Abstract
Herein, we have reviewed the role of glutamate, the major excitatory neurotransmitter in the brain, in a number of neurochemical, -physiological, and -behavioral processes mediating the development of alcohol dependence. The findings discussed include results from both preclinical as well as neuroimaging and postmortem clinical studies. Expression levels for a number of glutamate-associated genes and/or proteins are modulated by alcohol abuse and dependence. These changes in expression include metabotropic receptors and ionotropic receptor subunits as well as different glutamate transporters. Moreover, these changes in gene expression parallel the pharmacologic manipulation of these same receptors and transporters. Some of these gene expression changes may have predated alcohol abuse and dependence because a number of glutamate-associated polymorphisms are related to a genetic predisposition to develop alcohol dependence. Other glutamate-associated polymorphisms are linked to age at the onset of alcohol-dependence and initial level of response/sensitivity to alcohol. Finally, findings of innate and/or ethanol-induced glutamate-associated gene expression differences/changes observed in a genetic animal model of alcoholism, the P rat, are summarized. Overall, the existing literature indicates that changes in glutamate receptors, transporters, enzymes, and scaffolding proteins are crucial for the development of alcohol dependence and there is a substantial genetic component to these effects. This indicates that continued research into the genetic underpinnings of these glutamate-associated effects will provide important novel molecular targets for treating alcohol abuse and dependence.
1. ALCOHOLISM AND GENETICS
Over half of adult Americans have a family history of alcoholism or alcohol (ethanol) abuse and one in four Americans have had an alcohol use disorder (AUD) during their lifetime, costing the US economy an estimated $225 billion per year (Research Society on Alcoholism).1,2 AUDs continue to be ranked as the third leading cause of preventable death by the Centers for Disease Control and Prevention.3 Moreover, research supports a causal relationship between AUDs and at least 50 different medical conditions.4–6
The well-documented familial incidence of alcoholism indicates that heredity contributes significantly to a predisposition toward, and the development of, AUDs.7–9 In fact, family history positive (FHP) individuals are at a three- to sevenfold increased risk to develop alcoholism, relative to those who are family history negative (FHN).10 This genetic proposal has been confirmed by multiple gene studies (e.g., the Collaborative Study On the Genetics of Alcoholism (COGA), the Study of Addiction: Genes and Environment (SAGE), and the European research project on risk-taking behavior in teenagers (IMAGEN)) examining the association between diagnostic criteria for alcohol dependence/addiction and the presence of single nucleotide polymorphisms (SNPs) in alcohol-dependent individuals.11–17
Similar to humans, different lines of heterogeneous stock rats display a wide-range of ethanol-consumption levels.18 The very early work by Williams and coworkers,19 as well as Mardones and coworkers,20 resulted in the hypothesis that ethanol intake in rodents is also under substantial genetic control. From their early work and that of four other international sites, bidirectional selective breeding has resulted in at least five high alcohol–consuming versus their respective low alcohol–consuming rat lines.21 One of these selectively bred high alcohol–consuming rat lines is the alcohol-preferring P rat. Essentially, starting from a closed colony of Wistar rats, the highest alcohol drinkers were mated together and the lowest alcohol drinkers were mated together, which resulted in the P and NP lines, respectively.21,22 The selectively bred alcohol-preferring P rat meets all of the criteria put forth for a valid animal model23,24 of alcoholism.21,22 It also meets the more recently proposed criteria including relapse-like,25 binge-like,21,22,26,27 and early/adolescent-onset of excessive drinking, which results in blood alcohol concentrations (BACs) greater than the threshold (80 mg%) of NIAAA’s28 definition for binge drinking.26,29 By the nature of selective breeding, P rats represent multigenerational FHP subjects and their counterparts, the selectively bred alcohol-nonpreferring NP rats, represent multigenerational-FHN individuals. Regarding the point about representing FHP individuals, the P rat has some neurochemical, neuro-physiological, and behavioral characteristics similar to those seen clinically in FHP individuals.21,26,30
Some of these neurochemical characteristics of P rats involve the gluta-matergic system and these findings parallel clinical findings in both FHP individuals and chronic alcoholics. The present minireview will focus on the central glutamatergic system and its role in alcohol dependence. The basic structure and function of the glutamatergic system will be outlined; as it pertains to its activity in the brain, its receptors, and transporters as well as the excitatory synapse. A summary of the existing literature on how ethanol affects the structure and function of the central glutamatergic system will be presented in the context of both preclinical and clinical research. A synopsis of the genetic influence on the development and/or expression of alcohol dependence will be described. An overview of the current knowledge regarding how the ethanol–glutamate interaction affects gene and/or protein expression will be presented, again from both preclinical and clinical perspectives. This will be followed by a compilation of our findings with the P rat as it pertains to innate differences in gene and/or protein expression, relative to its NP counterpart, as well as ethanol’s modulation of gene and/or protein expression in subregions of the mesocorticolimbic reward circuitry. This minireview will then be concluded with some closing thoughts on some limitations observed in the existing literature.
2. CENTRAL GLUTAMATERGIC ACTIVITY
The amino acid glutamate is the primary excitatory neurotransmitter in the central nervous system (CNS). Therefore, it is not surprising that glutamate receptors are located throughout the brain (see Fig. 1 for glutamatergic projections). In addition, given the ubiquitous distribution of glutamate and its receptors, its function as the primary excitatory neurotransmitter is crucial for many processes, especially those mediating neuroplasticity, learning and memory.31–33 Glutamate interacts with both metabotropic mGlu1–mGlu8 (Grm1–Grm8 = mGluR1–mGluR8) and ionotropic receptors, which include those that can bind to N-methyl-D-aspartate (NMDA) (subunits: GluN1 (Grin1 = NR1); GluN2a–GluN2d (Grin2a–Grin2d = Nr2a–Nr2d); GluN3a–GluN3b (Grin3a–Grin3b = NR3a–NR3b), those that can bind to α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA) subunits: GluA1–GluA4 (Gria1–Gria4 = GluR1–GluR4) or kainite subunits GluK1–GluK4 (Grik1–Grik4 = GluR5–GluR7 + KA1–KA2); these nomenclatures reflect IUPHAR, HUGO, and “old” symbols respectively.34–36 Due to glutamate’s role in excitotoxicity, extracellular glutamate must be tightly controlled.34,37,38 This is accomplished, for the most part, by multiple glutamate transporters.34 The human excitatory amino acid transporter 2 (EAAT2) and its rodent analog glutamate transporter 1 (GLT1) appear to be the main transporters performing this function centrally.34,37
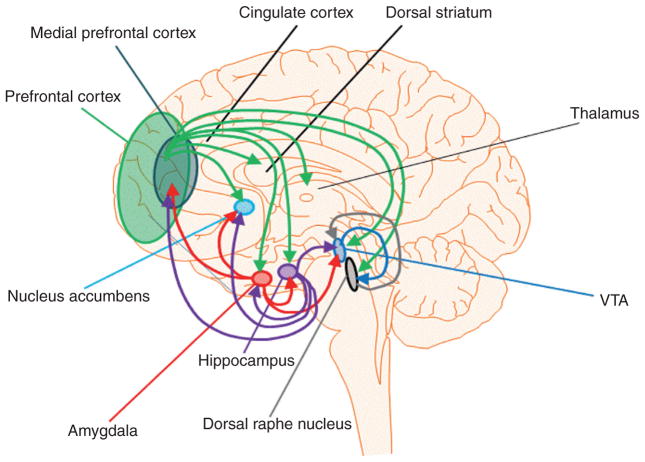
Figure 1.
A simplified diagram depicting glutamatergic projections of the mesocorticolimbic, extended amygdala, and brainstem reward neurocircuitry. The online version includes color coding of these projections; with green representing the PFC/mPFC, red representing the amygdala, purple representing the hippocampus, blue representing the VTA, and gray representing the DRN.
2.1 Metabotropic Glutamate Receptors (Fig. 2)
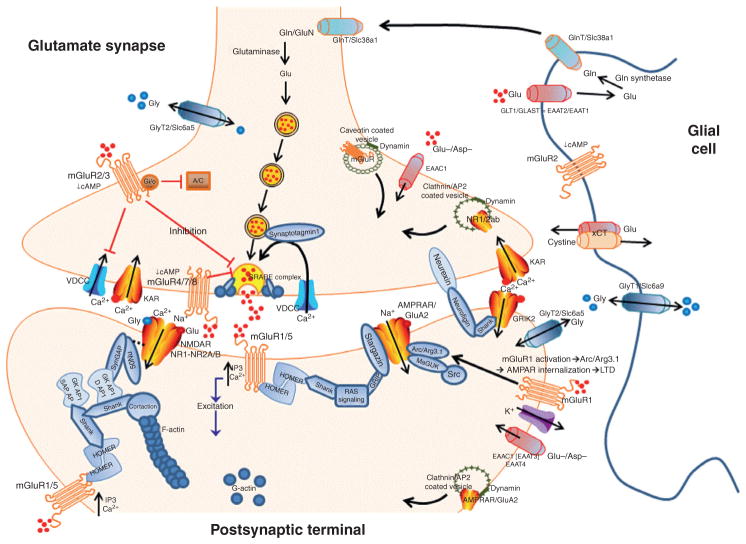
Figure 2.
A simplified diagram of a prototypic glutamatergic synapse in the brain. The figure depicts some of the intra-, inter-, and extracellular activities of glutamate-associated plasticity. Abbreviations: AC, adenylate cyclase; AP2, adaptor protein 2; Arc, activity-regulated cytoskeleton-association protein; Asp, aspartate; GKAP/DAP1, disks large associated protein 1 (part of PSD); Gln or GluN, glutamine; GlnT, glutamine transporter; GLT, glutamate transporter; Glu, glutamate; Gly, glycine; GlyT, glycine transporter; LTD, long-term depression; nNOS, neuronal nitric oxide synthase; Src, a tyrosine kinase; VDCC, voltage dependent calcium channel. Other abbreviations are in the text and tables.
The mGlu receptors are G-protein-coupled protein receptors (GPCRs) located at the neuronal synapse, extrasynaptically as well as on glial cells (Fig. 2). These receptors are divided into three groups. Group I mGluRs (mGlu1 and mGlu5) are predominately postsynaptic and engage in slow excitatory neurotransmission; Group II mGluRs (mGlu2 and mGlu3) are predominately presynaptic, with some postsynaptic and glial localization (Fig. 2) where they engage in slow inhibitory neurotransmission; and Group III mGluRs (mGlu4, mGlu6, mGlu7, and mGlu8) are generally restricted to the presynaptic terminal and, like Group II mGluRs, engage in slow inhibitory neurotransmission. The mesocorticolimbic and associated reward circuitry express high levels of mGlu1, mGlu2, mGlu3, and/or mGlu5, notably in the Acb, caudate nucleus, cortex, lateral septum, dorsal striatum, amygdala, and hippocampus.39–43
2.2 Ionotropic Glutamate Receptors (Fig. 2)
Ionotropic glutamate receptors are ligand-gated ion channels involved in fast excitatory transmission in the CNS. There are several types of glutamate ionotropic receptors including AMPA, kainate, and NMDA-receptor subunits. Most ionotropic glutamate receptors are located postsynaptically, although some are located presynaptically and on glia cells as well (Fig. 2). Similar to the mGlu receptors, they are found throughout the brain including cortical regions, hippocampus, amygdala, basal ganglia, midbrain, hind-brain, and brainstem nuclei.44–49 A recent functional addition to the iono-tropic glutamate receptors are the delta1 and delta2 subunits symbolized by Glud1 and Glud2. In the past these subunits were considered orphans and not functional but more recent findings indicate that they are indeed functional, by modulating LTD and prepulse inhibition of the acoustic startle response (sensorimotor gating), and their localization is not restricted to the cerebellum but includes cortical and limbic regions as well.50–54
2.3 Glutamate Transporters and Carriers (Fig. 2)
As discussed earlier, excessive glutamate in the synapse (e.g., the addiction-related hyperglutamatergic state) can lead to excitotoxicity and neuronal death. Thus, glutamate uptake/transport from the synapse and perisynapse is required to prevent the plasticity associated with addiction to become excitotoxicity.34,38 There are several transporters that regulate extracellular glutamate levels including the EAAT1 (glutamate aspartate transporter: GLAST or Slc1a3), EAAT2 (GLT1), EAAT3 (excitatory amino acid carrier 1: EAAC1 or Slc1a1), EAAT4 (Slc1a6), and EAAT5; where EAAT indicates the human homolog.55–57 There are also intracellular vesicular glutamate transporters (VGLUT1–3 of the Slc17 family of genes) that mediate the uptake of glutamate into synaptic vesicles. Intracellular glutamate carriers, other than the vGLUTs, include the Slc25a family of genes. Since the NMDAR NR1 subunit contains the glycine-binding site, it is important to recognize the role of bidirectional glycine transporters (GlyT1 which is primarily glial and GlyT2 which is primarily neuronal) at the excitatory synapse (GlyTs are represented by gene families Slc6a and Slc7a). Given the increased interest in N-acetyl-cysteine’s role in mental health, it is equally important to recognize the cystine–glutamate exchanger’s (xCT = Slc7a11) role in reversing damage induced by excessive extracellular glutamate. The xCT, generally located on glial cells, takes up cystine and releases glutamate molecules. Cystine is then converted to cysteine, which is used to synthesize glutathione and other proteins. Glutathione is a key antioxidant and important in reversing neuronal damage induced by excitotoxicity and oxidative stress.58
2.4 Glutamate Synthesis and Metabolism (Fig. 2)
Another biologic method for preventing excessive glutamate in the synapse is glutamate’s synthesis from glutamine intracellularly.34 Glutamine can be transported into and out of the synapse without inducing neurotoxicity. The metabolism of glutamate to glutamine occurs primarily in glial cells via the enzyme glutamine synthetase. Glutamine is then transported (glutamine transporter (GlnT), which is common to both glia and neurons and encoded by the Slc38a gene family) out of the glial cell. In turn, glutamine is transported out of the perisynapse and into the excitatory presynaptic compartment. Glutamine is then converted to glutamate by glutaminase and transported to the synaptic membrane, or shunted into GABA synthesis via glutamate decar-boxylase (GAD). Glutamate is also metabolized by glutamate dehydrogenase–yielding alpha-ketoglutarate, which enters the TCA/Krebs cycle.
2.5 Glutamate and the Postsynaptic Density (Fig. 2)
The postsynaptic density (PSD) is a cytoskeletal specialization that is located beneath the postsynaptic membrane and directly contiguous with the presyn-aptic “active zone” of excitatory synapses. Glutamate receptors, synaptic proteins, scaffolding proteins, kinases, and other downstream-signaling proteins are located within this PSD. There are several scaffolding proteins within the PSD including membrane-associated guanylate kinases (MAGUKs), Shanks, and Homers.59–61 Scaffolding proteins can be defined as molecules binding at least two other signaling proteins together. These scaffolding proteins are crucial for synaptic plasticity (e.g., learning and memory) by (1) acting as platforms where signaling molecules can assemble; (2) localizing signaling molecules at specific intracellular sites; (3) coordinating positive and negative feedback signals to modify intra- and extra-cellular signaling pathways; as well as (4) protecting these signaling pathways from inactivation, generally by preventing and/or disrupting phosphorylation.62–64 In general, scaffolding proteins act as signaling proteins for neuromodulator receptors and anchor these receptors (e.g., glutamate receptors) to the synaptic membrane.65 Given their role in receptor anchoring, dysregulation of these scaffolding proteins can lead to a number of neurological diseases.66
MAGUKs are expressed widely throughout the central nervous system. They are the scaffolding proteins closest to the surface of the postsynaptic membrane and they contain multiple PDZ domains.67 The PSD-95 protein is one of the most studied MAGUK scaffolding proteins and is involved in postsynaptic stability as well as excitatory receptor insertion.68 PSD-95 binds to numerous proteins associated with AMPAR and NMDAR complexes. Schnell et al.69 found that interaction between PSD-95 and the AMPA receptor-interacting protein, Stargazin, determines the density of AMPARs at the synapse and through this interaction can regulate synaptic maturation.70 The PSD-95 anchors NMDARs to the postsynaptic membrane and it acts as a signaling scaffold mediating the activation of neuronal nitric oxide synthase (nNOS) by calcium–calmodulin activity following, for example, entry of calcium through NMDAR channels.71 Differences between AMPA and NMDA receptors influence on synaptic events is due, in part, to their respective cytosolic C-terminal binding sites to the PSD-95/ discs large/zona occludens-1 (PDZ) domain-containing scaffolding proteins.72–74 The PDZ domain’s function is to regulate protein–protein interactions by binding to the C-terminus of each respective target protein; thus, highlighting its crucial role in neuroplasticity, dendritic growth, and dendritic arborization.68,75
The ionotropic NMDA and AMPA receptors are primarily concentrated in the PSD but there are distinct differences in synaptic regulation of these receptors.76,77 Additionally, NMDARs initiate synaptic plasticity by interacting with other components of the PSD.78–82 Similarly, AMPARs are involved in rapid synaptic transmission and these receptors cycle on and off in a manner tightly controlled by neuronal activity/plasticity in the PSD.78–82 The recycling (i.e., insertion and removal) of these AMPARs at the synapse modulates synaptic efficiency and thus, like NMDARs, influence learning and memory.76,77
Considerable evidence indicates the crucial role of Shank and Homer proteins in neuroplasticity, as well as alcohol and drug dependence.83–89 Shank and the Homer scaffolding proteins are located deeper within the PSD than the MAGUKs (i.e., toward the cytoplasm).70 Three genes encode for Shank proteins (Shank1, Shank2, and Shank3) and Shank scaffolding proteins bind to neuroligins and neurexins, as well as NMDAR complexes in the PSD.70 Shanks are capable of binding other scaffolding molecules within the PSD (e.g., Homers) and thus are capable of linking mGlu receptors, as well as AMPA and kainate ionotropic receptors to NMDARs. Therefore, scaffolding proteins present in the PSD mediate a number of plasticity-associated events through reorganization of PSD-associated proteins, regulation of membrane protein trafficking and activity, as well as the maintenance of associated epithelial cell polarity and morphology.90–92 Moreover, the connection between Shank proteins and neuroligins/neur-exins indicates that the PSD may bridge with the “active zone” of the presynaptic terminal. The Homer family of scaffolding proteins is encoded by 3 genes (Homer1, Homer2, and Homer3). Homer scaffolding proteins interact with the C-terminus of Group I mGluRs, bind to Shank/PSD-95/ NMDA-receptor complexes, and can also interact with a number of downstream effectors of mGlu1/5 including: IP3 receptors, diacylglycerol lipase-2, and PI3K enhancer (PIKE). Homer proteins are best characterized for their role in regulating mGlu1/5 trafficking, PSD localization, and signaling of mGlu1/5 and NMDA receptors, but are also critical in the regulation of actin and dendritic morphology.93 Furthermore, through their ability to associate with Shank, Homers facilitate cross-talk between mGlu1/5 and NMDA receptors and the integration of their calcium-dependent intracel-lular events underpinning synaptic plasticity.94
3. CENTRAL GLUTAMATE ACTIVITY AND ALCOHOL DEPENDENCE
3.1 Alcohol’s Effects on Glutamate Activity and Extracellular Levels
Substantial preclinical evidence indicates that glutamatergic activity mediates natural as well as alcohol- and drug-associated reward through direct and indirect interactions with other neurotransmitter/neuromodulatory systems within the mesocorticolimbic, extended amygdala, and associated reward neurocircuitry (Fig. 1).95–102 A key hypothesis that has received considerable attention postulates that the mesocorticolimbic and extended amygdala reward circuits, in the presence of alcohol, lose homeostasis between excitatory and inhibitory transmission and revert to a hyperglutamatergic/hyper-excitatory state resulting in the development and expression of alcohol/drug dependence.37,98,102–107
In general, ethanol consumption and/or exposure to low or moderate doses of ethanol elevate glutamatergic transmission and/or extracellular levels of glutamate in the nucleus accumbens (Acb)108; Acb shell (AcbSh)88,89,109–111; basolateral amygdala (BLA)112; cortex113; Hippocampus114,115; ventral teg-mental area (VTA)116; and posterior VTA (pVTA).110,117 It has also been shown that genetics influence ethanol-induced increases in extracellular glutamate within the Acb and/or PFC, such that rats with a predisposition for higher ethanol intake (P and Lewis rats) display greater elevations in glutamate relative to rats with a predisposition for lower ethanol intake (NP and F344 rats).118,119 Interestingly, similar relations have been observed between alcohol-preferring versus nonpreferring inbred strains of mice.86,120 In addition, these elevations in glutamatergic activity can be conditioned to the environment in which the animal had access to ethanol, with glutamatergic increases seen in the Acb core (AcbCo) or basolateral Amyg (BLA).103,121,122
A recent comprehensive preclinical study provides an excellent example of glutamate’s role in the development and expression of alcohol dependence.123 Griffin and coworkers123 evaluated whether free-choice ethanol access would increase Acb extracellular glutamate levels and found that dependence-induced (chronic intermittent access via ethanol-vapor chambers) ethanol-drinking doubled Acb glutamate levels over those seen in nondependent mice. Moreover, these authors reported that this doubling of glutamate was observed a week later indicating that these increases in glutamate activity were not dependent upon ethanol withdrawal itself. Consistent with prior neuropharmacologic results in studies of low versus high alcohol–consuming inbred mice,120 Griffin etal.123 showed that pharmacologic elevation of glutamate in the Acb, with a pan-glutamate-reuptake inhibitor (Threo-beta-benzyloxyaspartate, TBOA), increased the ethanol intake of nondependent mice to the levels observed in dependent animals, with TBOA also increasing ethanol intake further in the dependent mice. Also consistent with the results of Kapasova and Szumlinski,120 when the mGlu2/3 autoreceptor agonist LY379268 was microinjected into the Acb to lower glutamate levels, ethanol intake was decreased in both the dependent and nondependent mice.123 These latter results parallel earlier work indicating that manipulations of extracellular glutamate, including mGlu2/3 activation, actively regulate ethanol intake in multiple animal models of alcoholism (discussed later in the chapter).
Finally, evidence for altered glutamate neurotransmission within the previously mentioned brain regions, as well as the anterior cingulate cortex (ACC), has been reported in clinical studies of alcohol-dependent individuals as well.124–131 For example, a proton magnetic resonance spectroscopy (MRS) study examining the role of hippocampal glutamate in major depression and risky alcohol drinking revealed that elevated glutamate levels in the hippocampus were directly associated with both the presence of major depression and self-reported risky drinking.132 These authors noted that the major depression and risky drinking group did not differ from the control group in age-of-first alcohol use, Alcohol Use Disorders Identification Test (AUDIT) survey scores or smoking behavior; but, this group did have significantly more FHP individuals (approximately six-fold) indicating a possible confound. Another recent MRS study provided support for differences in glutamate activity of FHP versus FHN individuals.133 These authors reported that glutamate/glutamine ratios increased significantly between adolescence and emerging adulthood in FHN, but not FHP, individuals. This suggests that having a familial history of AUDs may genetically predispose an individual for abnormal developmental changes in glutamatergic neurotransmission across periadolescence.133
3.2 Metabotropic Glutamate Receptors and Alcohol
It has been shown that ethanol-binge drinking by mice upregulates mGlu1/ 5-Homer2 signaling in several mesocorticolimbic structures including the Acb83,84 and the central amygdala (CeA)85 and activation of either or both of these Group I mGluRs is required for mice to manifest binge-drinking behavior.83–85
Several studies have examined the effects of mGlu5 antagonists on operant ethanol self-administration behaviors in P rats as well. Systemic administration of the mGlu5 antagonist 2-methyl-6-(phenylethynyl)pyridine (MPEP) can reduce operant ethanol self-administration,134 reduce ethanol breakpoint without affecting sucrose breakpoint or locomotor activity,135 and block the repeated alcohol-deprivation effect (ADE).134 The effects of MPEP in rats were extended to mice by demonstrating that this mGlu5 antagonist interfered with the acquisition and maintenance of ethanol drinking by C57BL/6J mice as well,136,137 which appears to depend upon a protein kinase C-epsilon (PKC-epsilon) pathway.138 A subsequent study using P rats139 examined the effects of systemic MPEP on the extracellular signal-regulated kinase (ERK1/2) pathway,140 which is downstream of mGlu5 and implicated in addiction. MPEP attenuated cue-induced reinstatement of ethanol-seeking behavior, which was associated with decreased phosphorylated (p)ERK1/2 immunoreactivity (IR) in the BLA, but not CeA, and AcbSh, but not AcbCo.139 These findings support a role for ERK1/2 phosphorylation in the BLA and AcbSh in mediating cue-induced reinstatement of ethanol-seeking behavior. A third study from this laboratory141 confirmed a role for mGlu5 within the AcbCo in ethanol self-administration; such that local application of MPEP into the AcbCo reduced ethanol operant responding without affecting locomotor activity, sucrose or water responding. In contrast, MPEP infused into the dorsomedial caudate nucleus or mPFC did not alter operant ethanol self-administration.141 In a study that examined the effects of mGlu5 antagonist 3-[(2-methyl-1,3-tia-zol-4-yl)ethynyl]-pyridine (MTEP) on ethanol self-administration by two high alcohol–consuming rat lines (inbred P (iP) and Fawn-Hooded (FH)) reported that MTEP significantly decreased intake.142 However, these authors also reported that MTEP induced mild sedative effects in iP but not FH rats. Together, these findings indicate that mGlu5 receptors play an important role in regulating different aspects of alcoholism-related behavior in both rat and mouse models.
Other mGluR ligands have also been tested for their effects on excessive ethanol drinking or ethanol reinforcement. Earlier studies revealed that systemic pretreatment with the selective mGlu1 antagonist, (−)-ethyl (7E)-7-hydroxyimino-1,7a-dihydrocyclopropa[b]chromene-1a-carboxylate (CPCCOEt), produced inconsistent effects on operant ethanol self-administration in P rats134 versus C57BL/6J mice,137 such that intra-Acb infusions of CPCCOEt were unable to alter binge drinking in mice.83 These inconsistent effects of CPCCOEt on measures of alcohol intake likely reflect its relative insolubility. For instance, the more soluble, highly selective, mGlu1 antagonist JNJ 16259685 lowers operant ethanol self-administration and ethanol breakpoint in P rats when administered systemically135,143 and reduces binge drinking when infused into the AcbSh of mice.144 However, systemic JNJ 16259685 pretreatment has nonselective effects in that it also reduces locomotor activity,135,143 which may reflect the high abundance of mGlu1 receptors in the cerebellum and their effects on its control of motor movement.134,135,143
Studies have also examined the effects of targeting mGlu2/3 receptors within the contexts of operant ethanol self-administration. When given systemically, the mGlu2/3 antagonist LY341495 did not alter operant responding for ethanol by P rats,134 although systemic pretreatment with the mGlu2/3 receptor agonist LY404039 reduced ethanol-seeking and -relapse-like behavior.145 However, these latter authors reported that LY404039 did not alter the maintenance of operant ethanol self-administration in these animals.145 Interestingly, the local application of the mGlu2/3 agonist LY379268 in the AcbCo was sufficient to reduce operant ethanol self-administration by P rats,141 in a manner akin to the aforementioned studies of mice drinking under free-access conditions in the home cage.120,123 While understudied, the effects of mGlu2/3 agonists appear to depend upon the route of administration or the experimental procedures employed. However, as observed with mGlu1 antagonists,134,135,143 the effect of intra-AcbCo mGlu2/3 agonism on operant ethanol drinking by P rats is also not specific due to effects on locomotor activity.141 Other lines of evidence support an important role for mGlu2 receptors in drug addiction as well.97,102,146
3.3 Ionotropic Glutamate Receptors and Alcohol
As noted earlier, ionotropic glutamate receptors play an important part in the development of alcohol abuse and dependence, with many of ethanol’s effects mediated by these receptors. For instance, Enoch and coworkers147 as well as Jin and coworkers148 have reported that numerous ionotropic glutamate receptor subunit expression levels are significantly altered in chronic alcoholics and cocaine addicts, relative to control samples. Nevertheless, there has been limited behavioral–pharmacology research on the involvement of ionotropic receptors in excessive ethanol intake by P rats. Pretreatment with aniracetam, a selective positive allosteric modulator of AMPA receptors, increased operant responding for ethanol and cue-induced reinstatement of operant responding for ethanol by P rats, while not altering locomotor activity or operant responding for sucrose.149 These authors confirmed the involvement of AMPA receptors by demonstrating that the AMPA receptor antagonist 6,7-dinitroquinoxaline-2,3-dione blocked aniracetam-induced increases in ethanol self-administration. Besides the P rat, the selectively bred ALKO alcohol-accepting (high ethanol-consuming rat line from Finland) has also been used to assess the role of AMPA/kainate receptors in excessive ethanol intake, with systemic administration of the antagonist CNQX significantly reducing operant ethanol-seeking behavior by these rats.150 In addition, systemic administration of the mixed NMDA/glycine receptor antagonist L-701,324 also significantly reduces operant ethanol-seeking behavior in AA rats.150 While MTEP is an mGlu5 receptor antagonist and decreases ethanol self-administration, it also decreases mRNA expression for both Glua2 and Glun1 in the cingulate cortex of iP and FH rats.142 Thus, the effects of mGlu-receptor activity on ethanol intake are paralleled by its regulation of ionotropic glutamate receptor subunit gene expression underscoring the interaction of these two classes of glutamate receptors. Although there has been limited research on ionotropic glutamate receptors regarding alcohol- and/or drug-intake, -seeking, etc.151; there is clear evidence that NMDA and AMPA receptors are affected by ethanol, which in turn affects neuroplasticity, learning and memory.31,32,95,151,152
3.4 Glutamate Transporters/Carriers and Alcohol
Chronic ethanol-drinking or ethanol exposure significantly reduces glutamate uptake in the brain through downregulation of glutamate transporter, or antiporter, expression153–156 and reductions in glutamate transporter expression have been confirmed in postmortem evaluation of brains from alcoholics.128 Recent studies have examined the effects of modulating GLT-1, glial EAAT2, and the cystine–glutamate antiporter (xCT) on ethanol intake. Ceftriaxone, a beta-lactam antibiotic, increases glutamate reuptake by upregulating GLT1 expression.157 Given the general hyperglutamatergic state in alcohol/drug dependence, it is not surprising that this compound also decreases ethanol consumption, ethanol dependence (ED)-associated withdrawal signs, and withdrawal-associated escalation of ethanol intake in P rats.158–162 These authors reported a consistent upregulation of GLT1 expression in the Acb and mPFC, which was negatively associated with the observed reductions in ethanol consumption. Moreover, these authors have shown that ceftriaxone, or its analogs, significantly reduces ethanol intake and reverses chronic ethanol-induced downregulation of GLT1 expression in the mPFC, Acb, Amyg, and hippocampus, as well as reversing downregulation of xCT in some brain regions.153,163–166
Given that chronic ethanol induces excitotoxicity and oxidative stress, it is noteworthy that ethanol-induced increases in Slc7a11 (the protein product being xCT, the cystine–glutamate antiporter) are not dependent upon exci-totoxicity or oxidative stress.167 These authors reported that ethanol itself can inhibit octamer-binding transcription factor 1’s (OCT-1) repression of the Slc7a11 promoter in vitro, which in turn elevates Slc7a11 transcription. This increase in xCT would putatively increase the import of cystine into glial cells, where it would be converted to cysteine and subsequently into gluta-thione resulting in increased glutathione and decreased neurotoxicity.58 Regarding intracellular glutamate transport, an early study168 examined the effects of continuous ethanol exposure versus exposure interspersed with repeated deprivations on these vesicular glutamate transporters. These authors reported that repeated deprivations increased vGLUT2-immunos-tained terminals in the AcbSh compared to the water control group. However, ethanol exposure did not alter the level of vGLUT1-immunos-tained terminals in this brain region. These results suggest that the presence of multiple withdrawal episodes preferentially increases vGLUT2 expression in glutamate terminals in the AcbSh of P rats.168
3.5 Glutamate-Associated Enzyme Activity and Alcohol
Early work revealed that chronic ethanol exposure decreased glutamine synthetase while increasing glutamate and GAD in the cortex of rats.169 A more recent preclinical study found decreased GAD-67 expression levels, in the BLA, 2 months after 3 weeks of ethanol-diet initiated in adulthood, but not adolescence.170 Another study reported that chronic ethanol consumption decreased glutamine synthetase in the striatum (dorsal vs. ventral was not delineated) but not cortex of rats.171 A contemporary study also reported reduced glutamine synthetase in the brain, although the area of the brain was not identified, after chronic ethanol consumption, which started at the beginning of adolescence.172 A postmortem study indicated that glutamine synthetase was downregulated in the hippocampus of alcoholics without hepatic pathology.173 These consistent reductions in glutamine synthetase following ethanol exposure or consumption suggest the presence of astro-cytic pathology and, by extension, increased neurotoxicity. Regarding glutamate dehydrogenase, which metabolizes glutamate; adolescent binge-like drinking by rats resulted in a 40% decrease in hippocampal glutamate dehydrogenase 1, which was not seen in rats that received the same protocol during adulthood.174 Given this finding, it is noteworthy that ethanol inhibits NMDA excitation and LTP to a greater extent in hippocampal slices from adolescent versus adult rats.175,176 Therefore, significant differences in ethanol’s effects on glutamatergic activity occur across periadolescence and adulthood (see later in the chapter).
3.6 The Postsynaptic Density and Alcohol
Recent research indicates that alcohol and/or drugs of abuse have a profound developmental effect on the PSD as well174,177,178; such that, similar to earlier reports on the vulnerability of the adolescent hippocampus to alcohol and/or drug exposure,175,176 there is a differential effect of binge-like ethanol exposure between adolescent and adult rats. Risher et al.177 reported that adolescent intermittent ethanol (AIE) exposure in rats reduced PSD-95 expression levels in the hippocampus, leading to the retention of immature-like dendritic spine phenotypes into adulthood. There was also a reduction in the number of VGlut1/PSD-95 and VGlut1/SAP102 (another MAGUK) colocalized synaptic puncta and these effects were driven by decreases in PSD-95 and SAP102 density with no effect on presynaptic VGlut1 expression levels.177 In contrast, chronic intermittent ethanol (CIE) during adulthood178 did not alter PSD-95 expression in the hippocampus as a whole. However, these authors indicated that adult CIE could alter dendritic complexity in a subregion-specific manner, with a partial return to basal levels after protracted abstinence.178 Taken together these studies suggest that the PSD-95, and glutamate activity, may be more vulnerable to ethanol-induced changes during adolescence than during adulthood and that adolescent ethanol-induced changes in PSD-95 may interfere with the maturation of dendritic spines. Similarly, considerable evidence indicates a crucial role for Shank and Homer proteins in neuroplasticity as well as alcohol and drug dependence.86–89,179 For example, significant increases in both AcbCo and CeA Homer2a/b expression levels were seen 24 h after the removal of ethanol from chronically drinking P rats.180
4. GLUTAMATE-ASSOCIATED GENETIC VARIATIONS AND ALCOHOLISM
An early Genome Wide Association Study (GWAS) followed by a Gene Set Enrichment Analysis (GSEA) found that when gene variations were analyzed for grouping, neuronal signaling genes dominated other associations with an individual’s level of response to alcohol and glutamate was the primary neurotransmitter system implicated.181 These authors also noted that FHP individuals show an altered level of response to alcohol and ketamine (an NMDA antagonist), thus confirming a genetic risk for alcoholism and altered glutamatergic function.181,182 Similarly, a pathway analysis of variants in 130 addiction-related candidate genes confirmed a significant role for glutamate signaling in alcohol dependence, with the odds ratio of mGlu1-rs2300620 (>1.6) exceeding that of any other significant gene variant.183 A contemporary study using pathway analysis revealed the NMDA-dependent AMPA-trafficking cascade centered on the gene encoding the multiple PDZ domain protein (Mpdz) was significantly associated with alcohol dependence in a subset of the SAGE study.184
Regarding particular gene variants, polymorphisms of Glun2a, NR2a subunit of the NMDA receptor, are significantly associated with being an FHP individual, an early onset of risky drinking during adolescence as well as the maximum number of drinks in adulthood, with this association having been replicated in a second large sample.185 A subsequent study also found that a polymorphism in the promoter region of Glun2a is significantly associated with alcohol dependence, with this finding replicated in a second sample.186 A variant of another ionotropic glutamate receptor subunit, Gluk3, was found to be associated with alcohol withdrawal–associated delirium tremens, but not seizures, in a German cohort.187 Another study reported that a polymorphism in Gluk1 is significantly associated with alcohol dependence in Caucasians, although many of the alcohol-dependent, but none of the control, subjects were polysubstance users, which may represent a confound in this study.188
Following earlier work indicating theta event-related oscillations (EROs) are associated with the P300 event-related potential (ERP), whose amplitude is negatively associated with FHP,189,190 a Family Based Association Test (FBAT) was carried out on a subset of the COGA sample.191 These authors reported that multiple polymorphisms of the mGlu8 gene are significantly associated with theta EROs and a genetic risk to develop alcoholism.191 A subsequent study by this group confirmed that polymorphisms of the mGlu8 gene are associated with a genetic risk to develop alcohol dependence.192 In another recent study, the mGlu3-rs6465084 polymorphism was found to be significantly associated with alcohol dependence in a male Han Chinese cohort.193
A recent study examining the association between polymorphisms in vesicular glutamate transporters and severe alcoholism in a female Swedish cohort indicated a nominally significant association with Slc17a6-rs2290045.194 A study evaluating associations between alcohol dependence and GAD genes (Gad1 and Gad2), the enzyme responsible for conversion of glutamate to GABA, in a subset of the Irish Affected Sib Pair Study of Alcohol Dependence sample reported that two polymorphisms in Gad1 were associated with initial sensitivity to alcohol and a different polymorphism in Gad1 was associated with age at alcohol dependence onset.195 An analysis of the relationship between Gad1 variants and alcohol dependence in an Italian cohort revealed that the Gad1-rs11542313 polymorphism was significant.196 An earlier study evaluating polymorphisms of Gad2 indicated that the functional promoter Gad2-243A>G variant was significantly associated with alcohol dependence in Russian but not European American males.197 A contemporary study found that three polymorphisms of Gad1, but no polymorphisms of Gad2, were significantly associated with alcohol dependence in Han Taiwanese men.198 In a recent genome-wide DNA methylation study, it was reported that methylation of Gad1 is significantly associated with alcohol dependence in a cohort of Han Chinese men, suggesting epigenetic effects of chronic alcohol abuse.199 These results indicate that polymorphisms in Gad1 are significantly associated with alcohol dependence, or related phenotypes, in multiple populations around the world.
5. ALCOHOL AND GLUTAMATE GENE EXPRESSION
Extending the previously mentioned behavioral pharmacologic and neuropharmacologic studies are the results of a very comprehensive study by Meinhardt and coworkers,200 demonstrating that: (1) in ethanol-dependent rats, glutamate-related gene changes were primarily seen in the mPFC, rather than the Acb or amygdala; (2) within the mPFC of ethanol-dependent rats, downregulation of the NMDA-receptor subunits Glun2a and Glun2b, as well as mGlu2, but not mGlu3, and egr1 (early growth response protein 1: Zif268, a transcription factor involved in neuroplasticity and vesicular exo-cytosis at excitatory synapses) were significant in the infralimbic mPFC only, with mGlu2 displaying the greatest reduction; (3) the AcbSh receives its glutamatergic projections primarily from the infralimbic mPFC, according to retrograde tracing, and these projection neurons displayed significant ethanol-dependence-associated downregulation of mGlu2, Egr2, and Egr4; (4) while basal glutamate in the AcbSh did not differ between ethanol-dependent and control rats, peripheral administration of the mGlu2/3 agonist LY379268 significantly reduced extracellular glutamate in the AcbSh of control but not ethanol-dependent rats, suggesting a downregulation of mGlu2/3 function; (5) ethanol-dependent rats displayed greater relapse (almost twice as many responses) and progressive ratio than controls; (6) lentivial knockdown of mGlu2 in the infralimbic mPFC significantly attenuated cue-induced reinstatement of responding in ethanol-dependent but not nondependent rats; (7) mGlu2 knockdown rats did not differ from control rats in operant relapse responding, operant responding for sweetened condensed milk, or locomotor activity in an open-field test; and (8) a RT-qPCR analysis of postmortem ACC samples revealed that alcoholics had significantly less mGlu2 mRNA than their respective controls. An early postmortem study revealed that individuals who had abused nicotine, but not alcohol, displayed greater expression of Slc17a6 and Slc17a7 vesicular glutamate transporters in the VTA.201 However, in individuals who abused both nicotine and alcohol, these increases in Slc17a6 and Slc17a7 were significantly reduced.201
In another study, Enoch and coworkers147 reported that hippocampal glutamate receptor subunit gene expression levels differed between alcoholics, cocaine addicts, and healthy controls. Specifically, Glua4, Gluk3, and mGlu4 expression was significantly higher in alcoholics, relative to both controls and cocaine addicts; Glun2b expression was higher in both alcoholics and cocaine addicts, relative to control levels; and Glun2d and mGlu3 was upregulated in alcoholics while being downregulated in cocaine addicts, relative to control levels.147 Substantial changes of glutamate receptor subunit gene expression in the hippocampal dentate gyrus and to a lesser extent orbital frontal cortex, but not the dorsal-lateral prefrontal cortex, of alcoholics versus healthy controls was confirmed in another recent postmortem study.148 In particular, the data from the dentate gyrus revealed that alcoholics had higher expression levels of Glua2, Glua3, Gluk2, Gluk3, Gluk5, Glun1, Glun2a, Glun2c, Glun2d, and Glun3a; whereas only Glun3a expression was higher than controls in the orbital frontal cortex.148
6. ALCOHOL-ASSOCIATED CHANGES IN GENE/PROTEIN EXPRESSION OF P RATS
An examination of protein expression changes in subregions of the Acb and Amyg of chronic ethanol-drinking P rats revealed at 24-h withdrawal that GluN2a expression levels were increased, whereas GluN2b expression levels were decreased in the AcbSh.180 These authors also reported that Homer2a/b, mGlu1, mGlu5, GluN2a, and GluN2b expression levels were all consistently increased in the AcbCo and CeA.180 To test the hypothesized genetic role for the mGlu2 receptor in alcohol dependence, a recent RNA and exome sequencing study revealed that a SNP which creates a stop codon in the mGlu2 gene is present in P, but not NP, rats.202 This stop codon results in the absence of mGlu2 receptors, impaired gluta-matergic synaptic transmission, and altered levels of multiple genes associated with synaptic function. These authors also examined F2 rats from a PxNP–NPxP cross and found that mGlu2 expression levels were significantly and inversely related to ethanol consumption, with decreases in mGlu2 and mGlu2 receptor expression associated with significant increases in ethanol-drinking behavior.202 Similarly, mGlu2 knockout mice display significantly greater ethanol consumption and preference than their wild-type counterparts.202 As mGlu2 receptors function as autoreceptors, the results indicate an inverse relationship between mGlu2 gene/protein expression and ethanol intake, which is consistent with the aforementioned hypothesis that excessive drinking phenotypes are associated with a hyperglutamatergic state.
Our laboratories have published a number studies that examined central gene and/or protein expression profiles of P rats.180,203–215 Here we highlight alcohol-associated glutamate-related genes whose expression levels have been identified as significantly changed by ethanol self-administration in P rats, or innately different between P and NP rats, in brain regions implicated in alcohol reward and reinforcement (Fig. 1).
6.1 Metabotropic Glutamate Receptor Expression Differences (Table 1)
Table 1.
Metabotropic Glutamate Receptor (mGluR) Gene (mGlu) Expression Differences Between P and NP Rats (i.e., Fold-Change (F-C) P vs. NP) or Changes Induced by Ethanol Consumption in P Rats, Relative to Ethanol-Naive P Rats (i.e., F-C Ethanol (E) vs. Control (C))
| Gene ID | Gene Names | F-C P vs. NP | F-C E vs. C | Age of Rat | Brain Region |
|---|---|---|---|---|---|
| mGlu7 | Glutamate receptor, metabotropic 7 (Grm7: mGluR7) | 1.37 | Adolescent | CeA | |
| mGlu1 | Glutamate receptor, metabotropic 1 (Grm1: mGluR1) | 2.06 | Adolescent | DRN | |
| mGlu4 | Glutamate receptor, metabotropic 4 (Grm4: mGluR4) | 2.98 | Adolescent | DRN | |
| mGlu6 | Glutamate receptor, metabotropic 6 (Grm6: mGluR6) | −1.70 | Adolescent | DRN | |
| mGlu1 | Glutamate receptor, metabotropic 1 (Grm1: mGluR1) | 1.39 | Adult | AcbSh | |
| mGlu1 | Glutamate receptor, metabotropic 1 (Grm1: mGluR1) | −1.19 | Adult | AcbSh | |
| mGlu2 | Glutamate receptor, metabotropic 2 (Grm2: mGluR2) | −1.46 | Adult | AcbSh | |
| mGlu3 | Glutamate receptor, metabotropic 3 (Grm3: mGluR3) | −1.22 | Adult | AcbSh | |
| mGlu4 | Glutamate receptor, metabotropic 4 (Grm4: mGluR4) | −1.39 | Adult | AcbSh | |
| mGlu5 | Glutamate receptor, metabotropic 5 (Grm5: mGluR5) | −1.30 | Adult | AcbSh | |
| mGlu7 | Glutamate receptor, metabotropic 7 (Grm7: mGluR7) | −1.38 | Adult | AcbSh | |
| mGlu8 | Glutamate receptor, metabotropic 8 (Grm8: mGluR8) | −1.33 | Adult | AcbSh | |
| mGlu2 | Glutamate receptor, metabotropic 2 (Grm2: mGluR2) | −1.84 | Adult | CeA | |
| mGlu3 | Glutamate receptor, metabotropic 3 (Grm3: mGluR3) | −1.24 | Adult | CeA |
Age of rat refers to whether the P rats had access to ethanol during (peri-)adolescence (postnatal days (PNDs) 30–50) or during adulthood>PND75. Brain regions published thus far: CeA, DRN, and AcbSh. To facilitate distinction between directions of expression (e.g., up- vs. downregulation), downregulation, or lower level F-Cs are in italics and are below the findings of upregulation or higher-level F-Cs for that age and brain region.
A recent study revealed that periadolescent binge-ethanol consumption, using our drinking-in-the-dark—multiple-scheduled-access model,27,29 resulted in a general elevation of mGlu1 and mGlu4 (twice or three times that of controls, respectively) in the dorsal raphe nucleus (DRN) and mGlu7 (~1.4-fold increase) in the CeA.213 However, there was a significant decrease (~1.7-fold decrease) in mGlu6 expression within the DRN as well. Given that mGlu1, for the most part, is excitatory and the others inhibitory, this suggests a balance between ethanol-induced up- and downregulation of gene expression for these metabotropic receptors.
mGlu2 and mGlu3 expression levels were significantly lower in the AcbSh and CeA of adult ethanol-naive P versus NP rats.209,211 As discussed earlier, Zhou and coworkers202 reported a stop-codon polymorphism of mGlu2 in P, but not present in NP rats that appear to predispose them to high ethanol consumption. And, multiple studies134,135,141,143,145 have shown that mGlu2/3 agonists, presumably acting at the presynaptic autoreceptor, block ethanol-self-administration, -seeking, and -relapse behavior in P rats. Noteworthy is the fact that mGlu4, mGlu5, mGlu7, and mGlu8 (all are involved with inhibitory activity, generally at the presynaptic terminal) are also significantly lower in the AcbSh of P versus NP rats. Gene expression for only one mGluR, mGlu1, was ~1.4-fold higher in the AcbSh of P versus NP rats, which is a finding consistent with prior results from inbred C57BL/6J versus DBA2/J mice.86 Regarding the effects of ethanol, operant ethanol self-administration by adult P rats significantly reduced mGlu1 (~20% decrease) expression levels in the AcbSh, relative to ethanol-naive P rats.214 Given mGlu1’s basal elevation, relative to NP rats, it is possible that ethanol self-administration could reverse these levels; although, it is also possible for mGlu1 protein levels to be elevated by ethanol with mRNA levels responding in the opposite direction. Regarding this hypothesis, our laboratory has shown that extended (6 months) ethanol drinking by P rats does indeed increase mGlu1, and mGlu5, receptor protein expression in the AcbCo and CeA.180 Again, previous work with P rats found that an mGlu1 antagonist significantly reduced operant ethanol self-administration and breakpoint, although motor activity appeared to be affected as well.135,143 In general, these findings indicate that the effects of ethanol self-administration on Group1mGluR mRNA expression can be distinguished from effects upon protein expression and/or that different ethanol-drinking experiences/ procedures elicit distinct changes in receptor mRNA/protein expression within Acb subregions, with more protracted drinking regimens eliciting changes within the more dorsal AcbCo.
Overall, adolescent binge-like ethanol drinking upregulated two of the three mGlu receptors identified as significantly changed. In the AcbSh of adult animals, P rats had only one of the seven mGlu genes identified as significantly greater than that seen in NP rats. It is noteworthy that the one gene that had higher expression levels in the AcbSh of adult P rats was downregulated following ethanol self-administration. Both genes identified as significantly different in the CeA of adult rats were Group II (mGlu2 and mGlu3) metabotropic receptors and were lower in P versus NP rats.
6.2 Ionotropic Glutamate Receptor Expression Differences (Table 2)
Table 2.
Ionotropic Glutamate Receptor Subunit Gene Expression Differences Between P and NP Rats (i.e., Fold-Change (F-C) P vs. NP) or Changes Induced by Ethanol Consumption in P Rats, Relative to Ethanol-Naive P Rats (i.e., F-C Ethanol (E) vs. Control (C))
| Gene ID | Gene Names | F-C P vs. NP | F-C E vs. C | Age of Rat | Brain Region |
|---|---|---|---|---|---|
| Glua3 | Glutamate receptor, ionotropic (Gria3: AMPA3) | 1.29 | Adolescent | DRN | |
| Glua4 | Glutamate receptor, ionotropic (Gria4: AMPA4) | 1.31 | Adolescent | DRN | |
| Glud1 | Glutamate receptor, ionotropic (Grid1: delta1) | 1.20 | Adolescent | DRN | |
| Glud2 | Glutamate receptor, ionotropic (Grid2: delta2) | 2.31 | Adolescent | DRN | |
| Glua3 | Glutamate receptor, ionotropic (Grik3: kainate3) | 1.35 | Adolescent | DRN | |
| Glun2c | Glutamate receptor, ionotropic NMDA2c (Grin2c: NR2c) | 2.61 | Adolescent | DRN | |
| Glun2b | Glutamate receptor, ionotropic, NMDA2b (Grin2b: NR2b) | −1.56 | Adolescent | DRN | |
| Glun2d | Glutamate receptor, ionotropic, NMDA2d (Grin2d: NR2d) | −1.40 | Adolescent | DRN | |
| Glun3a | Glutamate receptor, ionotropic, NMDA3a (Grin3a: NR3a) | −1.39 | Adolescent | DRN | |
| Glunl1a | Glutamate receptor, ionotropic, NMDA1a like (Grinl1a: NR1a like) | −1.32 | Adolescent | DRN | |
| Glua1 | Glutamate receptor, ionotropic (Gria1: AMPA1) | 1.15 | Adult | AcbSh | |
| Glua4 | Glutamate receptor, ionotropic (Gria4: AMPA4) | 1.26 | Adult | AcbSh | |
| Glud2ip | Glutamate receptor, ionotropic (Grid2ip: delta2 interacting protein) | 3.49 | Adult | AcbSh | |
| Gluk1 | Glutamate receptor, ionotropic (Grik1: kainate1) | 1.73 | Adult | AcbSh | |
| Glun1 | Glutamate receptor, ionotropic, NMDA1 (Grin1: NR1) | 1.09 | Adult | AcbSh | |
| Glua1 | Glutamate receptor, ionotropic (Gria1: AMPA1) | −1.32 | Adult | AcbSh | |
| Glua2 | Glutamate receptor, ionotropic (Gria2: AMPA2) | −1.21 | Adult | AcbSh | |
| Glua2 | Glutamate receptor, ionotropic (Gria2: AMPA2) | −1.16 | Adult | AcbSh | |
| Glua3 | Glutamate receptor, ionotropic (Gria3: AMPA3) | −1.52 | Adult | AcbSh | |
| Glua3 | Glutamate receptor, ionotropic (Gria3: AMPA3) | −1.19 | Adult | AcbSh | |
| Gluk2 | Glutamate receptor, ionotropic (Grik2: kainate2) | −1.33 | Adult | AcbSh | |
| Glua1 | Glutamate receptor, ionotropic (Gria1: AMPA1) | 1.36 | Adult | CeA | |
| Glua2 | Glutamate receptor, ionotropic (Gria2: AMPA2) | 1.34 | Adult | CeA | |
| Glua3 | Glutamate receptor, ionotropic (Gria3: AMPA3) | 1.57 | Adult | CeA | |
| Gluk2 | Glutamate receptor, ionotropic (Grik2: kainate2) | 1.14 | Adult | CeA | |
| Gluk5 | Glutamate receptor, ionotropic (Grik5: kainate5) | 1.30 | Adult | CeA | |
| Glun3a | Glutamate receptor, ionotropic, NMDA3a (Grin3a: NR3a) | 1.23 | Adult | CeA |
Refer to Table 1 for details.
Ionotropic glutamate receptor subunits Glua3, Glua4, and Gluk3 were all elevated in the DRN following periadolescent binge-ethanol drinking, but not to the same extent as mGlu1 and mGlu4.213 Gene expression changes were also observed for the “orphan” ionotropic glutamate delta receptor subunits Glud1 and Glud2. Regarding Glud2, there was a 3.5-fold increase in Glud2ip (delphilin) after periadolescent binge drinking as well;213 Glud2ip is a scaffolding protein for Glud2. To some extent, this parallels increases in expression levels of mGlu7, in the CeA, induced by periadolescent binge-ethanol drinking. In the periadolescent binge-drinking P rats, the GluN subunits were generally downregulated, with only the Glun2c subunit being upregulated.213
In adult P rats, whereas metabotropic glutamate receptor gene expression levels were consistently lower than those of NP rats; this was not true for Glua and Gluk gene expression,209,211 which was mixed. Glua1, Glua2, Glua3, and Gluk3 expression levels were all lower in the AcbSh of P versus NP rats; whereas Glua4 and Gluk1 expression levels were higher in the AcbSh of P versus NP rats.211 Ethanol-binge drinking by adult P rats reversed the ~30% deficit of Glua1 in the AcbSh through a 15% increase, relative to ethanol-naive controls.208 However, operant ethanol self-administration by adult P rats significantly reduced Glua2 and Glua3 expression levels, versus ethanol-naive controls, in the AcbSh.214 Thus, in spite of AcbSh deficits in Glua2 and Glua3 of adult P versus NP rats,211 operant ethanol self-administration appears to exacerbate this condition with further decreases in expression levels.
Surprisingly, despite no adult CeA Glua, Gluk, or Glun gene expression differences between P and NP rats; ethanol-binge drinking by adult P rats elevated Glua1, Glua2, Glua3, Gluk2, Gluk5, and Glun3a expression levels, with no significant downregulation of ionotropic glutamate receptor genes in this region.208
Overall, periadolescent binge-like drinking decreased gene expression for certain NMDA receptor subunits in the DRN, but increased particular AMPA, delta, and kainate receptor subunits within this region as well. Ethanol self-administration by adult P rats did not downregulate any NMDA receptor subunits in the AcbSh but did upregulate an AMPA, delta, and NMDA-receptor subunit.
6.3 Expression Differences for Glutamate Transporters, Enzymes, and Postsynaptic Density (Table 3)
Table 3.
Gene Expression, for Ancillary Proteins of the Excitatory Synapse, Differences Between P and NP Rats (i.e., Fold-Change (F-C) P vs. NP) or Changes Induced by Ethanol Consumption in P Rats, Relative to Ethanol-Naive P Rats (i.e., F-C Ethanol (E) vs. Control (C))
| Gene ID | Gene Names | F-C P vs. NP | F-C E vs. C | Age of Rat | Brain Region |
|---|---|---|---|---|---|
| Homer3 | Homer homolog 3 | 1.15 | Adolescent | CeA | |
| Slc1a1 | Solute carrier family 1 (neuronal high affinity glutamate transporter), member 1 | 1.14 | Adolescent | CeA | |
| Dlg1/Sap97 | Discs, large homolog 1 (Sap97, AMPAR trafficking) | 1.28 | Adolescent | DRN | |
| Homer3 | Homer homolog 3 | 3.75 | Adolescent | DRN | |
| Mpp4 | Membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) | 1.68 | Adolescent | DRN | |
| Mpp6 | Membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) | 1.23 | Adolescent | DRN | |
| Pdlim5 | PDZ and LIM domain 5 | 1.28 | Adolescent | DRN | |
| Shank1 | SH3 and multiple ankyrin repeat domains 1 | 1.41 | Adolescent | DRN | |
| Shank2 | SH3 and multiple ankyrin repeat domains 2 | 1.79 | Adolescent | DRN | |
| Shank3 | SH3 and multiple ankyrin repeat domains 3 | 1.82 | Adolescent | DRN | |
| Slc1a3 | Solute carrier family 1 (glial glutamate transporter), member 3 | 1.70 | Adolescent | DRN | |
| Slc1a6 | Solute carrier family 1 (glial glutamate transporter), member 6 | 2.79 | Adolescent | DRN | |
| Slc17a7/ Vglut1 | Solute carrier family 17, member 7 (vesicular glutamate transporter) | 5.06 | Adolescent | DRN | |
| Slc25a18 | Solute carrier family 25 (mitochondrial glutamate carrier), member 18 | 1.45 | Adolescent | DRN | |
| Slc25a22 | Solute carrier family 25 (mitochondrial glutamate carrier), member 22 | 1.30 | Adolescent | DRN | |
| Tjp3 | Tight junction protein 3 (zona occludens 3) | 2.41 | Adolescent | DRN | |
| Mpp5 | Membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) | −1.27 | Adolescent | DRN | |
| Slc17a6/ Vglut2 | Solute carrier family 17, member 6 (vesicular glutamate transporter) | −1.82 | Adolescent | DRN | |
| Slc17a8/ Vglut3 | Solute carrier family 17, member 8 (vesicular glutamate transporter) | −1.97 | Adolescent | DRN | |
| Dlg1/Sap97 | Discs, large homolog 1 (Sap97, AMPAR trafficking) | 1.28 | Adult | AcbSh | |
| Dlg2/Psd93 | Discs, large homolog 2 (Chapsyn-110/PSD-93 a MAGUK) | 1.24 | Adult | AcbSh | |
| Gls2 | Glutaminase 2 | 1.40 | Adult | AcbSh | |
| Glul | Glutamate-ammonia ligase (glutamine synthase) | 1.14 | Adult | AcbSh | |
| Homer1 | Homer homolog 1 | 3.49 | Adult | AcbSh | |
| Mpdz | Multiple PDZ domain protein | 1.22 | Adult | AcbSh | |
| Mpp6 | Membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) | 1.71 | Adult | AcbSh | |
| Shank1 | SH3 and multiple ankyrin repeat domains 1 | 1.26 | Adult | AcbSh | |
| Shank1 | SH3 and multiple ankyrin repeat domains 1 | 1.19 | Adult | AcbSh | |
| Shank2 | SH3 and multiple ankyrin repeat domains 2 | 1.17 | Adult | AcbSh | |
| Slc6a9 | Solute carrier family 6 (glycine transporter), member 9 | 1.40 | Adult | AcbSh | |
| Slc7a10 | Solute carrier family 7 (glycine transporter), member 10 (glial) | 1.42 | Adult | AcbSh | |
| Slc17a6/ Vglut2 | Solute carrier family 17, member 6 (vesicular glutamate transporter) | 2.25 | Adult | AcbSh | |
| Slc25a13/ Citrin | Solute carrier family 25 (mitochondrial glutamate carrier), member 13 | 1.23 | Adult | AcbSh | |
| Dlg1/Sap97 | Discs, large homolog 1 (Sap97, AMPAR trafficking) | −1.09 | Adult | AcbSh | |
| Dlg2/Psd93 | Discs, large homolog 2 (Chapsyn-110/PSD-93 a MAGUK) | −1.17 | Adult | AcbSh | |
| Dlg2/Psd93 | Discs, large homolog 2 (Chapsyn-110/PSD-93 a MAGUK) | −1.38 | Adult | AcbSh | |
| Dlg4/Psd95 | Discs, large homolog 4 (PSD95 or Sap90) | −1.25 | Adult | AcbSh | |
| Gls | Glutaminase | −1.35 | Adult | AcbSh | |
| Homer1 | Homer homolog 1 | −1.68 | Adult | AcbSh | |
| Mpp4 | Membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) | −1.08 | Adult | AcbSh | |
| Mpp7 | Membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) | −1.58 | Adult | AcbSh | |
| Pdlim7 | PDZ and LIM domain 7 | −1.25 | Adult | AcbSh | |
| Shank1 | SH3 and multiple ankyrin repeat domains 1 | −1.14 | Adult | AcbSh | |
| Shank2 | SH3 and multiple ankyrin repeat domains 2 | −1.22 | Adult | AcbSh | |
| Shank3 | SH3 and multiple ankyrin repeat domains 3 | −1.30 | Adult | AcbSh | |
| Slc1a3 | Solute carrier family 1 (glial glutamate transporter), member 3 | −1.30 | Adult | AcbSh | |
| Cask | Calcium/calmodulin-dependent serine protein kinase (MAGUK family) | 1.14 | Adult | CeA | |
| Homer1 | Homer homolog 1 | 2.19 | Adult | CeA | |
| Pdlim3 | PDZ and LIM domain 3 | 1.28 | Adult | CeA | |
| Slc25a22 | Solute carrier family 25 (mitochondrial glutamate carrier), member 22 | 1.12 | Adult | CeA | |
| Gls2 | Glutaminase 2 | −1.17 | Adult | CeA | |
| Homer2 | Homer homolog 2 | −1.16 | Adult | CeA | |
| Mpp3 | Membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3) | −1.45 | Adult | CeA | |
| Tgm2 | Transglutaminase 2, C polypeptide | −1.30 | Adult | CeA | |
| Glul | Glutamate-ammonia ligase (glutamine synthetase) | 1.08 | Adult | VTA | |
| Dlg2/Psd93 | Discs, large homolog 2 (Chapsyn-110/PSD-93 a MAGUK) | −1.17 | Adult | VTA | |
| Mpp6 | Membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) | −1.10 | Adult | VTA | |
| Pdlim7 | PDZ and LIM domain 7 | −1.12 | Adult | VTA | |
| Tgm2 | Transglutaminase 2, C polypeptide | −1.38 | Adult | VTA |
Age of rat refers to whether the P rats had access to ethanol during (peri-)adolescence (PNDs, 30–50) or during adulthood>PND75. Brain regions published thus far: CeA, DRN, AcbSh, and VTA. To facilitate distinction between directions of expression (e.g., up- vs. downregulation), downregulation or lower level F-Cs are in italics and are below the findings of upregulation or higher-level F-Cs for that age and brain region.
One of the most striking findings is that periadolescent binge-like drinking significantly downregulated gene expression for only three ancillary proteins (Mpp5, a MAGUK subfamily member, by ~30%; as well asVglut2 and Vglut3 by ~2-fold in the DRN); while upregulating expression levels for Homer3 and Slc1a1 in the CeA along with nine genes from the PSD (e.g., Homer3 ~4-fold and Tjp3 ~2.5-fold), as well as all three Shanks, two glutamate transporters (e.g., Slc1a6 ~3-fold), and two glutamate carriers in the DRN.213
In the adult AcbSh, P rats had lower gene expression of Homer1 than NP rats.211 However, operant self-administration of ethanol by adult P rats upregulated Homer1 by 3.5-fold in the AcbSh.214 These data are interesting as studies of rodents with free-access to ethanol83–85,89,180 or rodents injected repeatedly with alcohol86,88 have consistently detected increases in Homer2 protein expression within Acb and amygdala structures, without detecting significant changes in Homer1 protein expression. This raises the possibility that ethanol-induced changes in Homer1 mRNA/protein expression may depend upon nonpharmacologic factors associated with the act of ethanol taking, which has recently been demonstrated to be the case with respect to intravenous cocaine taking.216 Again in the AcbSh, P rats had higher expression levels of Shank1, whereas NP rats had higher expression levels of Shank2 and Shank3.211 Our data suggest a dissociation between the effects of home-cage ethanol drinking which decreased expression of Shank1 by ~15%204 as opposed to operant ethanol self-administration which increased expression levels of Shank1 by ~20%214 in the AcbSh. Some support for this dissociation can be deduced from the fact that operant ethanol self-administration also increased Shank2 expression levels by ~ 20% in the AcbSh.214
In the adult CeA, operant ethanol self-administration upregulated Homer1 expression levels by over two-fold; although operant ethanol self-administration also downregulated Homer2 expression by ~20%, relative to controls.214 This latter result is peculiar given that chronic free-choice access to ethanol upregulates CeA Homer2 protein expression in P rats180 and C57BL/6J mice;85 which, again, may be due to nonpharmacologic factors related to the operant-conditioning procedures employed in the mRNA study.
Glycine transporter expression levels in the adult AcbSh were higher and glutamate transporter expression levels were lower in P rats compared to NP rats.211 Additionally, whenever ethanol intake modified membrane glutamate transporter expression levels, once each in the adult or adolescent CeA and multiple times in the adolescent DRN, it was always for an increase relative to control levels.208,213 However, when examining vesicular glutamate transporter expression levels, Vglut1 was increased by five-fold, whereas Vglut2 and Vglut3 were reduced by ~two-fold in the DRN following peri-adolescent binge-like drinking.213 Also, in the adult AcbShVglut2 expression levels were more than two-fold higher than that seen in NP rats.211
In general, adolescent binge-like drinking upregulated 14 of the 17 glutamate ancillary genes identified as significantly changed, suggesting that ethanol induces substantial increases in DRN glutamatergic activity during this stage of development. Of the genes identified as significantly different between P and NP rats, in the AcbSh of adult animals, half were higher and half were lower. In the CeA (9 of 17) and VTA (4 of 5), adult P rats generally had lower expression levels of glutamate ancillary genes than their NP counterparts. Regarding ethanol exposure, 6 of the 10 genes identified as significantly different relative to ethanol-naive controls were upregulated in the AcbSh of adult P rats. In ethanol-drinking/self-administering adult P rats, two of the five genes identified as significant in the CeA were downregulated. Overall, these findings of glutamate transporter and cytoskeleton/scaffolding protein gene expression level changes induced by ethanol parallel the existing literature indicating that ethanol exposure alters glutamate clearance from the synapse and induces neuro-plastic changes in the PSD.
7. CONCLUSIONS
The findings discussed in this review include results from both pre-clinical as well as neuroimaging and postmortem clinical studies. Expression levels for a number of glutamate-associated genes and/or proteins are modulated by alcohol abuse and dependence. These changes in expression include metabotropic receptors and ionotropic receptor subunits as well as different glutamate transporters. Moreover, these changes in gene expression parallel pharmacologic manipulation of these same receptors and transporters. Some of these gene expression changes may have predated alcohol abuse and dependence, because a number of glutamate-associated polymorphisms are related to a genetic predisposition to develop alcohol dependence. Other glutamate-associated polymorphisms have been linked to age at the onset of alcohol-dependence and/or initial level of response/sensitivity to alcohol. Finally, findings of innate and/or ethanol-induced glutamate-associated gene expression differences/changes observed in a genetic animal model of alcoholism, the P rat, are highlighted. Overall, the existing literature indicates that changes in receptors, transporters, enzymes, and scaffolding proteins are crucial for the development of alcohol dependence and there is a substantial genetic component to these effects.
This review reveals that there are presently key limitations to our understanding of glutamate’s role in the development of alcohol dependence and the impact that genetics has on this process. First, there are no studies examining glutamate-associated gene and/or protein expression changes across the juvenile, adolescent, emerging adult, and full adult stages of development. This information is crucial given the fact that the risky drinking age-of-onset is inversely associated with the probability of developing alcohol dependence (i.e., earlier onset leads to higher risk of developing alcoholism). And, while there is some evidence for an association between being a FHP individual and initiating risky drinking at a younger age, findings comparing the effects of ethanol on glutamate function between FHP individuals/models and FHN individuals/models are very limited. Second, studies thus far have been limited to gross examinations of regions and/or subregions of major structures in the central reward neu-rocircuitry. Also related to this point, a third limitation is the lack of publications examining multiple regions (i.e., putative circuits) within a single study. Despite these limitations, substantial progress has been made with new targets for medications development/screening identified, such as the GLT1/EAAT2 glutamate transporter or PKC-epsilon’s modulation of mGlu5 activity. Nevertheless, the research community still has much to do in unraveling the role of glutamate-associated genes in the development of alcohol dependence, especially as it relates to pharmacogenomics and personalized pharmacologic interventions.
Acknowledgments
This work was supported in part by AA13522 to RLB; AA020396 to RLB; AA020892 to HJE; AA016650 to KKS from the National Institutes of Health (NIH)/National Institute on Alcohol Abuse and Alcoholism (NIAAA). The views expressed in this manuscript are completely those of the authors and do not necessarily reflect the views of the funding agencies at the National Institutes of Health.
References
- 1.Research Society on Alcoholism. Impact of Alcoholism and Alcohol Induced Disease on America. Austin, TX: Research Society on Alcoholism; 2009. [Google Scholar]
- 2.Research Society on Alcoholism. Impact of Alcoholism and Alcohol Induced Disease on America. Austin, TX: Research Society on Alcoholism; 2015. [Google Scholar]
- 3.Mokdad A, Marks J, Stroup D, Gerberding J. Actual causes of death in the United States, 2000. JAmMedAssoc. 2004;291:1238–1245. doi: 10.1001/jama.291.10.1238. [DOI] [PubMed] [Google Scholar]
- 4.Rehm J. The risks associated with alcohol use and alcoholism. Alcohol Res Health. 2011;34:135–143. [PMC free article] [PubMed] [Google Scholar]
- 5.Rehm J, Room R, Graham K, Monteiro M, Gmel G, Sempos CT. The relationship of average volume of alcohol consumption and patterns of drinking to burden of disease—an overview. Addict. 2003;98:1209–1228. doi: 10.1046/j.1360-0443.2003.00467.x. [DOI] [PubMed] [Google Scholar]
- 6.Reed T, Page WF, Viken RJ, Christian JC. Genetic predisposition to organ-specific endpoints of alcoholism. AlcoholClinExpRes. 1996;20:1528–1533. doi: 10.1111/j.1530-0277.1996.tb01695.x. [DOI] [PubMed] [Google Scholar]
- 7.Cloninger CR. Neurogenetic adaptive mechanisms in alcoholism. Science. 1987;236:410–416. doi: 10.1126/science.2882604. [DOI] [PubMed] [Google Scholar]
- 8.Cotton NS. The familial incidence of alcoholism. JStudAlcohol. 1979;40:89–116. doi: 10.15288/jsa.1979.40.89. [DOI] [PubMed] [Google Scholar]
- 9.Schuckit MA. Genetic aspects of alcoholism. AnnEmergMed. 1986;15:991–996. doi: 10.1016/s0196-0644(86)80117-2. [DOI] [PubMed] [Google Scholar]
- 10.Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, VanEerdewegh P, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, Porjesz B, Li T-K, Conneally PM, Nurnberger JI, Jr, Tischfield JA, Crowe R, Cloninger CR, Wu W, Shears S, Carr K, Crose C, Willig C, Begleiter H. Genome-wide search for genes affecting the risk for alcohol dependence. AmJMedGenet. 1998;81:207–215. [PubMed] [Google Scholar]
- 11.Agrawal A, Hinrichs AL, Dunn G, Bertelsen S, Dick DM, Saccone SF, Saccone NL, Grucza RA, Wang JC, Cloninger CR, Edenberg HJ, Foroud T, Hesselbrock V, Kramer J, Bucholz KK, Kuperman S, Nurnberger JI, Porjesz B, Schuckit MA, Goate AM, Bierut LJ. Linkage scan for quantitative traits identifies new regions of interest for substance dependence in the Collaborative Study on the Genetics of Alcoholism (COGA) sample. DrugAlcoholDepend. 2008;93:12–20. doi: 10.1016/j.drugalcdep.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edenberg HJ, Foroud T. Genetics and alcoholism. Nat Rev Gastroenterol Hepatol. 2013;10:487–494. doi: 10.1038/nrgastro.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapoor M, Wang J-C, Wetherill L, Le N, Bertelsen S, Hinrichs AL, Budde J, Agrawal A, Bucholz K, Dick D, Harari O, Hesselbrock V, Kramer J, Nurnberger JI, Rice J, Saccone N, Schuckit M, Tischfield J, Porjesz B, Edenberg HJ, Bierut L, Foroud T, Goate A. A meta-analysis of two genome-wide association studies to identify novel loci for maximum number of alcoholic drinks. HumGenet. 2013;132:1141–1151. doi: 10.1007/s00439-013-1318-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levey DF, Le-Niculescu H, Frank J, Ayalew M, Jain N, Kirlin B, Learman R, Winiger E, Rodd Z, Shekhar A, Schork N, Kiefe F, Wodarz N, Muller-Myhsok B, Dahmen N, Nothen M, Sherva R, Farrer L, Smith AH, Kranzler HR, Rietschel M, Gelernter J, Niculescu AB Consortium GESGASA. Genetic risk prediction and neurobiological understanding of alcoholism. TranslPsychiatry. 2014;4:e391. doi: 10.1038/tp.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rietschel M, Treutlein J. The genetics of alcohol dependence. Ann NY Acad Sci. 2013;1282:39–70. doi: 10.1111/j.1749-6632.2012.06794.x. [DOI] [PubMed] [Google Scholar]
- 16.Wong CCY, Schumann G. Genetics of addictions: strategies for addressing heterogeneity and polygenicity of substance use disorders. PhilosTrans R Soc Lond B Biol Sci. 2008;363:3213–3222. doi: 10.1098/rstb.2008.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan J, Aliev F, Webb BT, Kendler KS, Williamson VS, Edenberg HJ, Agrawal A, Kos MZ, Almasy L, Nunberger JI, Schuckit MA, Kramer JR, Rice JP, Kuperman S, Goate AM, Tischfield JA, Porjesz B, Dick DM. Using genetic information from candidate gene and genome-wide association studies in risk prediction for alcohol dependence. AddictBiol. 2014;19:708–721. doi: 10.1111/adb.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richter CP, Campbell KH. Alcohol taste thresholds and concentrations of solutions preferred by rats. Science. 1940;9:507–508. doi: 10.1126/science.91.2369.507. [DOI] [PubMed] [Google Scholar]
- 19.Williams RJ, Berry LJ, Beerstecher E., Jr Individual metabolic patterns, alcoholism, genotrophic diseases. ProcNatlAcadSciUSA. 1949;35:265–271. doi: 10.1073/pnas.35.6.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mardones J, Segovia-Riquelme N. Thirty-two years of selection of rats by ethanol preference: UChA and UChB strains. NeurobehavToxicolTeratol. 1983;5:171–178. [PubMed] [Google Scholar]
- 21.Bell RL, Sable HJK, Colombo G, Hyytia P, Rodd ZA, Lumeng L. Animal models for medications development targeting alcohol abuse using selectively bred rat lines: neu-robiological and pharmacological validity. PharmacolBiochemBehav. 2012;103:119–155. doi: 10.1016/j.pbb.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McBride WJ, Rodd ZA, Bell RL, Lumeng L, Li T-K. The alcohol-preferring (P) and high-alcohol-drinking (HAD) rats—animal models of alcoholism. Alcohol. 2014;48:209–215. doi: 10.1016/j.alcohol.2013.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cicero TJ. A critique of animal analogues of alcoholism. In: Majchrowicz E, Noble EP, editors. Biochemistry and Pharmacology of Ethanol. Vol. 2. New York: Plenum Press; 1979. pp. 533–560. [Google Scholar]
- 24.Lester D, Freed EX. Criteria for an animal model of alcoholism. Pharmacol Biochem Behav. 1973;1:103–107. doi: 10.1016/0091-3057(73)90062-2. [DOI] [PubMed] [Google Scholar]
- 25.McBride WJ, Li T-K. Animal models of alcoholism: neurobiology of high alcohol-drinking behavior in rodents. CritRevNeurobiol. 1998;12:339–369. doi: 10.1615/critrevneurobiol.v12.i4.40. [DOI] [PubMed] [Google Scholar]
- 26.Bell RL, Franklin KM, Hauser SR, Engleman EA. Next stop dependence. Binge drinking on the road to alcoholism: preclinical findings on its neurobiology from rat animal models. In: Harris SB, editor. Binge Eating and Binge Drinking:Psychological, Social and Medical Implications. New York: Nova Science Publishers; 2013. pp. 1–60. [Google Scholar]
- 27.Bell RL, Rodd ZA, Engleman EA, Toalston JE, McBride WJ. Scheduled access alcohol drinking by alcohol-preferring (P) and high alcohol-drinking (HAD) rats: modeling adolescent and adult binge-like drinking. Alcohol. 2014;48:225–234. doi: 10.1016/j.alcohol.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.National Institute on Alcohol Abuse and Alcoholism (NIAAA) National Advisory Council. NIAAA Council approves definition of binge drinking. NIAAANewsl. 2004;3:5. [Google Scholar]
- 29.Bell RL, Rodd ZA, Smith RJ, Toalston JE, Franklin KM, McBride WJ. Modeling binge-like ethanol drinking by peri-adolescent and adult P rats. Pharmacol Biochem Behav. 2011;100:90–97. doi: 10.1016/j.pbb.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crabbe JC, Bell RL, Ehlers CL. Human and laboratory rodent low response to alcohol: is better consilience possible? Addict Biol. 2010;15:125–144. doi: 10.1111/j.1369-1600.2009.00191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henley JM, Wilkinson KA. AMPA receptor trafficking and the mechanisms underlying synaptic plasticity and cognitive aging. DialoguesClinNeurosci. 2013;15:11–27. doi: 10.31887/DCNS.2013.15.1/jhenley. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morris RGM. NMDA receptors and memory encoding. Neuropharmacology. 2013;74:32–40. doi: 10.1016/j.neuropharm.2013.04.014. [DOI] [PubMed] [Google Scholar]
- 33.Warburton EC, Barker GRI, Brown MW. Investigations into the involvement of NMDA mechanisms in recognition memory. Neuropharmacology. 2013;74:41–47. doi: 10.1016/j.neuropharm.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Danbolt NC. Glutamate uptake. ProgNeurobiol. 2001;65:1–105. doi: 10.1016/s0301-0082(00)00067-8. [DOI] [PubMed] [Google Scholar]
- 35.Niciu MJ, Kelmendi B, Sanacora G. Overview of glutamatergic neurotransmission in the nervous system. PharmacolBiochemBehav. 2012;100:656–664. doi: 10.1016/j.pbb.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. Glutamate receptor ion channels: structure, regulation, and function. PharmacolRev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sari Y. Role of glutamate transporter 1 in the attenuation of alcohol intake. Front Neurosci. 2014;8:200. doi: 10.3389/fnins.2014.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Qin ZH. Molecular and cellular mechanisms of excitotoxic neuronal death. Apoptosis. 2010;15:1382–1402. doi: 10.1007/s10495-010-0481-0. [DOI] [PubMed] [Google Scholar]
- 39.Abe T, Sugihara H, Nawa H, Shigemoto R, Mizuno N, Nakanishi S. Molecular characterization of a novel metabotropic glutamate receptor mGluR5 coupled to inositol phosphate/Ca2-signal transduction. J Biol Chem. 1992;267:13361–13368. [PubMed] [Google Scholar]
- 40.Fotuhi M, Sharp AH, Glatt CE, Hwang PM, von Krosigk M, Snyder SH, Dawson TM. Differential localization of phosphoinositide-linked metabotropic glutamate receptor (mGluR1) and the inositol 1,4,5-trisphosphate receptor in rat brain. J Neurosci. 1993;13:2001–2012. doi: 10.1523/JNEUROSCI.13-05-02001.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shigemoto R, Nomura S, Ohishi H, Sugihara H, Nakanishi S, Mizuno N. Immunohistochemical localization of a metabotropic glutamate receptor, mGluR5, in the rat brain. Neurosci Lett. 1993;163:53–57. doi: 10.1016/0304-3940(93)90227-c. [DOI] [PubMed] [Google Scholar]
- 42.Romano C, Sesma MA, McDonald CT, O’Malley K, Van den Pol AN, Olney JW. Distribution of metabotropic glutamate receptor mGluR5 immunoreactivity in rat brain. JCompNeurol. 1995;355:455–469. doi: 10.1002/cne.903550310. [DOI] [PubMed] [Google Scholar]
- 43.Tamaru Y, Nomura S, Mizuno N, Shigemoto R. Distribution of metabotropic glutamate receptor mGluR3 in the mouse CNS: differential location relative to pre- and postsynaptic sites. Neuroscience. 2001;106:481–503. doi: 10.1016/s0306-4522(01)00305-0. [DOI] [PubMed] [Google Scholar]
- 44.Bischoff S, Barhanin J, Bettler B, Mulle C, Heinemann S. Spatial distribution of kainate receptor subunit mRNA in the mouse basal ganglia and ventral mesencephalon. JComp Neurol. 1997;379:541–562. doi: 10.1002/(sici)1096-9861(19970324)379:4<541::aid-cne6>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 45.Isaac JTR, Nicoll RA, Malenka RC. Silent glutamatergic synapses in the mammalian brain. CanJPhysiolPharmacol. 1997;77:735–737. [PubMed] [Google Scholar]
- 46.Monaghan DT, Buller AL. Anatomical, pharmacological, and molecular diversity of native NMDA receptor subtypes. In: Collingridge GL, Watkins JC, editors. The NMDA Receptor. 2. New York: Oxford University Press; 1994. pp. 158–176. [Google Scholar]
- 47.Petralia RS, Wang Y-X, Wenthold RJ. Histological and ultrastructural localization of the kainite receptor subunits, KA2 and GluR6/7, in the rat nervous system using selective antipeptide antibodies. JComp Neurol. 1994;349:85–110. doi: 10.1002/cne.903490107. [DOI] [PubMed] [Google Scholar]
- 48.Tarazi FI, Baldessarini RJ. Regional localization of dopamine and ionotropic glutamate receptor subtypes in striatolimbic brain regions. JNeurosci Res. 1999;55:401–410. doi: 10.1002/(SICI)1097-4547(19990215)55:4<401::AID-JNR1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 49.Van Bockstaele EJ, Colago EEO. Ultrastructural localization of the kainite selective glutamate receptor in noradrenergic perikarya and dendrites of the nucleus locus coeruleus in the rat brain. BrainRes. 1996;732:223–231. doi: 10.1016/0006-8993(96)00569-0. [DOI] [PubMed] [Google Scholar]
- 50.Ady V, Perroy J, Tricoire L, Piochon C, Dadak S, Chen X, Dusart I, Fagni L, Lambolez B, Levenes C. Type 1 metabotropic glutamate receptors (mGlu1) trigger the gating of GluD2 delta glutamate receptors. EMBORep. 2014;15:103–109. doi: 10.1002/embr.201337371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kohda K, Kakegawa W, Matsuda S, Yamamoto T, Hirano H, Yuzaki M. The delta2 glutamate receptor gates long-term depression by coordinating interactions between two AMPA receptor phosphorylation sites. Proc Natl Acad Sci USA. 2013;110:E948–E957. doi: 10.1073/pnas.1218380110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Konno K, Matsuda K, Nakamoto C, Uchigashima M, Miyazaki T, Yamasaki M, Sakimura K, Yuzaki M, Watanabe M. Enriched expression of GluD1 in higher brain regions and its involvement in parallel fiber-interneuron synapse formation in the cerebellum. JNeurosci. 2014;34:7412–7424. doi: 10.1523/JNEUROSCI.0628-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Orth A, Tapken D, Hollman M. The delta subfamily of glutamate receptors: characterization of receptor chimeras and mutants. EurJNeurosci. 2013;37:1620–1630. doi: 10.1111/ejn.12193. [DOI] [PubMed] [Google Scholar]
- 54.Swerdlow NR, Shilling PD, Breier M, Trim RS, Light GA, Marie RS. Fronto-temporal-mesolimbic gene expression and heritable differences in amphetamine-disrupted sensorimotor gating in rats. Psychopharmacology. 2012;224:349–362. doi: 10.1007/s00213-012-2758-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wadiche JI, Amara SG, Kavanaugh MP. Ion fluxes associated with excitatory amino acid transport. Neuron. 1995;15:721–728. doi: 10.1016/0896-6273(95)90159-0. [DOI] [PubMed] [Google Scholar]
- 56.Arriza JL, Eliasof S, Kavanaugh MP, Amara SG. Excitatory amino acid transporter 5, a retinal glutamate transporter coupled to a chloride conductance. ProcNatlAcadSciUSA. 1997;94:4155–4160. doi: 10.1073/pnas.94.8.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tanaka K. Functions of glutamate transporters in the brain. Neurosci Res. 2000;37:15–19. doi: 10.1016/s0168-0102(00)00104-8. [DOI] [PubMed] [Google Scholar]
- 58.Patten AR, Brocardo PS, Sakiyama C, Wortman RC, Noonan A, Gil-Mohapel J, Christie BR. Impairments in hippocampal synaptic plasticity following prenatal ethanol exposure are dependent on glutathione levels. Hippocampus. 2013;23:1463–1475. doi: 10.1002/hipo.22199. [DOI] [PubMed] [Google Scholar]
- 59.Sala C, Roussignol G, Meldolesi J, Fagni L. Key role of the postsynaptic density scaffold proteins Shank and Homer in the functional architecture of Ca2+ homeostasis at dendritic spines in hippocampal neurons. JNeurosci. 2005;25:4587–4592. doi: 10.1523/JNEUROSCI.4822-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sheng M, Kim E. The Shank family of scaffold proteins. J Cell Sci. 2000;113:1851–1856. doi: 10.1242/jcs.113.11.1851. [DOI] [PubMed] [Google Scholar]
- 61.O’Connor EC, Bariselli S, Bellone C. Synaptic basis of social dysfunction: a focus on postsynaptic proteins linking group-I mGluRs with AMPARs and NMDARs. EurJ Neurosci. 2014;39:1114–1129. doi: 10.1111/ejn.12510. [DOI] [PubMed] [Google Scholar]
- 62.Funke L, Dakoji S, Bredt DS. Membrane-associated guanylate kinases regulate cell adhesion and plasticity at cell junctions. Annu RevBiochem. 2005;74:219–245. doi: 10.1146/annurev.biochem.74.082803.133339. [DOI] [PubMed] [Google Scholar]
- 63.Garbett D, Bretscher A. The surprising dynamics of scaffolding proteins. MolBiolCell. 2014;25:2315–2319. doi: 10.1091/mbc.E14-04-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shaw AS, Filbert EL. Scaffold proteins and immune-cell signaling. Nat Rev Immunol. 2009;9:47–56. doi: 10.1038/nri2473. [DOI] [PubMed] [Google Scholar]
- 65.Bard L, Sainlos M, Bouchet D, Cousins S, Mikasova L, Breillat C, Stephenson FA, Imperiali B, Choquet D, Groc L. Dynamic and specific interaction between synaptic NR2-NMDA receptor and PDZ proteins. Proc Natl Acad Sci USA. 2010;107:19561–19566. doi: 10.1073/pnas.1002690107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bockaert J, Perroy J, Bécamel C, Marin P, Fagni L. GPCR interacting proteins (GIPs) in the nervous system: roles in physiology and pathologies. Annu Rev PharmacolToxicol. 2010;50:89–109. doi: 10.1146/annurev.pharmtox.010909.105705. [DOI] [PubMed] [Google Scholar]
- 67.Tao-Cheng JH, Yang Y, Reese TS, Dosemeci A. Differential distribution of Shank and GKAP at the postsynaptic density. PLoSOne. 2015;10:e0118750. doi: 10.1371/journal.pone.0118750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Béïque JC, Lin DT, Kang MG, Aizawa H, Takamiya K, Huganir RL. Synapse-specific regulation of AMPA receptor function by PSD-95. Proc Natl Acad Sci USA. 2006;103:19535–19540. doi: 10.1073/pnas.0608492103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schnell E, Sizemore M, Karimzadegan S, Chen L, Bredt DS, Nicoll RA. Direct interactions between PSD-95 and stargazin control synaptic AMPA receptor number. ProcNatlAcad SciUSA. 2002;99:13902–13907. doi: 10.1073/pnas.172511199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen J, Yu S, Fu Y, Li X. Synaptic proteins and receptors defects in autism spectrum disorders. FrontCellNeurosci. 2014;8:276. doi: 10.3389/fncel.2014.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jaffrey SR, Snowman AM, Eliasson MJL, Cohen NA, Snyder SH. CAPON: a protein associated with neuronal nitric oxide synthase that regulates its interaction with PSD95. Neuron. 1998;20:115–124. doi: 10.1016/s0896-6273(00)80439-0. [DOI] [PubMed] [Google Scholar]
- 72.Garner CC, Nash J, Huganir RL. PDZ domains in synapse assembly and signaling. TrendsCellBiol. 2000;10:274–280. doi: 10.1016/s0962-8924(00)01783-9. [DOI] [PubMed] [Google Scholar]
- 73.Kornau HC, Seeburg PH, Kennedy MB. Interaction of ion channels and receptors with PDZ domain proteins. CurrOpin Neurobiol. 1997;7:368–373. doi: 10.1016/s0959-4388(97)80064-5. [DOI] [PubMed] [Google Scholar]
- 74.Ziff EB. Enlightening the postsynaptic density. Neuron. 1997;19:1163–1174. doi: 10.1016/s0896-6273(00)80409-2. [DOI] [PubMed] [Google Scholar]
- 75.El-Husseini AE, Schnell E, Chetkovich DM, Nicoll RA, Bredt DS. PSD-95 involvement in maturation of excitatory synapses. Science. 2000;290:1364–1368. [PubMed] [Google Scholar]
- 76.Malenka RC, Nicoll RA. Long-term potentiation—a decade of progress? Science. 1999;285:1870–1874. doi: 10.1126/science.285.5435.1870. [DOI] [PubMed] [Google Scholar]
- 77.Malinow R, Mainen ZF, Hayashi Y. LTP mechanisms: from silence to four-lane traffic. CurrOpin Neurobiol. 2000;10:352–357. doi: 10.1016/s0959-4388(00)00099-4. [DOI] [PubMed] [Google Scholar]
- 78.Beattie EC, Carroll RC, Yu X, Morishita W, Yasuda H, von Zastrow M, Malenka RC. Regulation of AMPA receptor endocytosis by a signaling mechanism shared with LTD. NatNeurosci. 2000;3:1291–1300. doi: 10.1038/81823. [DOI] [PubMed] [Google Scholar]
- 79.Ehlers MD. Reinsertion or degradation of AMPA receptors determined by activity-dependent endocytic sorting. Neuron. 2000;28:511–525. doi: 10.1016/s0896-6273(00)00129-x. [DOI] [PubMed] [Google Scholar]
- 80.Lin JW, Ju W, Foster K, Lee SH, Ahmadian G, Wyszynski M, Wang YT, Sheng M. Distinct molecular mechanisms and divergent endocytotic pathways of AMPA receptor internalization. NatNeurosci. 2000;3:1282–1290. doi: 10.1038/81814. [DOI] [PubMed] [Google Scholar]
- 81.Lüscher C, Xia H, Beattie EC, Carroll RC, von Zastrow M, Malenka RC, Nicoll RA. Role of AMPA receptor cycling in synaptic transmission and plasticity. Neuron. 1999;24:649–658. doi: 10.1016/s0896-6273(00)81119-8. [DOI] [PubMed] [Google Scholar]
- 82.Tomita S, Nicoll RA, Bredt DS. PDZ protein interactions regulating glutamate receptor function and plasticity. JCellBiol. 2001;153:F19–F24. doi: 10.1083/jcb.153.5.f19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cozzoli DK, Goulding SP, Zhang P-W, Xiao B, Hu J-H, Ary AW, Obara I, Rahn A, Abou-Ziab H, Tyrrel B, Marini C, Yoneyama N, Metten P, Snelling C, Dehoff M, Crabbe JC, Finn DA, Klugmann M, Worley PF, Szumlinski KK. Binge drinking up-regulated accumbens mGluR5-Homer2-PI3K signaling: functional implications for alcoholism. JNeurosci. 2009;29:8655–8668. doi: 10.1523/JNEUROSCI.5900-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cozzoli DK, Courson J, Caruana AL, Miller BW, Thompson AB, Wroten M, Zhang PW, Xiao B, Hu J-H, Klugmann M, Metten P, Worley PW, Crabbe JC, Szumlinski KK. Accumbens shell metabotropic glutamate receptor 5-associated signaling regulates binge alcohol drinking: evidence from drinking-in-the-dark studies. Alcohol Clin Exp Res. 2012;36:1623–1633. doi: 10.1111/j.1530-0277.2012.01776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cozzoli DK, Courson J, Wroten MG, Greentree DI, Lum EN, Campbell RR, Thompson AB, Worley PF, Jonquieres G, Klugmann M, Finn DA, Szumlinski KK. Binge alcohol drinking by mice requires intact Group1 metabotropic glutamate receptor signaling within the central nucleus of the amygdala. Neuropsychopharmacology. 2014;39:435–444. doi: 10.1038/npp.2013.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Goulding SP, Obara I, Lominac KD, Gould AT, Miller BW, Klugmann M, Szumlinski KK. Accumbens Homer2-mediated signaling: a factor contributing to mouse strain differences in alcohol drinking? GenesBrainBehav. 2011;10:111–126. doi: 10.1111/j.1601-183X.2010.00647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Szumlinski KK, Dehoff MH, Kang SH, Frys KA, Lominac KD, Rohrer J, Griffin W, III, Klugmann M, Toda S, Champtiaux NP, Berry T, Shealy S, During M, Middaugh LD, Worley PF, Kalivas PW. Homer proteins regulate vulnerability to cocaine. Neuron. 2004;43:401–413. doi: 10.1016/j.neuron.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 88.Szumlinski KK, Lominac KD, Oleson EB, Walker JK, Mason A, Dehoff MH, Klugmann M, Cagle S, Welt K, During MT, Worley PF, Middaugh LD, Kalivas PW. Homer2 is necessary for ethanol-induced neuroplasticity. J Neurosci. 2005;25:7054–7061. doi: 10.1523/JNEUROSCI.1529-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Szumlinski KK, Ary AW, Lominac KD, Klugmann M, Kippin TE. Accumbens Homer2 over-expression facilitates alcohol-induced neuroplasticity in C57BL/6J mice. Neuropsychopharmacology. 2008;33:1365–1378. doi: 10.1038/sj.npp.1301473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Harris BZ, Lim WA. Mechanism and role of PDZ domains in signaling complex assembly. JCellSci. 2001;114:3219–3231. doi: 10.1242/jcs.114.18.3219. [DOI] [PubMed] [Google Scholar]
- 91.Jani K, Schöck F. Zasp is required for the assembly of functional integrin adhesion sites. JCellBiol. 2007;179:1583–1597. doi: 10.1083/jcb.200707045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Maday S, Anderson E, Chang HC, Shorter J, Satoh A, Sfakianos J, Fölsch H, Anderson JM, Walther Z, Mellman I. A PDZ-binding motif controls basolateral targeting of syndecan-1 along the biosynthetic pathway in polarized epithelial cells. Tra/c. 2008;9:1915–1924. doi: 10.1111/j.1600-0854.2008.00805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shiraishi-Yamaguchi Y, Furuichi T. The Homer family of proteins. Genome Biol. 2007;8:206–212. doi: 10.1186/gb-2007-8-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Worley PF, Zeng W, Huang G, Kim JY, Shin DM, Kim MS, Yuan JP, Kiselyov K, Muallem S. Homer proteins in Ca2+ signaling by excitable and non-excitable cells. Cell Calcium. 2007;42:363–371. doi: 10.1016/j.ceca.2007.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chandler LJ. Ethanol and brain plasticity: receptors and molecular networks of the postsynaptic density as targets of ethanol. PharmacolTher. 2003;99:311–326. doi: 10.1016/s0163-7258(03)00096-2. [DOI] [PubMed] [Google Scholar]
- 96.Grace AA, Floresco SB, Goto Y, Lodge DJ. Regulation of firing of dopaminergic neurons and control of goal-directed behaviors. Trends Neurosci. 2007;30:220–227. doi: 10.1016/j.tins.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 97.Kalivas PW, Lalumiere RT, Knackstedt L, Shen H. Glutamate transmission in addiction. Neuropharmacology. 2009;56(suppl 1):169–173. doi: 10.1016/j.neuropharm.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Koob GF. Alcoholism: allostasis and beyond. AlcoholClinExpRes. 2003;27:232–243. doi: 10.1097/01.ALC.0000057122.36127.C2. [DOI] [PubMed] [Google Scholar]
- 99.McBride WJ. Central nucleus of the amygdala and the effects of alcohol and alcohol-drinking behavior in rodents. PharmacolBiochemBehav. 2002;71:509–515. doi: 10.1016/s0091-3057(01)00680-3. [DOI] [PubMed] [Google Scholar]
- 100.McCool BA. Ethanol modulation of synaptic plasticity. Neuropharmacology. 2011;61:1097–1108. doi: 10.1016/j.neuropharm.2010.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Steketee JD, Kalivas PW. Drug wanting: behavioral sensitization and relapse to drug-seeking behavior. PharmacolRev. 2011;63:348–365. doi: 10.1124/pr.109.001933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Spanagel R. Alcoholism: a systems approach from molecular physiology to addictive behavior. PhysiolRev. 2009;89:649–705. doi: 10.1152/physrev.00013.2008. [DOI] [PubMed] [Google Scholar]
- 103.Gass JT, Olive MF. Glutamatergic substrates of drug addiction and alcoholism. Biochem Pharmacol. 2008;75:218–265. doi: 10.1016/j.bcp.2007.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Quintero GC. Role of nucleus accumbens glutamatergic plasticity in drug addiction. Neuropsychiatric DisTreat. 2013;9:1499–1512. doi: 10.2147/NDT.S45963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rao PSS, Bell RL, Engleman EA, Sari Y. Targeting glutamate uptake to treat alcohol use disorders. FrontNeurosci. 2015;9:144. doi: 10.3389/fnins.2015.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Reissner KJ, Kalivas PW. Using glutamate homeostasis as a target for treating addictive disorders. BehavPharmacol. 2010;21:514–522. doi: 10.1097/FBP.0b013e32833d41b2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vengeliene V, Bilbao A, Molander A, Spanagel R. Neuropharmacology of alcohol addiction. BrJPharmacol. 2008;154:299–315. doi: 10.1038/bjp.2008.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lallemand F, Ward RJ, de Witte P, Vernanck P. Binge-drinking +/− chronic nicotine administration alters extracellular glutamate and arginine levels in the nucleus accum-bens of adult male and female Wistar rats. AlcoholAlcohol. 2011;46:373–382. doi: 10.1093/alcalc/agr031. [DOI] [PubMed] [Google Scholar]
- 109.Carlezon WA, Wise RA. Microinjections of phencyclidine (PCP) and related drugs into nucleus accumbens shell potentiate medial forebrain bundle brain stimulation reward. Psychopharmacology. 1996;128:413–420. doi: 10.1007/s002130050151. [DOI] [PubMed] [Google Scholar]
- 110.Ding ZM, Rodd ZA, Engleman EA, Bailey JA, Lahiri DK, McBride WJ. Alcohol drinking and deprivation alter basal extracellular glutamate concentrations and clearance in the mesolimbic system of alcohol-preferring (P) rats. Addict Biol. 2013;18:297–306. doi: 10.1111/adb.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Szumlinski KK, Diab ME, Friedman R, Henze LM, Lominac KD, Bowers MS. Accumbens neurochemical adaptations produced by binge-like alcohol consumption. Psychopharmacology. 2007;190:415–431. doi: 10.1007/s00213-006-0641-7. [DOI] [PubMed] [Google Scholar]
- 112.Floyd DW, Jung KY, McCool BA. Chronic ethanol ingestion facilitates N-methyl-D aspartate receptor function and expression in rat lateral/basolateral amygdala neurons. JPharmacolExpTher. 2003;307:1020–1029. doi: 10.1124/jpet.103.057505. [DOI] [PubMed] [Google Scholar]
- 113.Chandler LJ, Newsom H, Sumners C, Crews F. Chronic ethanol exposure potentiates NMDA excitotoxicity in cerebral cortical neurons. JNeurochem. 1993;60:1578–1581. doi: 10.1111/j.1471-4159.1993.tb03326.x. [DOI] [PubMed] [Google Scholar]
- 114.Chefer V, Meis J, Wang G, Kuzmin A, Bakalkin G, Shippenberg T. Repeated exposure to moderate doses of ethanol augments hippocampal glutamate neurotransmission by increasing release. Addict Biol. 2010;16:229–237. doi: 10.1111/j.1369-1600.2010.00272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ward RJ, Colivicchi MA, Allen R, Schol F, Lallemand F, de Witte P, Ballini C, Corte LD, Dexter D. Neuro-inflammation induced in the hippocampus of ‘binge drinking’ rats may be mediated by elevated extracellular glutamate content. J Neurochem. 2009;111:1119–1128. doi: 10.1111/j.1471-4159.2009.06389.x. [DOI] [PubMed] [Google Scholar]
- 116.Xiao C, Shao XM, Olive MF, Griffin WC, III, Li K-Y, Krnjevic K, Zhou C, Ye J-H. Ethanol facilitates glutamatergic transmission to dopamine neurons in the ventral tegmental area. Neuropsychopharmacology. 2009;34:307–318. doi: 10.1038/npp.2008.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ding ZM, Engleman EA, Rodd ZA, McBride WJ. Ethanol increases glutamate neu-rotransmission in the posterior ventral tegmental area of female Wistar rats. AlcoholClin ExpRes. 2012;36:633–640. doi: 10.1111/j.1530-0277.2011.01665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.McBride WJ, Murphy JM, Lumeng L, Li T-K. Effects of ethanol on monoamine and amino acid release from cerebral cortex slices of the alcohol-preferring P line of rats. AlcoholClinExpRes. 1986;10:205–208. doi: 10.1111/j.1530-0277.1986.tb05072.x. [DOI] [PubMed] [Google Scholar]
- 119.Selim M, Bradburry CW. Effect of ethanol on extracellular 5-HT and glutamate in the nucleus accumbens and prefrontal cortex: comparison between the Lewis and Fischer 344 rat strains. BrainRes. 1996;716:157–164. doi: 10.1016/0006-8993(95)01385-7. [DOI] [PubMed] [Google Scholar]
- 120.Kapasova Z, Szumlinski KK. Strain differences in alcohol induced neurochemical plasticity: a role for accumbens glutamate in alcohol intake. Alcohol Clin Exp Res. 2008;32:617–631. doi: 10.1111/j.1530-0277.2008.00620.x. [DOI] [PubMed] [Google Scholar]
- 121.Gass JT, Sinclair CM, Cleva RM, Widholm JJ, Olive MF. Alcohol-seeking behavior is associated with increased glutamate transmission in basolateral amygdala and nucleus accumbens as measured by glutamate-oxidase-coated biosensors. Addict Biol. 2010;16:215–228. doi: 10.1111/j.1369-1600.2010.00262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nevo I, Hamon M. Neurotransmitter and neuromodulatory mechanisms involved in alcohol abuse and alcoholism. NeurochemInt. 1995;26:305–342. doi: 10.1016/0197-0186(94)00139-l. [DOI] [PubMed] [Google Scholar]
- 123.Griffin WC, III, Haun HL, Hazelbaker CL, Ramachandra VS, Becker HC. Increased extracellular glutamate in the nucleus accumbens promotes excessive ethanol drinking in ethanol dependent mice. Neuropsychopharmacology. 2014;39:707–717. doi: 10.1038/npp.2013.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bauer J, Pedersen A, Scherbaum N, Bening J, Patschke J, Kugel H, Heindel W, Arolt V, Ohrmann P. Craving in alcohol-dependent patients after detoxification is related to glutamatergic dysfunction in the nucleus accumbens and the anterior cingulate cortex. Neuropsychopharmacology. 2013;38:1401–1408. doi: 10.1038/npp.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ende G, Hermann D, Demirakca T, Hoerst M, Tunc-Skarka N, Weber-Fahr W, Wichert S, Rabinstein J, Frischknecht U, Mann K, Vollstädt-Klein S. Loss of control of alcohol use and severity of alcohol dependence in non-treatment-seeking heavy drinkers are related to lower glutamate in frontal white matter. Alcohol Clin Exp Res. 2013;37:1643–1649. doi: 10.1111/acer.12149. [DOI] [PubMed] [Google Scholar]
- 126.Hermann D, Weber-Fahr W, Sartorius A, Hoerst M, Frischknecht U, Tunc-Skarka N, Perreau-Lenz S, Hansson AC, Krumm B, Kiefer F, Spanagel R, Mann K, Ende G, Sommer WH. Translational magnetic resonance spectroscopy reveals excessive central glutamate levels during alcohol withdrawal in humans and rats. Biol Psychiatry. 2012;71:1015–1021. doi: 10.1016/j.biopsych.2011.07.034. [DOI] [PubMed] [Google Scholar]
- 127.Karkkainen O, Kupila J, Hakkinen M, Laukkanen V, Tupala E, Kautiainen H, Tiihonen J, Storvik M. AMPA receptors in post-mortem brains of Cloninger type 1 and 2 alcoholics: a whole-hemisphere autoradiography study. Psychiatry Res Neuroimaging. 2013;214:429–434. doi: 10.1016/j.pscychresns.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 128.Kryger R, Wilce PA. The effects of alcoholism on the human basolateral amygdala. Neuroscience. 2010;167:361–371. doi: 10.1016/j.neuroscience.2010.01.061. [DOI] [PubMed] [Google Scholar]
- 129.Kupila J, Karkkainen O, Laukkanen V, Tupala E, Tiihonen J, Storvik M. mGluR1/5 receptor densities in the brains of alcoholic subjects: a whole hemisphere autoradiography study. PsychiatryRes. 2013;212:245–250. doi: 10.1016/j.pscychresns.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 130.Laukkainen V, Karkkainen O, Kupila J, Kautiainen H, Tiihonen J, Storvik M. Increased metabotropic glutamate 2/3 receptor binding in the perigenual anterior cingulate cortex of Cloninger type 2 alcoholics: a whole-hemisphere autoradiography study. AlcoholAlcohol. 2015;50:62–67. doi: 10.1093/alcalc/agu081. [DOI] [PubMed] [Google Scholar]
- 131.Thoma R, Mullins P, Ruhl D, Monnig M, Yeo RA, Caprihan A, Bogenschutz M, Lysne P, Tonigan S, Kalyanam R, Gasparovic C. Perturbation of the glutamate–glutamine system in alcohol dependence and remission. Neuropsychopharmacology. 2011;36:1359–1365. doi: 10.1038/npp.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hermens DF, Chitty KM, Lee RSC, Tickell A, Haber PS, Naismith SL, Hickie IB, Lagopoulas J. Hippocampal glutamate is increased and associated with risky drinking in young adults with major depression. JA¡ect Disord. 2015;186:95–98. doi: 10.1016/j.jad.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 133.Cohen-Gilbert JE, Sneider JT, Crowley DJ, Rosso IM, Jensen JE, Silveri MM. Impact of family history of alcoholism on glutamine/glutamate ratio in anterior cingulate cortex in substance-naive adolescents. Dev Cogn Neurosci. 2015 doi: 10.1016/j.dcn.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Schroeder JP, Overstreet DH, Hodge CW. The mGluR5 antagonist MPEP decreases operant ethanol self-administration during maintenance and after repeated alcohol deprivations in alcohol-preferring (P) rats. Psychopharmacology (Berl) 2005;179:262–270. doi: 10.1007/s00213-005-2175-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Besheer J, Faccidomo S, Grondin JJ, Hodge CW. Regulation of motivation to self-administer ethanol by mGluR5 in alcohol-preferring (P) rats. Alcohol Clin Exp Res. 2008;32:209–221. doi: 10.1111/j.1530-0277.2007.00570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hodge CW, Miles MF, Sharko AC, Stevenson RA, Hillmann JR, Lepoutre V, Besheer J, Schroeder JP. The mGluR5 antagonist MPEP selectively inhibits the onset and maintenance of ethanol self-administration in C57BL/6J mice. Psychopharmacology. 2006;183:429–438. doi: 10.1007/s00213-005-0217-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lominac KD, Kapasova Z, Hannun RA, Patterson C, Middaugh LD, Szumlinski KK. Behavioral and neurochemical interactions between Group 1 mGluR antagonists and ethanol: potential insight into their anti-addictive properties. Drug Alcohol Depend. 2006;85:142–156. doi: 10.1016/j.drugalcdep.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 138.Olive MF, McGeehan AJ, Kinder JR, McMahon T, Hodge CW, Janak PH, Messing RO. The mGluR5 antagonist 6-methyl-2-(phenylethynyl)pyridine decreases ethanol consumption via a protein kinase C-epsilon dependent mechanism. J Pharmacol Exp Ther. 2005;67:349–355. doi: 10.1124/mol.104.003319. [DOI] [PubMed] [Google Scholar]
- 139.Schroeder JP, Spanos M, Stevenson JR, Besheer J, Salling M, Hodge CW. Cue-induced reinstatement of alcohol-seeking behavior is associated with increased ERK1/2 phosphorylation in specific limbic brain regions: blockade by the mGluR5 antagonist MPEP. Neuropharmacology. 2008;55:546–554. doi: 10.1016/j.neuropharm.2008.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zamora-Martinez ER, Edwards S. Neuronal extracellular signal-regulated kinase (ERK) activity as marker and mediator of alcohol and opioid dependence. Front Integr Neurosci. 2014;8:24. doi: 10.3389/fnint.2014.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Besheer J, Grondin JJ, Cannady R, Sharko AC, Faccidomo S, Hodge CW. Metabotropic glutamate receptor 5 activity in the nucleus accumbens is required for the maintenance of ethanol self-administration in a rat genetic model of high alcohol intake. BiolPsychiatry. 2010;67:812–822. doi: 10.1016/j.biopsych.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cowen MS, Djouma E, Lawrence AJ. The metabotropic glutamate 5 receptor antagonist 3-[(2-methyl-1,3-thiazol-4-yl)ethynyl]-pyridine reduces ethanol self-administration in multiple strains of alcohol-preferring rats and regulates olfactory glutamatergic systems. JPharmacolExpTher. 2005;315:590–600. doi: 10.1124/jpet.105.090449. [DOI] [PubMed] [Google Scholar]
- 143.Besheer J, Faccidomo S, Grondin JJ, Hodge CW. Effects of mGlu1-receptor blockade on ethanol self-administration in inbred alcohol-preferring rats. Alcohol. 2008;42:13–20. doi: 10.1016/j.alcohol.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lum EN, Campbell RR, Rostock C, Szumlinski KK. mGluR1 within the nucleus accumbens regulates alcohol intake in mice under limited-access conditions. Neuropharmacology. 2014;79:679–687. doi: 10.1016/j.neuropharm.2014.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rodd ZA, McKinzie DL, Bell RL, McQueen VK, Murphy JM, Schoepp DD, McBride WJ. The metabotropic glutamate 2/3 receptor agonist LY404039 reduces alcohol-seeking but not alcohol self-administration in alcohol-preferring (P) rats. Behav Brain Res. 2006;171:207–215. doi: 10.1016/j.bbr.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 146.Cannella N, Halbout B, Uhrig S, Evrard L, Corsi M, Corti C, Deroche-Gamonet V, Hansson AC, Spanagel R. The mGluR2/3 agonist LY379268 induced anti-reinstatement effects in rats exhibiting addition-like behavior. Neuropsycholpharmacology. 2013;38:2048–2056. doi: 10.1038/npp.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Enoch M-A, Rosser AA, Zhou Z, Mash DC, Yuan Q, Goldman D. Expression of glutamatergic genes in healthy humans across 16 brain regions; altered expression in the hippocampus after chronic exposure to alcohol or cocaine. Genes Brain Behav. 2014;13:758–768. doi: 10.1111/gbb.12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jin Z, Bhandage AK, Bazov I, Kononenko O, Bakalkin G, Korpi ER, Birnir B. Selective increases of AMPA, NMDA, and kainite receptor subunit mRNAs in the hippocampus and orbitofrontal cortex but not in prefrontal cortex of human alcoholics. Front Cell Neurosci. 2014;8:11. doi: 10.3389/fncel.2014.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cannady R, Fisher KR, Durant B, Besheer J, Hodge CW. Enhanced AMPA receptor activity increases operant alcohol self-administration and cue-induced reinstatement. AddictBiol. 2013;18:54–65. doi: 10.1111/adb.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bäckström P, Hyytiä P. Ionotropic glutamate receptor antagonists modulate cue-induced reinstatement of ethanol-seeking behavior. AlcoholClinExpRes. 2004;28:558–565. doi: 10.1097/01.alc.0000122101.13164.21. [DOI] [PubMed] [Google Scholar]
- 151.Watterson LR, Olive MF. Are AMPA receptor positive allosteric modulators potential pharmacotherapeutics for addiction? Pharmaceuticals. 2014;7:29–45. doi: 10.3390/ph7010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Krystal JH, Petrakis IL, Mason G, Trevisan L, D’Souza DC. N-methyl-D-aspartate glutamate receptors and alcoholism: reward, dependence, treatment, and vulnerability. PharmacolTher. 2003;99:79–94. doi: 10.1016/s0163-7258(03)00054-8. [DOI] [PubMed] [Google Scholar]
- 153.Alhaddad H, Kim NT, Aal-Aaboda M, Althobaiti YS, Leighton J, Boddu SH, Wei Y, Sari Effects of MS-153 on chronic ethanol consumption and GLT1 modulation of glutamate levels in male alcohol-preferring rats. FrontBehavNeurosci. 2014;8:366. doi: 10.3389/fnbeh.2014.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Melendez RI, Hicks MP, Cagle SS, Kalivas PW. Ethanol exposure decreases glutamate uptake in the nucleus accumbens. AlcoholClinExpRes. 2005;29:326–333. doi: 10.1097/01.alc.0000156086.65665.4d. [DOI] [PubMed] [Google Scholar]
- 155.Sari Y, Sreemantula SN. Neuroimmunophilin GPI-1046 reduces ethanol consumption in part through activation of GLT1 in alcohol-preferring rats. Neuroscience. 2012;227:327–335. doi: 10.1016/j.neuroscience.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sari Y, Franklin KM, Alazizi A, Rao PS, Bell RL. Effects of ceftriaxone on the acquisition and maintenance of ethanol drinking in peri-adolescent and adult female alcohol-preferring (P) rats. Neuroscience. 2013;241:229–238. doi: 10.1016/j.neuroscience.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Rothstein JD, Van Kammen M, Levey AI, Martin LJ, Kuncl RW. Selective loss of glial glutamate transporter GLT-1 in amyotrophic lateral sclerosis. AnnNeurol. 1995;38:73–84. doi: 10.1002/ana.410380114. [DOI] [PubMed] [Google Scholar]
- 158.Abulseoud OA, Camsari UM, Ruby CL, Kasasbeh A, Choi S, Choi DS. Attenuation of ethanol withdrawal by ceftriaxone-induced upregulation of glutamate transporter EAAT2. Neuropsychopharmacology. 2014;39:1674–1684. doi: 10.1038/npp.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Das SC, Yamamoto BK, Hristov AM, Sari Y. Ceftriaxone attenuates ethanol drinking and restores extracellular glutamate concentration through normalization of GLT-1 in nucleus accumbens of male alcohol-preferring rats. Neuropharmacology. 2015;97:67–74. doi: 10.1016/j.neuropharm.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Qrunfleh AM, Alazizi A, Sari Y. Ceftriaxone, a beta-lactam antibiotic, attenuates relapse-like ethanol-drinking behavior in alcohol-preferring rats. J Psychopharmacol. 2013;27:541–549. doi: 10.1177/0269881113482529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sari Y, Sakai M, Weedman JM, Rebec GV, Bell RL. Ceftriaxone, a beta-lactam antibiotic, reduces ethanol consumption in alcohol-preferring rats. Alcohol Alcohol. 2011;46:239–246. doi: 10.1093/alcalc/agr023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sari Y, Sreemantula SN, Lee MR, Choi DS. Ceftriaxone treatment affects the levels of GLT1 and ENT1 as well as ethanol intake in alcohol-preferring rats. J Mol Neurosci. 2013;51:779–787. doi: 10.1007/s12031-013-0064-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Aal-Aaboda M, Alhaddad H, Osowik F, Nauli SM, Sari Y. Effects of (R)-(−)-5-methyl-1 nicotinoyl-2-pyrazoline on glutamate transporter 1 and cysteine/glutamate exchanger as well as ethanol drinking behavior in male, alcohol-preferring rats. J Neurosci Res. 2015;93:930–937. doi: 10.1002/jnr.23554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Alasmari F, Abuhamdah S, Sari Y. Effects of ampicillin on cystine/glutamate antiporter and glutamate transporter 1 isoforms as well as ethanol drinking in male P rats. Neurosci Lett. 2015;600:148–152. doi: 10.1016/j.neulet.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Goodwani S, Rao PSS, Bell RL, Sari Y. Amoxicillin and amoxicillin/clavulanate reduce ethanol intake and increase GLT-1 expression as well as AKT phosphorylation in mesocorticolimbic regions. BrainRes. 2015;1622:397–408. doi: 10.1016/j.brainres.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Rao PSS, Goodwani S, Bell RL, Wei Y, Boddu SH, Sari Y. Effects of ampicillin, cefazolin and cefoperazone treatments on GLT1 expressions in the mesocorticolimbic system and ethanol intake in alcohol-preferring rats. Neuroscience. 2015;295:164–174. doi: 10.1016/j.neuroscience.2015.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lin X, Yang H, Zhang H, Zhou L, Guo Z. A novel transcription mechanism activated by ethanol: induction of Slc7a11 gene expression via inhibition of the DNA-binding activity of transcriptional repressor octamer-binding transcription factor 1 (OCT-1) JBiolChem. 2013;288:14815–14823. doi: 10.1074/jbc.M113.466565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhou FC, Sahr RN, Sari Y, Behbahani K. Glutamate and dopamine synaptic terminals in extended amygdala after 14-week chronic alcohol drinking in inbred alcohol-preferring rats. Alcohol. 2006;39:39–49. doi: 10.1016/j.alcohol.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 169.Rani VA, Nadiger HA, Marcus SR, Chandrakala MV, Sadasivudu B. Acute and short term effects of ethanol on the metabolism of glutamaic acid and GABA in rat brain. NeurochemRes. 1985;10:297–306. doi: 10.1007/BF00964575. [DOI] [PubMed] [Google Scholar]
- 170.Falco AM, Bergstrom HC, Bachus SE, Smith RF. Persisting changes in basolateral amygdala mRNAs after chronic ethanol consumption. PhysiolBehav. 2009;96:169–173. doi: 10.1016/j.physbeh.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 171.Bondy SC, Guo SX. Regional selectivity in ethanol-induced pro-oxidant events within the brain. BiochemPharmacol. 1995;49:69–72. doi: 10.1016/0006-2952(94)00442-o. [DOI] [PubMed] [Google Scholar]
- 172.Babu PP, Kumari R, Vemuri MC. Differential changes in cell morphology, macromo-lecular composition and membrane protein profiles of neurons and astrocytes in chronic ethanol treated rats. MolCellBiochem. 1994;130:29–40. doi: 10.1007/BF01084265. [DOI] [PubMed] [Google Scholar]
- 173.Matsumoto I. Proteomics approach in the study of the pathophysiology of alcohol-related brain damage. AlcoholAlcohol. 2009;44:171–176. doi: 10.1093/alcalc/agn104. [DOI] [PubMed] [Google Scholar]
- 174.Hargreaves GA, Quinn H, Kashem MA, Matsumoto I, McGregor IS. Proteomic analysis demonstrates adolescent vulnerability to lasting hippocampal changes following chronic alcohol consumption. AlcoholClinExpRes. 2009;33:86–94. doi: 10.1111/j.1530-0277.2008.00814.x. [DOI] [PubMed] [Google Scholar]
- 175.Swartzwelder HS, Wilson WA, Tayyeb MI. Age-dependent inhibition of long-term potentiation by ethanol in immature versus mature hippocampus. AlcoholClinExpRes. 1995;19:1480–1485. doi: 10.1111/j.1530-0277.1995.tb01011.x. [DOI] [PubMed] [Google Scholar]
- 176.Swartzwelder HS, Wilson WA, Tayyeb MI. Differential sensitivity of NMDA receptor-mediated synaptic potentials to ethanol in immature versus mature hippocampus. AlcoholClinExpRes. 1995;19:320–323. doi: 10.1111/j.1530-0277.1995.tb01509.x. [DOI] [PubMed] [Google Scholar]
- 177.Risher ML, Fleming RL, Risher WC, Miller KM, Klein RC, Wills T, Acheson SK, Moore SD, Wilson WA, Eroglu C, Swartzwelder HS. Adolescent intermittent alcohol exposure: persistence of structural and functional hippocampal abnormalities into adulthood. AlcoholClinExpRes. 2015;39:989–997. doi: 10.1111/acer.12725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Staples MC, Kim A, Mandyam CD. Dendritic remodeling of hippocampal neurons is associated with altered NMDA receptor expression in alcohol dependent rats. MolCell Neurosci. 2015;65:153–162. doi: 10.1016/j.mcn.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Klugmann M, Szumlinski KK. Targeting Homer genes using AAV: lessons learned from behavioural and neurochemical studies. BehavPharmacol. 2008;19:485–500. doi: 10.1097/FBP.0b013e32830c369f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Obara I, Bell RL, Goulding SP, Reyes CM, Larson LA, Ary AW, Truitt WA, Szumlinski KK. Differential effects of chronic ethanol consumption and withdrawal on homer/ glutamate receptor expression in subregions of the accumbens and amygdala of P rats. AlcoholClinExpRes. 2009;33:1924–1934. doi: 10.1111/j.1530-0277.2009.01030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Joslyn G, Ravindranathan A, Brush G, Schuckit M, White RL. Human variation in alcohol response is influenced by variation in neuronal signaling genes. AlcoholClinExp Res. 2010;34:800–812. doi: 10.1111/j.1530-0277.2010.01152.x. [DOI] [PubMed] [Google Scholar]
- 182.Petrakis IL, Limoncelli D, Gueorguieva R, Jatlow P, Boutros NN, Trevisan L, Gelernter J, Krystal JH. Altered NMDA glutamate receptor antagonist response in individuals with a family vulnerability to alcoholism. AmJPsychiatry. 2004;161:1776–1782. doi: 10.1176/ajp.161.10.1776. [DOI] [PubMed] [Google Scholar]
- 183.Reimers MA, Riley BP, Kalsi G, Kertes DA, Kendler KS. Pathway based analysis of genotypes in relation to alcohol dependence. PharmacogenomicsJ. 2012;12:342–348. doi: 10.1038/tpj.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Karpyak VM, Geske JR, Colby CL, Mrazek DA, Biernaka JM. Genetic variability in the NMDA-dependent AMPA trafficking cascade is associated with alcohol dependence. AddictBiol. 2011;17:798–806. doi: 10.1111/j.1369-1600.2011.00338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Schumann G, Johann M, Frank J, Preuss U, Dahmen N, Laucht M, Rietschel M, Rujescu D, Lourdusamy A, Clarke T-K, Krause K, Dyer A, Depner M, Wellek S, Treutlein J, Szegedi A, Giegling I, Cichon S, Blomeyer D, Heinz A, Heath S, Lathrop M, Wodarz N, Soyka M, Spanagel R, Mann K. Systematic analysis of gluta-matergic neurotransmission genes in alcohol dependence and adolescent risky drinking behavior. ArchGenPsychiatry. 2008;65:826–838. doi: 10.1001/archpsyc.65.7.826. [DOI] [PubMed] [Google Scholar]
- 186.Domart M-C, Benyamina A, Lemoine A, Bourgain C, Blecha L, Debuire B, Reynaud M, Saffroy R. Association between a polymorphism in the promotor of a glutamate receptor subunit gene (GRIN2A) and alcoholism. AddictBiol. 2011;17:783–785. doi: 10.1111/j.1369-1600.2011.00321.x. [DOI] [PubMed] [Google Scholar]
- 187.Preuss UW, Zill P, Koller G, Bondy B, Hesselbrock V, Soyka M. Ionotropic glutamate receptor gene GRIK3 ser310ala functional polymorphism is related to delirium tremens in alcoholics. PharmacogenomicsJ. 2006;6:34–41. doi: 10.1038/sj.tpj.6500343. [DOI] [PubMed] [Google Scholar]
- 188.Kranzler HR, Gelernter J, Anton RF, Arias AJ, Herman A, Zhao H, Burian L, Covault J. Association of markers in the 3′ region of the GluR5 kainate receptor subunit gene to alcohol dependence. AlcoholClinExpRes. 2009;33:925–930. doi: 10.1111/j.1530-0277.2009.00913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Begleiter H, Porjesz B, Reich T, Edenberg HJ, Goate A, Blangero J, Almasy L, Foroud T, Van Eerdewegh P, Polich J, et al. Quantitative trait loci analysis of human event-related brain potentials: P3 voltage. Electroenceph Clin Neurophysiol. 1998;108:244–250. doi: 10.1016/s0168-5597(98)00002-1. [DOI] [PubMed] [Google Scholar]
- 190.O’Connor S, Morzorati S, Christian JC, Li T-K. Heritable features of the auditory oddball event-related potential: peaks, latencies, morphology and topography. ElectroencephClinNeurophysiol. 1994;92:115–125. doi: 10.1016/0168-5597(94)90052-3. [DOI] [PubMed] [Google Scholar]
- 191.Chen ACH, Tang Y, Rangaswamy M, Wang JC, Almasy L, Foroud T, Edenberg HJ, Hesselbrock V, Nurnberger JI, Jr, Kuperman S, O’Connor SJ, Schuckit MA, Bauer LO, Tischfield J, Rice JP, Bierut L, Goate A, Porjesz B. Association of single nucleotide polymorphisms in a glutamate receptor gene [GRM8] with theta power of event-related oscillations and alcohol dependence. AmJMedGenetPartB. 2008;150B:359–368. doi: 10.1002/ajmg.b.30818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Long EC, Aliev F, Wang J-C, Edenberg HJ, Nurnberger JI, Jr, Hesselbrock V, Porjesz B, Dick DM. Further analyses of genetic association between GRM8 and alcohol dependence symptoms among young adults. JStudAlcoholDrugs. 2015;76:414–418. doi: 10.15288/jsad.2015.76.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Xia X, Wu Z, Ma D, Tang C, Liu L, Xin F, Zhu D, Hu J. Association of single-nucleotide polymorphisms in a metabotropic glutamate receptor GRM3 gene subunit to alcohol-dependent male subjects. AlcoholAlcohol. 2014;49:256–260. doi: 10.1093/alcalc/agu004. [DOI] [PubMed] [Google Scholar]
- 194.Comasco E, Hallman J, Wallen-Mackenzie A. Haplotype tag single nucleotide polymorphism analysis of the vesicular glutamate transporter (VGLUT) genes in severely alcoholic women. PsychiatryRes. 2014;219:403–405. doi: 10.1016/j.psychres.2014.05.052. [DOI] [PubMed] [Google Scholar]
- 195.Kuo P-H, Kalsi G, Prescott CA, Hodgkinson CA, Goldman D, Alexander J, van den Oord EJ, Chen X, Sullivan PF, Patterson DG, Walsh D, Kendler KS, Riley BP. Associations of glutamate decarboxylase genes with initial sensitivity and age-at-onset of alcohol dependence in the Irish Affected Sib Pair Study of Alcohol Dependence. DrugAlcoholDepend. 2009;101:80–87. doi: 10.1016/j.drugalcdep.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Terranova C, Tucci M, Forza G, Barzon L, Palu G, Ferrara SD. Alcohol dependence and glutamate decarboxylase gene polymorphisms in an Italian male population. Alcohol. 2010;44:407–413. doi: 10.1016/j.alcohol.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 197.Lappalainen J, Krupitsky E, Kranzler HR, Luo X, Remizov M, Pchelina S, Taraskina A, Zvartau E, Rasanen P, Makikyro T, Somberg LK, Krystal JH, Stein MB, Gelernter J. Mutation screen of the GAD2 gene and association study of alcoholism in three populations. AmJMedGenetPartB. 2007;144B:183–192. doi: 10.1002/ajmg.b.30377. [DOI] [PubMed] [Google Scholar]
- 198.Loh E-W, Lane H-Y, Chen C-H, Chang P-S, Ku L-W, Wang KHT, Cheng ATA. Glutamate decarboxylase genes and alcoholism in Han Taiwanese men. AlcoholClinExp Res. 2006;30:1817–1823. doi: 10.1111/j.1530-0277.2006.00218.x. [DOI] [PubMed] [Google Scholar]
- 199.Zhao R, Zhang R, Li W, Liao Y, Tang J, Miao Q, Hao W. Genome-wide DNA methylation patterns in discordant sib pairs with alcohol dependence. Asia Paci¢c Psychiatry. 2013;5:39–50. doi: 10.1111/appy.12010. [DOI] [PubMed] [Google Scholar]
- 200.Meinhardt MW, Hansson AC, Perreau-Lenz S, Bauder-Wenz C, Stahlin O, Heilig M, Harper C, Drescher KU, Spanagel R, Sommer WH. Rescue of infralimbic mGluR2 deficit restores control over drug-seeking behavior in alcohol dependence. J Neurosci. 2013;33:2794–2806. doi: 10.1523/JNEUROSCI.4062-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Flatscher-Bader T, Zuvela N, Landis N, Wilce PA. Smoking and alcoholism target genes associated with plasticity and glutamate transmission in the human ventral tegmental area. HumMolGenet. 2008;17:38–51. doi: 10.1093/hmg/ddm283. [DOI] [PubMed] [Google Scholar]
- 202.Zhou Z, Karlsson C, Liang T, Xiong W, Kimura M, Tapocik JD, Yuan Q, Barbier E, Feng A, Flanigan M, Augier E, Enoch MA, Hodgkinson CA, Shen PH, Lovinger DM, Edenberg HJ, Heilig M, Goldman D. Loss of metabotropic glutamate receptor 2 escalates alcohol consumption. ProcNatlAcadSciUSA. 2013;110:16963–16968. doi: 10.1073/pnas.1309839110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Bell RL, Kimpel MW, Rodd ZA, Strother WN, Bai F, Peper CL, Mayfield RD, Lumeng L, Crabb DW, McBride WJ, Witzmann FA. Protein expression changes in the nucleus accumbens and amygdala of inbred alcohol-preferring rats given either continuous or scheduled access to ethanol. Alcohol. 2006;40:3–17. doi: 10.1016/j.alcohol.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 204.Bell RL, Kimpel MW, McClintick JN, Strother WN, Carr LG, Liang T, Rodd ZA, Mayfield RD, Edenberg HJ, McBride WJ. Gene expression changes in the nucleus accumbens of alcohol-preferring rats following chronic ethanol consumption. PharmacolBiochem Behav. 2009;94:131–147. doi: 10.1016/j.pbb.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Edenberg HJ, Strother WN, McClintick JN, Tian H, Stephens M, Jerome RE, Lumeng L, Li T-K, McBride WJ. Gene expression in the hippocampus of inbred alcohol-preferring (iP) and -nonpreferring (iNP) rats. GenesBrainBehav. 2005;4:20–30. doi: 10.1111/j.1601-183X.2004.00091.x. [DOI] [PubMed] [Google Scholar]
- 206.Kimpel MW, Strother WN, McClintick JN, Carr LG, Liang T, Edenberg HJ, McBride WJ. Functional gene expression differences between inbred alcohol-preferring and -nonpreferring rats in five brain regions. Alcohol. 2007;41:95–132. doi: 10.1016/j.alcohol.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.McBride WJ, Schultz JA, Kimpel MW, McClintick JN, Wang M, You J, Rodd ZA. Differential effects of ethanol in the nucleus accumbens shell of alcohol-preferring (P), alcohol non-preferring (NP) and Wistar rats: a proteomic study. Pharmacol Biochem Behav. 2009;92:304–313. doi: 10.1016/j.pbb.2008.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.McBride WJ, Kimpel MW, Schultz JA, McClintick JN, Edenberg HJ, Bell RL. Changes in gene expression in regions of the extended amygdala of alcohol-preferring rats after binge-like alcohol drinking. Alcohol. 2010;44:171–183. doi: 10.1016/j.alcohol.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.McBride WJ, Kimpel MW, McClintick JN, Ding ZM, Hyytia P, Colombo G, Edenberg HJ, Lumeng L, Bell RL. Gene expression in the ventral tegmental area of 5 pairs of rat lines selectively bred for high or low ethanol consumption. Pharmacol Biochem Behav. 2012;102:275–285. doi: 10.1016/j.pbb.2012.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.McBride WJ, Kimpel MW, McClintick JN, Ding ZM, Hauser SR, Edenberg HJ, Bell RL, Rodd ZA. Changes in gene expression within the ventral tegmental area following repeated excessive binge-like alcohol drinking by alcohol-preferring (P) rats. Alcohol. 2013;47:367–380. doi: 10.1016/j.alcohol.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.McBride WJ, Kimpel MW, McClintick JN, Ding Z-M, Hyytia P, Colombo G, Liang T, Edenberg HJ, Lumeng L, Bell RL. Gene expression within the extended amygdala of 5 pairs of rat lines selectively bred for high or low ethanol consumption. Alcohol. 2013;47:517–529. doi: 10.1016/j.alcohol.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.McBride WJ, Kimpel MW, McClintick JN, Ding Z-M, Edenberg HJ, Liang T, Rodd ZA, Bell RL. Changes in gene expression within the extended amygdala following binge-like alcohol drinking by adolescent alcohol-preferring (P) rats. PharmacolBiochem Behav. 2014;117:52–60. doi: 10.1016/j.pbb.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.McClintick JN, McBride WJ, Bell RL, Ding Z-M, Liu Y, Xuei X, Edenberg HJ. Gene expression changes in serotonin, GABA-A receptors, neuropeptides and ion channels in the dorsal raphe nucleus of adolescent alcohol-preferring (P) rats following binge-like alcohol drinking. PharmacolBiochemBehav. 2015;129:87–96. doi: 10.1016/j.pbb.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Rodd ZA, Kimpel MW, Edenberg HJ, Bell RL, Strother WN, McClintick JN, Carr LG, Liang T, McBride WJ. Differential gene expression in the nucleus accumbens with ethanol self-administration in inbred alcohol-preferring rats. Pharmacol Biochem Behav. 2008;89:481–498. doi: 10.1016/j.pbb.2008.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Witzmann FA, Li J, Strother WN, McBride WJ, Hunter L, Crabb DW, Lumeng L, Li T-K. Innate differences in protein expression in the nucleus accumbens and hippocampus of inbred alcohol-preferring and -nonpreferring rats. Proteomics. 2003;3:1335–1344. doi: 10.1002/pmic.200300453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Gould AT, Sacramento AD, Wroten MG, Miller BW, Klugmann M, Ben-Shahar O, Szumlinski KK. Extended access to intravenous cocaine imbalances ventromedial pre-frontal cortex Homer1 versus Homer2 expression: implications for relapse. AddictBiol. 2015;20:148–157. doi: 10.1111/adb.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]