Abstract
Automated data collection is an integral part of modern workflows in single particle electron microscopy (EM) research. This review surveys the software packages available for automated single particle EM data collection. The degree of automation at each stage of data collection is evaluated, and the capabilities of the software packages are described. Finally, future trends in automation are discussed.
Keywords: single particle electron microscopy, data collection, automation, target acquisition, software packages, high throughput
Introduction
The field of single particle electron microscopy (EM) is undergoing astonishing changes in achievement of higher resolution, brought about by a combination of technological advances and improved specimen preparation. This ‘resolution revolution’ as termed by Kühlbrandt [1] shows no signs of abating: so far this year a total of eight structures [2–9] surpassed the 3 Å barrier [10] for single particle EM reconstruction for the first time, with many more expected soon.
As a result of these technology breakthroughs, there has been a surge in the popularity of single particle EM. Automation is important in both lowering the barrier for entry for new scientists into the field as well as freeing up the time of experienced scientists during data collection. The topic of automation in EM has a long history – early work by the Agard [11], Baumeister [12,13] and Brisson groups [14] in the 1990s contributed to semi-automated systems to control electron microscopes and manipulate downstream data in single particle EM, electron tomography and electron crystallography.
The purpose of this review is to survey the state of automation in data collection of single particle EM. Each sequential step in the collection pipeline is addressed, and the program suites available will be discussed in the context of the pipeline. Data processing will not be covered here, nor will electron tomography and electron crystallography related software. The goal of this review is to provide new researchers in the field of single particle EM with an overview of the technical advantages and challenges faced in automation of EM data collection.
Automation in data collection
In this review, an automated data collection package will be defined as a software system that is able to interface with both the camera and the microscope and can collect multiple images of an EM grid without user input. The automation software must also be able to perform autofocusing and determine the eucentric height of the specimen without intervention in order to image multiple locations on the grid. Semi-automated packages will refer to software systems that have some but not all of these capabilities. Automation software packages that are discussed are listed in Tables 1 and 2.
Table 1.
Software packages available for automated single particle EM Data collection
| Software package | Latest version | Capabilities | Supported equipment | Reference(s) | Number of citationsa |
|---|---|---|---|---|---|
| Academic packages | |||||
| SerialEM | 2015 | Single particle, RCT/OT, tomography |
|
[15] | 449 |
| Leginon | 2014 | Single particle, RCT/OT, tomography, electron crystallography |
|
[16] | 87 |
| [17] | 148 | ||||
| [18] | 328 | ||||
| UCSFImage4 | 2013 | Single particle |
|
[19] | 0 |
| TOM2 | 2012 | Single particle, tomography |
|
[20] | 21 |
| SAM | 2008 | Single particle |
|
[21] | 18 |
| JAMES | 2006 | Single particle |
|
[22] | 9 |
| AutoEMation | 2005 | Single particle |
|
[23] | 51 |
| AutoEM | 2003 | Single particle |
|
[24] | 42 |
| [25] | 52 | ||||
| Commercial packages | |||||
| EM-TOOLS | 2015 | Single particle, tomography |
|
– | – |
| EPU | 2014 | Single particle, RCT/OT |
|
– | – |
| JADAS | 2009 | Single particle |
|
[26] | 36 |
a As of 15 May 2015.
Table 2.
Specific capabilities of software packages available for automated single particle EM data collection
| Software package | Auto targeting on grids with regular hole pattern | Auto targeting on grids with lacey carbon | MSI level customizable by user | Ability to discriminate holes based on ice thickness | Breath-first tree traversal work flow (Queuing) | Additional hardware (computer or GPU card) required | Built-in remote data monitoring |
|---|---|---|---|---|---|---|---|
| Academic packages | |||||||
| SerialEM | Yes | No information | Yes | No | Yes | No | No |
| Leginon | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| UCSFImage4 | Yes | No | No | No | No | Yes | No |
| TOM2 | Yes | No information | Yes | No | No information | Yes | No |
| SAM | No | No | No | No | No | No information | No |
| JAMES | No information | No | No | No | No | No information | No |
| AutoEMation | Yes | No | No | Yes | No | No information | No |
| AutoEM | No information | No | No | Yes | No | No information | No |
| Commercial packages | |||||||
| EM-TOOLS | Yes | No | No information | No information | No information | No information | No |
| EPU | Yes | Yes | No | Yes | Yes | No | No |
| JADAS | Yes | No | Yes | Yes | No | No information | No |
Grid compatibility
The first variable that an automation software package must adjust to is the nature of the EM grid itself. EM grids for negative stain typically have a uniform layer of carbon on which the sample is placed – the area available for imaging would be the entire square demarcated by the grid bars. For cryo-EM, holey carbon or holey gold grids [27] are typically used. These grids have regular arrays of holes within the film that must be targeted during imaging. The most commonly used commercial holey carbon grids are Quantifoil [28] and C-flat [29,30]. The diameter of the holes and distance between holes can be customized, and the automation software has to adapt to these differences. All automation software packages are able to deal with imaging on these conventional substrates.
Other types of less regular grids are also sometimes used, including irregular holes or lacey carbon grids, with the latter preferred when dealing with samples that tend to adhere to carbon surfaces and thus creating difficulty in distributing sample across the holes of conventional holey carbon grids. These non-conventional grids require the software to be able to identify noncircular holes and select areas within these holes, as has been accomplished by Nicholson et al. [31] using Leginon and also in EM-TOOLS. More recently, regular grids have been used to load multiple samples [32,33] for the purposes of screening, and Leginon can be used in a semi-automated mode to image these multiple targets.
Microscope alignment
An important aspect of successful automation is the alignment of the microscope. The purpose of this alignment is to minimize lens aberrations, maximize image quality and provide convenience of operation [34,35]. Alignments are usually done starting from the electron source and continuing sequentially down to the projector lens. Most alignments are stable and do not need to be adjusted by the microscopist during routine data collection [18,36]. Gun and condenser alignments are independent of the sample and thus typically do not need to be changed during the imaging process. Alignments that do need to be regularly checked and refined are termed direct alignments on FEI microscopes [36] and other manufacturers have similar alignment routines. No automation software has thus far been developed to fully replace a trained microscopist in aligning the microscope for data collection, although significant progress has been made with making this process semi-automated. Automation has the advantage, in some instances, of providing more accuracy for certain alignments than an operator can achieve [37]. In the following sections, we discuss automation of direct alignments based on the type of aberration that must be corrected.
On-axis aberration: out-of-focus
Image focus is adjusted by changing the strength of the objective lens. Defocus refers to the amount of displacement of the imaging plane from the focused position (Fig. 1). Most automation packages (AutoEM, AutoEMation, EPU, JADAS, Leginon, SerialEM, TOM2) make use of an autofocusing technique called the beam-tilt-induced image displacement (BID) method first described by Koster et al. [39]. In this method, the electron beam is tilted, and displacement of the image is recorded. The amplitude of the displacement is mathematically related to the extent of defocus through the following equation, taken from Koster et al. [39]:
| (1) |
where D is the image displacement, M is the magnification, β is the beam tilt angle, Δf is the defocus, Cs is the objective lens spherical aberration, ω is the spatial frequency of the sample and λ is the wavelength of the electron beam.
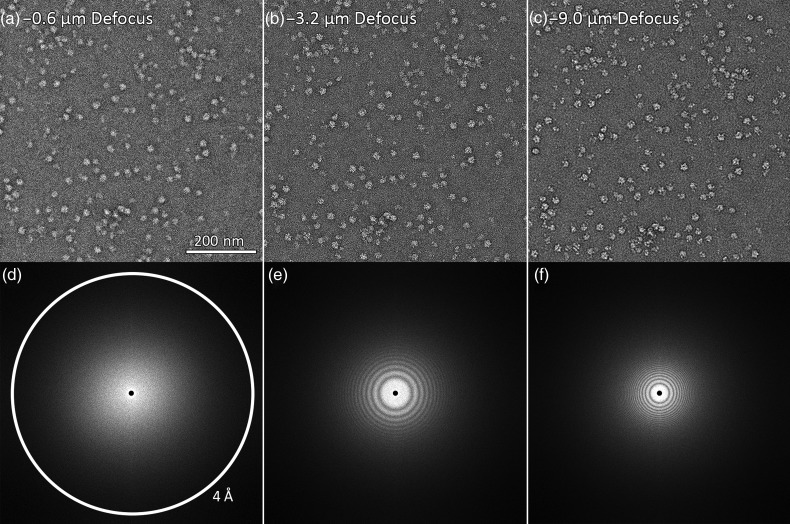
Fig. 1.
Negative stain electron micrographs of the same field at different defocus values. (a) is closest to focus, followed by (b) and (c) with sequentially higher defocus values. The corresponding Fourier transforms of each micrograph are shown (d–f). Defocus values were estimated using CTFFind3 [38]. Images had a pixel size of 1.83 Å and were taken with a 200 kV microscope with Cs of 2.0 mm.
A second technique that can be used for focusing is the diffractogram method first described by Saxton et al. [40]. An image is acquired and the computed power spectrum is used to calculate the defocus (Fig. 2d–f). Krivanek and Maloney [41] have also described using this method to allow for computer-controlled automated focus correction on a CCD camera. Both JADAS [26] and UCSFImage4 [19] have implemented this method.
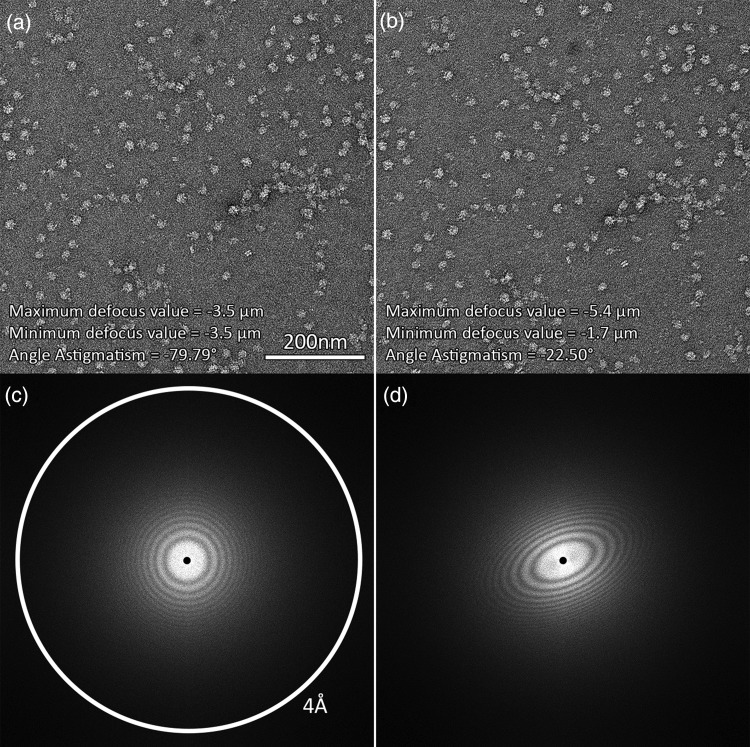
Fig. 2.
Comparison of non-astigmatic (a) and strongly astigmatic (b) micrographs. Their corresponding Fourier transforms are shown below (c and d). Images had a pixel size of 1.83 Å and were taken with a 200-kV microscope with Cs of 2.0 mm.
One other method for focusing has been described in the literature [37] whereby the focus is determined by finding the image with the least contrast, but this is not currently used by any of the automation packages described in this review.
The BID method for autofocusing is the most widely used for several reasons. It is very efficient – only one untilted and one beam tilt image are required to determine the defocus – whereas the diffractogram method requires two or more diffractograms in order to compensate for the ambiguity and errors present during imaging [39]. The BID method works well at low magnifications [39] and unlike the other two methods [37] is largely specimen independent, except in cases where there is interference from the diffraction from crystalline support substrates like gold grids [27]. The BID method's main drawback is its lack of dose efficiency [39] – the area illuminated during BID is generally damaged by radiation and not suitable for further imaging.
Off-axis aberration: astigmatism
Two-fold astigmatism in an image arises when two orthogonally propagated electron rays are focused to two different locations by the lens (Fig. 2) [37]. Thus, it can be corrected automatically using the focusing methods described above. More complex forms of astigmatism, such as 3-fold astigmatism, will compromise image quality if higher than 1.5 Å resolution is a required goal [42,43]. These effects cannot be reliably corrected by current automated data collection software, but a manual alignment is expected to be stable for periods well in excess of a year.
Off-axis aberration: coma
If a lens has coma, off-axis illumination will not be focused to a point. Minimizing the effect of the coma is important for high-resolution imaging as highlighted by Zemlin et al. [44] and Glaeser et al. [45] because the phase error caused by coma increases with the third power of the spatial frequency. The equation for the phase error is given below, taken from Glaeser et al. [45]:
| (2) |
where θ is the beam tilt angle, Cs is the spherical aberration, λ is the wavelength of the electrons, s is the spatial frequency, is the unit vector in the beam tilt direction and is the unit vector in the spatial frequency vector direction.
To minimize the effect of coma on a conventional electron microscope, the beam must be aligned with the optical axis so that the average beam tilt angle is zero. Under these conditions, the phase error due to coma is zero as calculated using Eq. (2). Two methods can be used to achieve this so-called coma-free alignment [45]. The first method is to slowly wobble the beam back and forth about an angle of ∼10 mrad. This is done in both the x and y axes, and the live image is observed either on a screen or a camera. If the beam is tilted in the corresponding axis to begin with, a positive tilt will result in an image of different defocus compared with a negative tilt of the same magnitude. Only when the beam is parallel to the optical axis will a positive and negative tilt give an image of the same defocus and astigmatism [45]. The similarity of the two images is usually judged by eye, thus making this method rather qualitative, but it has the advantage of speed.
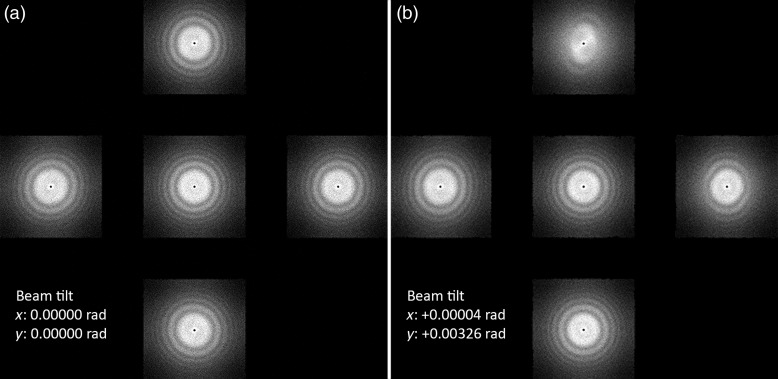
The second method uses Zemlin tableaus [44]. A series of images using different beam tilts, positive and negative, are acquired in the x and y tilt directions (and possibly also the intermediate axes between them) and Fourier transforms are calculated of the acquired images, as shown in Fig. 3. The concept is similar to the first method, except comparing the transformed images is more quantitative, allowing the microscopist to obtain a more accurate coma-free alignment. Zemlin tableaus are implemented in the Leginon package [18], but only in a semi-automated manner, as the user has to manually adjust the beam tilt using the Leginon interface until the Zemlin tableaus look optimal. No single particle automation software that we are aware of so far offers a fully automated coma-free alignment.
Fig. 3.
Zemlin tableaus acquired with coma-free alignment (a) and without coma-free alignment (b). The calculated resolution limit (based on Eq. (2)) for the aberration seen in (b) and corrected in (a) is ∼0.55 nm, given a resolution limit cutoff at 45° phase error, using a 200 kV microscope with Cs of 2.0 mm.
One other way to eliminate the coma effect would be to adjust the Cs value to zero in the equation for phase error, and this has been achieved by hardware in Cs corrected Titan Krios microscopes [4]. The ability to generate Zemlin tableaus is still required as the microscopist has to now adjust the Cs corrector instead of the beam tilt in order to eliminate coma and other aberrations.
Given that no software is so far able to automatically correct for coma, it is important to know how long a coma-free alignment remains stable, in order to determine if this will be a factor limiting the unsupervised duration of a high-resolution single particle data collection session. Based on work by Uhlemann and Haider [46] on a spherical aberration-corrected EM, the change in coma within a 12-h experimental collection period violates an acceptable threshold of at 1.4 Å resolution and needs to be corrected about once every 3 h. Considering that phase error increases with the cube of the resolution, a 2.5 Å resolution threshold is predicted to be breached only after 17 h, indicating the need to consider automatic coma-free alignment only during very long-duration collection of high resolution data.
Finally, it is important to note that coma-free alignment can only eliminate axial-coma effects; off-axis coma can still corrupt the image if the beam is not parallel across the field of view [45]. No program automatically checks this so ensuring a parallel beam is an essential step during manual microscope alignment. The magnitude of this effect can be determined by the scope lens geometry [45] or estimated from diffractograms from different parts of the camera sensor using a known average beam tilt.
Calibrations and corrections
A consequence of relying on automation software to interface with the microscope is the need to calibrate the microscope adjustments within the software environment. This is essential to ensure that the program can adjust the microscope parameters accurately. Broad categories for calibrations include image shift, stage shift, pixel size, autofocusing, beam shift, beam tilt, flat field, CCD sensitivity, beam intensity (I0) and dose [15,18,36]. Fortunately of all these calibrations, only image shift offsets and I0 calibration (required for EPU) have to be updated prior to starting each data collection session.
Image shift offsets are determined at each magnification and define the corrective image shifts required to be applied when moving between different magnification presets so as to keep the same image features in the center of view. This procedure is called ‘image shift calibration’ by EPU [36], ‘preset image shift alignment’ by Leginon [18] and ‘mag IS offsets’ by SerialEM [15]. These calibrations are done by first picking an identifiable feature visible at both high and low magnifications while at eucentric height. Thereafter, an image is taken at each magnification and the microscopist centers the feature manually each time, and the offset is then stored by the automation software.
I0 refers to the intensity of electron illumination without a sample present [36]. This calibration is used as a reference for the illumination that would pass through an empty hole in a cryo-EM grid, thus allowing the automation software to identify holes with a suitable thickness of ice.
Usually before each imaging session, dark field and bright field images are acquired with the sample removed from the beam path. These are used for flat field corrections of subsequently acquired images, allowing for the correction of the difference in sensitivity for each pixel on the detector. In addition, the automation or camera software may exclude bad pixels and suppress spikes in the image due to stray X-rays [18,23].
Target acquisition
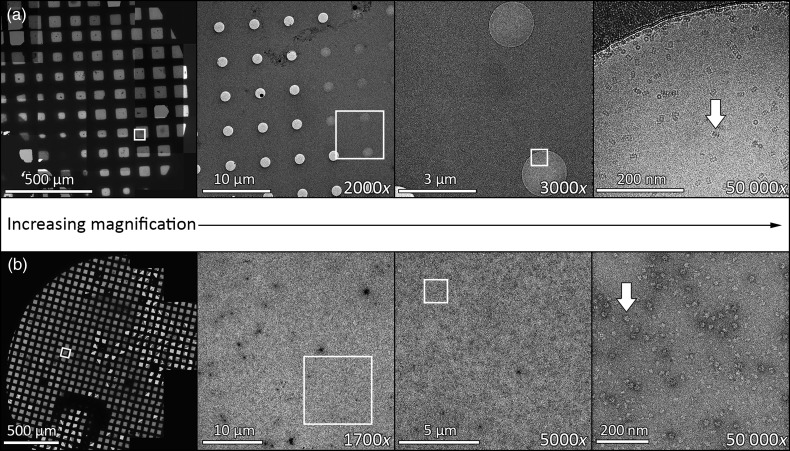
Target acquisition is a multi-scale process – starting from the lowest magnification, suitable regions to image are identified, and this process is repeated with sequentially higher magnification until the final desired magnification is reached (Fig. 4). Automation software streamlines this process.
Fig. 4.
MSI procedure employed in data collection software for both cryo T20S proteasome (a) and negative stain 50S ribosome (b) samples. The white boxes indicate the area where an image is acquired at subsequent higher magnification and the white arrows point to a single particle of the respective proteins.
The first step in the multi-scale imaging (MSI) process is to image the entire grid and ascertain the location and quality of potential squares available for data collection. Many automation software packages (EM-TOOLS, EPU, JADAS, Leginon, SerialEM, TOM2) provide the option of automatically collecting such a map of the entire grid, often referred to as an atlas.
With an atlas for reference, suitable squares for imaging are selected. For negative stain grids, good squares are unbroken and not too thickly covered by the stain so particles are visible (Fig. 5). For vitrified samples (Fig. 5), good squares need to be unbroken, not too thickly covered by ice (which will adversely affect contrast and result in excessive variation in Z-height of particles), and not so thin as to exclude the particles completely [47]. Automated picking of good squares for cryo-EM entails checking ice thickness by estimating the transmittance of electrons [18,20,23,26]. In order to eliminate holes containing uneven ice or some contamination, some programs such as AutoEMation and Leginon also offer the option to reject holes that have a measured transmittance beyond a certain mean or standard deviation threshold [18,23].
Fig. 5.
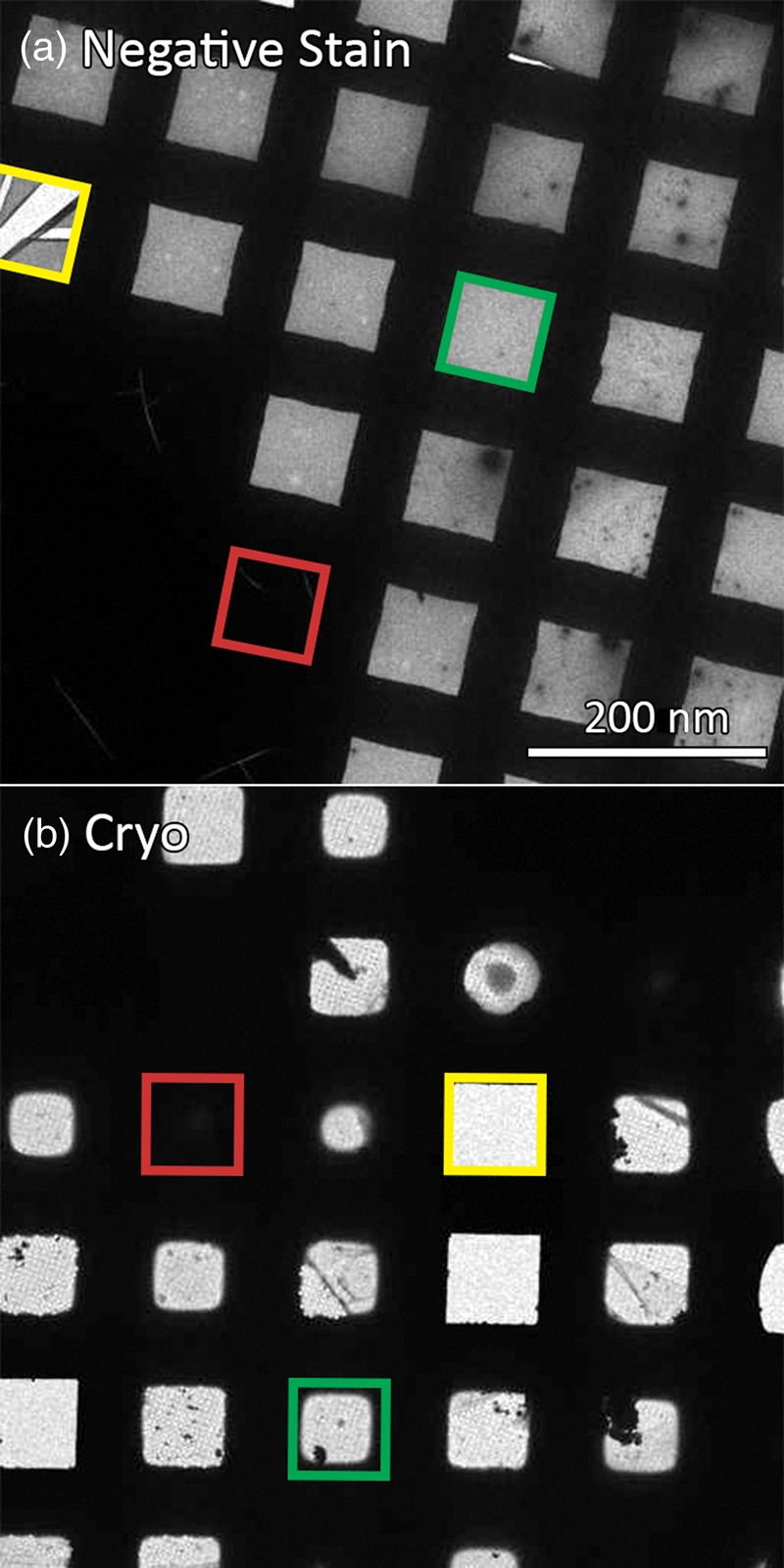
Variation of squares in a typical negative stain (a) and cryo-EM (b) grid. An example of a broken square (yellow), a square with too thick ice/stain (red) and a potentially good square (green) are shown in the respective colors above.
If the grid has a regular array of holes, the automation software can be programed to identify them in a variety of ways. AutoEMation requires a template created at the initialization stage and makes use of the correlation map between micrographs and the template to pinpoint the holes. JADAS similarly marks holes via a template-based hole matching algorithm. In Leginon, the correlation peaks from these are filtered with a loose lattice defined by the microscopist to account for slight distortion. In SerialEM, a grid of points is defined by the microscopist to indicate where the holes are. In UCSFImage4, the microscopist sets two vectors that define the location of neighboring holes in a regularly spaced lattice. The spacing and extent of the grid is also defined. EPU on the other hand requires users to define the size of a hole, and the spacing between two holes, before it automatically populates the array of holes. For all of these software packages, the automatically picked holes are also subject to ice thickness criteria defined by transmittance as mentioned in the previous paragraph.
Many automation packages also offer an option of selecting imaging areas manually in case the automated selection fails consistently on a challenging grid; indeed this is the only mode of operation for SAM [21]. While using this method of imaging area selection, the automation software may collect images at the higher magnifications breadth-first (i.e. collect all images at one magnification first before proceeding to the next higher magnification) or depth-first (i.e. image one area from low to high magnification, before moving to next area). The former method offers the advantage of requiring user interaction only at the beginning of the workflow. Software packages that provide breadth-first tree traversal at all points of the workflow include Leginon and SerialEM.
Once the imaging areas are selected, either automatically or manually, the software proceeds through a fixed workflow before exposing the target. First, eucentric height is adjusted, followed by autofocusing by one of the methods mentioned above. Once focus has been determined, a set defocus value is applied. The applied defocus value can be randomized across a range (Leginon, SerialEM, TOM2, JADAS), sequentially increased across a range (AutoEMation) or be selected from a fixed list of values (EPU, UCSFImage4). Once focus has been adjusted, an image (or movie series) of the target is acquired using a specified dose.
Centering the target for the final high magnification exposure imaging can be achieved by two methods: using the beam and image shifts or using the mechanical stage movement. In the beam and image shift method, deflectors bring multiple selected targets on the grid to the center of the viewing area for acquisition by the camera. The method is rapid and accurate but, depending on microscope optics, may introduce beam tilt at the specimen level. Alternatively, the stage can be physically moved to center each target. The former method has the advantage of speed as there is no need to either move the stage (which is slow and inaccurate) or wait for the stage to settle after movement, while the latter has the potential advantage of providing the highest resolution data as no coma is introduced by the beam tilt. Where a choice of targeting method is available (such as in EPU, Leginon and SerialEM), users should estimate the beam tilt induced by the former method either using a Zemlin tableau or observing the beam displacement in the diffraction mode, and then weigh the substantial extra time needed for targeting (up to a minute) against the resolution loss based on Eq. (2).
One problem that must be dealt with during data collection is the stage drift, which is especially pronounced immediately after physically shifting the stage. Typically, stage drift reduces over time as the stage settles. If exposures are acquired when the stage drift is too large, the focus may fail, the image will be blurred and high resolution information will be lost. In order to ameliorate this problem, software like SAM [21] pauses for 40 s after the goniometer movement and before collection to allow for the drift to stabilize. Other automation software such as AutoEM [23,24,25], EPU [36], Leginon [18] and SerialEM [15] provide the option of drift monitoring, whereby the drift of the stage is checked before exposure. If the drift is above a pre-defined threshold, the program waits for the stage to settle. If the drift does not fall below the threshold after a pre-defined period, the program skips the area and moves to the next target. The frequency of the drift check can be adjusted. AutoEM [24,25], AutoEMation [23] and Leginon [18] can intelligently select the order of target acquisition to minimize distance traveled, which may minimize stage drift. JADAS has the option for active compensation of drift by controlling a physical piezo device [26].
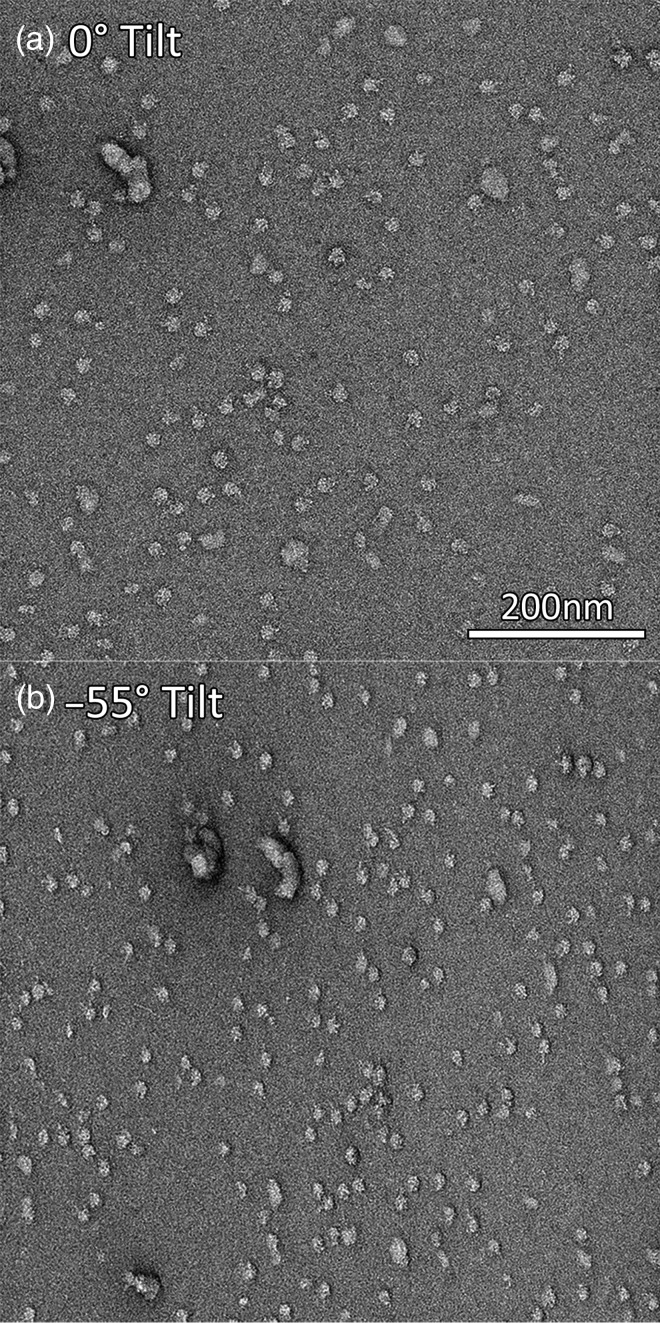
Tilt pair collection
Tilt data collection is an important tool for single particle EM for two main reasons: first, for random conical tilt (RCT) and orthogonal tilt (OT) reconstructions of single-particle samples and second for tilt pair validation [48] of the final 3D reconstruction model. A series of tilt series images separated by small angle increments have long been collected on a routine basis using EM tomographic software [12,15,49,50]. However, RCT tilt pair data collection requires correlation of image pairs with a much larger tilt difference (typically 45–60° as seen in Fig. 6) and consequently with a much larger distortion between the image pairs. Aligning these two images presents technical challenges, especially in the absence of supplementary fiducial markers like gold particles. EPU has a prototypic implementation of tilt pair collection, while Leginon [50] and SerialEM [15] have functional RCT and OT data collection capabilities.
Fig. 6.
Electron micrographs of negative stain samples at 0° tilt (a) and −55° tilt (b).
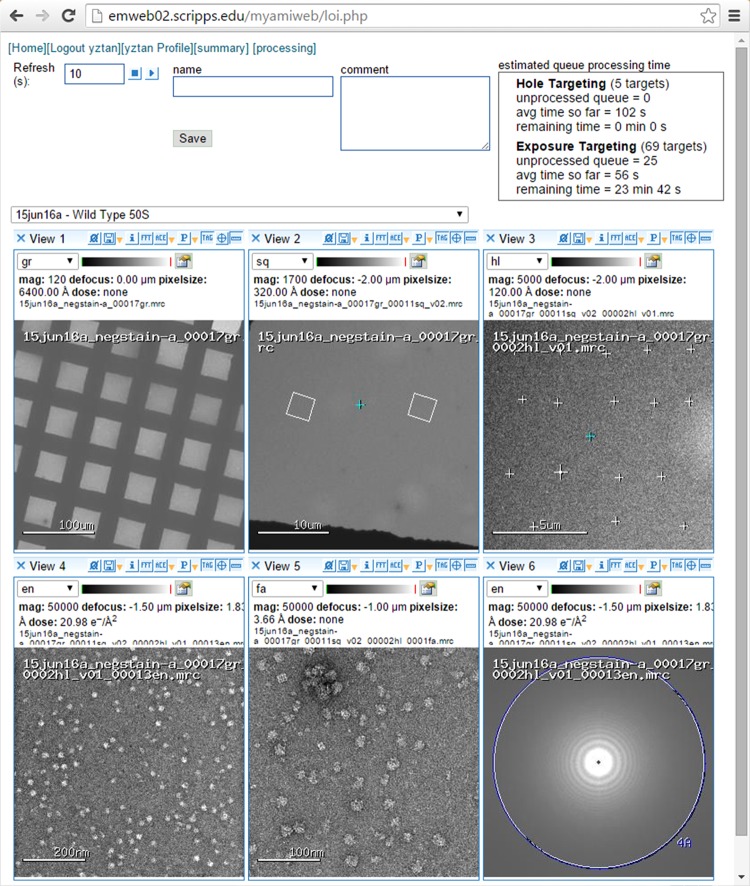
Monitoring of collection
Once the imaging targets are selected, automated data collection can begin and continue with no further user intervention, save for the need to refill the liquid nitrogen of the cryo-stage and cold-finger, if necessary. Sporadic user monitoring of collection is however good practice and sometimes necessary for challenging samples. Leginon [18] has a specialized monitoring interface called Leginon observer interface (LOI) as illustrated in Fig. 7. This interface allows users to see the most recently acquired images and get an update on the number of targets imaged and number of images remaining in the queue. It also provides an estimate of the time left for data collection. The LOI is a web-based interface, allowing for the convenient monitoring of the experiment remotely, either by a computer or smart phone. SerialEM [15] also has an interface for modifying the order of targets for tilt series collection.
Fig. 7.
Remote monitoring of data collection by LOI.
Automation can enable data collection to proceed unsupervised for long periods, up to several days [51]. One potential problem that could occur is the buildup of contamination on the grid as the vacuum in the column is not perfect. Cheng et al. [51] have shown that for a Tecnai F20, using a CCD in a well-controlled environment and good vacuum, the buildup occurs at a rate of 1.3 Å per hour – not a major limiting factor for data collection from one to several days. Experience also shows (Cheng, unpublished data) that for the latest generation of instruments like the Titan Krios, contamination is usually not a limitation unless there are extraneous circumstances like microscope leaks or malfunctions.
Hardware compatibility
Data collection software suites are just part of a chain of tools that electron microscopists use, and their degree of integration with other tools determines the ease with which the user can conduct their experiments (Table 1). Many automation packages were optimized for specific instrumentation available in the labs that developed the software – JAMES for example was designed around two JEOL microscopes [22]. Some of these packages also took advantage of microscopes and cameras that already had vendor programs for controlling them in place – SAM [21] accesses FEI's Tecnai User Interface and tools in Gatan's Digital Micrograph in its target selection and saving. The advantage of this design philosophy is that less effort is required to develop the automation package as ready-made parts are incorporated. The disadvantages are that the system has limited portability and might become outdated with equipment upgrades or as vendor software changes. For example, JAMES [22] used FASTEM on older JEOL microscopes for control. The latest series of JEOL microscopes no longer supports that software, resulting in the development of JADAS [26].
Other automation software use an object-orientated approach to deal with each hardware component, allowing for greater compatibility. In TOM2 [20], the hardware abstraction layer (HAL) deals with individual hardware components. Leginon [18] and SerialEM [15] have also been built with this approach, allowing them to be compatible with multiple brands of microscopes and detectors (Table 1).
Commercial software programs may have a different agenda from academic developments – most commercial programs are required only to support the company's equipment, but can be compatible with non-competing hardware. For example, EM-TOOLS supports FEI, Hitachi, JEOL, Philips and Zeiss microscopes, but only supports the company's TVIPS cameras.
Various automation packages also have the additional ability to adjust to physical add-ons present on microscopes. The energy filter, an important module for tomography, can be controlled by software like Leginon [18], SerialEM [15] and TOM2 [20]. Oster et al. [52] have used the Leginon framework to develop a positioning module for electrostatic physical phase plates on Zeiss microscopes.
Software compatibility
Downstream from data collection is data processing, and packages that are able to offer connection to their data processing counterparts are extremely helpful. JADAS is integrated with the EMEN2 [53] database, which doubles-up as a lab notebook that records experimental parameters and data collection variables. Leginon records almost all experimental parameters into a MySQL database during data collection [18] and is interfaced with Appion [54], which is a data processing pipeline wrapping a large variety of EM tools and software packages.
Some automation packages also allow users to program their own modules. SerialEM has a built-in macro ability [15] while Leginon is open-source [18] and modules have been extended by users [31,52,55].
Combining automated and manual procedures during data collection
Although automated data collection has many potential benefits, it is, after all, only one of many tools available to a microscopist – knowing when and where to use automated, semi-automated or manual data collection can help increase efficiency and productivity.
Screening of samples is a crucial first step in any single particle EM project – many parameters such as buffer conditions, protein concentration, type of negative stain, type of grid and freezing conditions need to be optimized. During this phase, fully automated data collection would not be the most efficient process for several reasons. First, fully automated data collection would necessitate correction of all aberrations and stabilization of drift before each image is collected. During the screening process, such aberrations can be well tolerated as a moderate resolution and moderate quality image is sufficient to determine parameters such as particle concentration or ice thickness. Implementing aberration correction and wait time for drift stabilization would add unnecessary delay to the screening process. Second, encouraging the microscopist to be closely involved in data collection during screening has the benefit of ensuring they observe grid conditions not usually recorded during automated data collection. For example, a microscopist operating manually might observe that many of the grid squares are broken and be able to pursue upstream problems – like whether the entire batch of grids was defective, or whether the grids should be more delicately handled during the preparation process.
Nevertheless, despite the advantages of manual input during screening, some level of automated data collection is still helpful. For example, when a good condition is found during screening, the microscopist can readily switch to an automated collection mode to quickly obtain a dataset for particle picking and initial 2D classification for further and more quantitative evaluation. Once sample and grid preparation conditions are optimized, the full power of automated data collection can then be unleashed.
One group of users who may benefit from a reduced repertoire of automated options is novices. Being presented with a complex data collection schema and multiple options may be acceptable, or even desirable, for an experienced microscopist, but may easily overwhelm a beginner. Often when new users start out in the field, they may only be interested in simply visualizing their protein and their immediate goal is usually not a subnanometer reconstruction. Leginon is one example of an automated data collection program that has the choice of hiding advanced options from novice users and providing users with imaging profiles that contain a reduced set of control nodes. As long as an experienced microscopist has already aligned the microscope, the new user can mainly focus on a straightforward data collection and not be overwhelmed by numerous menu options.
One observation from our own experience is that at a facility including a mixture of expert and occasional users, consistent use of an automated data collection package has the benefit of maintaining the microscopes in a stable condition for all. This is because the requirements for microscope alignments when using automated focusing and targeting are more stringent than those that might be acceptable to an inexperienced human operator. Thus, problems with microscope optics or alignment become immediately evident and quickly corrected. Finally, the use of a standard data collection software suite ensures that the microscope is used in a very consistent and routine manner, enhancing the overall stability of the microscope over time.
Future evolution of automated data collection
The main advantage of automated data collection is the ability to obtain reproducible results with less human effort. An additional benefit is that collection of very large datasets is now more feasible. Currently, collection of hundreds of thousands [56] or even millions of particles [4] has become the norm, enabled by automated data collection. This explosion in data collection rates allows the resolution limit to be pushed further [3,5–7] and also permits many more interesting biological questions to be addressed [57], such as the structural analysis of rare complexes and the understanding of minor conformational states [58,59].
As mentioned earlier, microscope alignment and aberration correction is an important aspect for achieving high resolution imaging [60,61]. Many of these aberration corrections have now been automated (focus) or semi-automated (astigmatism and coma), but more development is required to fully automate this process [62], given that the aberration-corrected state of the microscope has a limited lifetime [46].
One potential area for development of automation software in single particle EM is real-time feedback on data collection. Jonić et al. [63] have developed an algorithm to generate better power spectra for cryo-EM images, thereby allowing images to be discarded due to defects caused by astigmatism, drift, charging or even signal loss. Vargas et al. [64] added to this idea by creating FASTDEF, a method to very quickly estimate the contrast transfer function (CTF) of an image, taking between 11 and 20 s per image. Recently, Zhang [65] has pushed this limit further by developing GCTF, which is able to generate CTF estimates for micrographs within fractions of a second, thanks to GPU acceleration and a quick one-dimensional search plus two-dimensional refinement protocol. In the Appion pipeline, the cross-correlation coefficient (CC) between the CTF estimated for the image and a 1-D radially averaged power spectrum is automatically calculated [66] and was used by Campbell et al. [3] to select the optimal micrographs for further analysis. This kind of information could potentially be fed back into the automation software to improve focusing, astigmatism and to deduce areas of the grid which could potentially provide the highest spatial resolution images, while the grid is in the microscope. Similarly, a history of the CTF value and its calculated figure of merit, the quality and quantity of particles selected, as well as selected image target locations can be used to counteract situations of persistent unsupervised automation errors such as focusing and targeting failure. Automation software should also be capable of monitoring physical conditions, such as room temperature or vacuum status. This is especially important when using automation, as the user is no longer physically present and needs to be alerted to problems when they arise. Brunner and Resch [67] have developed software called MoniTEM that monitors the microscope and network status, and messages users if problems occur. Fellmann et al. [68] devised a temperature monitoring system tailored for EM facilities. The latest FEI Titan Krios microscopes are also provided with a health and monitoring system. Integration of these physical condition monitors with automation software could aid in reproducibility and reduce downtime during collection and may also be used to identify issues that limit image quality. Finally, the eventual aim of EM automation will be to make the entire process from grid preparation to 3D structure determination fully automated, in a similar fashion to the automated pipelines developed for protein crystallography [69,70]. Automated data processing is already becoming a reality with software that can acquire EM images and process them into electron density maps semi-automatically, such as Appion [54], EMAN2 [71] and Relion [72]. Work is also underway to develop automation in other parts of the EM pipeline [62], perhaps most urgently in the preparation of grids [33,73]. With commercial companies such as FEI, Gatan, JEOL and TVIPS also actively developing automation software at the same time as academic developers, we anticipate that automation methods will continue to mature rapidly and ultimately provide a high-throughput single particle EM pipeline readily available to the entire scientific community.
Conflict of interest
The authors declare that no competing interests exist.
Funding
This study was supported by National Institutes of Health, National Institute of General Medical Sciences [GM103310 to A.C., C.S.P. and B.C.] Simons Foundation [349247 to C.S.P. and B.C.]; Agency for Science, Technology and Research Singapore [to Y.Z.T.]. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
Many thanks to Joey Davis for the wild-type 50S ribosome sample and Christoph Wigge for the T20S proteasome sample and images. Thanks to everyone in the Automated Molecular Imaging Group, including David Veesler, Jana Albrecht and Melody Campbell for their help and feedback, and especially to Dmitry Lyumkis for invaluable comments on the draft. Thanks to Bill Anderson for technical help, especially with EPU. Thanks as well to experts who responded to our queries via email, namely Chris Booth, Dieter Typke, Ingo Daberkow, Jeffrey Lengyel, Nicolas Coudray, Voigt Andreas, and Xueming Li.
References
- 1.Kühlbrandt W. (2014) The resolution revolution. Science 343: 1443–1444. [DOI] [PubMed] [Google Scholar]
- 2.Bartesaghi A, Merk A, Banerjee S, Matthies D, Wu X, Milne J L, Subramaniam S (2015) 2.2 Å resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitor. Science 348: 1147–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell M G, Veesler D, Cheng A, Potter C S, Carragher B (2015) 2.8 Å resolution reconstruction of the Thermoplasma acidophilum 20S proteasome using cryo-electron microscopy. Elife 4: e06380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fischer N, Neumann P, Konevega A L, Bock L V, Ficner R, Rodnina M V, Stark H (2015) Structure of the E. coli ribosome-EF-Tu complex at <3 Å resolution by Cs-corrected cryo-EM. Nature 520: 567–570. [DOI] [PubMed] [Google Scholar]
- 5.Grant T, Grigorieff N (2015) Measuring the optimal exposure for single particle cryo-EM using a 2.6Å reconstruction of rotavirus VP6. Elife 4: e06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang J, Pentelute B L, Collier R J, Zhou Z H (2015) Atomic structure of anthrax protective antigen pore elucidates toxin translocation. Nature 521: 545–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.dos Passos D O, Lyumkis D (2015) CryoEM structure of a eukaryotic large 60S ribosomal subunit at 2.9Å resolution from 75,000 single particles. J. Struct. Biol. 192: 235–244. [DOI] [PubMed] [Google Scholar]
- 8.Spear J, Noble A J, Xie Q, Sousa D R, Chapman M S, Stagg S M (2015) The influence of frame alignment with dose compensation on the quality of single particle reconstructions. J. Struct. Biol. 192: 196–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu X, Jiang J, Sun J, Zhou Z H (2015) A putative ATPase mediates RNA transcription and capping in a dsRNA virus. eLife 4: e07901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bai X C, McMullan G, Scheres S H (2015) How cryo-EM is revolutionizing structural biology. Trends Biochem. Sci. 40: 49–57. [DOI] [PubMed] [Google Scholar]
- 11.Chen H, Hughes D D, Chan T, Sedat J W, Agard D A (1996) IVE (image visualization environment): a software platform for all three-dimensional microscopy applications. J. Struct. Biol. 116: 56–60. [DOI] [PubMed] [Google Scholar]
- 12.Dierksen K, Typke D, Hegerl R, Koster A J, Baumeister W (1992) Towards automatic electron tomography. Ultramicroscopy. 40: 71–87. [Google Scholar]
- 13.Dierksen K, Typke D, Hegerl R, Baumeister W (1993) Towards automatic electron tomography: II. Implementation of autofocus and low-dose procedures. Ultramicroscopy 49: 109–120. [Google Scholar]
- 14.Oostergetel G T, Keegstra W, Brisson A (1998) Automation of specimen selection and data acquisition for protein electron crystallography. Ultramicroscopy 74: 47–59. [Google Scholar]
- 15.Mastronarde D N. (2005) Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152: 36–51. [DOI] [PubMed] [Google Scholar]
- 16.Potter C S, Chu H, Frey B, Green C, Kisseberth N, Madden T J, Miller K L, Nahrstedt K, Pulokas J, Reilein A, Tcheng D, Weber D, Carragher B (1999) Leginon: a system for fully automated acquisition of 1000 electron micrographs a day. Ultramicroscopy 77: 153–161. [DOI] [PubMed] [Google Scholar]
- 17.Carragher B, Kisseberth N, Kriegman D, Milligan R A, Potter C S, Pulokas J, Reilein A (2000) Leginon: an automated system for acquisition of images from vitreous ice specimens. J. Struct. Biol. 132: 33–45. [DOI] [PubMed] [Google Scholar]
- 18.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter C S, Carragher B (2005) Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151: 41–60. [DOI] [PubMed] [Google Scholar]
- 19.Li X, Zheng S, Agard A D, Cheng Y (2015) Asynchronous data acquisition and on-the-fly analysis of dose fractionated cryoEM images by UCSFImage. J. Struct. Biol. 192: 174–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korinek A, Beck F, Baumeister W, Nickell S, Plitzko J M (2011) Computer controlled cryo-electron microscopy – TOM2 a software package for high-throughput applications. J. Struct. Biol. 175: 394–405. [DOI] [PubMed] [Google Scholar]
- 21.Shi J, Williams D R, Stewart P L (2008) A Script-Assisted Microscopy (SAM) package to improve data acquisition rates on FEI Tecnai electron microscopes equipped with Gatan CCD cameras. J. Struct. Biol. 164: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marsh M P, Chang J T, Booth C R, Liang N L, Schmid M F, Chiu W (2007) Modular software platform for low-dose electron microscopy and tomography. J. Microsc. 228: 384–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lei J, Frank J (2005) Automated acquisition of cryo-electron micrographs for single particle reconstruction on an FEI Tecnai electron microscope. J. Struct. Biol. 150: 69–80. [DOI] [PubMed] [Google Scholar]
- 24.Zhang P, Beatty A, Milne J L, Subramaniam S (2001) Automated data collection with a Tecnai 12 electron microscope: applications for molecular imaging by cryomicroscopy. J. Struct. Biol. 135: 251–261. [DOI] [PubMed] [Google Scholar]
- 25.Zhang P, Borgnia M J, Mooney P, Shi D, Pan M, O'Herron P, Mao A, Brogan D, Milne J L S, Subramaniam S (2003) Automated image acquisition and processing using a new generation of 4K × 4 K CCD cameras for cryo electron microscopic studies of macromolecular assemblies. J. Struct. Biol. 143: 135–144. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Nakamura N, Shimizu Y, Liang N, Liu X, Jakana J, Marsh M P, Booth C R, Shinkawa T, Nakata M, Chiu W (2009) JADAS: a customizable automated data acquisition system and its application to ice-embedded single particles. J. Struct. Biol. 165: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russo C J, Passmore L A (2014) Electron microscopy: ultrastable gold substrates for electron cryomicroscopy. Science 346: 1377–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ermantraut E, Wohlfart K, Tichelaar W (1998) Perforated support foils with pre-defined hole size, shape and arrangement. Ultramicroscopy 74: 75–81. [Google Scholar]
- 29.Grassucci R A, Taylor D J, Frank J (2007) Preparation of macromolecular complexes for cryo-electron microscopy. Nat. Protoc. 2: 3239–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quispe J, Damiano J, Mick S E, Nackashi D P, Fellmann D, Ajero T G, Carragher B, Potter C S (2007) An improved holey carbon film for cryo-electron microscopy. Microsc. Microanal. 13: 365–371. [DOI] [PubMed] [Google Scholar]
- 31.Nicholson W V, White H, Trinick J (2010) An approach to automated acquisition of cryoEM images from lacey carbon grids. J. Struct. Biol. 172: 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castro-Hartmann P, Heck G, Eltit J M, Fawcett P, Samso M (2013) The ArrayGrid: a methodology for applying multiple samples to a single TEM specimen grid. Ultramicroscopy. 135: 105–112. [DOI] [PubMed] [Google Scholar]
- 33.Mulligan S K, Speir J A, Razinkov I, Cheng A, Crum J, Jain T, Duggan E, Liu E, Nolan J, Carragher B, Potter C S (2015) Multiplexed TEM specimen preparation and analysis of plasmonic nanoparticles. Microsc. Microanal. 21: 1017–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bozzola J J, Russell L D (1981) Electron Microscopy: Principles and Techniques for Biologists. (Jones & Bartlett, USA: ). [Google Scholar]
- 35.Rodenburg J M. (2004) Understanding transmission electron microscope alignment: a tutorial. Microsc. Anal. 18: 9–11. [Google Scholar]
- 36.Janus M, Voigt A (2014) EPU Software User‘s Guide (FEI Company, Hillsboro: ). [Google Scholar]
- 37.Koster A J, de Ruijter W J (1992) Practical autoalignment of transmission electron microscopes. Ultramicroscopy 40: 89–107. [Google Scholar]
- 38.Mindell J A, Grigorieff N (2003) Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142: 334–347. [DOI] [PubMed] [Google Scholar]
- 39.Koster A J, Van Den Bos A, Van Der Mast K D (1987) An autofocus method for a TEM. Ultramicroscopy 21: 209–222. [Google Scholar]
- 40.Saxton W O, Smith D J, Erasmus S (1983) Procedures for focusing, stigmating and alignment in high resolution electron microscopy. J. Microsc. 130: 187–201. [Google Scholar]
- 41.Krivanek O L, Mooney P E (1993) Applications of slow-scan CCD cameras in transmission electron microscopy. Ultramicroscopy 49: 95–108. [Google Scholar]
- 42.Ishizuka K. (1994) Coma-free alignment of a high-resolution electron microscope with three-fold astigmatism. Ultramicroscopy 55: 407–418. [DOI] [PubMed] [Google Scholar]
- 43.Overwijk M H F, Bleeker A J, Thust A (1997) Correction of three-fold astigmatism for ultra-high-resolution TEM. Ultramicroscopy 67: 163–170. [Google Scholar]
- 44.Zemlin F, Weiss K, Schiske P, Kunath W, Herrmann K H (1978) Coma-free alignment of high resolution electron microscopes with the aid of optical diffractograms. Ultramicroscopy 3: 49–60. [Google Scholar]
- 45.Glaeser R M, Typke D, Tiemeijer P C, Pulokas J, Cheng A (2011) Precise beam-tilt alignment and collimation are required to minimize the phase error associated with coma in high-resolution cryo-EM. J. Struct. Biol. 174: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uhlemann S, Haider M (1998) Residual wave aberrations in the first spherical aberration corrected transmission electron microscope. Ultramicroscopy. 72: 109–119. [Google Scholar]
- 47.Agard D, Cheng Y, Glaeser R M, Subramaniam S (2014) Chapter two – single-particle cryo-electron microscopy (Cryo-EM): progress, challenges, and perspectives for further improvement. Adv. Imag. Elect. Phys. 185: 113–137. [Google Scholar]
- 48.Henderson R, Chen S, Chen J Z, Grigorieff N, Passmore L A, Ciccarelli L, Rubinstein J L, Crowther R A, Stewart P L, Rosenthal P B (2011) Tilt-pair analysis of images from a range of different specimens in single-particle electron cryomicroscopy. J. Mol. Biol. 413: 1028–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koster A J, Chen H, Sedat J W, Agard D A (1992) Automated microscopy for electron tomography. Ultramicroscopy 46: 207–227. [DOI] [PubMed] [Google Scholar]
- 50.Yoshioka C, Pulokas J, Fellmann D, Potter C S, Milligan R A, Carragher B (2007) Automation of random conical tilt and orthogonal tilt data collection using feature-based correlation. J. Struct. Biol. 159: 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng A, Fellmann D, Pulokas J, Potter C S, Carragher B (2006) Does contamination buildup limit throughput for automated cryoEM? J. Struct. Biol. 154: 303–311. [DOI] [PubMed] [Google Scholar]
- 52.Oster M, Frindt N, Schröder R R (2012) Automating phase plate assisted phase contrast microscopy with Leginon and Zeiss TEMs. EMC2012.
- 53.Rees I, Langley E, Chiu W, Ludtke S J (2013) EMEN2: an object oriented database and electronic lab notebook. Microsc. Microanal. 19: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lander G C, Stagg S M, Voss N R, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau P W, Lyumkis D, Potter C S, Carragher B (2009) Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166: 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lasala R, Coudray N, Abdine A, Zhang Z, Lopez-Redondo M, Kirshenbaum R, Alexopoulos J, Zolnai Z, Stokes D L, Ubarretxena-Belandia I (2015) Sparse and incomplete factorial matrices to screen membrane protein 2D crystallization. J. Struct. Biol. 189: 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Urnavicius L, Zhang K, Diamant A G, Motz C, Schlager M A, Yu M, Patel N A, Robinson C V, Carter A P (2015) The structure of the dynactin complex and its interaction with dynein. Science. 347: 1441–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Glaeser R M, Lee J, Typke D (2006) Advantages and objectives of high-throughput data collection in single-particle cryo-EM. Microsc Microanal. 12: 84–85. [Google Scholar]
- 58.Bai X, Rajendra E, Yang G, Shi Y, Scheres S (2015) Sampling the conformational space of the catalytic subunit of human γ-secretase. bioRxiv. 025890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lyumkis D, Oliveira dos Passos D, Tahara E B, Webb K, Bennett E J, Vinterbo S, Potter C S, Carragher B, Joazeiro C A (2014) Structural basis for translational surveillance by the large ribosomal subunit-associated protein quality control complex. Proc. Natl. Acad. Sci. USA. 111: 15981–15986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zemlin F. (1979) A practical procedure for alignment of a high resolution electron microscope. Ultramicroscopy 4: 241–245. [Google Scholar]
- 61.Ishizuka K, Shirota K (1996) Voltage-center and coma-free alignment for high-resolution electron microscopy. Ultramicroscopy 62: 9–13. [DOI] [PubMed] [Google Scholar]
- 62.Lyumkis D, Moeller A, Cheng A, Herold A, Hou E, Irving C, Jacovetty E L, Lau P, Mulder A M, Pulokas J, Quispe J D, Voss N R, Potter C S, Carragher B (2010) Chapter fourteen – automation in single-particle electron microscopy: connecting the pieces. Method. Enzymol. 483: 291–338. [DOI] [PubMed] [Google Scholar]
- 63.Jonić S, Sorzano C O S, Cottevieille M, Larquet E, Boisset N (2007) A novel method for improvement of visualization of power spectra for sorting cryo-electron micrographs and their local areas. J. Struct. Biol. 157: 156–167. [DOI] [PubMed] [Google Scholar]
- 64.Vargas J, Oton J, Marabini R, Jonic S, de la Rosa-Trevin J M, Carazo J M, Sorzano C O (2013) FASTDEF: fast defocus and astigmatism estimation for high-throughput transmission electron microscopy. J. Struct. Biol. 181: 136–148. [DOI] [PubMed] [Google Scholar]
- 65.Zhang K. (2015) Gctf: real-time CTF determination and correction. bioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sheth L K, Piotrowski A L, Voss N R (2015) Visualization and quality assessment of the contrast transfer function estimation. J. Struct. Biol. 192: 222–234. [DOI] [PubMed] [Google Scholar]
- 67.Brunner M J, Resch G P (2009) Automated monitoring to reduce electron microscope downtime. Ultramicroscopy 109: 1389–1392. [DOI] [PubMed] [Google Scholar]
- 68.Fellmann D, Bañez R, Carragher B, Potter C S (2006) Temperature monitoring of an EM environment. Microsc. Today 14: 24–29. [Google Scholar]
- 69.Tsai Y, McPhillips S E, Gonzalez A, McPhillips T M, Zinn D, Cohen A E, Feese M D, Bushnell D, Tiefenbrunn T, Stout C D, Ludaescher B, Hedman B, Hodgson K O, Soltis S M (2013) AutoDrug: fully automated macromolecular crystallography workflows for fragment-based drug discovery. Acta Crystallogr. D Biol. Crystallogr. 69: 796–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Elsliger M A, Deacon A M, Godzik A, Lesley S A, Wooley J, Wuthrich K, Wilson I A (2010) The JCSG high-throughput structural biology pipeline. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66: 1137–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tang G, Peng L, Baldwin P R, Mann D S, Jiang W, Rees I, Ludtke S J (2007) EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157: 38–46. [DOI] [PubMed] [Google Scholar]
- 72.Scheres S H. (2015) Semi-automated selection of cryo-EM particles in RELION-1.3. J. Struct. Biol. 189: 114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Coudray N, Hermann G, Caujolle-Bert D, Karathanou A, Erne-Brand F, Buessler J L, Daum P, Plitzko J M, Chami M, Mueller U, Kihl H, Urban J, Engel A, Rémigy H W (2011) Automated screening of 2D crystallization trials using transmission electron microscopy: a high-throughput tool-chain for sample preparation and microscopic analysis. J. Struct. Biol. 173: 365–374. [DOI] [PubMed] [Google Scholar]