Abstract
Background
Although genetic factors are risk factors for schizophrenia, some environmental factors are thought to be required for the manifestation of disease. Epigenetic mechanisms regulate gene functions without causing a change in the nucleotide sequence of DNA. Brain-derived neurotrophic factor (BDNF) is a neurotrophin that regulates synaptic transmission and plasticity. It has been suggested that BDNF may play a role in the pathophysiology of schizophrenia. It is established that methylation status of the BDNF gene is associated with fear learning, memory, and stressful social interactions. In this study, we aimed to investigate the DNA methylation status of BDNF gene in patients with schizophrenia.
Material/Methods
The study included 49 patients (33 male and 16 female) with schizophrenia and 65 unrelated healthy controls (46 male and 19 female). Determination of methylation pattern of CpG islands was based on the principle that bisulfite treatment of DNA results in conversion of unmethylated cytosine residues into uracil, whereas methylated cytosine residues remain unmodified. Methylation-specific PCR was performed with primers specific for either methylated or unmethylated DNA.
Results
There was no significant difference in methylated or un-methylated status for BDNF promoters between schizophrenia patients and controls. The mean duration of illness was significantly lower in the hemi-methylated group compared to the non-methylated group for BDNF gene CpG island-1 in schizophrenia patients.
Conclusions
Although there were no differences in BDNF gene methylation status between schizophrenia patients and healthy controls, there was an association between duration of illness and DNA methylation.
MeSH Keywords: Brain-Derived Neurotrophic Factor; DNA Methylation; Epigenesis, Genetic; Schizophrenia and Disorders with Psychotic Features
Background
Schizophrenia is a multifactorial disorder, and genetic and environmental factors are involved in its etiology [1]. Although genetic factors are risk factors for schizophrenia, it is believed that some environmental factors are required for the manifestation of disease [2]. Epigenetic mechanisms regulate gene functions without changing the nucleotide sequence of DNA [3]. These regulations are reversible, and the most commonly studied epigenetic mechanisms are DNA methylation and histone modification [3,4]. DNA methylation and epigenetic mechanisms are associated with psychiatric disorders, such as depression, psychotic disorders, post-traumatic stress disorder, autism, eating disorders, and substance dependence [5–7]. Epigenetic factors have an important role in gene and environment interactions, and environmental factors influence the genomic expressions through epigenetic mechanisms [8]. Epigenetic mechanisms are associated with many environmental factors, including nutrition, maternal care and behavior, hormones and drugs, early life experiences, and environmental agents in early development stages [9,10]. DNA methylation is caused by coupling of a methyl group to CpG sites with the DNA methyltransferase enzyme [11]. A normal level of DNA methylation is required for controlling genomic expressions [12]. Disease-related hypermethylation is thought to be associated with decreased RNA levels and hypomethylation is associated with increased RNA levels [13].
An animal study reported that animals showed schizophrenia-like behaviors and molecular changes, such as significant decrease in BDNF mRNA variants, increase in DNA methyltransferase1 (DNMT1), and ten-eleven translocase methylcytosine dioxygenase1 (TET1), no changes in histone deacetylases, histone methyltransferases and demethylases, or methyl CpG binding protein 2 in mice exposed to prenatal stress. This and several other studies show that environmental factors, such as prenatal stress, influence brain development and cause schizophrenia by epigenetic mechanisms, especially in early stage of brain development [14–16].
Brain-derived neurotrophic factor (BDNF) is a neurotrophin that regulates synaptic transmission and plasticity, and it has a role in proliferation, differentiation, survival, and death of neuronal and non-neuronal cells [17]. It has been suggested that BDNF plays a role in the pathophysiology of schizophrenia [18]. A schizophrenia animal model induced by prenatal stress showed increased DNA methylation of the BDNF gene and reduced BDNF transcription [14]. Genetic studies show an association between BDNF and schizophrenia [19]. Reduced BDNF levels in the prefrontal cortex, serum and CSF samples of patients with schizophrenia have been reported [20–23]. BDNF protein level and mRNA expression are lower in the prefrontal cortex of schizophrenia patients [20,24]. However, other studies showed higher BDNF levels in serum of schizophrenia patients [25,26] and there are conflicting results regarding BDNF levels in the hippocampus [27,28]. Postmortem studies have shown increased BDNF levels in the parietal cortex and frontal cortex, but decreased levels in the temporal cortex and occipital cortex [29]. Another post-mortem study showed abnormalities of DNA methylation in brains of patients with schizophrenia [30]. Methylation status of the BDNF gene is associated with fear learning, memory, and stressful social interactions. DNA methylation has a role in regulation of the BDNF gene in schizophrenia [31–33] and DNA methylation and demethylation may be used as targets for antipsychotic therapy [14].
In this study we aimed to investigate the DNA methylation status of the BDNF gene in patients with schizophrenia. We report the methylation status of 2 CpG islands of the BDNF gene that are located on the promoter region and intron 3-exon 4 boundary, as shown in Figure 1.
Figure 1.
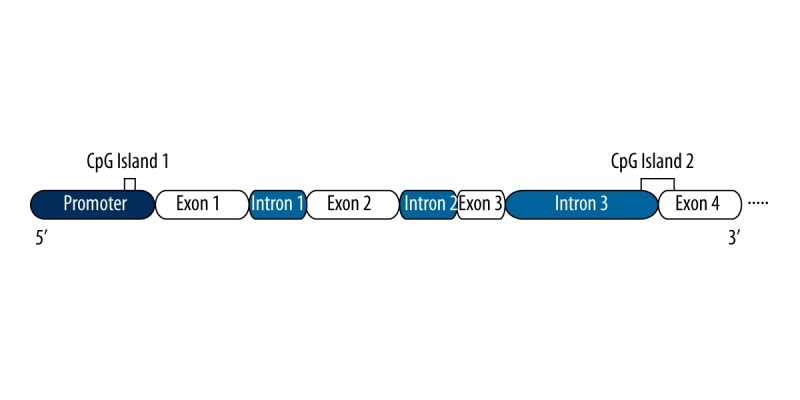
Schematic representation of BDNF gene. Only four exons are shown. The CpG islands amplified are idicated by orange lines.
Material and Methods
The study included 49 patients (aged 35.31±10.35 years, 33 male and 16 female) with schizophrenia and 65 unrelated healthy controls (aged 35.18±9.05 years, 46 male and 19 female). Mean duration of illness was 11.6±8.7 (min: 1, max: 36) years, mean PANSS total score was 66.3±30.2 (min: 30, max: 129), and mean CGI-S score was 3.9±2.16 (min: 1, max: 7). All patients were on medication. Thirty-two patients received atypical antipsychotic, 1 patient received typical and atypical antipsychotic combination, 11 patients received RLAI and atypical antipsychotic combination, and 7 patients received zuclopenthixol or flupenthixol depot and atypical antipsychotic combination. Volunteers in the control group had no personal or familial history of schizophrenia. Individuals with known major health problems, including diabetes and malignancies, were excluded from the study. Patients were diagnosed with schizophrenia according to the 4th edition of the Diagnostic and Statistical Manual for Mental Disorders (DSM-IV) [34]. Patients and controls were of the same ethnic origin and from the same geographical area (south-eastern Turkey). The severity of schizophrenia symptoms in the patients was evaluated using the Positive and Negative Syndrome Scale (PANSS) and the Clinical Global Impression Severity Scale (CGI-S) [35,36]. Physical and neurological examinations were performed in each of the patients and controls. The study was approved by the local ethics committee, informed consent was obtained from all participants, and the study was conducted in accordance with the Declaration of Helsinki.
DNA was extracted from blood samples by using the salt-chloroform method. Determination of methylation pattern of CpG islands was based on the principle that bisulfite treatment of DNA results in conversion of unmethylated cytokine residues into uracil, whereas methylated cytokine residues remain unmodified. Methylation-specific PCR was performed with primers specific for either methylated or unmethylated DNA. The primers were designed for 2 different CpG islands in the BDNF promoter (Figure 1). We treated 100 ng DNA samples with EpiMark Bisulfite Conversion Kit (Catalog No. E3318, New England Biolabs), in accordance with the manufacturer’s standard instructions. Primer sequences were: BDNF_1_MFw: 5′-GTAGTTTTCGTAGGATGAGGAAGC-3′ and BDNF_1_MRv: 5′-AATATAAATTAACAACCCCGATACG-3′ for the methylated (M) reaction and BDNF_1_UFw: 5′-GTAGTTTTTGTAGGATGAGGAAGTG-3′ and BDNF_1_URv 5′-TATAAATTAACAACCCCAATACACA-3′ for the unmethylated (U) reaction. BDNF_2_MFw: 5′-ATGATAGCGTACGTTAAGGTATCGT-3′, BDNF_2_MRv: 5′-AACGAAAAACTCCATTTAATCTCG-3′ and BDNF_2_UFw: 5′-ATATGATAGTGTATGTTAAGGTATTGT-3′, BDNF_2_URv: 5′-ACAAAAAACTCCATTTAATCTCAAC-3′. PCR mixture contained 10 ng of modified DNA, 2.0 mmol each of dATP, dGTP, dCTP, and dTTP, 1.0 pmol each of primer, 2.0 mmol MgCl2, 1× reaction buffer, and 1.0 U Taq polymerase. The PCR conditions were: 1 cycle at 95°C for 1 min, 12 cycles at 95°C for 10 s, at 54°C to 48°C (touch-down) for 40 s, at 72°C for 60 s, then 25 cycles at 95°C for 10 s, at 52°C for 40 s, at 72°C for 60 s, and 1 cycle at 72°C for 5 min. Amplified PCR products were electrophoresed on 2% agarose gels, visualized by staining with ethidium bromide, and illuminated under UV light. Positive control sample (Fermentas, Cat. No: SD1131) was utilized as hypermethylated.
Statistical analyses
The collected data were analyzed using SPSS version 18.0 (SPSS Inc., Chicago, IL, USA). Both descriptive and analytical statistics were used. Chi-square/Fisher’s exact test was used to compare categorical variables. The t test was used to compare continuous variables.
Results
The mean ages (F=1.306, p=0.947) and the sex distribution (x2=0.154, DF=1, p=0.695) of the study groups were similar. For BDNF gene CpG island-1, 3.9% (n=2) was methylated, 68.6% (n=35) was hemi-methylated, and 27.4% (n=14) was unmethylated in he patient group. There were no methylated subjects, 67.7% (n=44) were hemi-methylated, and 32.3% (n=21) were unmethylated in the control group (Figure 2). There were no significant difference in methylated or unmethylated status for each area between schizophrenia patients and controls (p>0.05). The comparisons of the methylated or unmethylated status for each area according to the study groups are presented in Table 1. When patients were compared with clinical parameters (duration of the illness, CGI-S, PANSS scores, BMI, and methylated or unmethylated status for each area) we did not find any significant difference (p>0.05). There were no differences in DNA methylation status of BDNF gene CpG island-1 and CpG island-2 regions with respect to sex or age between the schizophrenia patients and the control group (p>0.05). The mean duration of illness was significantly lower in the hemi-methylated group compared to non-methylated group for BDNF gene CpG island-1 in schizophrenia patients (p=0.017). However, there was no difference in mean duration of illness between the hemi-methylated group compared to the non-methylated group for BDNF gene CpG island-2 in schizophrenia patients (p>0.05). There were no differences in median CGI-S PANSS total and subscale scores between hemi-methylated group compared to non-methylated group for BDNF gene CpG island-1 and CpG island-2 regions in schizophrenia patients (p>0.05).
Figure 2.
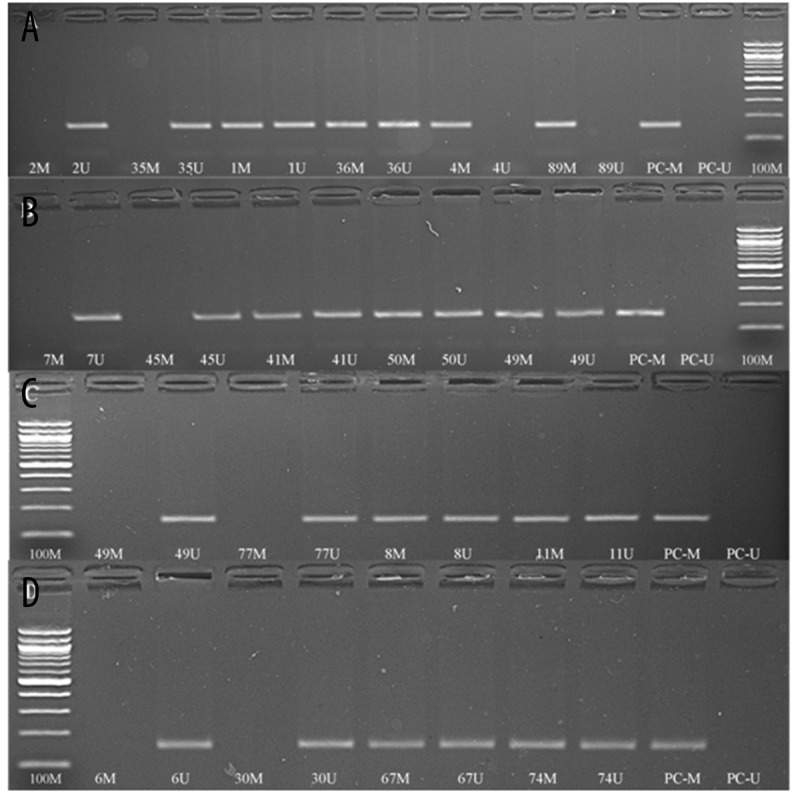
Methylation-specific PCR results of some randomly selected sample patients with schizophrenia and control groups, analyzed with 2% agarose gel electrophoresis followed by ethidium bromide staining. A: Methylation pattern of some patients in CpG island-1 from BDNF gene. B: Methylation pattern of some controls in CpG island-1 from BDNF gene. C: Methylation pattern of some patients in CpG island-2 from BDNF gene. D: Methylation pattern of some controls in CpG island-2 from BDNF gene. PC: Positive control; M: methylated; U: unmethylated; 100M: 100 bp DNA ladder.
Table 1.
Methylation status of BDNF gene with schizophrenia patients and healthy controls.
| Schizophrenia patients | Healthy controls (n= 65) | p | |
|---|---|---|---|
| CpG island 1 (n=49) | 0.669* | ||
| Hemi-methylated | 35 (71.4%) | 44 (67.7%) | |
| Unmethylated | 14 (28.6%) | 21 (32.3%) | |
| CpG island 2 (n=51) | 1.00** | ||
| Hemi-methylated | 2 (3.9%) | 2 (3.1%) | |
| Unmethylated | 49 (96.1%) | 63 (96.9%) |
Chi-square test;
Fisher’s exact test.
Discussion
We found that there was no difference in methylation status of 2 regions of the BNDF gene between schizophrenia patients and healthy controls, suggesting that the genetic component of schizophrenia is very complex. Genetic studies have reported that some genes are associated with schizophrenia. The most likely candidate genes are: GABAergic genes (RELN and GAD1), dopaminergic genes (DAT and DRD2, MB-COMT), 5-HT2A receptor (HTR2A) gene, and BDNF [33,37]. It is suggested that down-regulation of GABAergic genes such as RELN and GAD1 are related to hypermethylation of their gene promoters, causing GABAergic dysfunction; moreover, it plays a role in schizophrenia [38–40]. However, in another study, decreased methylation of the GAD1promoter and lower levels ofGAD1mRNA in schizophrenia patients were found [41]. A postmortem study performed in patients with schizophrenia showed increased methylation of RELN gene and decreased RELN gene expression [38]. However, another study shows no detectable DNA methylation in RELN promoter region [42]. Another study showed no or less methylation in RELN gene until puberty, while increased levels of methylation were found after puberty [43]. 5-HT2A receptor (HTR2A) gene is a serotonergic system gene and in a study it was found that methylation of the HTR2A gene promoter correlated with HTR2A expression levels. However, another study found no difference between schizophrenia patients compared to controls in HTR2A methylation [44,45]. Epigenetic regulations of genes influencing the dopaminergic system, such as COMT, DRD2, and DAT genes, have been investigated in schizophrenia. It was shown that there was a hypomethylation in MB-COMT gene and it was suggested that this methylation is a risk factor for schizophrenia [46]. However, another study was found no difference in COMT promoter methylation between schizophrenia patients and controls [47]. It was found that there were no differences in DRD2 and DAT gene methylation between schizophrenia patients and controls [48,49].
There are few studies about methylation status of the BNDF gene in schizophrenia patients. A postmortem study found that there were no differences in BDNF gene exon I, III, VI, and IX regions methylation status between schizophrenia patients and controls, but the same study fond reduced DNA methylation in the Val66Met SNP in exon IX [50]. There are 2 other studies on BDNF gene methylation with peripheral blood samples of schizophrenia patients – one study showed reduced BDNF promoter IV methylation, and the other one showed increased BDNF promoter I methylation in schizophrenia patients compared to controls [48]. Another study with peripheral blood samples of Japanese schizophrenia patients found that there was a small but significant increase in methylation in the BDNF 1 promoter, and the same study suggested that epigenetic differences in peripheral blood might be a potential biomarker and reflect the pathophysiology of schizophrenia [51]. A review by Mitchelmore and Gede suggested that BDNF had a central role in neuronal development and neuroplasticity, and dysregulation of the BDNF gene was associated with several psychiatric and neurologic disorders. In addition, epigenetic mechanisms that regulate BDNF gene expression could be used in diagnosis, prognoses, and as biomarkers of several psychiatric disorders [37]. In the present study we found that there were no associations between DNA methylation status of BDNF gene CpG island-1 and CpG island-2 regions with respect to sex or age. However, another study showed that the methylation status of BDNF promoter was associated with sex – there was more prominent methylation in male patients with schizophrenia.
The mean duration of illness was significantly shorter in the hemi-methylated group compared to the non-methylated group for BDNF gene CpG island-1 in schizophrenia patients. To the best of our knowledge there is no published data about the association between duration of illness and DNA methylation in schizophrenia. A review of DNA hypomethylation and human diseases showed that DNA methylation plays a role in neuronal cell survival and maturation; the balance of the methylation level is important for neuronal survival and hypomethylation causes abnormalities in neuronal function [52]. Although the mechanism is not clear, a study mentioned that DNA hypomethylation may cause cell death by apoptosis [53]. Considering these data, it appears that reduced methylation ratios with duration of disease may cause the neuronal dysfunction by altering neuronal survival, suggesting that there is an association between duration of illness and DNA methylation.
We hypothesized that there was a defect in the DNA methylation mechanisms that affect the BDNF gene, and that there is an association between DNA methylation and schizophrenia. However, there was only an association between duration of illness and DNA methylation. As mentioned above, there are inconsistent results in methylation studies. Several other factors, such as patients receiving antipsychotics and analyzing methylation in blood, may affect the methylation status. A study that examined the effect of antipsychotics on DNA methylation found that clozapine and quetiapine reduced the DNA methylation, but there are no similar effects of haloperidol and risperidone on DNA methylation status [54]. Another study in animals showed that clozapine reduces BDNF methylation and that this increases social interaction [55,56].
There are some limitations of this study. First, we analyzed DNA methylation in peripheral blood, which does not directly reflect the central nervous system. Second, this is a naturalistic and cross-sectional study and patients were continuing their medication. Therefore, receiving antipsychotics may have affected the methylation status. DNA methylation is a dynamic and reversible process, and many other environmental factors also affect methylation. BDNF methylation may be different in different populations and environments [57]. For these reasons, examining the methylation status and protein levels of BDNF gene at certain intervals throughout the treatment period in the same patients may be more valuable.
Conclusions
Although there were no differences in BDNF gene methylation status between schizophrenia patients and healthy controls, there was an association between duration of illness and DNA methylation. Further studies with more patients are necessary, and the limitations referred to in this paper should be considered.
Footnotes
Conflicting interests
The authors declare no conflicting interests.
Source of support: Departmental sources
References
- 1.Tsuang M. Schizophrenia: genes and environment. Biol Psychiatry. 2000;47(3):210–20. doi: 10.1016/s0006-3223(99)00289-9. [DOI] [PubMed] [Google Scholar]
- 2.Fuller Torrey E, Yolken RH. Familial and genetic mechanisms in schizophrenia. Brain Res Brain Res Rev. 2000;31(2):113–17. doi: 10.1016/s0165-0173(99)00028-4. [DOI] [PubMed] [Google Scholar]
- 3.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–54. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 4.Rutten BPF, Mill J. Epigenetic mediation of environmental influences in major psychotic disorders. Schizophr Bull. 2009;35(6):1045–56. doi: 10.1093/schbul/sbp104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Favalli G, Li J, Belmonte-de-Abreu P, Wong AHC, Daskalakis ZJ. The role of BDNF in the pathophysiology and treatment of schizophrenia. J Psychiatr Res. 2012;46(1):1–11. doi: 10.1016/j.jpsychires.2011.09.022. [DOI] [PubMed] [Google Scholar]
- 6.Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nat Rev Neurosci. 2007;8(5):355–67. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- 7.Unternaehrer E, Luers P, Mill J, et al. Dynamic changes in DNA methylation of stress-associated genes (OXTR, BDNF) after acute psychosocial stress. Transl Psychiatry. 2012;2(8):e150. doi: 10.1038/tp.2012.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bagot RC, Meaney MJ. Epigenetics and the biological basis of gene x environment interactions. J Am Acad Child Adolesc Psychiatry. 2010;49(8):752–71. doi: 10.1016/j.jaac.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 9.Aguilera O, Fernández AF, Muñoz A, Fraga MF. Epigenetics and environment: a complex relationship. J Appl Physiol. 2010;109(1):243–51. doi: 10.1152/japplphysiol.00068.2010. [DOI] [PubMed] [Google Scholar]
- 10.Meaney MJ. Epigenetics and the biological definition of gene x environment interactions. Child Dev. 2010;81(1):41–79. doi: 10.1111/j.1467-8624.2009.01381.x. [DOI] [PubMed] [Google Scholar]
- 11.Bird AP. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980;8(7):1499–504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366(6453):362–65. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 13.Akbarian S. The molecular pathology of schizophrenia – focus on histone and DNA modifications. Brain Res Bull. 2010;83(3):103–7. doi: 10.1016/j.brainresbull.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong E, Dzitoyeva SG, Matrisciano F, et al. Brain-derived neurotrophic factor epigenetic modifications associated with schizophrenia-like phenotype induced by prenatal stress in mice. Biol Psychiatry. 2015;77(6):589–96. doi: 10.1016/j.biopsych.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai SJ. Central N-acetyl aspartylglutamate deficit: a possible pathogenesis of schizophrenia. Med Sci Monit. 2005;11(9):HY39–45. [PubMed] [Google Scholar]
- 16.Fruntes V, Limosin F. Schizophrenia and viral infection during neurodevelopment: a pathogenesis model? Med Sci Monit. 2008;14(6):RA71–77. [PubMed] [Google Scholar]
- 17.Lu B. BDNF and activity-dependent synaptic modulation. Learn Mem. 2003;10(2):86–98. doi: 10.1101/lm.54603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Angelucci F, Brene S, Mathe AA. BDNF in schizophrenia, depression and corresponding animal models. Mol Psychiatry. 2005;10(4):345–52. doi: 10.1038/sj.mp.4001637. [DOI] [PubMed] [Google Scholar]
- 19.Szekeres G, Juhász A, Rimanóczy Á, et al. The C270T polymorphism of the brain-derived neurotrophic factor gene is associated with schizophrenia. Schizophr Res. 2003;65(1):15–18. doi: 10.1016/s0920-9964(02)00505-4. [DOI] [PubMed] [Google Scholar]
- 20.Issa G, Wilson C, Terry AV, Jr, Pillai A. An inverse relationship between cortisol and BDNF levels in schizophrenia: data from human postmortem and animal studies. Neurobiol Dis. 2010;39(3):327–33. doi: 10.1016/j.nbd.2010.04.017. [DOI] [PubMed] [Google Scholar]
- 21.Jindal RD, Pillai AK, Mahadik SP, et al. Decreased BDNF in patients with antipsychotic naive first episode schizophrenia. Schizophr Res. 2010;119(1):47–51. doi: 10.1016/j.schres.2009.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pillai A, Kale A, Joshi S, et al. Decreased BDNF levels in CSF of drug-naive first-episode psychotic subjects: correlation with plasma BDNF and psychopathology. Int J Neuropsychopharmacol. 2010;13(04):535–39. doi: 10.1017/S1461145709991015. [DOI] [PubMed] [Google Scholar]
- 23.Toyooka K, Asama K, Watanabe Y, et al. Decreased levels of brain-derived neurotrophic factor in serum of chronic schizophrenic patients. Psychiatry Res. 2002;110(3):249–57. doi: 10.1016/s0165-1781(02)00127-0. [DOI] [PubMed] [Google Scholar]
- 24.Weickert CS, Hyde TM, Lipska BK, et al. Reduced brain-derived neurotrophic factor in prefrontal cortex of patients with schizophrenia. Mol Psychiatry. 2003;8(6):592–610. doi: 10.1038/sj.mp.4001308. [DOI] [PubMed] [Google Scholar]
- 25.Gama CS, Andreazza AC, Kunz M, et al. Serum levels of brain-derived neurotrophic factor in patients with schizophrenia and bipolar disorder. Neurosci Lett. 2007;420(1):45–48. doi: 10.1016/j.neulet.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 26.Reis HJ, Nicolato R, Barbosa IG, et al. Increased serum levels of brain-derived neurotrophic factor in chronic institutionalized patients with schizophrenia. Neurosci Lett. 2008;439(2):157–59. doi: 10.1016/j.neulet.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 27.Ray MT, Weickert CS, Wyatt E, Webster MJ. Decreased BDNF, trkB-TK+ and GAD67 mRNA expression in the hippocampus of individuals with schizophrenia and mood disorders. J Psychiatry Neurosci. 2011;36(3):195–203. doi: 10.1503/jpn.100048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi M, Shirakawa O, Toyooka K, et al. Abnormal expression of brain-derived neurotrophic factor and its receptor in the corticolimbic system of schizophrenic patients. Mol Psychiatry. 2000;5(3):293–300. doi: 10.1038/sj.mp.4000718. [DOI] [PubMed] [Google Scholar]
- 29.Durany N, Michel T, Zöchling R, et al. Brain-derived neurotrophic factor and neurotrophin 3 in schizophrenic psychoses. Schizophr Res. 2001;52(1):79–86. doi: 10.1016/s0920-9964(00)00084-0. [DOI] [PubMed] [Google Scholar]
- 30.Veldic M, Caruncho HJ, Liu WS, et al. DNA-methyltransferase 1 mRNA is selectively overexpressed in telencephalic GABAergic interneurons of schizophrenia brains. Proc Natl Acad Sci. 2004;101(1):348–53. doi: 10.1073/pnas.2637013100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu B, Martinowich K, editors. NIH Public Access. 2008. Cell biology of BDNF and its relevance to schizophrenia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28(42):10576–86. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roth TL, Lubin FD, Sodhi M, Kleinman JE. Epigenetic mechanisms in schizophrenia. Biochim Biophys Acta. 2009;1790(9):869–77. doi: 10.1016/j.bbagen.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.American Psychiatric A. Diagnostic and statistical manual of mental disorders: DSM-IV-TR®. American Psychiatric Pub; 2000. [Google Scholar]
- 35.Guy W. Clinical global impression scale. The ECDEU Assessment Manual for Psychopharmacology-Revised Volume DHEW Publ No ADM 76. 1976;338:218–22. [Google Scholar]
- 36.Kay SR, Flszbein A, Opfer LA. The positive and negative syndrome scale (PANSS) for schizophrenia. Schizophr Bull. 1987;13(2):261–76. doi: 10.1093/schbul/13.2.261. [DOI] [PubMed] [Google Scholar]
- 37.Mitchelmore C, Gede L. Brain derived neurotrophic factor: epigenetic regulation in psychiatric disorders. Brain Res. 2014;1586:162–72. doi: 10.1016/j.brainres.2014.06.037. [DOI] [PubMed] [Google Scholar]
- 38.Abdolmaleky HM, Cheng KH, Russo A, et al. Hypermethylation of the reelin (RELN) promoter in the brain of schizophrenic patients: a preliminary report. Am J Med Genet B Neuropsychiatr Genet. 2005;134B(1):60–66. doi: 10.1002/ajmg.b.30140. [DOI] [PubMed] [Google Scholar]
- 39.Costa E, Chen Y, Dong E, et al. GABAergic promoter hypermethylation as a model to study the neurochemistry of schizophrenia vulnerability. Expert Rev Neurother. 2009;9(1):87–98. doi: 10.1586/14737175.9.1.87. [DOI] [PubMed] [Google Scholar]
- 40.Grayson DR, Jia X, Chen Y, et al. Reelin promoter hypermethylation in schizophrenia. Proc Natl Acad Sci USA. 2005;102(26):9341–46. doi: 10.1073/pnas.0503736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang HS, Akbarian S. GAD1 mRNA expression and DNA methylation in prefrontal cortex of subjects with schizophrenia. PloS One. 2007;2(8):e809. doi: 10.1371/journal.pone.0000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tochigi M, Iwamoto K, Bundo M, et al. Methylation status of the reelin promoter region in the brain of schizophrenic patients. Biol Psychiatry. 2008;63(5):530–33. doi: 10.1016/j.biopsych.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 43.Lintas C, Persico AM. Neocortical RELN promoter methylation increases significantly after puberty. Neuroreport. 2010;21(2):114–18. doi: 10.1097/WNR.0b013e328334b343. [DOI] [PubMed] [Google Scholar]
- 44.De Luca V, Viggiano E, Dhoot R, et al. Methylation and QTDT analysis of the 5-HT2A receptor 102C allele: analysis of suicidality in major psychosis. J Psychiatr Res. 2009;43(5):532–37. doi: 10.1016/j.jpsychires.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 45.Polesskaya OO, Aston C, Sokolov BP. Allele C-specific methylation of the 5-HT2A receptor gene: Evidence for correlation with its expression and expression of DNA methylase DNMT1. J Neurosci Res. 2006;83(3):362–73. doi: 10.1002/jnr.20732. [DOI] [PubMed] [Google Scholar]
- 46.Abdolmaleky HM, Cheng KH, Faraone SV, et al. Hypomethylation of MB-COMT promoter is a major risk factor for schizophrenia and bipolar disorder. Hum Mol Genet. 2006;15(21):3132–45. doi: 10.1093/hmg/ddl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dempster EL, Mill J, Craig IW, Collier DA. The quantification of COMT mRNA in post mortem cerebellum tissue: diagnosis, genotype, methylation and expression. BMC Med Genet. 2006;7(1):10. doi: 10.1186/1471-2350-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kordi-Tamandani DM, Sahranavard R, Torkamanzehi A. DNA methylation and expression profiles of the brain-derived neurotrophic factor (BDNF) and dopamine transporter (DAT1) genes in patients with schizophrenia. Mol Biol Rep. 2012;39(12):10889–93. doi: 10.1007/s11033-012-1986-0. [DOI] [PubMed] [Google Scholar]
- 49.Zhang A-p, Yu J, Liu J-x, et al. The DNA methylation profile within the 5′-regulatory region of DRD2 in discordant sib pairs with schizophrenia. Schizophr Res. 2007;90(1–3):97–103. doi: 10.1016/j.schres.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 50.Mill J, Tang T, Kaminsky Z, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet. 2008;82(3):696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ikegame T, Bundo M, Sunaga F, et al. DNA methylation analysis of BDNF gene promoters in peripheral blood cells of schizophrenia patients. Neurosci Res. 2013;77(4):208–14. doi: 10.1016/j.neures.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 52.Hutnick LK, Golshani P, Namihira M, et al. DNA hypomethylation restricted to the murine forebrain induces cortical degeneration and impairs postnatal neuronal maturation. Hum Mol Genet. 2009;18(15):2875–88. doi: 10.1093/hmg/ddp222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69(6):915–26. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 54.Guidotti A, Auta J, Chen Y, et al. Epigenetic GABAergic targets in schizophrenia and bipolar disorder. Neuropharmacology. 2011;60(7):1007–16. doi: 10.1016/j.neuropharm.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gavin DP, Akbarian S. Epigenetic and post-transcriptional dysregulation of gene expression in schizophrenia and related disease. Neurobiol Dis. 2012;46(2):255–62. doi: 10.1016/j.nbd.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 56.Matrisciano F, Dong E, Gavin DP, et al. Activation of group II metabotropic glutamate receptors promotes DNA demethylation in the mouse brain. Mol Pharmacol. 2011;80(1):174–82. doi: 10.1124/mol.110.070896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma W, Zhou X, Ji H, et al. Population difference in the association of BDNF promoter methylation with mild cognitive impairment in the Xinjiang Uygur and Han populations. Psychiatry Res. 2015;229(3):926–32. doi: 10.1016/j.psychres.2015.07.017. [DOI] [PubMed] [Google Scholar]


