Abstract
Aberrant expression of miRNAs is critically implicated in cancer initiation and progression. Therapeutic approaches focused on regulating miRNAs are therefore a promising approach for treating cancer. Antisense oligonucleotides, miRNA sponges, and CRISPR/Cas9 genome editing systems are being investigated as tools for regulating miRNAs. Despite the accruing insights in the use of these tools, delivery concerns have mitigated clinical application of such systems. In contrast, little attention has been given to the potential of small molecules to modulate miRNA expression for cancer therapy. In these years, many researches proved that small molecules targeting cancer-related miRNAs might have greater potential for cancer treatment. Small molecules targeting cancer related miRNAs showed significantly promising results in different cancer models. However, there are still several obstacles hindering the progress and clinical application in this area. This review discusses the development, mechanisms and application of small molecules for modulating oncogenic miRNAs (oncomiRs). Attention has also been given to screening technologies and perspectives aimed to facilitate clinical translation for small molecule-based miRNA therapeutics.
Keywords: miRNA, small molecule miRNA inhibitors, oncomiR, cancer therapy
Graphical Abstract

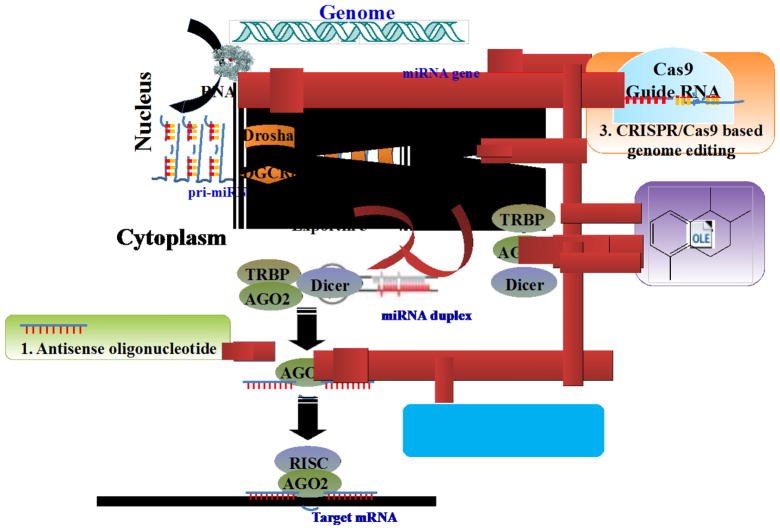
Strategies to modulate miRNA expression. i) antisense oligonucleotide (antagomir), ii) miRNA sponges, iii) CRISPR/Cas9-base genome editing, and iv) small molecule inhibitor of miRNA (SMIR)
1. Introduction
MicroRNAs (miRNAs) are single stranded small non-coding RNAs (21–23 nucleotides) that have emerged as regulators of gene expression by hindering translation and triggering degradation of target mRNA post-transcriptionally.1–3 miRNAs play a crucial role in the initiation and development of a variety of human cancers with numerous studies reporting aberrant miRNA expression as characteristic signatures.4–6 miRNAs are not only deregulated in cancers, but are also acting as oncogenes or tumor suppressors. Oncogenic miRNAs (oncomiRs) function by either inhibiting tumor suppressor genes or genes responsible for promoting apoptosis or cell differentiation and are normally upregulated in cancer (Table 1). The corresponding antagomirs, also known as anti-miRNAs, are used to suppress these oncomiRs to inhibit tumor growth. In contrast, tumor suppressor miRNAs are downregulated in cancers.5,7–9 These miRNAs function by inhibiting oncogenes or genes that hinder apoptosis or cell differentiation. For instance, miR-143 and miR-145 are both downregulated in colon cancer while miR-99 is overexpressed in pancreatic cancer.10 The widespread involvement of miRNAs across human cancers suggests their utility as new ideal therapeutic targets. RNA inhibition agents such as antisense oligonucleotides and miRNA sponges have been used to restore miRNA balance in cancer networks by inhibiting overexpressed oncomiRs. Nonetheless, intrinsic challenges associated with these systems hinder their clinical translation. Obstacles include potential off-target effects, compromised tissue specific delivery, poor cellular uptake and in vivo instability. Although numerous delivery systems were developed for animal work, most of them including nanoparticles and liposomes have been proved to be ineffective or toxic for clinical use. In this regard, small molecule modulators of miRNA function are potentially better therapeutic candidates since they can be more easily delivered and more stable to overcome serum degradation.
Table 1.
Examples of oncomiRs for miRNA inhibition treatment
| OncomiRs | Target genes | References |
|---|---|---|
| miR-21 | PDCD4, PTEN, BCL2, TPM1, RECK, | 14–18 |
| miR-17-92 cluster | PTEN, Bim | 19,20 |
| miR-221/222 | p27, TIMP2, DKK2 | 21–23 |
| miR-155 | DMTF1, annexin 7, LKB1, E2F2, GABA receptor | 24–28 |
| miR-223 | PAX6, Stathmin1, FBXW7/hCdc4 | 29–31 |
| miR-214 | PTEN, p53 | 32,33 |
| miR-191 | C/EBPβ, checkpoint kinase 2 | 34,35 |
| miR-25 | CDKN1C, LATS2, RECK | 36–38 |
First small molecule inhibitor of miRNA was developed by Gumireddy et al. for inhibition of miR-21.11 In this study, a luciferase reporter plasmid was constructed for screening and diazobenzene was finally selected as a potent compound. Since then, numbers of miRNA inhibitors targeting oncomiRs have been identified using high throughput screening or in silico sequence-based design.12 Several potent oncomiRs have been selected as potent targets for small molecule miRNA inhibitor development. Moreover, fresh approaches such as the construction of small molecule-miRNA networks for a variety of cancers are being examined as alternative ways to fast track the drug discovery process.13 Meanwhile, the mechanisms about miRNA inhibition by small molecules are elucidated. These promising findings are sparking a renaissance in developing small molecule modulators of miRNAs for cancer therapy.
In this review, we discuss the rationale and therapeutic strategies for targeting miRNAs responsible for cancer initiation and progression. We describe the development, mechanisms and applications of small molecules for modulating cancer related miRNAs. We also discuss the potential and pitfalls of small molecule modulators of miRNAs for treating cancer with emphasis on their delivery technologies to facilitate their clinical translation into cancer therapeutics.
2. miRNAs as valid Drug Targets in Cancer Therapy
Drug target selection remains a bottleneck in the quest for potent anticancer therapeutics. The current paradigm where drugs are designed to target proteins is flawed for several reasons. Since cancer is a complex process involving multiple factors and multistep processes, the efficacy of anticancer agents designed to target single therapeutic protein is often sub-optimal in cancer therapy. Although combination therapy, in which more than one targets are addressed, yields better therapeutic outcomes compared to single drug treatment, it is typically costly, associated with detrimental drug-drug interactions and involves complicated treatment regimens. Considering their dysregulated expression in cancer compared to normal tissues, miRNAs are regarded as high value drug targets for cancer therapy and targeting their expression can change cancer phenotype.39–41 miRNA-based therapeutics are attractive alternatives to protein based cancer therapy since single oncomiR always downregulates multiple anticancer genes while tumor suppressor miRNA downregulates multiple oncogenes. For example, miR-221 has an oncogenic function by targeting Bmf, a proapoptotic BH3-only protein, to inhibit cell apoptosis.42 In addition, miR-221 enhances cell migration and make cancer cell more aggressive by targeting PTEN and TIMP3.43 Let-7g suppresses tumor cell proliferation by targeting c-Myc44 and COL1A2.45 Meanwhile, Bcl-xL, an anti-apoptotic member of the Bcl-2 family, is identified as a target of let-7g to induce cell apoptosis.46 Consequently, miRNAs are highly efficient regulators of cellular processes pertinent for normal and malignant homeostasis.47–49
One rationale for miRNA-based therapeutics elegantly described by Garzon et al., is the notion of cancer networks being miRNA wired.49 The “miRNA wired cancer network” hypothesis suggests miRNAs to be the code that maintains required connection between all genes and protein networks in normal cells. As a result, normal tissues can be thoroughly characterized and miRNA expression patterns established as a coding blueprint. It might then be possible to compare this blueprint to miRNA expression patterns in cancerous tissue. Therapeutic approaches can be developed to “reboot” the cancerous tissue by restoring the miRNA patterns to the default settings observed in the normal tissue. Clearly, such a therapeutic strategy involves targeting more than just a single miRNA, gene or protein. It may involve simultaneous inhibition and replacement of more than one miRNAs. From an implementation standpoint, it might be tempting to dispose the “miRNA wired cancer network” hypothesis due to its potential complexity. Another instructive argument might be that there is no universal miRNA blueprint for normal tissues. Nonetheless, since aberrant expression of single miRNA may affect hundreds of proteins,50 reprogramming cancer network may be more feasible using miRNAs compared to proteins.
3. miRNA-based Therapeutic Strategies
For many cancers, oncogenic miRNAs are overexpressed while tumor suppressor miRNAs are downregulated. Therefore, two miRNA-based therapeutic strategies used are: (1) miRNA inhibition for addressing oncogenic miRNAs and (2) miRNA replenishment for tumor suppressor miRNAs. Similar therapeutic molecules such as oligonucleotides and small molecules may be employed in both approaches to either directly inhibit miRNAs or indirectly by targeting specific genes or transcription factors which modulate specific miRNA expression.
3.1. miRNA Inhibition
In contrast to the diminished levels of tumor suppressive miRNAs, a number of oncomiRs are overexpressed in cancer cells and directly target tumor suppressor genes. The current strategies (Fig. 1) to ablate oncomiRs include: (1) antisense oligonucleotide (ASOs, also known as antagomirs or antimiRs), which includes cholesterol-conjugated antimiRs, locked nucleic acid (LNA) antimiRs and tiny LNA antimiRs; (2) miRNA sponges which contain multiple tandem binding sites to target miRNA; (3) CRISPR/Cas9 based genome editing which modify the genome of cancer cells; and (4) small molecule inhibitors of miRNAs. ASOs are single stranded RNA molecules complementary to the target miRNAs that function as competitive inhibitors by obstructing their interaction with target miRNAs; LNA antimiRs, in which an extra methylene bridge connecting the 2′-O atom and the 4′-C atom “locks” the ribose, exhibit higher thermal stability and superior hybridization efficiency with their miRNA targets; tiny LNA antimiRs are 8nt long LNA-modified ASOs designed to target the 5′-seed region of miRNAs; miRNA sponges use transgenic overexpression of RNA molecules harboring complementary binding sites to a miRNA of interest to block the function of a given miRNA or a miRNA family; CRISPR/Cas9 is a novel technique for editing genomes by delivering Cas9 protein and guide RNAs into target cells; SMIRs are small molecule chemical compounds that interfere with miRNA biogenesis or maturation.
Figure 1.
Schemes of miRNA generation and the inhibition effect of antisense oligonucleotide, miRNA sponges, CRISPR/Cas 9 genome editing, and small molecule inhibitor of miRNA (SMIR). miRNA is transcribed by RNA polymerase II into primary transcripts pri-miRNA (~1 to 3 kb long). This pri-miRNA undergoes further processing by the ribonucleases Drosha and DiGeorge syndrome critical region gene 8 (DGCR8) complex in the nucleus, thereby resulting in a hairpin intermediate pre-miRNA (~70–100 nucleotides long) which then transported to the cytoplasm via exportin 5. In the cytoplasm, the pre-miRNA is processed by another ribonuclease, Dicer, into a mature double strand miRNA (~18–25 nucleotides long). After strand separation, the guide strand or mature miRNA is incorporated into an RNA-induced silencing complex (RISC) complex target the 3′-UTR region of mRNAs, resulting is a decreased level of targeted protein, while the passenger strand is commonly degraded. Antisense oligonucleotide and miRNA sponges work on mature miRNA while CRISPR/Cas 9 genome editing works on genome. SMIR works on almost every stage of miRNA biogenesis.
3.1.1 Antisense Oligonucleotides
The demonstration that oncomiRs are upregulated in cancer (Table 1) provides a rationale to investigate the use of antisense oligonucleotides to inhibit their expression. Antisense oligonucleotides work as competitive inhibitors of miRNAs, presumably by annealing to the mature miRNA guide strand and inducing degradation or stoichiometric duplex formation. However, the unmodified antisense oligonucleotides are degraded before reaching their targets. Thus, researchers introduced modifications to the chemical structure of oligonucleotides to increase stability, binding affinity and specificity. Among these modifications, the introduction of 2′-O-methyl groups contributes to nuclease resistance and improved binding affinities to RNA.51 Oligonucleotides with 2′-O-methyl groups have been proven to be effective inhibitors of miRNA expression in several cancer cell lines. As a proof of concept, Krutzfeldt et al. developed chemically modified (2-OMe-modified nucleotides, with a phosphorothioate linkage), cholesterol-conjugated single stranded RNA analogues (named ‘antagomirs’), complementary to miR-122, which is abundant in the liver.52 These antagomirs were injected into the tail vein of mice, and specific targeting of miR-122 in the liver was observed after 24 h. The silencing of endogenous miRNAs by this method was specific, efficient and long lasting, and the effects were observed even after 23 days post-injection. Gene expression and bioinformatics analysis of the whole transcriptome from antagomir-treated animals revealed that the 3′-UTRs of upregulated transcripts were strongly enriched in miR-122 recognition motifs, whereas downregulated genes were depleted of these motifs. To improve the binding affinity, LNA nucleotides were further developed. By “locking” the molecule with the methylene bridge, LNA oligonucleotides displayed unprecedented hybridization affinity toward complementary single stranded RNA and complementary single- or double stranded DNA.53,54 In addition, they displayed excellent mismatch discrimination and high aqueous solubility. LNA antimiRs have been used successfully in several in vitro studies to knock down specific miRNA expression.55,56 Studies in mice using LNA antimiR have shown the feasibility and high efficiency of this approach.57,58 Elmen and colleagues examined whether combining LNA antimiR with phosphorothioate modifications could improve delivery of the compounds and silence miR-122 in mice without requiring additional chemical modifications.59 This research suggests that LNA antimiRs are able to effectively silence their targets at much lower doses than cholesterol-based oligonucleotides. The simple systemic delivery of an unconjugated LNA antimiR-122 has also been shown to effectively antagonize liver-expressed miR-122 in non-human primates.60
3.1.2. miRNA Sponges
miRNA sponges are competitive inhibitors expressed from strong promoters, containing multiple, tandem binding sites to a miRNA of interest or miRNA family.61 The binding sites in these miRNA sponges are either perfectly antisense or contain mismatches in the middle position 9–12, which probably induce more stable interaction with miRNA, including miRNA complexed with Ago2. Normal miRNA sponge constructs contain four to ten binding sites separated by a few nucleotides as more binding sites increased the possibility of sponge RNA degradation.62 In recent years, miRNA sponges have been proved effective in several cell lines, including non-small long cancer cell63, embryonic neural stem cell64 and B cell lymphoma.65 To achieve stable miRNA sponge activity, several groups express the transgene from chromosomal integration and thereby perform long-term miRNA loss of function studies.66–68 The applications of miRNA sponges are to elucidate the role of miRNA in differentiation pathways69,70 and to mimic the downregulation of specific miRNA in certain diseases.71,72 For in vivo studies, viral vectors are used to deliver sponge constructs to tissue in mice73,74 while stable germline miRNA sponge expression was achieved in Drosophila using Gal4-UAS system.75
3.1.3. CRISPR/Cas9-based Genome Editing
Ishino et al. first discovered clustered regularly interspaced short palindromic repeats (CRISPR) in Escherichia coli more than 30 years ago.76 CRISPR is characterized as cell immune system that confers resistance to foreign genetic elements such as plasmids by cleaving target DNA sequence via a short RNA molecule and the endonuclease Cas9.77 Nowadays, CRISPR has been proven to be an efficient genome editing method, which includes a Cas9 protein and CRISPR RNA complex. CRISPR RNA guides the complex to a complementary sequence in the target DNA and destroys it to perform gene silencing. Several researches have been published about utilizing CRISPR/Cas9 system to perform genome editing in cultured mammalian cells.78–80 A GFP-tagged CRISPR/Cas9 imaging system was also developed to monitor telomeres and coding genes in living cells.81 As a novel gene silencing technique, researchers are concentrating to explore the application of CRISPR/Cas9 system for inhibiting miRNAs. For instance, Zhao et al. reported construction of sequence specific CRISPR/Cas9 based miRNA inhibitor to downregulate miR-17-92 cluster and miR-21, two typical oncomiRs, in vitro.82 Xiao et al. co-injected Cas9 mRNA and guide RNA into one cell-stage zebrafish embryos and obtained chromosomal deletions and inversions.83
3.2. miRNA Replenishment
miRNA replenishment therapy involves restoring of downregulated tumor suppressor miRNAs in cancer cells. Loss of tumor suppressor miRNAs causes hyperactivation of multiple cancer pathways which facilitates cancer cell proliferation, inhibits apoptosis and promotes tumor-forming ability of cancer stem cells. Introducing miRNA mimics reestablishes tumor suppressor status in cancer cells thereby hindering tumorigenesis. miRNA mimics only need to be delivered to the cytoplasm to be active and available technologies for siRNA delivery are sufficient. Compared to miRNA inhibition therapy, miRNA replacement therapy is only now being explored and may at first appear counter intuitive. However, several advantages validate its application since miRNAs are generally downregulated in cancer9,84 and miRNA mimics do not generate off-target effect once delivered.85 For example, Sun et al. reported the role of miR-1280 in suppressing melanoma by regulating Src, which acts as proto-oncogenes by mediating tumor proliferation and invasion.86 Intratumoral injection of miR-1280 complexed with the siPORT transfection reagent significantly suppressed melanoma progression in vivo. Tiwari et al. proved that oral squamous cell carcinoma cells with overexpressed miR-125a showed reduced proliferation and invasion since estrogen-related receptor α was significantly downregulated.87 For combination therapy using an anti-tumor drug and a tumor suppressor miRNA, Mittal et al. reported that combining gemcitabine and miR-205 significantly inhibited tumor growth in a subcutaneous pancreatic cancer model.88 Kumar et al. demonstrated that co-delivery of GDC-0449 and let-7b effectively decreased tumor cell proliferation with increased cell apoptosis by inhibiting hedgehog pathway.89 Recently, several small molecule modulators of tumor suppressor miRNAs were reported to restore the levels of tumor suppressor miRNAs and inhibited tumor growth in animal models.
3.3. Challenges of Non-Small Molecule miRNA Therapeutics
Non-small molecule miRNA have the potential to be an efficient method for miRNA inhibition. Babar et al. demonstrated that systemic delivery of anti-miR-155 formulated in nanoparticles resulted in rapid regression of lymphadenopathy.90 Silencing of miR-21 through miRNA inhibitor markedly antagonized B-cell lymphoma tumor growth in mouse model.91 As discussed before, LNA-modified miRNA inhibitor showed even higher efficiency of miRNA inhibition in mouse and non-human primates.60 However, there are still several crucial obstacles that need to be overcome. Currently, 2588 mature human miRNA sequences are registered in miRBase, while around 200 of them have sufficiently high expression for targeting. As discussed before, most of antisense oligonucleotides are perfect complementary to their targets with chemical modifications to improve binding affinity and stability. However, these miRNA inhibitors may not distinguish between miRNAs within the same family, causing off-target effects.92 Although the off-target effect of miRNA sponges are not reported yet, miRNA sponges always exhibit different degrees of inhibition in different contexts and it is still challenging to evaluate the degree of miRNA silencing under a sponge treatment.62 Furthermore, chemical modifications have been observed to induce sequence-independent toxicity in vivo.93 Although previously described phosphorothioate-modified antisense oligonucleotide showed significant miRNA silencing effect in non-human primate, it was shown to activate C5 complement and induce a transient decrease in peripheral white blood cell counts.94 For LNA-modified oligonucleotides, Swayze et al. reported that they had significant hepatotoxicity as measured by serum transaminase activity as well as body weight during preclinical animal tests.95 Compared to antisense oligonucleotides and miRNA sponges, CRISPR-Cas9 causes permanent genome alterations. However, its off-target effect has not been well-recognized and accurately profiled when applied in gene therapy, which significantly limits its clinical application. Moreover, this genome editing method still needs to be optimized since commonly used Cas9 gene derived from Streptococcus pyogenes is too big to be transduced, leading to less than 20% genome editing efficiency in vitro.96
Another issue which hinders the clinical development of non-small molecular miRNA inhibitors is the delivery-related concerns. Depending on the diseases and targets, people need to carefully consider and design the delivery systems to achieve optimized clinical effects. Organs, which are more accessible and responsible for metabolism and excretion including liver, kidney, and spleen, have shown exciting results for antisense oligonucleotide delivery. For example, Hatakeyama et al. encapsulated anti-miRNA oligonucleotides into pH-sensitive liposome and reduced the level of miR-122 in mouse liver.97 Kriegel et al. demonstrated that intravenously delivered LNA modified anti-miR382 blocked miR-382 expression and significantly reduced kidney medullary fibrosis.98 For hard-to-reach tissues such as solid tumors, people developed target delivery systems (less than 100 nm) including a target ligand, hydrophilic membrane, and positive charged and hydrophobic core to overcome off-target effects on normal tissues. Nevertheless, it is still difficult to ensure an effective dose reaching and entering the appropriate target cells.99 For example, tumor vessels exhibit high permeability, high hydraulic conductivity, and high interstitial pressure that slow down the diffusion and convection of nanoparticles within the tissue.100 Gilleron et al. recently reported that only 1–2% of siRNAs escaped from endosome degradation after delivering with lipid nanoparticles using an imaging-based fluorescence and electron microscopy,101 suggesting the delivery system facilitating the release of oligonucleotides may considerably decrease the effective dose in vivo. Theoretically, liposome or nanoparticle based non-viral delivery system can be used to deliver miRNA sponges or CRISPR/Cas9 based genome editing systems. However, these systems generally suffer from low gene delivery efficiency, especially for in vivo studies. Interestingly, the delivery of recombinant Cas9 protein instead of Cas9 gene can achieve a genome editing efficiency as high as 79%.102 Currently, most miRNA sponges or CRISPR/Cas9 are using viral vectors for in vitro and in vivo gene delivery due to the high delivery efficacy, which probably explained why much of current efforts have been addressed on modifying viral vectors for safe and effective clinical applications.103
4. Small Molecule miRNA Therapeutic Agents
Due to the above challenges of non-small molecule miRNA inhibitors, it would be promising to develop small molecular weight drugs to target specific miRNAs and inhibit their activities (named SMIR). Actually, miRNAs have long been neglected as promising drug targets due to their structural flexibility and highly electronegative surface.104 Furthermore, poor understanding of miRNA X-Ray crystallography or NMR structure as well as the limited availability of miRNA-Dicer or RISC complex structure makes the design of small molecule inhibitor of miRNA much more difficult.105 These might be the reasons why the first reported SMIR by Gumireddy et al.11 and most following designs were based on non-specific selection assay. For the first SMIR, they selected miR-21 as the target miRNA, which is overexpressed in various cancers including breast, ovarian, and lung cancer.106,107 Lentiviral vector encoding complementary sequences of miR-21 and downstream luciferase reporter gene was constructed for HeLa cell transduction and non-specific compound selection. As a result, diazobenzene was selected for further modification since 251% increase of luciferase signal was detected relative to untreated cells. Currently, with the advancement of RNA (or RNA and protein complex) structure simulation software, high throughput virtual screening are performed to select SMIR according to RNA secondary and tertiary structures although vector based non-specific screening is still playing a crucial role for SMIR selection. On the other hand, several studies were carried out to elucidate the inhibitory mechanism of SMIRs. In the following sub-sections, we have first reviewed the current SMIR screening methods (Fig. 2), followed by discussion of their inhibitory mechanisms.
Figure 2.
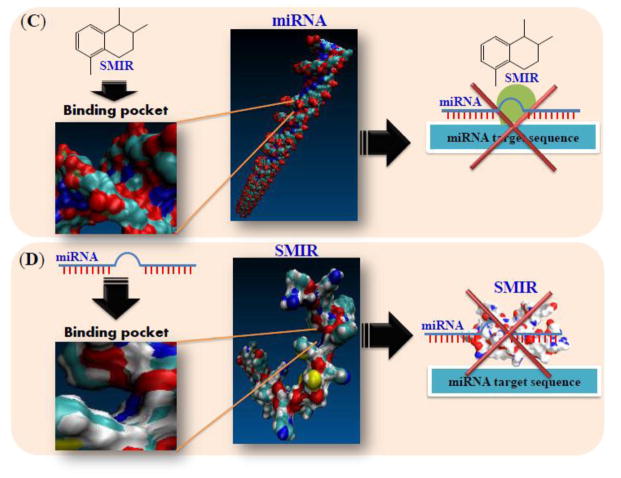
Mechanisms of (A) luciferase/GFP based screening. Effective SMIR binds mature miRNA and further inhibit the binding of this miRNA to miRNA target sequence, leading to the expression of Luciferase/GFP. (B) Molecular beacon-based screening. Effective SMIR binds mature miRNA and further inhibit the activity of Dicer. The 5′-fluorophore (F) and 3′-quencher (Q) attach to each other and no signal can be detected. (C) structure-based design. Effective SMIR binds to the binding pocket of certain miRNA and further prevent this miRNA to bind miRNA target sequence. (D) Peptides or peptoids screening. Effective SMIR (peptides or peptoids) binds specific miRNA and further prevent this miRNA to bind miRNA target sequence. SMIR, small molecule inhibitor of miRNA.
4.1 Luciferase (or GFP)-based Screening
Luciferase-based vectors, which contain a complementary sequence or control sequence of target miRNA linked with downstream luciferase reporter gene, are widely used for SMIR screening. After cloning into lentiviral vectors, they are transduced into culture cells in which target miRNA is highly expressed. These gene modified cells are thereby able to determine the efficacy of potent SMIRs. With the presence of effective SMIRs, less target miRNA is available for binding the complementary sequence and luciferase gene is expressed as a result. Thus, the more effective SMIR, the more luciferase signals will be detected. Young et al.12 improved their previous screening methods11 using psiCHECK-2 vector, which co-expressed both Renilla and firefly luciferase to normalize the signal. The potent inhibitors they found for miR-122 induced 773±38% and 1251±125% increase in the relative luciferase signal. Connelly et al.108 used similar psiCHECK-miR-122 vector for the same miRNA to screen SMIRs using Huh7 cell line and found benzothiazole was a potent SMIR for miR-122. Bose et al.109 used a modified psiCHECK-2-prohibitin vector, where prohibitin was a miR-27a inhibitor, to select SMIR for miR-27a in MCF-7 cell line. They found luciferase signals were significantly increased under the treatment of amikacin, streptomycin, tobramycin, and neomycin at the concentration of 20 μM. Bose et al.110 used another pEZX-MT01 plasmid, which co-express luciferase and PDCD4, a known target of miR-21, to screen SMIRs for miR-21 in MCF-7 cell line. Streptomycin was identified as the most potent target and was characterized as a direct miR-21 inhibitor docking with pre-miR-21 at a region close to the terminal loop. Tan et al. also used luciferase reporter system to prove and evaluate a specific SMIR for miR-1.111 Luciferase reporter system resulting in an increased luciferase signal in the presence of an effective SMIR excludes false-positive caused by compound toxicity, which may occur in an assay based on a decreased signal. These studies need to be carried out using cell line, which is probably modified by lentiviral vectors containing reporter assay plasmid according to previous studies. Similarly, this luciferase reporter-based screening method can be used to select specific compounds promoting tumor suppressor miRNA activity. Xiao et al.112 constructed miR-34a reporter vector using Huh7 cell line and found Rubone, which effectively inhibited luciferase activity, was a potent miR-34a promoter. Young et al.12 reported an effective miR-122 modulator selected by luciferase assay and further demonstrated by RT-PCR. These miRNA promoting compounds were named small molecule modulator of miRNA (SMMR). Compared to SMIR selected by luciferase reporter system, small molecule modulator of miRNA needs further evaluation to exclude false-positive phenomenon caused by toxicity since this compound decrease luciferase activity.
Recently, another similar GFP-based screening was also developed for SMIR screening. To screen a general miRNA inhibitor, a cell line stably expressing lenti-GFP and lenti-shGFP was developed. A compound was considered potent SMIR if green fluorescence was increased. To screen a SMIR for specific miRNA, EGFP reporter gene expression was under the control of specific miRNA through its complementary sequence present at the 3′ UTR. Using this GFP based screening assay, Shum et al.113 obtained 6 potent miR-21 inhibitors and 6-hydroxy-DL-DOPA was characterized as the most potent SMIR. This GFP-based screening is another method for screening SMMR as reported by Shan et al.114 In their study, enoxacin, which decreased EGFP expression and enhanced GAPDH gene silencing, was selected as a small molecule modulator of miRNA. Melo et al.115 further reported the anticancer activity of enoxacin, which acted by enhancing TAR RNA-binding protein 2-mediated miRNA processing.
4.2. Molecular Beacon-based Screening
Fluorescent beacons are usually hairpin shaped oligonucleotides which contain a 5′-fluorophore and a 3′-quencher, along with a miRNA targeting sequence (anti-miRNA sequence) in the loop. Davies et al.116 first described the design of a fluorescent beacon and forecasted its potent application for screening SMIRs. In a hairpin shape, the base pair would bring the fluorophore and quencher closely, leading to quenching of the fluorescence. Thus, a Dicer-dependent increase in the fluorescence would be detected since mature miRNA is generated from Dicer-mediated hydrolysis, resulting in a dissociation of the fluorophore and quencher, and an increase of fluorescence. In the presence of a Dicer inhibitor or ligand of pre-miRNA, Dicer activity would be inhibited and the beam showed a lack of fluorescence increase. Vo et al.117 used this fluorescent beacon system to select Dicer inhibitor to inhibit biogenesis of oncogenic miR-372 and miR-373. In their study, Neomycin appeared to be the best aminoglycoside for Dicer inhibition and thereby be used for further modification. Bose et al.109 modified this fluorescent beacon system since it was reported that the cleavage of miRNA by human Dicer depended on both 5′ and 3′-ends.118,119 Therefore, the addition of quencher and fluorophore might affect the activity of Dicer and obscure the results. They overcame these drawbacks by using a DNA molecular beacon with a 5′ fluorophore and a 3′ quencher. This newly designed beacon was independent of pre-miRNA and the loop was complementary to mature target miRNA sequence after cleavage by Dicer. This beacon was further used for SMIR of miR-27a and neomycin was found the most potent compound, which was also demonstrated by luciferase-based vector screening. Recently, there were several other screening methods, such as fluorescence polarization screening assay reported by Tan et al.120 and catalytic enzyme-linked click chemistry assay reported by Lorenz et al.121 Molecular beacon-based screening does not need to perform under cell line and it is not widely used as luciferase-based screening method. Since SMIR causes a decrease of fluorescence in this molecular beacon system, further experiments need to be carried out to exclude false-positive caused by interfering with the fluorophore or quencher.
4.3. Structure-based Design
One difficulty that encountered in the drug development process is the high cost in the process of drug screening. With more accurate understanding of miRNA (or miRNA protein complex) structure and the simulation of binding affinity of SMIR to miRNA, in silico high-throughput screening is a promising technique to speed up the discovery of SMIRs and decrease the cost during the process. This computational approach is still challenging and needs further demonstration and recalibration to ensure the efficacy of screened compounds due to the flexible and complicated RNA tertiary structure. Shi et al.122 reported AC1MMYR2 as an inhibitor of Dicer-mediated biogenesis of miR-21 using MC-Fold/MC-Sym pipeline for RNA secondary and tertiary structure prediction. In their studies, AC1MMYR2 was demonstrated a specific miR-21 inhibitor, which repressed pri-miR-21 expression by approximately 50% after 6 h and inhibited tumor growth in an orthotopic tumor model. Velagapudi et al.123 reported a new method called Inforna for sequence-based design of SMIR to target pre-miRs. Inforna integrated a selection-based strategy (Two-Dimensional Combinatorial Screening; 2DCS),124 a statistical approach (Structure-Activity Relationship through Sequencing; StARTS),125,126 and the structural information about the RNA target of interest that identified RNA motifs that positively and negatively contributed to binding. After screening and optimization, they selected three compounds for miR-96 precursor, miR-210 precursor, and miR-182 precursor, respectively. The secondary structure was proved by enzymatic mapping assays and the downstream effect of miR-96 inhibitor was evaluated. Compared to traditional medicinal chemical approaches, Inforna provided a reliable prediction of SMIRs that was able to target specific miRNA.
4.4. Peptide or Peptoid Screening
Another category of SMIRs is peptides or peptoids, which are well reported for selective RNA binding. Herein, we introduce two peptide selection methods, peptoid microarrays and phage display selection. Chirayil et al.127 performed peptoid microarrays to identify specific ligands for RNA hairpin precursor of miR-21. In their studies, they used peptoid microarrays as the foundation for a system of RNA ligand discovery to screen a library of 7680 N-substituted oligoglycines. Among them, two compounds were shown to have specific binding affinity to the secondary structure of miR-21 precursor hairpin. After identifying the functional groups contributing to the affinity and specificity, a compound with dissociation constant of 1.9 μM for miR-21 precursor hairpin was created. However, this compound did not show detectable binding to the targeted hairpin loop in the presence of Mg2+ which is required for microprocessor activity. Thus, they further modified their screening strategy and found higher affinity compounds inhibiting microprocessor activity in vitro.128 The newly selected peptoid shows weaker binding affinity but has better selectivity of pri-miR-21 over pri-miR-16. A fusion phage is a filamentous virion displaying on its surface a foreign peptide fused to a coat protein.129 In one study, the library of this fusion phage may represent up to billions of peptides.130 If a phage displays a peptide which is a strong ligand of target miRNA, it can be eluted and the peptide sequences responsible for the binding are easily obtained by infecting the specific phage into bacteria and sequencing the relevant part of their viral DNAs.131 Using this method, Bose et al.132 reported that ‘ALWPPNLHAWVP’ was a potent peptide sequence for binding miR-21. After identifying the binding pocket of this peptide using a PEP-FOLD web server, they further demonstrated that this peptide suppressed tumor cell proliferation, invasion and migration by antiagonizing miR-21.
4.5. Mechanism of Small Molecule miRNA Therapeutic Agents
Deiters proposed three basic processing stages that present potent target for the activation or deactivation of miRNA function by small molecules: (1) the pre-transcriptional stage, (2) the transcription stage, and (3) the post-transcriptional stage.133 For pre-transcriptional stage, the biogenesis of miRNA is affected by miRNA gene copy number, mutations in miRNA gene, or histone deacetylation and hypermethylation of miRNA promoters.134,135 Scott et al. reported that 22 miRNAs were downregulated after treatment with LAQ824, a histone deacetylases inhibitor.136 In transcriptional stage, the transcription factors regulating the expression of miRNAs are potent target for SMIR. Recently, novel c-Myc inhibitors, JQ1, were developed for the treatment of hematopoietic malignancies137,138 and lymphoma.139 Directly interacting with Drosha gene promoter, c-Myc activates the transcription of Drosha, which promotes the biogenesis of miRNAs.140
Compared to the previous two stages, much more SMIRs were developed targeting post-transcriptional process because SMIR targeting previous two stages mostly inhibited the biogenesis of several miRNAs, significantly decreasing the efficacy and specificity for oncomiRs. Bose et al. discovered that streptomycin can efficiently repress miR-21 by binding to its precursor and interfering with its downstream process by Dicer.110 Velagapudi et al. used sequence-based design and found a SMIR specific for miR-96 by binding Dicer and Drosha.123 Murata et al. reported that Xanthone derivatives could inhibit miR-29a by targeting its secondary structure and suppressing Dicer binding.141 Shi et al. reported that AC1MMYR2 probably blocked the Dicer binding site on pre-miR-21 to prevent the cleavage of pre-miR-21 to the mature miRNA.122 The peptide miR-21 inhibitor developed by Bose et al. has a binding pocket for miR-21 and thus inhibit Dicer processing for miRNA maturation.132 Nevertheless, there are still general SMIR that inhibits several miRNAs in post-transcriptional process. Watashi et al.142 screened 530 compounds and discovered poly-L-lysine hydrobromide as a Dicer inhibitor and 3,6-diamino-10-methylacridinium chloride as an AGO2 inhibitor. Lünse et al. discovered an aptamer targeting the apical-loop domain of pri-miRNAs and modulating the maturation processing of miR-17, miR-18a, and miR-19.143 Tan et al. discovered aurintricarboxylic acid as a RISC loading inhibitor after searching thousands of compounds using a novel method based on fluorescence polarization.120 Enoxacin was demonstrated by Melo et al. to function by enhancing TAR RNA-binding protein 2-mediated miRNA processing, which was the only available mechanism study for SMMR.115
5. Promises and Challenges of Small Molecule miRNA Therapeutic Agents for treating Cancer
5.1. Therapeutic Potential of SMIR and Small Molecular Modulators of miRNA (SMMRs)
miRNAs are considered crucial factors in spectrums of human disease, especially cancer. In the past decade, various miRNAs have been reported to be associated with cancer development process. Drug discovery and development are always a time-consuming and expensive process, which significantly influences the therapeutic progress of cancer and other diseases, leading to the urgent need for new therapeutic alternatives. SMIRs and Small Molecular Modulators of miRNA (SMMRs) show another promising approach for the treatment of cancer due to its less time-consuming characteristic for drug development with reduced cost in the whole process. In addition, their exciting results as previously discussed further proved them to be an efficient tool for therapeutic use. JQ1, c-Myc inhibitor, significantly promoted differentiation, tumor regression, and improved survival in murine xenograft models of NUT midline cancinoma.137 AC1MMYR2 inhibited tumorigenesis and invasiveness in an orthotopic U87 glioma intracranial model.122 PLL, the Dicer inhibitor, and TPF, the AGO2 inhibitor, treatments suppressed tumorigenic activity of miR-93 over-expressed NIH3T3 cell lines when subcutaneously implanted into nude mice for tumor formation.142 The replenishment of miR-34a reported by Xiao et al. significantly inhibited the growth of hepatocellular carcinoma in xenograft mouse model.112
To yields better therapeutic outcomes compared to single drug treatment, combination therapy can be performed to address more than one pivotal target. Qian et al. reported the synergistic inhibition of human glioma cell lines with the combination of temozolomide and antisense oligonucleotide miR-21 inhibitor.144 Xu et al. reported that inhibition of miR-21 enhanced chemotherapeutic effect of cisplatin in non-small-cell lung cancer.145 Theoretically, we can use previously discussed SMIRs to target miR-21 for combination therapy with another small molecule drug. Recently, Yu et al. developed mesoporous silica nanoparticles for combination therapy by co-delivering SMIR and antagomir against miR-122 in hepatocellular carcinoma cells.146 In this research, they selected a previous published SMIR against miR-122 selected by Young et al.12 and miR-122 antagomir delivered by RGD-conjugated mesoporous silica nanoparticles. In vitro analysis showed that this SMIR and miR-122 antagomir significantly downregulated miR-122 expression in Huh 7 cells. To our knowledge, this is the only available combination therapy by using SMIR and antagomir for targeting cancer related miRNA. Compared to antisense oligonucleotide-based miRNA inhibitor, SMIR is more easily for systemic delivery using current drug delivery systems, including liposomes, micelles, and nanoparticles.
5.2. Pitfalls of Small Molecule miRNA Therapeutic Agents for treating Cancer
The challenges for development and application of SMIR and SMMR are searching for more potent compounds and the delivery issue. Based on previous researches, people are still far away from being able to efficiently design potent SMIRs and SMMRs with clear understanding of their inhibition mechanisms. According to recently developed SMIRs, we can conclude that what we did was only to discover the new application of previous drugs. Currently designed SMIRs were able to target only a small number of oncomiRs (Table 3). Furthermore, several crucial defects of the current screening strategies or structure-based design techniques cannot be ignored. For instance, molecular beacon-based screening needs further improvement to exclude the false positive which might be caused by the interaction with fluorophore and quencher. For structure based screening method, more powerful simulation software needs to be developed to accurately predict secondary and tertiary structure of target miRNA. In 2011, Paige reported RNA mimics of GFP and predicted its secondary structure using Mfold web-based software,147 whereas the correct structure was discovered by Huang et al.148 and Warner et al.149 in 2014. Thus, more effective screening methods and accurate structural and thermodynamic simulation on the interaction between miRNA and SMIR or SMMR are in urgent need. Furthermore, drug interaction study needs to be performed if combination therapy is used to improve therapeutic efficiency. We also need to design efficient drug delivery systems for SMIR and SMMR delivery.
Table 3.
Summary of current SMIRs for specific oncomiRs
| OncomiR | SMIRs | References |
|---|---|---|
| miR-21 | diazobenzene, streptomycin, 6-hydroxy-DL-DOPA, AC1MMYR2, peptoid and peptide as described in reference, | 11,110,113,122,127,128,132 |
| miR-122 | Benzothiazole based compounds | 108 |
| miR-96, miR-210, miR-182 | Compound 1, 2, 3 as described in reference | 123 |
| miR-27a | amikacin, streptomycin, tobramycin, and neomycin | 109 |
| miR-1 | Compound 14 as described in the reference | 111 |
| miR-372/373 | neomycin | 117 |
| miR-29a | Compound 5 as described in the reference | 141 |
| miR-17, miR-18a, and miR-19 | Aptamer 7 as described in the reference | 143 |
Since single miRNA may regulate several genes, the potent off-target effects are one of the major concerns for miRNA therapy. For example, miR-29 oligonucleotide mimics may act as anticancer drugs by targeting several oncogenic pathways including Mcl-1150 and CDK6.151 In contrast, miR-29 may promote tumor cell migration, invasion, and apoptotic resistance through direct targeting PTEN.152 Meanwhile, it also regulates osteoblast differentiation153 and immune inhibitory molecule expression.154 Thus, an efficient target delivery system is always used to deliver oligonucleotide-based miRNA or anti-miRNA mimics. Except for tissue targeting related off-target effect, there is another kind of off-target effect of SMIR need to be solved since single SMIR may target multiple miRNAs. For example, streptomycin can target both miR-21 and miR-27a, whereas neomycin can target both miR-27a and miR-372/373 (Table 3). Thus, a miRNA profiling study might be crucial to evaluate the specificity of a certain SMIR.
6. Conclusions and Future Perspectives
OncomiRs are exciting targets for drug development and cancer treatment. In the past decade, SMIR has been proven a novel and effective method for inhibiting oncomiRs. Compared to non-small molecule miRNA therapeutics, SMIR and SMMR are more easily systematically delivered. Furthermore, the development of non-small molecule miRNA therapeutic agents is always costly and the in vivo stability of these agents is another outstanding issue to be overcome. Thus, extensive work has been done to develop several promising methods for SMIR and SMMR discovery in recent years, including non-structure based screening and structure based design. There is significant improvement in the design of screening vectors and validating rational computational approaches. Moreover, peptide and peptoid based SMIRs were developed as another category for miRNA inhibition. However, we are still at the early stage of this area since outstanding challenges, including screening methods and simulation techniques, remain to be overcome. Overall, targeting miRNAs with SMIRs for cancer treatment constitutes a reasonable and evidence based strategy with strong potential and chance for success. The progress of screening techniques and computational stimulation may address bright future in this field.
Table 2.
Different mechanisms of current SMIRs
| Stage | SMIRs | Mechanism | Reference |
|---|---|---|---|
| Pre-transcription | LAQ824 | Inhibiting histone deacetylases | 136 |
| Transcription | JQ1 | Inhibiting c-Myc | 137–139 |
| Post-transcription | Streptomycin, xanthone derivatives, AC1MMYR2, poly-L-lysine hydrobromide, 3,6-diamino-10-methylacridinium chloride, aurintricarboxylic acid, Enoxacin | Binding pre-miRNA or pri-miRNA; binding Dicer, Drosha, or AGO2; inhibiting Dicer process or RISC loading, enhancing TAR RNA-binding protein 2-mediated miRNA processing | 110,115,120,122,123,132,141–143 |
Acknowledgments
The National Institutes of Health (1RO1EB017853 and 1R01GM113166), Fred, and Pamela Buffet Cancer Center and the faculty start-up fund are duly acknowledged for providing financial support for this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ambros V. MicroRNA pathways in flies and worms: Growth, death, fat, stress, and timing. Cell. 2003;113(6):673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Lai EC. microRNAs: Runts of the genome assert themselves. Curr Biol. 2003;13(23):R925–36. doi: 10.1016/j.cub.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 4.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10(10):704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 6.Iorio MV, Croce CM. Causes and consequences of microRNA dysregulation. Cancer J. 2012;18(3):215–222. doi: 10.1097/PPO.0b013e318250c001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Babashah S, Soleimani M. The oncogenic and tumour suppressive roles of microRNAs in cancer and apoptosis. Eur J Cancer. 2011;47(8):1127–1137. doi: 10.1016/j.ejca.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 8.Bandyopadhyay S, Mitra R, Maulik U, Zhang MQ. Development of the human cancer microRNA network. Silence. 2010;1(1):6-907X-1-6. doi: 10.1186/1758-907X-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 10.Michael MZ, O’Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1(12):882–891. [PubMed] [Google Scholar]
- 11.Gumireddy K, Young DD, Xiong X, Hogenesch JB, Huang Q, Deiters A. Small-molecule inhibitors of microrna miR-21 function. Angew Chem Int Ed Engl. 2008;47(39):7482–7484. doi: 10.1002/anie.200801555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Young DD, Connelly CM, Grohmann C, Deiters A. Small molecule modifiers of microRNA miR-122 function for the treatment of hepatitis C virus infection and hepatocellular carcinoma. J Am Chem Soc. 2010;132(23):7976–7981. doi: 10.1021/ja910275u. [DOI] [PubMed] [Google Scholar]
- 13.Jiang W, Chen X, Liao M, et al. Identification of links between small molecules and miRNAs in human cancers based on transcriptional responses. Sci Rep. 2012;2:282. doi: 10.1038/srep00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peacock O, Lee AC, Cameron F, et al. Inflammation and MiR-21 pathways functionally interact to downregulate PDCD4 in colorectal cancer. PLoS One. 2014;9(10):e110267. doi: 10.1371/journal.pone.0110267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu J, Zhang W, Lv Q, Zhu D. Overexpression of miR-21 promotes the proliferation and migration of cervical cancer cells via the inhibition of PTEN. Oncol Rep. 2015;33(6):3108–3116. doi: 10.3892/or.2015.3931. [DOI] [PubMed] [Google Scholar]
- 16.Wu Z, Lu H, Sheng J, Li L. Inductive microRNA-21 impairs anti-mycobacterial responses by targeting IL-12 and bcl-2. FEBS Lett. 2012;586(16):2459–2467. doi: 10.1016/j.febslet.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282(19):14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 18.Zhao W, Dong Y, Wu C, Ma Y, Jin Y, Ji Y. MiR-21 overexpression improves osteoporosis by targeting RECK. Mol Cell Biochem. 2015 doi: 10.1007/s11010-015-2404-4. [DOI] [PubMed] [Google Scholar]
- 19.Battistella M, Romero M, Castro-Vega LJ, et al. The high expression of the microRNA 17-92 cluster and its paralogs, and the downregulation of the target gene PTEN, is associated with primary cutaneous B-cell lymphoma progression. J Invest Dermatol. 2015;135(6):1659–1667. doi: 10.1038/jid.2015.27. [DOI] [PubMed] [Google Scholar]
- 20.Xiao C, Srinivasan L, Calado DP, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9(4):405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang YF, Wang F, Xiao JJ, et al. MiR-222 overexpression promotes proliferation of human hepatocellular carcinoma HepG2 cells by downregulating p27. Int J Clin Exp Med. 2014;7(4):893–902. [PMC free article] [PubMed] [Google Scholar]
- 22.Yang F, Wang W, Zhou C, et al. MiR-221/222 promote human glioma cell invasion and angiogenesis by targeting TIMP2. Tumour Biol. 2015 doi: 10.1007/s13277-014-3017-3. [DOI] [PubMed] [Google Scholar]
- 23.Li Q, Shen K, Zhao Y, et al. MicroRNA-222 promotes tumorigenesis via targeting DKK2 and activating the wnt/beta-catenin signaling pathway. FEBS Lett. 2013;587(12):1742–1748. doi: 10.1016/j.febslet.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Peng Y, Dong W, Lin TX, et al. MicroRNA-155 promotes bladder cancer growth by repressing the tumor suppressor DMTF1. Oncotarget. 2015 doi: 10.18632/oncotarget.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai ZK, Chen Q, Chen YB, et al. microRNA-155 promotes the proliferation of prostate cancer cells by targeting annexin 7. Mol Med Rep. 2015;11(1):533–538. doi: 10.3892/mmr.2014.2744. [DOI] [PubMed] [Google Scholar]
- 26.Lao G, Liu P, Wu Q, et al. Mir-155 promotes cervical cancer cell proliferation through suppression of its target gene LKB1. Tumour Biol. 2014;35(12):11933–11938. doi: 10.1007/s13277-014-2479-7. [DOI] [PubMed] [Google Scholar]
- 27.Li T, Yang J, Lv X, et al. miR-155 regulates the proliferation and cell cycle of colorectal carcinoma cells by targeting E2F2. Biotechnol Lett. 2014;36(9):1743–1752. doi: 10.1007/s10529-014-1540-3. [DOI] [PubMed] [Google Scholar]
- 28.D’Urso PI, D’Urso OF, Storelli C, et al. miR-155 is up-regulated in primary and secondary glioblastoma and promotes tumour growth by inhibiting GABA receptors. Int J Oncol. 2012;41(1):228–234. doi: 10.3892/ijo.2012.1420. [DOI] [PubMed] [Google Scholar]
- 29.Huang BS, Luo QZ, Han Y, Li XB, Cao LJ, Wu LX. microRNA-223 promotes the growth and invasion of glioblastoma cells by targeting tumor suppressor PAX6. Oncol Rep. 2013;30(5):2263–2269. doi: 10.3892/or.2013.2683. [DOI] [PubMed] [Google Scholar]
- 30.Kang W, Tong JH, Chan AW, et al. Stathmin1 plays oncogenic role and is a target of microRNA-223 in gastric cancer. PLoS One. 2012;7(3):e33919. doi: 10.1371/journal.pone.0033919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Guo Y, Liang X, et al. MicroRNA-223 functions as an oncogene in human gastric cancer by targeting FBXW7/hCdc4. J Cancer Res Clin Oncol. 2012;138(5):763–774. doi: 10.1007/s00432-012-1154-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao C, Sun W, Zhang P, et al. miR-214 promotes osteoclastogenesis by targeting pten/PI3k/akt pathway. RNA Biol. 2015;12(3):343–353. doi: 10.1080/15476286.2015.1017205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang F, Lv P, Liu X, Zhu M, Qiu X. microRNA-214 enhances the invasion ability of breast cancer cells by targeting p53. Int J Mol Med. 2015;35(5):1395–1402. doi: 10.3892/ijmm.2015.2123. [DOI] [PubMed] [Google Scholar]
- 34.Zhang XF, Li KK, Gao L, et al. miR-191 promotes tumorigenesis of human colorectal cancer through targeting C/EBPbeta. Oncotarget. 2015;6(6):4144–4158. doi: 10.18632/oncotarget.2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang YZ, Zhang J, Shao HY, Chen JP, Zhao HY. MicroRNA-191 promotes osteosarcoma cells proliferation by targeting checkpoint kinase 2. Tumour Biol. 2015 doi: 10.1007/s13277-015-3290-9. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J, Gong X, Tian K, et al. miR-25 promotes glioma cell proliferation by targeting CDKN1C. Biomed Pharmacother. 2015;71:7–14. doi: 10.1016/j.biopha.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 37.Feng S, Pan W, Jin Y, Zheng J. MiR-25 promotes ovarian cancer proliferation and motility by targeting LATS2. Tumour Biol. 2014;35(12):12339–12344. doi: 10.1007/s13277-014-2546-0. [DOI] [PubMed] [Google Scholar]
- 38.Zhao H, Wang Y, Yang L, Jiang R, Li W. MiR-25 promotes gastric cancer cells growth and motility by targeting RECK. Mol Cell Biochem. 2014;385(1–2):207–213. doi: 10.1007/s11010-013-1829-x. [DOI] [PubMed] [Google Scholar]
- 39.Ciafre SA, Galardi S, Mangiola A, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334(4):1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 40.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 41.Pichiorri F, Suh SS, Ladetto M, et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc Natl Acad Sci U S A. 2008;105(35):12885–12890. doi: 10.1073/pnas.0806202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gramantieri L, Fornari F, Ferracin M, et al. MicroRNA-221 targets bmf in hepatocellular carcinoma and correlates with tumor multifocality. Clin Cancer Res. 2009;15(16):5073–5081. doi: 10.1158/1078-0432.CCR-09-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garofalo M, Di Leva G, Romano G, et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16(6):498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Lan FF, Wang H, Chen YC, et al. Hsa-let-7g inhibits proliferation of hepatocellular carcinoma cells by downregulation of c-myc and upregulation of p16(INK4A) Int J Cancer. 2011;128(2):319–331. doi: 10.1002/ijc.25336. [DOI] [PubMed] [Google Scholar]
- 45.Ji J, Zhao L, Budhu A, et al. Let-7g targets collagen type I alpha2 and inhibits cell migration in hepatocellular carcinoma. J Hepatol. 2010;52(5):690–697. doi: 10.1016/j.jhep.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shimizu S, Takehara T, Hikita H, et al. The let-7 family of microRNAs inhibits bcl-xL expression and potentiates sorafenib-induced apoptosis in human hepatocellular carcinoma. J Hepatol. 2010;52(5):698–704. doi: 10.1016/j.jhep.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 47.Calin GA, Cimmino A, Fabbri M, et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci U S A. 2008;105(13):5166–5171. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang TC, Wentzel EA, Kent OA, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26(5):745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: Rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9(10):775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Budd WT, Seashols S, Weaver D, Joseph C, Zehner ZE. A networks method for ranking microRNA dysregulation in cancer. BMC Syst Biol. 2013;7 (Suppl 5):S3-0509-7-S5-S3. doi: 10.1186/1752-0509-7-S5-S3. Epub 2013 Dec 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dias N, Stein CA. Antisense oligonucleotides: Basic concepts and mechanisms. Mol Cancer Ther. 2002;1(5):347–355. [PubMed] [Google Scholar]
- 52.Krutzfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438(7068):685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 53.Petersen M, Nielsen CB, Nielsen KE, et al. The conformations of locked nucleic acids (LNA) J Mol Recognit. 2000;13(1):44–53. doi: 10.1002/(SICI)1099-1352(200001/02)13:1<44::AID-JMR486>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 54.Wengel J, Vester B, Lundberg LB, et al. LNA and alpha-L-LNA: Towards therapeutic applications. Nucleosides Nucleotides Nucleic Acids. 2003;22(5–8):601–604. doi: 10.1081/NCN-120021963. [DOI] [PubMed] [Google Scholar]
- 55.Galardi S, Mercatelli N, Giorda E, et al. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem. 2007;282(32):23716–23724. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- 56.Liang Z, Li Y, Huang K, Wagar N, Shim H. Regulation of miR-19 to breast cancer chemoresistance through targeting PTEN. Pharm Res. 2011;28(12):3091–3100. doi: 10.1007/s11095-011-0570-y. [DOI] [PubMed] [Google Scholar]
- 57.Di Martino MT, Gulla A, Gallo Cantafio ME, et al. In vitro and in vivo activity of a novel locked nucleic acid (LNA)-inhibitor-miR-221 against multiple myeloma cells. PLoS One. 2014;9(2):e89659. doi: 10.1371/journal.pone.0089659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang ZC, Li YY, Wang HY, et al. Knockdown of miR-214 promotes apoptosis and inhibits cell proliferation in nasopharyngeal carcinoma. PLoS One. 2014;9(1):e86149. doi: 10.1371/journal.pone.0086149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Elmen J, Lindow M, Silahtaroglu A, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36(4):1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Elmen J, Lindow M, Schutz S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452(7189):896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 61.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4(9):721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ebert MS, Sharp PA. MicroRNA sponges: Progress and possibilities. RNA. 2010;16(11):2043–2050. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kumar MS, Erkeland SJ, Pester RE, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105(10):3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rybak A, Fuchs H, Smirnova L, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10(8):987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 65.Bolisetty MT, Dy G, Tam W, Beemon KL. Reticuloendotheliosis virus strain T induces miR-155, which targets JARID2 and promotes cell survival. J Virol. 2009;83(23):12009–12017. doi: 10.1128/JVI.01182-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bonci D, Coppola V, Musumeci M, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14(11):1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 67.Gentner B, Schira G, Giustacchini A, et al. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat Methods. 2009;6(1):63–66. doi: 10.1038/nmeth.1277. [DOI] [PubMed] [Google Scholar]
- 68.Huang J, Zhao L, Xing L, Chen D. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells. 2010;28(2):357–364. doi: 10.1002/stem.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Papapetrou EP, Korkola JE, Sadelain M. A genetic strategy for single and combinatorial analysis of miRNA function in mammalian hematopoietic stem cells. Stem Cells. 2010;28(2):287–296. doi: 10.1002/stem.257. [DOI] [PubMed] [Google Scholar]
- 70.Barbato C, Ruberti F, Pieri M, et al. MicroRNA-92 modulates K(+) cl(−) co-transporter KCC2 expression in cerebellar granule neurons. J Neurochem. 2010;113(3):591–600. doi: 10.1111/j.1471-4159.2009.06560.x. [DOI] [PubMed] [Google Scholar]
- 71.Ma L, Reinhardt F, Pan E, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol. 2010;28(4):341–347. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Valastyan S, Reinhardt F, Benaich N, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137(6):1032–1046. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 73.Du C, Liu C, Kang J, et al. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009;10(12):1252–1259. doi: 10.1038/ni.1798. [DOI] [PubMed] [Google Scholar]
- 74.Care A, Catalucci D, Felicetti F, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13(5):613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 75.Loya CM, Lu CS, Van Vactor D, Fulga TA. Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nat Methods. 2009;6(12):897–903. doi: 10.1038/nmeth.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in escherichia coli, and identification of the gene product. J Bacteriol. 1987;169(12):5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wei C, Liu J, Yu Z, Zhang B, Gao G, Jiao R. TALEN or Cas9 - rapid, efficient and specific choices for genome modifications. J Genet Genomics. 2013;40(6):281–289. doi: 10.1016/j.jgg.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 78.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31(3):230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 79.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen B, Gilbert LA, Cimini BA, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/cas system. Cell. 2013;155(7):1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhao Y, Dai Z, Liang Y, et al. Sequence-specific inhibition of microRNA via CRISPR/CRISPRi system. Sci Rep. 2014;4:3943. doi: 10.1038/srep03943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xiao A, Wang Z, Hu Y, et al. Chromosomal deletions and inversions mediated by TALENs and CRISPR/cas in zebrafish. Nucleic Acids Res. 2013;41(14):e141. doi: 10.1093/nar/gkt464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6(6):590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39(5):673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 86.Sun V, Zhou WB, Nosrati M, et al. Antitumor activity of miR-1280 in melanoma by regulation of src. Mol Ther. 2015;23(1):71–78. doi: 10.1038/mt.2014.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tiwari A, Shivananda S, Gopinath KS, Kumar A. MicroRNA-125a reduces proliferation and invasion of oral squamous cell carcinoma cells by targeting estrogen-related receptor alpha: Implications for cancer therapeutics. J Biol Chem. 2014;289(46):32276–32290. doi: 10.1074/jbc.M114.584136. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 88.Mittal A, Chitkara D, Behrman SW, Mahato RI. Efficacy of gemcitabine conjugated and miRNA-205 complexed micelles for treatment of advanced pancreatic cancer. Biomaterials. 2014;35(25):7077–7087. doi: 10.1016/j.biomaterials.2014.04.053. [DOI] [PubMed] [Google Scholar]
- 89.Kumar V, Mondal G, Slavik P, Rachagani S, Batra SK, Mahato RI. Codelivery of small molecule hedgehog inhibitor and miRNA for treating pancreatic cancer. Mol Pharm. 2015;12(4):1289–1298. doi: 10.1021/mp500847s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Babar IA, Cheng CJ, Booth CJ, et al. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc Natl Acad Sci U S A. 2012;109(26):E1695–704. doi: 10.1073/pnas.1201516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li Y, Yan L, Zhang W, et al. miR-21 inhibitor suppresses proliferation and migration of nasopharyngeal carcinoma cells through down-regulation of BCL2 expression. Int J Clin Exp Pathol. 2014;7(6):3478–3487. [PMC free article] [PubMed] [Google Scholar]
- 92.Li Z, Yang CS, Nakashima K, Rana TM. Small RNA-mediated regulation of iPS cell generation. EMBO J. 2011;30(5):823–834. doi: 10.1038/emboj.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bennett CF, Swayze EE. RNA targeting therapeutics: Molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 94.Galbraith WM, Hobson WC, Giclas PC, Schechter PJ, Agrawal S. Complement activation and hemodynamic changes following intravenous administration of phosphorothioate oligonucleotides in the monkey. Antisense Res Dev. 1994;4(3):201–206. doi: 10.1089/ard.1994.4.201. [DOI] [PubMed] [Google Scholar]
- 95.Swayze EE, Siwkowski AM, Wancewicz EV, et al. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res. 2007;35(2):687–700. doi: 10.1093/nar/gkl1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xiao-Jie L, Hui-Ying X, Zun-Ping K, Jin-Lian C, Li-Juan J. CRISPR-Cas9: A new and promising player in gene therapy. J Med Genet. 2015;52(5):289–296. doi: 10.1136/jmedgenet-2014-102968. [DOI] [PubMed] [Google Scholar]
- 97.Hatakeyama H, Murata M, Sato Y, et al. The systemic administration of an anti-miRNA oligonucleotide encapsulated pH-sensitive liposome results in reduced level of hepatic microRNA-122 in mice. J Control Release. 2014;173:43–50. [PubMed] [Google Scholar]
- 98.Kriegel AJ, Liu Y, Cohen B, Usa K, Liu Y, Liang M. MiR-382 targeting of kallikrein 5 contributes to renal inner medullary interstitial fibrosis. Physiol Genomics. 2012;44(4):259–267. doi: 10.1152/physiolgenomics.00173.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li Z, Rana TM. Therapeutic targeting of microRNAs: Current status and future challenges. Nat Rev Drug Discov. 2014;13(8):622–638. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 100.Jain RK. Delivery of molecular and cellular medicine to solid tumors. Adv Drug Deliv Rev. 2012;64(Suppl):353–365. doi: 10.1016/j.addr.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gilleron J, Querbes W, Zeigerer A, et al. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat Biotechnol. 2013;31(7):638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 102.Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24(6):1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tay FC, Lim JK, Zhu H, Hin LC, Wang S. Using artificial microRNA sponges to achieve microRNA loss-of-function in cancer cells. Adv Drug Deliv Rev. 2015;81:117–127. doi: 10.1016/j.addr.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 104.del Monroig PC, Chen L, Zhang S, Calin GA. Small molecule compounds targeting miRNAs for cancer therapy. Adv Drug Deliv Rev. 2015;81:104–116. doi: 10.1016/j.addr.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang HW, Noland C, Siridechadilok B, et al. Structural insights into RNA processing by the human RISC-loading complex. Nat Struct Mol Biol. 2009;16(11):1148–1153. doi: 10.1038/nsmb.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65(14):6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 107.Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299(4):425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Connelly CM, Thomas M, Deiters A. High-throughput luciferase reporter assay for small-molecule inhibitors of microRNA function. J Biomol Screen. 2012;17(6):822–828. doi: 10.1177/1087057112439606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bose D, Jayaraj GG, Kumar S, Maiti S. A molecular-beacon-based screen for small molecule inhibitors of miRNA maturation. ACS Chem Biol. 2013;8(5):930–938. doi: 10.1021/cb300650y. [DOI] [PubMed] [Google Scholar]
- 110.Bose D, Jayaraj G, Suryawanshi H, et al. The tuberculosis drug streptomycin as a potential cancer therapeutic: Inhibition of miR-21 function by directly targeting its precursor. Angew Chem Int Ed Engl. 2012;51(4):1019–1023. doi: 10.1002/anie.201106455. [DOI] [PubMed] [Google Scholar]
- 111.Tan SB, Huang C, Chen X, et al. Small molecular inhibitors of miR-1 identified from photocycloadducts of acetylenes with 2-methoxy-1,4-naphthalenequinone. Bioorg Med Chem. 2013;21(20):6124–6131. doi: 10.1016/j.bmc.2013.04.058. [DOI] [PubMed] [Google Scholar]
- 112.Xiao Z, Li CH, Chan SL, et al. A small-molecule modulator of the tumor-suppressor miR34a inhibits the growth of hepatocellular carcinoma. Cancer Res. 2014;74(21):6236–6247. doi: 10.1158/0008-5472.CAN-14-0855. [DOI] [PubMed] [Google Scholar]
- 113.Shum D, Bhinder B, Radu C, et al. An image-based biosensor assay strategy to screen for modulators of the microRNA 21 biogenesis pathway. Comb Chem High Throughput Screen. 2012;15(7):529–541. doi: 10.2174/138620712801619131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shan G, Li Y, Zhang J, et al. A small molecule enhances RNA interference and promotes microRNA processing. Nat Biotechnol. 2008;26(8):933–940. doi: 10.1038/nbt.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Melo S, Villanueva A, Moutinho C, et al. Small molecule enoxacin is a cancer-specific growth inhibitor that acts by enhancing TAR RNA-binding protein 2-mediated microRNA processing. Proc Natl Acad Sci U S A. 2011;108(11):4394–4399. doi: 10.1073/pnas.1014720108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Davies BP, Arenz C. A homogenous assay for micro RNA maturation. Angew Chem Int Ed Engl. 2006;45(33):5550–5552. doi: 10.1002/anie.200601332. [DOI] [PubMed] [Google Scholar]
- 117.Vo DD, Staedel C, Zehnacker L, Benhida R, Darfeuille F, Duca M. Targeting the production of oncogenic microRNAs with multimodal synthetic small molecules. ACS Chem Biol. 2014;9(3):711–721. doi: 10.1021/cb400668h. [DOI] [PubMed] [Google Scholar]
- 118.Macrae IJ, Zhou K, Li F, et al. Structural basis for double-stranded RNA processing by dicer. Science. 2006;311(5758):195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 119.MacRae IJ, Zhou K, Doudna JA. Structural determinants of RNA recognition and cleavage by dicer. Nat Struct Mol Biol. 2007;14(10):934–940. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- 120.Tan GS, Chiu CH, Garchow BG, Metzler D, Diamond SL, Kiriakidou M. Small molecule inhibition of RISC loading. ACS Chem Biol. 2012;7(2):403–410. doi: 10.1021/cb200253h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lorenz DA, Song JM, Garner AL. High-throughput platform assay technology for the discovery of pre-microrna-selective small molecule probes. Bioconjug Chem. 2015;26(1):19–23. doi: 10.1021/bc500544v. [DOI] [PubMed] [Google Scholar]
- 122.Shi Z, Zhang J, Qian X, et al. AC1MMYR2, an inhibitor of dicer-mediated biogenesis of oncomir miR-21, reverses epithelial-mesenchymal transition and suppresses tumor growth and progression. Cancer Res. 2013;73(17):5519–5531. doi: 10.1158/0008-5472.CAN-13-0280. [DOI] [PubMed] [Google Scholar]
- 123.Velagapudi SP, Gallo SM, Disney MD. Sequence-based design of bioactive small molecules that target precursor microRNAs. Nat Chem Biol. 2014;10(4):291–297. doi: 10.1038/nchembio.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Disney MD, Labuda LP, Paul DJ, et al. Two-dimensional combinatorial screening identifies specific aminoglycoside-RNA internal loop partners. J Am Chem Soc. 2008;130(33):11185–11194. doi: 10.1021/ja803234t. [DOI] [PubMed] [Google Scholar]
- 125.Velagapudi SP, Seedhouse SJ, Disney MD. Structure-activity relationships through sequencing (StARTS) defines optimal and suboptimal RNA motif targets for small molecules. Angew Chem Int Ed Engl. 2010;49(22):3816–3818. doi: 10.1002/anie.200907257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Velagapudi SP, Seedhouse SJ, French J, Disney MD. Defining the RNA internal loops preferred by benzimidazole derivatives via 2D combinatorial screening and computational analysis. J Am Chem Soc. 2011;133(26):10111–10118. doi: 10.1021/ja200212b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chirayil S, Chirayil R, Luebke KJ. Discovering ligands for a microRNA precursor with peptoid microarrays. Nucleic Acids Res. 2009;37(16):5486–5497. doi: 10.1093/nar/gkp549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Diaz JP, Chirayil R, Chirayil S, Tom M, Head KJ, Luebke KJ. Association of a peptoid ligand with the apical loop of pri-miR-21 inhibits cleavage by drosha. RNA. 2014;20(4):528–539. doi: 10.1261/rna.042911.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Smith GP. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228(4705):1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 130.Scott JK, Smith GP. Searching for peptide ligands with an epitope library. Science. 1990;249(4967):386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- 131.Smith GP, Scott JK. Libraries of peptides and proteins displayed on filamentous phage. Methods Enzymol. 1993;217:228–257. doi: 10.1016/0076-6879(93)17065-d. [DOI] [PubMed] [Google Scholar]
- 132.Bose D, Nahar S, Rai MK, Ray A, Chakraborty K, Maiti S. Selective inhibition of miR-21 by phage display screened peptide. Nucleic Acids Res. 2015;43(8):4342–4352. doi: 10.1093/nar/gkv185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Deiters A. Small molecule modifiers of the microRNA and RNA interference pathway. AAPS J. 2010;12(1):51–60. doi: 10.1208/s12248-009-9159-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Scott GK, Mattie MD, Berger CE, Benz SC, Benz CC. Rapid alteration of microRNA levels by histone deacetylase inhibition. Cancer Res. 2006;66(3):1277–1281. doi: 10.1158/0008-5472.CAN-05-3632. [DOI] [PubMed] [Google Scholar]
- 137.Filippakopoulos P, Qi J, Picaud S, et al. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mertz JA, Conery AR, Bryant BM, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108(40):16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhao X, Lwin T, Zhang X, et al. Disruption of the MYC-miRNA-EZH2 loop to suppress aggressive B-cell lymphoma survival and clonogenicity. Leukemia. 2013;27(12):2341–2350. doi: 10.1038/leu.2013.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chen BJ, Wu YL, Tanaka Y, Zhang W. Small molecules targeting c-myc oncogene: Promising anti-cancer therapeutics. Int J Biol Sci. 2014;10(10):1084–1096. doi: 10.7150/ijbs.10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Murata A, Fukuzumi T, Umemoto S, Nakatani K. Xanthone derivatives as potential inhibitors of miRNA processing by human dicer: Targeting secondary structures of pre-miRNA by small molecules. Bioorg Med Chem Lett. 2013;23(1):252–255. doi: 10.1016/j.bmcl.2012.10.108. [DOI] [PubMed] [Google Scholar]
- 142.Watashi K, Yeung ML, Starost MF, Hosmane RS, Jeang KT. Identification of small molecules that suppress microRNA function and reverse tumorigenesis. J Biol Chem. 2010;285(32):24707–24716. doi: 10.1074/jbc.M109.062976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lunse CE, Michlewski G, Hopp CS, et al. An aptamer targeting the apical-loop domain modulates pri-miRNA processing. Angew Chem Int Ed Engl. 2010;49(27):4674–4677. doi: 10.1002/anie.200906919. [DOI] [PubMed] [Google Scholar]
- 144.Qian X, Ren Y, Shi Z, et al. Sequence-dependent synergistic inhibition of human glioma cell lines by combined temozolomide and miR-21 inhibitor gene therapy. Mol Pharm. 2012;9(9):2636–2645. doi: 10.1021/mp3002039. [DOI] [PubMed] [Google Scholar]
- 145.Xu L, Huang Y, Chen D, et al. Downregulation of miR-21 increases cisplatin sensitivity of non-small-cell lung cancer. Cancer Genet. 2014;207(5):214–220. doi: 10.1016/j.cancergen.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 146.Yu C, Qian L, Uttamchandani M, Li L, Yao SQ. Single-vehicular delivery of antagomir and small molecules to inhibit miR-122 function in hepatocellular carcinoma cells by using “smart” mesoporous silica nanoparticles. Angew Chem Int Ed Engl. 2015 doi: 10.1002/anie.201504913. [DOI] [PubMed] [Google Scholar]
- 147.Paige JS, Wu KY, Jaffrey SR. RNA mimics of green fluorescent protein. Science. 2011;333(6042):642–646. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Huang H, Suslov NB, Li NS, et al. A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore. Nat Chem Biol. 2014;10(8):686–691. doi: 10.1038/nchembio.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Warner KD, Chen MC, Song W, et al. Structural basis for activity of highly efficient RNA mimics of green fluorescent protein. Nat Struct Mol Biol. 2014;21(8):658–663. doi: 10.1038/nsmb.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mott JL, Kobayashi S, Bronk SF, Gores GJ. Mir-29 regulates mcl-1 protein expression and apoptosis. Oncogene. 2007;26(42):6133–6140. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhao JJ, Lin J, Lwin T, et al. microRNA expression profile and identification of miR-29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma. Blood. 2010;115(13):2630–2639. doi: 10.1182/blood-2009-09-243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wang C, Bian Z, Wei D, Zhang JG. miR-29b regulates migration of human breast cancer cells. Mol Cell Biochem. 2011;352(1–2):197–207. doi: 10.1007/s11010-011-0755-z. [DOI] [PubMed] [Google Scholar]
- 153.Li Z, Hassan MQ, Jafferji M, et al. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem. 2009;284(23):15676–15684. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Xu H, Cheung IY, Guo HF, Cheung NK. MicroRNA miR-29 modulates expression of immunoinhibitory molecule B7-H3: Potential implications for immune based therapy of human solid tumors. Cancer Res. 2009;69(15):6275–6281. doi: 10.1158/0008-5472.CAN-08-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]