Abstract
Predictive toxicology plays an important role in the assessment of toxicity of chemicals and the drug development process. While there are several well-established in vitro and in vivo assays that are suitable for predictive toxicology, recent advances in high-throughput analytical technologies and model systems are expected to have a major impact on the field of predictive toxicology. This commentary provides an overview of the state of the current science and a brief discussion on future perspectives for the field of predictive toxicology for human toxicity. Computational models for predictive toxicology, needs for further refinement and obstacles to expand computational models to include additional classes of chemical compounds are highlighted. Functional and comparative genomics approaches in predictive toxicology are discussed with an emphasis on successful utilization of recently developed model systems for high-throughput analysis. The advantages of three-dimensional model systems and stem cells and their use in predictive toxicology testing are also described.
Keywords: predictive toxicology, model systems, in silico approaches, genotoxicity
INTRODUCTION
A variety of well-established and validated in vitro and in vivo assays have been used successfully in predictive toxicology testing. The Ames test, mouse lymphoma assay, in vitro chromosome aberration test, in vitro micronucleus (MN) assay, and in vivo assays for toxicity, genotoxicity, carcinogenicity, and reproduction toxicity are some of the commonly used assays in predictive toxicology. While these assays have evolved and have been modified for specificity, sensitivity, and high-throughput capabilities, there are several inherent disadvantages that relate to predictive power, relevance to route of exposure, lack of complexity, and cell type specific effects that relate to mammalian tissues (in vitro assays), and species specific differences (rodent vs. human) that limit extrapolation to human toxicity.
Recent advances in high-throughput sequencing, computational biology, bioinformatics, and cell biology assay development have provided important avenues to predict the toxicity of chemicals. The newly developed approaches have the advantages relating to in silico approaches that circumvent laboratory assays for initial analysis, use of relevant target cells, ability to mimic physiological conditions, mechanism based predictive power, and systems biology-based analysis. Further development of predictive toxicology methodologies and the rational combination of such approaches is expected to be cost effective, accurate, and reduce time in the chemical product/drug development pipeline. This commentary reviews the recent developments and emerging areas in predictive toxicology that includes computational approaches, model systems, and genomic approaches and discusses the challenges in the field.
PREDICTIVE TOXICOLOGY—NEEDS AND CHALLENGES
The use of predictive methods in genetic toxicology has been around for over three decades. As a result, these methods have gained a great degree of success in predicting genotoxicity of a novel compound based on a surrogate set of information. Indeed, the Ames assay was developed as a surrogate to identify carcinogens, and thus, reduce the need for long and expensive in vivo experiments to determine carcinogenic risk from exposure to chemicals [Ames et al., 1975]. The late 1980s saw the rising development of computational methods to predict the Ames assay based on the chemical structure of a compound and since then numerous methods and models have been developed in this area. These computational approaches have now reached a stage of maturity and acceptance whereby they are being proposed for inclusion in an international regulatory guideline as part of a screening cascade to detect low-level genotoxic impurities in drug products. The ideal model is one that has both high specificity that correctly identifies true negatives with few false positives and high sensitivity, correctly identifying true positives with few false negatives. In reality, however, computational models tend to have either high sensitivity or high specificity but not both. This stems from an incomplete understanding of the effects of chemical substitutions on reactivity and/or a compound’s susceptibility to metabolic activation.
There are numerous ways in which computational methods can be applied in practice. At one end of the spectrum, computational models may be applied as a first pass filter to remove any obvious “bad actors” from a large set of molecules prior to further screening and investigation. In this scenario, computational models tend to focus on minimizing false positives and accepting the presence of false negatives, that is, compounds predicted to be negative that later turn out to be positive when tested, on the basis that these compounds will be detected prior to exposing human subjects to the chemical. At the opposite end of the spectrum, models may be used to detect potential hazards presented by exposure to low levels of a compound where the focus would be to minimize the number of false negatives and tolerate higher false-positive rates as this would minimize the health risks to humans from exposure.
APPLICATIONS TO GENOTOXICITY PREDICTION
Numerous computational models exist for predicting the mutagenic potential of a compound and the relative performances of these have been well-studied in the public literature [Naven et al., 2012]. Performance measures are, however, dataset-dependent and some models perform well for one set of compounds as they are well represented in the model’s training set but maybe less well when used on compound sets that are outside of the model’s experience, commonly referred to as its applicability domain. Some modeling approaches try to distinguish between active and inactive molecules by looking for features, either structural or chemical physical properties, that are more associated with activity than inactivity, therefore, a model may predict inactivity by inference from the absence of properties that cause activity. These approaches rely heavily on having enough examples in their training sets to establish with high confidence that absence of known activity is indeed synonymous with inactivity. This requires accurate definition of the applicability domain, a scientifically challenging and often subjective exercise, especially with systems that use expert-derived rules or structural alerts.
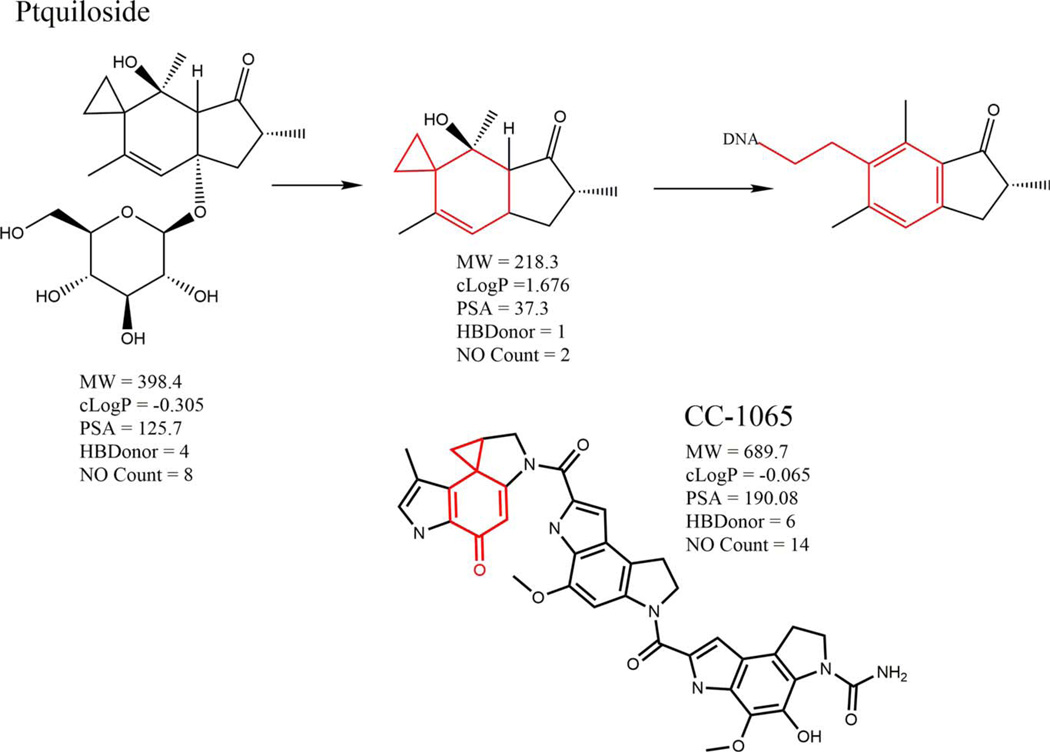
Extrapolation across compounds that contain a small functional DNA-reactive group is quite legitimate when the basic rules of chemical reactivity are upheld. These can span across compounds that would be considered quite dissimilar by conventional methods of measuring chemical similarity, for example, using knowledge of the mechanism behind the mutagenic activity of ptaquiloside to predict the mutagenic potential of CC-1065 and similar analogues (Fig. 1).
Fig. 1.
The mechanism of ptaquiloside genotoxicity and its extrapolation to CC-1065. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Similarly, extrapolating beyond the current descriptor space within a quantitative structure–activity relationship model may be perfectly legitimate in some cases, for example, where a particular descriptor has no bearing on the activity of the molecule. This, however, could be context-dependent and only be true for one class of compounds but not another. The problem of defining chemical similarity is especially challenging when the mechanism of action is chemical reactivity and, to date, no methods have been developed that adequately address and resolve this.
A further complication of defining applicability domains of models for mutagenicity prediction is that some compounds can cause mutagenicity through mechanisms other than direct DNA-reactivity (see below). This will undoubtedly introduce activity relationships with descriptors that are quite different from those where chemical reactivity is the primary mechanism of action. One solution to this problem lies in human intervention (expert opinion) when assessing the predictions from a computational platform. This human judgment is the only way to ensure that the model has either not over extrapolated from one compound to another (i.e., has not made a false-positive prediction) or has not been overly cautious and made a false-negative prediction.
A largely overlooked aspect of chemical entity/DNA binding is that which occurs in a noncovalent fashion, for example, by hydrogen bonding-associated intercalation or groove binding. Noncovalent binding has been the subject of a recent Special issue of mutation research [Snyder, 2007] within which the chemistry [Strekowski and Wilson, 2007] and the biology [Ferguson and Denny, 2007; Hoffmann et al., 2007; Nelson et al., 2007] of such interactions are discussed. In addition, the evidence in favor of a directed evolutionary process driving small molecule intercalation in a regulatory role [Hendry et al., 2007] and the involvement of topoisomerases in the genotoxicity of intercalating agents is presented. There seems little doubt that we are just beginning to appreciate the likely importance of noncovalent DNA interactions. This said, we do not have a lot of tools, either experimental or in silico that allow us to determine if a particular drug or chemical has the three-dimensional (3-D) structure and charge distribution that would facilitate intercalation or groove binding. Moreover, as much of the work in this field is still in its infancy, it has been understandably difficult for industry, in particular, to champion the development of methods to detect possible new classes of genotoxic compounds. Nevertheless, the absence of acceptable and compelling explanations for the genotoxicity (primarily clastogenicity) of over 80 marketed pharmaceuticals not possessing classically recognized structural alerts as first elucidated by Ashby et al. [1989], suggests that there must be other mechanisms of genotoxicity that we have yet to identify.
Without question, some of these discrepancies are explained by general or specific perturbances of DNA metabolic processes perhaps not even requiring direct chemical/DNA interaction, for example, inhibition of DNA polymerase, ribonucleotide reductase, histone processing, and DNA topoisomerase. But growing evidence suggests a central role for noncovalent DNA interaction. Early DNA docking studies coupled with cell-based bleomycin amplification assays in V79 and other cell systems [Snyder and Strekowski, 1999; Snyder et al., 2004] identified “atypically” structured molecules, that is, not classical fused tricyclics, that appeared to be clastogenic through intercalation into DNA. The use of catalytic inhibitors of topoisomerase II in these cell-based systems, further suggested that the clastogenicity was, in many cases, due to topoisomerase inhibition [Snyder, 2000; Snyder and Arnone, 2002]. More recently, over 1,350 launched drugs from the MDL drig data reports database from Accelrys (http://accelrys.com/products/databases/bioactivity/mddr.html) were docked with 10 different DNA sequences and two human topoisomerase II ATP binding sites using two independent programs, Autodock and Surflex, which model noncovalent DNA binding by charge and shape, respectively. These in silico studies confirmed the broad capability of a number of previously unexpected molecular scaffolds and chemical features to intercalate into DNA [Snyder et al., 2013]. Included among these scaffolds are first generation antihistamines, antiestrogens such as tamoxifen and benzimidazoles; structural features of apparent importance include N-dialkyl and N-ortho aryl ketones. The studies also revealed intriguing aspects of topoisomerase binding, specifically that of steroids and vitamin D analogues, which quite likely form the basis for the long appreciated but not understood class-specific genotoxicity of these drugs and chemicals.
Additional in silico and in vitro/in vivo studies must be directed toward confirmation and expansion of these findings and underlying mechanisms of genotoxicity. It is anticipated that noncovalent DNA interaction, particularly that resulting in perturbation of DNA topoisomerase activity, will be a major motif among nonclassical intercalating agents as it has been for classical fused tricyclics. Studies are underway evaluating the practical value of incorporation of these DNA docking scores into Leadscope Model Applier (http://www.leadscope.com). Extension of the proposed important structural features identified in the drug docking exercise to the much larger database would provide additional compelling reason to consider inclusion of noncovalent DNA binding data to in silico systems.
FUNCTIONAL AND COMPARATIVE GENOMICS: SYSTEMATIC SCREENING FOR GENES AND PATHWAYS INVOLVED IN HUMAN SUSCEPTIBILITY TO CHEMICAL EXPOSURES
Genetic variation likely underlies a significant proportion of the individual variation in human susceptibility to toxicants [Aldridge et al., 2003] by influencing processes such as metabolism, oxidative stress, DNA damage response, and repair. Characterization of this genetic variability, which is currently not well-understood, will enable more accurate chemical exposure risk assessment [Dorne, 2009; Guyton et al., 2009] and the identification of subgroups of individuals at greater risk of disease resulting from exposure to toxicants. The main approaches to identifying gene–environment interactions in toxicant-mediated disease are candidate gene association studies and genome-wide association studies (GWAS), which test for an association of a subset of genes or pathways or all genes, respectively, with a toxicant-related phenotypic outcome. These approaches are limited by insufficient knowledge of a priori genes (candidate approach) and a requirement for large exposed and control populations and associated expenses. Alternative approaches are, therefore, needed to identify human susceptibility genes.
Recently, innovative in vitro functional genomics testing systems have been developed, including a targeted approach assessing DNA damage and repair pathways in chicken DT40 B-lymphocytes [Yamazoe et al., 2004; Evans et al., 2010] and untargeted functional screening assays in yeast [North and Vulpe, 2010] and, more recently, in near-haploid human cells (KBM7) [Carette et al., 2009] and mouse haploid embryonic stem cells [ESC; Elling et al., 2011]. These systematic approaches use large-scale gene disruption or insertion techniques to generate null allele mutants in specific pathways, for example, DNA-repair-deficient clones in DT40 cells, or in all nonessential genes, for example, parallel deletion analysis [PDA] in yeast and insertional mutagenesis in human haploid cells. Genes that are essential for cell survival are easily identified by comparing cellular proliferation in wild type and mutant cells exposed to toxic chemicals. The yeast and haploid cell systems are high-throughput, cell survival bioassays. Comparative genomic and computational analyses are used to identify corresponding human susceptibility genes. A novel functional genomics approach has been developed that combines functional screening in yeast and human haploid cells with targeted follow up analyses in mammalian cells, to identify human susceptibility genes involved in arsenic, benzene, and formaldehyde toxicity, as detailed in the following section. It is envisioned that these types of approaches will enable the discovery of susceptibility genes and associated toxicity mechanisms, as an alternative or complement to candidate gene association studies and GWAS.
DNA-REPAIR PATHWAYS IN CHICKEN DT40 B-LYMPHOCYTES
The DT40 cell line, established from chicken B-lymphocytes and conditionally null for essential genes, was used to establish a unique higher eukaryotic system for comprehensive reverse genetic analysis of gene function [Yamazoe et al., 2004]. Isogenic mutant clones of all known DNA damage response pathways are available [Buerstedde and Takeda, 1991]. Its remarkably stable phenotype and karyotype, long S phase and lack of a G1/S checkpoint, make the DT40 system useful for testing DNA damaging agents. Ridpath et al. [2007] reported hypersensitivity to plasma levels of formaldehyde, a carcinogen and recently classified leukemogen, in DT40 mutants deficient in the BRCA/FANC and homologous recombination (HR) pathways. Recently, using human lymphoblast cell models of FANCD2 deficiency (PD20 cells) and sufficiency (PD20-D2 cells), it was shown that FANCD2 protein and the Fanconi anemia pathway are essential to protect human lymphoblastoid cells against formaldehyde toxicity [Ren et al., 2013].
Using a quantitative and high-throughput DT40 screening assay, Ji et al. [2009] screened genotoxic environmental contaminants and found that sodium meta-arsenite induced at least two types of damage: chromosomal breaks and UV photoproduct-like DNA lesions. Their assay measured cellular proliferation during exposure of cells to chemical compounds, offered enhanced sensitivity through the use of genetically defined and phenotypically characterized mutants defective in DNA repair pathways and minimization of false-negative outcomes through the use of DNA repair proficient wild type cells as a negative control [Ji et al., 2009; Evans et al., 2010]. Despite these assay improvements, the DT40 system is limited by the fact that it is a genetic and not a genomic screen.
YEAST PDA APPROACH
Saccharomyces cerevisiae is a good model for human and higher eukaryote disease and toxicity testing, as yeast has functional orthologues of many human disease genes. PDA is a powerful technique that allows for the quantitative analysis of the fitness of every deletion strain, representing all nonessential genes, simultaneously [North and Vulpe, 2010]. This approach has been used to identify genes involved in susceptibility to toxicity and prioritize Superfund chemicals including arsenic [Jo et al., 2009a,b] and benzene metabolites [North et al., 2011, 2014]. Through comparative genomic and computational analysis, several candidate human susceptibility genes were identified for potential analysis in single nucleotide polymorphism association studies [Lan et al., 2009]. Several such candidate genes were evaluated for their roles in human susceptibility to toxicity to specific compounds in exposed human cells using RNAi [Galvan et al., 2008; Jo et al., 2009a,b; Ren et al., 2009].
Arsenic
N6AMT1 (N-6 adenine-specific DNA methyltransferase 1, putative) and MYST1 (MYST histone acetyltransferase 1) are genes that play an important role in arsenic methylation and histone acetylation, respectively. Results from studies in yeast [Jo et al., 2009a,b] and validation in mammalian cells [Ren et al., 2011] suggested that N6AMT1 and MYST1 are candidate human susceptibility genes involved in arsenic toxicity. Studies are undergoing for investigating the biochemical roles of these two proteins in in vitro studies and examining the role of N6AMT1 polymorphisms in susceptibility to lung cancer associated with arsenic exposure in DNA samples from a case control study conducted in Chile. In fact, a very recent study has reported that N6AMT1 polymorphisms were associated with arsenic methylation in Andean women, independent of AS3MT [Harari et al., 2013].
Benzene
Yeast genes and pathways that modulate the cellular toxicity of three of the phenolic metabolites of benzene, hydroquinone (HQ), catechol and 1,2,4-benzenetriol have been identified [North et al., 2011]. Benzene metabolites generate oxidative and cytoskeletal stress and tolerance requires correct regulation of iron homeostasis and the vacuolar ATPase. It was found that IRA2, a yeast orthologue of the human tumor suppressor gene neurofibromin (NF1) and a modulator of Ras signaling, was required for tolerance to HQ [North et al., 2011]. A follow up study found that HQ toxicity is modulated by Ras signaling and that NF1 knockout mice exhibited both increased levels of DNA damage in erythroid progenitors and increased proliferation of CFU-GM progenitors on exposure to HQ, compared to wild type mice, which together could increase risk of myeloid disease [North et al., 2014].
Formaldehyde
de Graaf et al. [2009] used an alternative quantitative yeast screening methodology than the system employed by [North and Vulpe, 2010] to screen for genes involved in formaldehyde toxicity. They identified genes in the DNA repair and tolerance pathways that confer formaldehyde resistance under acute and chronic exposure conditions. Following chronic low-dose exposure, strains containing deletions in genes mediating HR showed the greatest sensitivity. Following acute formaldehyde exposure, repair and/or tolerance of DNA-protein crosslinks was reportedly mediated by nucleotide excision repair without the accumulation of double-strand breaks.
HAPLOID SCREENING SYSTEMS
Human Haploid Cell Line
Though genome-wide, the yeast screening approach is restricted to identifying mammalian genes with yeast homologues or orthologues. Recently, a human near-haploid cell (KBM7)-based insertional mutagenesis system that conceptually parallels the yeast system and screens directly for human susceptible genes was developed from a derivative of a chronic myeloid leukemia cell line [Carette et al., 2009]. Furthermore, a library of null mutants for most nonessential genes, that is, genes that are nonessential to the survival of the cells under normal conditions, (KBM7-Mu) was created. This system can identify genes whose insertional disruption allows cells to survive and proliferate in a selective environment. Using this approach, Carette et al. focused on host–pathogen biology and identified genes involved in susceptibility to influenza infection and diphtheria toxin and exotoxin A cytotoxicity.
Mouse Haploid ESC
A mouse ESC screening system is being established that will combine the power of a haploid genome with the pluripotency of ESC to identify susceptibility to toxicants in relevant cell types, such as hematopoietic stem and progenitor cells in the case of leukemia, at a genomic scale as described by Elling et al. [2011] and others [Leeb and Wutz, 2011; Leeb et al., 2012; Li et al., 2012; Yang et al., 2012; Zhang and Teng, 2013]. Elling et al. [2011] reported the generation of haploid mouse ESC lines, which carry 20 chromosomes, express stem cell markers, and develop into all germ layers in vitro and in vivo, from parthenogenetic embryos. They also developed a reversible mutagenesis protocol that allows saturated genetic recessive screens and results in homozygous alleles.
The selection of additional genes from the chicken, yeast and human haploid screening systems for validation in mammalian cell lines and knockout mice and in human population studies is continuing and is expanding the experimental models that are available. Additionally, a mouse ESC screening system is being established. Together, the screening approaches described here have the potential to identify human genes involved in susceptibility to toxicants and to elucidate mechanisms of toxicity.
3-D MODELS IN PREDICTIVE TESTING FOR HUMAN TOXICITY
Standard 2-D cell cultures are most commonly used in current in vitro toxicology testing strategies. Extrapolating from cell cultures to the in vivo situation can be challenging, however, since p53 deficient cell lines, cell lines lacking DNA repair and normal cells are often used, exposures in 2-D cultures are nonphysiological, natural cell–cell and cell–matrix interactions are missing, and exogenous metabolic activation is typically added that is not representative of normal metabolism. Chemicals that are positive in in vitro assays are typically tested further in in vivo assays which are costly, time consuming, and not consistent with the 3Rs: Refine, Reduce, Replace, to eliminate/reduce animal tests. For some chemicals, in vivo testing is not permitted, such as by the seventh Amendment to the EU Cosmetics Directive: ban on in vivo genotoxicity testing March 2009. Due to the limitations of 2-D cell cultures, and limits on in vivo assays, 3-D tissue constructs for toxicity testing are receiving increased interest.
3-D tissue constructs demonstrate “in vivo-like” behavior for key parameters like cell viability, proliferation, differentiation, morphology, metabolism gene, and protein expression, as well as metabolic function. 3-D tissue constructs can be very simple (e.g., epidermal skin models with only one cell type) to very complex (e.g., vascularized human liver model). Generally the more complex models will be more in vivo-like, with the downside being that they become more difficult to handle and more expensive.
3-D skin models have been successfully established for testing corrosivity, irritation, and genotoxicity. Skin models are particularly relevant since skin has the highest exposure to many chemicals, drugs, including cosmetics. 3-D skin models represent more relevant exposure/toxicokinetics/penetration versus in vitro cell cultures and more relevant metabolism compared to the S9 typically added to in vitro cell cultures. A MN assay in the 3-D human reconstructed EpiDerm™ skin model (reconstructed skin micronucleus) has been developed, which is currently part of a global prevalidation project sponsored by the Cosmetics European Association (formerly COLIPA), and the European Center for Validation of Alternative Methods (ECVAM) [Curren et al., 2006; Hu et al., 2009; Mun et al., 2009; Dahl et al., 2011]. Results to date demonstrate international interlaboratory and interexperimental reproducibility of the assay [Aardema et al., 2010] and its utility for chemicals that require metabolic activation [Aardema et al., 2013]. The 3-D skin MN assay is robust and has recently been established as a GLP assay at Bio-Reliance Corporation. The use of reconstructed skin [RS] models for genotoxicity assessment of dermally applied cosmetics was recently described [Pfuhler et al., 2010].
A 3-D skin Comet assay is also under development [Reus et al., 2013]. Genotoxicity results in other 3-D tissue models including EpiOcular, Cornea-FT, EpiVaginal, EpiVaginal-FT, Epi-Airway, and EpiOral, has been presented at various meetings by Yulia Kaluzhny, MatTek Corporation, Ashland, MA. In another skin model, Stratatech demonstrated UV-induction of thymidine dimers [Rasmussen et al., 2010]. Because of the clear benefits, 3-D tissue models will increasingly be used in toxicology testing to identify potential hazards in a variety of target tissues, to investigate toxicological mechanisms and to help extrapolate from in vitro cell cultures to results in animals and humans.
STEM CELLS IN PREDICTIVE TESTING FOR HUMAN TOXICITY
Toxicity during drug development represents between 30–40% of the drug attrition rate [Kola and Landis, 2004; Mahajan and Gupta, 2010]. Despite a better understanding of biological systems and improved technology, this rate does not appear to have declined over years. There may be many reasons for this. For example, the requirement to use animal models to predict a response in human patients does not usually correlate. The use of human cell lines as targets rather than primary cell targets can lead to false conclusions. Potential toxicity may also be missed due to lack of knowledge and/or understanding of the biology and physiology involved in how a system is regulated and/or failure to measure toxicity correctly due to incorrect assays.
Recently, there has been an effort to use new cell models to predict toxicity. These models incorporate the use of stem cells [Sison-Young et al., 2012]. There is a considerable amount of “hype” involving the use of stem cells. That “hype” concerns so-called nondefinitive stem cell systems that include ESC, induced pluripotent stem (iPS) cells and even primordial germ cells. These are “nondefinitive” systems because they have the potential to produce cells of virtually any of the primary, definitive cell systems. To do this, however, these nondefinitive stem cells must first produce stem cells of the “definitive” system that will allow for the production of functionally, mature end cells of that system. Definitive stem cell systems can be divided into continuously and partially proliferating systems. These systems are “definitive” because they are essentially “locked” into producing functionally, mature end cells that represent a specific organ or tissue. Partially proliferating stem cell systems include, but are not limited to, the production of hepatocytes, renal cells, lung cells, cardiomyocytes, insulin-producing cells, and neural/neuronal cells. Continuously proliferating stem cell systems include lympho-hematopoietic cells, gastrointestinal cells, cells of the reproductive organs, hair and skin cells, epithelial cells of the eye, and even mesenchymal stem/stromal cells (MSC). Technology has now been able to “unlock” cells from a primary, definitive system, return them to a pluripotential stem cell status (iPS), and then reprogram them into cells of a completely different lineage [Yamanaka, 2007; Aksoy and Stanton, 2013]. Such is the case, for example, with iPS-derived cardiomyocytes and hepatocytes.
Although considerable effort has gone into demonstrating that ES- or iPS-derived cells are similar to their fresh, primary counterparts, and that they can be produced in bulk with consistent quality, it is unlikely that these end cells will help reduce the drug attrition rate. However, the use of stem cells as the targets for toxicity focuses attention at the source and at a level in cellular development that has only been possible in a rare number of instances. Human ES cells as a target cell model for predicting, for example, developmental toxicity has yet to be conclusively demonstrated [Riebeling et al., 2012]. Conversely, use of primary, definitive hematopoietic stem cells has not only been shown to be highly predictive of the “global” response of the system as a whole and the effect of agents on individual cell lineages [Rich and Hall, 2005] but also a high concordance between in vitro and in vivo results can also be expected [Olaharski et al., 2009]. This system can be used as a model to illustrate how predictive in vitro stem cell toxicology could be used to reduce drug attrition rates during drug development.
Predictive in vitro stem cell toxicology is the ability to predict and identify potential in vivo toxic effects to a biological system before the system becomes partially or completely damaged causing a life-threatening situation. It follows that to achieve this predictive ability, not only must a thorough knowledge-base of the biological system be available, but a good understanding of the properties of the stem cells that give rise to this system must be known. Unlike nondefinitive stem cell systems, primary, definitive stem cells represent only a very small population (<0.01%) within the tissue or organ. The small number of definitive stem cells does not usually allow them to be morphologically identified. As a consequence, the ability of stem cells to proliferate is used as an identifying functional property. Indeed, stem cells exhibit the greatest proliferation ability and potential of all cells in the body. Within the stem cell compartment of any definitive system, the stem cells can be characterized by different degrees of proliferation potential. The greater the proliferation potential, the more primitive the stem cell population. It is these properties that were used to develop the hemotoxicity assays via luminescence output (HALO) predictive hemotoxicity platform, an in vitro, high throughput and fully validated screening assay system capable of detecting up to seven different hematopoietic stem cell populations from at least eight species (human, nonhuman primate, horse, pig, sheep, dog, rat, and mouse) [Rich and Hall, 2005].
All too often, however, the focus is not on stem cell toxicity, but on downstream cell population toxicity. For example, anticancer, anti-inflammatory, and antiviral drugs and many other agents cause neutropenia. However, just testing the response of primitive progenitor cells, in this case granulocyte-macrophage progenitor cells, often does not provide the whole toxicity story. Only a very small aspect of the total toxicity can be addressed when a single lineage is studied. This is because the majority of agents affect more than one cell lineage. When this occurs, the target is not the differentiation lineage, but the common stem cells that feed into the different lineages.; the effect may occur at different levels, but the net result can be quite different and more significant to that expected from toxicity to a single lineage [Harper and Rich, 2013]. This is a potentially more dangerous situation. The stem cells usually exhibit greater sensitivity to potential cytotoxic agents than any of their downstream-derived cells. Cytotoxicity at the stem cell level can result in the complete eradication of both the lymphopoietic and hematopoietic systems. This is why stem cell toxicity testing is so important; it not only provides a “global” view of how the system will respond to an insult and predict the effect on mature population(s), but also provides invaluable information regarding the status of the cells that are responsible for maintaining the system.
The predictive hemotoxicity platform as well as other in vitro toxicity platforms, such as those for immune cells (ImmunoGlo-Tox HT), MSC (MSCGlo-Tox HT), and other primary stem cells as well as ESC and iPS cells (XVPrime-Tox HT), all incorporate the ability to multiplex with other assay systems (e.g., flow cytometry, gene expression analysis, and mechanism of action) providing more information on potential toxicity using the same sample. However, they also have the capability to compare the response of primary normal tissue with primary diseased tissue or even matched samples, thereby providing the ability to estimate the therapeutic index of a drug. More recently, the HALO platform has been adapted to measure both cytotoxicity and genotoxicity, using a flow cytometric MN assay, for hematopoietic stem cells. This provides the first example of a genotoxicity assay specifically developed for a definitive stem cell system. Thus, the ability to measure both cytotoxicity and genotoxicity in stem cells not only increases the already highly predictive capability of the assay platform, but also provides the potential to predict tumorgenicity within a specific biological system.
CONCLUSIONS AND FUTURE PERSPECTIVES
The well-established and currently used in vitro and in vivo assays have been instrumental in our understanding of the toxicity of chemicals and their mode of action in humans. These assays have provided the foundation for the development and evolution of computational approaches that are capable of predicting the toxicity of chemicals and chemical classes. Further, refinement of computational methods to include a wider range of mode of action variables are needed to expand their utility in predicting toxicity of chemicals. Coincidentally, recent developments in high-throughput assays coupled with novel in vitro model systems and genomic analyses have provided us the opportunity of interrogating mechanisms of toxicity on a systems biology level. The possibility of analyzing pathways of toxicity at the molecular level has enormous implications for the future development of computational models that can predict toxicity based on measurable molecular and cellular readouts (gene expression, proliferation, and so forth) and extrapolating to chemical structures and classes. In addition to the advances in computational approaches and high-throughput assay protocols, in vitro 3-D model systems that are able to closely mimic physiological conditions and cell types have also been developed and are being validated. Such models provide an important avenue for surrogate analysis and prediction of toxicity of chemicals that can possibly replace the in vivo testing for at least a certain class of compounds (e.g., cosmetics). Complementing the aforementioned models and approaches, the successful development and use of definitive stem cell systems from humans (as well as other species) to predict cytotoxicity and genotoxicity is expected to provide additional insights on the effects of chemicals within the stem cell compartment.
In summary, a variety of emerging computational approaches, high-throughput screening assays and model systems are beginning to provide a comprehensive and clinically relevant data on toxicity. Further, refinement and integration of these approaches will be important step toward strengthening our ability to predict the toxicity of chemicals.
Acknowledgments
This commentary originates from the symposium on “Emerging Approaches in Predictive Toxicology” at the 43rd Annual Meeting of the Environmental Mutagenesis and Genomics Society, held in Bellevue, WA from September 8–12, 2012 that discussed the state of the current science, need and challenges in predictive toxicology testing.
Footnotes
AUTHOR CONTRIBUTIONS
LZ and CMM wrote the section on functional and comparative genomics; GN and RDS wrote the section on computational methods; INR wrote the section on stem cells; MJA, RS and SP wrote the section on 3-D models and VS wrote the abstract, introduction and conclusions besides managing and editing the commentary.
REFERENCES
- Aardema MJ, Barnett BC, Khambatta Z, Reisinger K, Ouedraogo-Arras G, Faquet B, Ginestet AC, Mun GC, Dahl EL, Hewitt NJ, et al. International prevalidation studies of the EpiDerm 3D human reconstructed skin micronucleus [RSMN] assay: transferability and reproducibility. Mutat Res. 2010;701:123–131. doi: 10.1016/j.mrgentox.2010.05.017. [DOI] [PubMed] [Google Scholar]
- Aardema MJ, Barnett BB, Mun GC, Dahl EL, Curren RD, Hewitt NJ, Pfuhler S. Evaluation of chemicals requiring metabolic activation in the EpiDerm 3D human reconstructed skin micronucleus [RSMN] assay. Mutat Res. 2013;750:40–49. doi: 10.1016/j.mrgentox.2012.08.009. [DOI] [PubMed] [Google Scholar]
- Aksoy I, Stanton LW. Pluripotency-regulating networks provide basis for reprogramming. Curr Mol Med. 2013;13:695–706. doi: 10.2174/1566524011313050002. [DOI] [PubMed] [Google Scholar]
- Aldridge JE, Gibbons JA, Flaherty MM, Kreider ML, Romano JA, Levin ED. Heterogeneity of toxicant response: sources of human variability. Toxicol Sci. 2003;76:3–20. doi: 10.1093/toxsci/kfg204. [DOI] [PubMed] [Google Scholar]
- Ames BN, McCann J, Yamasaki E. Methods for detecting carcinogens and mutagens with the Salmonella/mammalian-microsome mutagenicity test. Mutat Res. 1975;31:347–364. doi: 10.1016/0165-1161(75)90046-1. [DOI] [PubMed] [Google Scholar]
- Ashby J, Tennant RW, Zeiger E, Stasiewicz S. Classification according to chemical structure, mutagenicity to Salmonella and level of carcinogenicity of a further 42 chemicals tested for carcinogenicity by the U.S. National Toxicology Program. Mutat Res. 1989;223:73–103. doi: 10.1016/0165-1218(89)90037-2. [DOI] [PubMed] [Google Scholar]
- Buerstedde JM, Takeda S. Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell. 1991;67:179–188. doi: 10.1016/0092-8674(91)90581-i. [DOI] [PubMed] [Google Scholar]
- Carette JE, Guimaraes CP, Varadarajan M, Park AS, Wuethrich I, Godarova A, Kotecki M, Cochran BH, Spooner E, Ploegh HL, et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 2009;326:1231–1235. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- Curren RD, Mun GC, Gibson DP, Aardema MJ. Development of a method for assessing micronucleus induction in a 3D human skin model [EpiDerm] Mutat Res. 2006;607:192–204. doi: 10.1016/j.mrgentox.2006.04.016. [DOI] [PubMed] [Google Scholar]
- Dahl EL, Curren R, Barnett BC, Khambatta Z, Reisinger K, Ouedraogo G, Faquet B, Ginestet AC, Mun G, Hewitt NJ, et al. The reconstructed skin micronucleus assay [RSMN] in EpiDerm: detailed protocol and harmonized scoring atlas. Mutat Res. 2011;720:42–52. doi: 10.1016/j.mrgentox.2010.12.001. [DOI] [PubMed] [Google Scholar]
- de Graaf B, Clore A, McCullough AK. Cellular pathways for DNA repair and damage tolerance of formaldehyde-induced DNA-protein crosslinks. DNA Repair [Amst] 2009;8:1207–1214. doi: 10.1016/j.dnarep.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorne JL. Metabolism, variability and risk assessment. Toxicology. 2009;268:156–164. doi: 10.1016/j.tox.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Elling U, Taubenschmid J, Wirnsberger G, O’Malley R, Demers SP, Vanhaelen Q, Shukalyuk AI, Schmauss G, Schramek D, Schnuetgen F, et al. Forward and reverse genetics through derivation of haploid mouse embryonic stem cells. Cell Stem Cell. 2011;9:563–574. doi: 10.1016/j.stem.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans TJ, Yamamoto KN, Hirota K, Takeda S. Mutant cells defective in DNA repair pathways provide a sensitive high-throughput assay for genotoxicity. DNA Repair [Amst] 2010;9:1292–1298. doi: 10.1016/j.dnarep.2010.09.017. [DOI] [PubMed] [Google Scholar]
- Ferguson LR, Denny WA. Genotoxicity of non-covalent interactions: DNA intercalators. Mutat Res. 2007;623:14–23. doi: 10.1016/j.mrfmmm.2007.03.014. [DOI] [PubMed] [Google Scholar]
- Galvan N, Lim S, Zmugg S, Smith MT, Zhang L. Depletion of WRN enhances DNA damage in HeLa cells exposed to the benzene metabolite, hydroquinone. Mutat Res. 2008;649:54–61. doi: 10.1016/j.mrgentox.2007.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyton KZ, Kyle AD, Aubrecht J, Cogliano VJ, Eastmond DA, Jackson M, Keshava N, Sandy MS, Sonawane B, Zhang L, et al. Improving prediction of chemical carcinogenicity by considering multiple mechanisms and applying toxicogenomic approaches. Mutat Res. 2009;681:230–240. doi: 10.1016/j.mrrev.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Harari F, Engstrom K, Concha G, Colque G, Vahter M, Broberg K. N-6-adenine-specific DNA methyltransferase 1 [N6AMT1] polymorphisms and arsenic methylation in Andean women. Environ Health Perspect. 2013;121:797–803. doi: 10.1289/ehp.1206003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper H, Rich IN. Stem cell predictive hemotoxicology. In: Alimoghaddam K, editor. Stem Cell Biology in Normal Life and Diseases. Croatia: InTech, Rijeka; 2013. [Google Scholar]
- Hendry LB, Mahesh VB, Bransome ED, Jr, Ewing DE. Small molecule intercalation with double stranded DNA: implications for normal gene regulation and for predicting the biological efficacy and genotoxicity of drugs and other chemicals. Mutat Res. 2007;623:53–71. doi: 10.1016/j.mrfmmm.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Hoffmann GR, Gessner GS, Hughes JF, Ronan MV, Sylvia KE, Willett CJ. Modulation of the genotoxicity of bleomycin by amines through noncovalent DNA interactions and alteration of physiological conditions in yeast. Mutat Res. 2007;623:41–52. doi: 10.1016/j.mrfmmm.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Hu T, Kaluzhny Y, Mun GC, Barnett B, Karetsky V, Wilt N, Klausner M, Curren RD, Aardema MJ. Intralaboratory and interlaboratory evaluation of the EpiDerm 3D human reconstructed skin micronucleus [RSMN] assay. Mutat Res. 2009;673:100–108. doi: 10.1016/j.mrgentox.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Ji K, Kogame T, Choi K, Wang X, Lee J, Taniguchi Y, Takeda S. A novel approach using DNA-repair-deficient chicken DT40 cell lines for screening and characterizing the genotoxicity of environmental contaminants. Environ Health Perspect. 2009;117:1737–1744. doi: 10.1289/ehp.0900842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo WJ, Loguinov A, Wintz H, Chang M, Smith AH, Kalman D, Zhang L, Smith MT, Vulpe CD. Comparative functional genomic analysis identifies distinct and overlapping sets of genes required for resistance to monomethylarsonous acid [MMAIII] and arsenite [AsIII] in yeast. Toxicol Sci. 2009a;111:424–436. doi: 10.1093/toxsci/kfp162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo WJ, Ren X, Chu F, Aleshin M, Wintz H, Burlingame A, Smith MT, Vulpe CD, Zhang L. Acetylated H4K16 by MYST1 protects UROtsa cells from arsenic toxicity and is decreased following chronic arsenic exposure. Toxicol Appl Pharmacol. 2009b;241:294–302. doi: 10.1016/j.taap.2009.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov. 2004;3:711–715. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- Lan Q, Zhang L, Shen M, Jo WJ, Vermeulen R, Li G, Vulpe C, Lim S, Ren X, Rappaport SM, et al. Large-scale evaluation of candidate genes identifies associations between DNA repair and genomic maintenance and development of benzene hematotoxicity. Carcinogenesis. 2009;30:50–58. doi: 10.1093/carcin/bgn249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeb M, Wutz A. Derivation of haploid embryonic stem cells from mouse embryos. Nature. 2011;479:131–134. doi: 10.1038/nature10448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeb M, Walker R, Mansfield B, Nichols J, Smith A, Wutz A. Germline potential of parthenogenetic haploid mouse embryonic stem cells. Development. 2012;139:3301–3305. doi: 10.1242/dev.083675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Shuai L, Wan H, Dong M, Wang M, Sang L, Feng C, Luo GZ, Li T, Li X, et al. Androgenetic haploid embryonic stem cells produce live transgenic mice. Nature. 2012;490:407–411. doi: 10.1038/nature11435. [DOI] [PubMed] [Google Scholar]
- Mahajan R, Gupta K. Food and drug administration’s critical path initiative and innovations in drug development paradigm: Challenges, progress, and controversies. J Pharm Bioallied Sci. 2010;2:307–313. doi: 10.4103/0975-7406.72130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun GC, Aardema MJ, Hu T, Barnett B, Kaluzhny Y, Klausner M, Karetsky V, Dahl EL, Curren RD. Further development of the EpiDerm 3D reconstructed human skin micronucleus [RSMN] assay. Mutat Res. 2009;673:92–99. doi: 10.1016/j.mrgentox.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Naven RT, Greene N, Williams RV. Latest advances in computational genotoxicity prediction. Expert Opin Drug Metab Toxicol. 2012;8:1579–1587. doi: 10.1517/17425255.2012.724059. [DOI] [PubMed] [Google Scholar]
- Nelson SM, Ferguson LR, Denny WA. Non-covalent ligand/DNA interactions: minor groove binding agents. Mutat Res. 2007;623:24–40. doi: 10.1016/j.mrfmmm.2007.03.012. [DOI] [PubMed] [Google Scholar]
- North M, Vulpe CD. Functional toxicogenomics: mechanism-centered toxicology. Int J Mol Sci. 2010;11:4796–4813. doi: 10.3390/ijms11124796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North M, Tandon VJ, Thomas R, Loguinov A, Gerlovina I, Hubbard AE, Zhang L, Smith MT, Vulpe CD. Genome-wide functional profiling reveals genes required for tolerance to benzene metabolites in yeast. PLoS One. 2011;6:e24205. doi: 10.1371/journal.pone.0024205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North M, Shuga J, Fromowitz M, Shannon KM, Zhang L, Smith MT, Vulpe CD. Modulation of ras signaling alters the toxicity of hydroquinone, a benzene metabolite and component of cigarette smoke. BMC Cancer. 2014;14:6. doi: 10.1186/1471-2407-14-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaharski AJ, Uppal H, Cooper M, Platz S, Zabka TS, Kolaja KL. In vitro to in vivo concordance of a high throughput assay of bone marrow toxicity across a diverse set of drug candidates. Toxicol Lett. 2009;188:98–103. doi: 10.1016/j.toxlet.2009.03.012. [DOI] [PubMed] [Google Scholar]
- Pfuhler S, Kirst A, Aardema M, Banduhn N, Goebel C, Araki D, Costabel-Farkas M, Dufour E, Fautz R, Harvey J, et al. A tiered approach to the use of alternatives to animal testing for the safety assessment of cosmetics: genotoxicity. A COLIPA analysis. Regul Toxicol Pharmacol. 2010;57:315–324. doi: 10.1016/j.yrtph.2010.03.012. [DOI] [PubMed] [Google Scholar]
- Rasmussen C, Gratz K, Liebel F, Southall M, Garay M, Bhattacharyya S, Simon N, Vander Zanden M, Van Winkle K, Pirnstill J, et al. The StrataTest[R] human skin model, a consistent in vitro alternative for toxicological testing. Toxicol In Vitro. 2010;24:2021–2029. doi: 10.1016/j.tiv.2010.07.027. [DOI] [PubMed] [Google Scholar]
- Ren X, Lim S, Smith MT, Zhang L. Werner syndrome protein, WRN, protects cells from DNA damage induced by the benzene metabolite hydroquinone. Toxicol Sci. 2009;107:367–375. doi: 10.1093/toxsci/kfn254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Aleshin M, Jo WJ, Dills R, Kalman DA, Vulpe CD, Smith MT, Zhang L. Involvement of N-6 adenine-specific DNA methyltransferase 1 [N6AMT1] in arsenic biomethylation and its role in arsenic-induced toxicity. Environ Health Perspect. 2011;119:771–777. doi: 10.1289/ehp.1002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Ji Z, McHale CM, Yuh J, Bersonda J, Tang M, Smith MT, Zhang L. The impact of FANCD2 deficiency on formaldehyde-induced toxicity in human lymphoblastoid cell lines. Arch Toxicol. 2013;87:189–196. doi: 10.1007/s00204-012-0911-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reus AA, Reisinger K, Downs TR, Carr CJ, Zeller A, Corvi R, Krul CA, Pfuhler S. Comet assay in reconstructed 3D human epidermal skin models—investigation of intra- and inter-laboratory reproducibility with coded chemicals. Mutagenesis. 2013;28:709–720. doi: 10.1093/mutage/get051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich IN, Hall KM. Validation and development of a predictive paradigm for hemotoxicology using a multifunctional bioluminescence colony-forming proliferation assay. Toxicol Sci. 2005;87:427–441. doi: 10.1093/toxsci/kfi250. [DOI] [PubMed] [Google Scholar]
- Ridpath JR, Nakamura A, Tano K, Luke AM, Sonoda E, Arakawa H, Buerstedde JM, Gillespie DA, Sale JE, Yamazoe M, et al. Cells deficient in the FANC/BRCA pathway are hypersensitive to plasma levels of formaldehyde. Cancer Res. 2007;67:11117–11122. doi: 10.1158/0008-5472.CAN-07-3028. [DOI] [PubMed] [Google Scholar]
- Riebeling C, Hayess K, Peters AK, Steemans M, Spielmann H, Luch A, Seiler AE. Assaying embryotoxicity in the test tube: current limitations of the embryonic stem cell test [EST] challenging its applicability domain. Crit Rev Toxicol. 2012;42:443–464. doi: 10.3109/10408444.2012.674483. [DOI] [PubMed] [Google Scholar]
- Sison-Young RL, Kia R, Heslop J, Kelly L, Rowe C, Cross MJ, Kitteringham NR, Hanley N, Park BK, Goldring CE. Human pluripotent stem cells for modeling toxicity. Adv Pharmacol. 2012;63:207–256. doi: 10.1016/B978-0-12-398339-8.00006-9. [DOI] [PubMed] [Google Scholar]
- Snyder RD. Use of catalytic topoisomerase II inhibitors to probe mechanisms of chemical-induced clastogenicity in Chinese hamster V79 cells. Environ Mol Mutagen. 2000;35:13–21. [PubMed] [Google Scholar]
- Snyder RD. Assessment of atypical DNA intercalating agents in biological and in silico systems. Mutat Res. 2007;623:72–82. doi: 10.1016/j.mrfmmm.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Snyder RD, Arnone MR. Putative identification of functional interactions between DNA intercalating agents and topoisomerase II using the V79 in vitro micronucleus assay. Mutat Res. 2002;503:21–35. doi: 10.1016/s0027-5107(02)00028-3. [DOI] [PubMed] [Google Scholar]
- Snyder RD, Strekowski L. Enhancement of bleomycin-induced micronucleus formation in V79 cells as a rapid and sensitive screen for non-covalent DNA-binding compounds. Mutat Res. 1999;444:181–192. doi: 10.1016/s1383-5718(99)00080-7. [DOI] [PubMed] [Google Scholar]
- Snyder RD, Ewing DE, Hendry LB. Evaluation of DNA intercalation potential of pharmaceuticals and other chemicals by cell-based and three-dimensional computational approaches. Environ Mol Mutagen. 2004;44:163–173. doi: 10.1002/em.20036. [DOI] [PubMed] [Google Scholar]
- Snyder RD, Holt PA, Maguire JM, Trent JO. Prediction of noncovalent Drug/DNA interaction using computational docking models: studies with over 1350 launched drugs. Environ Mol Mutagen. 2013;54:668–681. doi: 10.1002/em.21796. [DOI] [PubMed] [Google Scholar]
- Strekowski L, Wilson B. Noncovalent interactions with DNA: an overview. Mutat Res. 2007;623:3–13. doi: 10.1016/j.mrfmmm.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Yamanaka S. Strategies and new developments in the generation of patient-specific pluripotent stem cells. Cell Stem Cell. 2007;1:39–49. doi: 10.1016/j.stem.2007.05.012. [DOI] [PubMed] [Google Scholar]
- Yamazoe M, Sonoda E, Hochegger H, Takeda S. Reverse genetic studies of the DNA damage response in the chicken B lymphocyte line DT40. DNA Repair [Amst] 2004;3:1175–1185. doi: 10.1016/j.dnarep.2004.03.039. [DOI] [PubMed] [Google Scholar]
- Yang H, Shi L, Wang BA, Liang D, Zhong C, Liu W, Nie Y, Liu J, Zhao J, Gao X, et al. Generation of genetically modified mice by oocyte injection of androgenetic haploid embryonic stem cells. Cell. 2012;149:605–617. doi: 10.1016/j.cell.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Zhang S, Teng Y. Powering mammalian genetic screens with mouse haploid embryonic stem cells. Mutat Res. 2013;741–742:44–50. doi: 10.1016/j.mrfmmm.2013.01.002. [DOI] [PubMed] [Google Scholar]