Abstract
Development of blood cells through hematopoiesis occurs in the bone marrow (BM), and can be adversely impacted by various substances and/or conditions ranging from known therapeutic, intentionally administered xenobiotics to unintentional food additives and exposure to environmental chemicals. The principles underlying the techniques for evaluating toxicity to BM progenitors (erythroid, myeloid, and lymphoid) exploit changes in the normal hematopoietic process, biochemical cell surface and intracellular markers, as well as components of the BM microenvironment. Toxicological investigations following in vivo exposures of mice or in vitro exposures of mouse primary BM cell cultures allow the assessment of the developmental and functional integrity of BM cells, cell population shifts, and adverse biochemical effects due to toxicity. Colony forming unit (CFU) assays and flow cytometry are indispensable techniques in these toxicity studies.
Keywords: Bone Marrow Toxicology, Hematopoiesis, Progenitor Cell Differentiation, CFU Assays, Surface and Intracellular Staining
INTRODUCTION
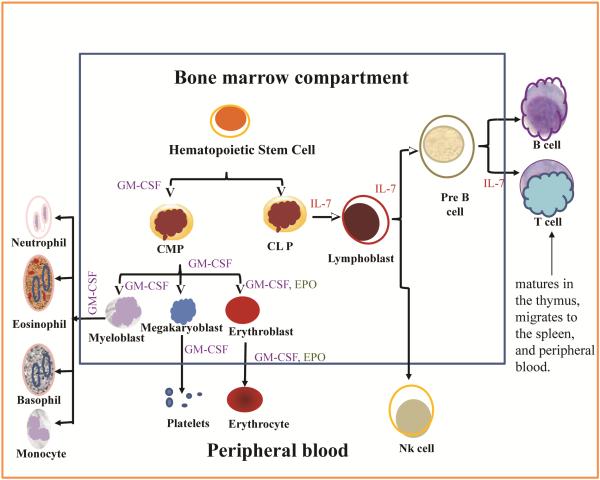
All blood cells are derived from a pluripotent hematopoietic stem cell which differentiates through several stages in the bone marrow (Figure 1). The BM vasculature and rich blood supply predispose the BM to toxicity by substances that may enter the body (Iverson, 1997). Exposure to environmental chemicals (Burchiel et al, 1988, Ezeh et al, 2014, 2015, N’jai et al, 2010), known therapeutic drugs and chemotherapy (Sommadossi and Carlisle, 1987), alcohol (Scharf and Aul, 1988), and protein malnutrition (Fantuzzi and Faggioni, 2000, Cunha et al, 2013), are associated with measurable toxicity to the hematopoietic process and progenitor cells in the BM. The most important role of BM progenitors is the regulation of hematopoiesis and immune function through the generation of cells for oxygen utilization, hemostasis, as well as innate, and adaptive immune responses. CFU assays are used to determine the specific lineage of progenitor cells that are affected by a test chemical or adverse condition and also to assess functionality of such lineage. Changes in the numbers of specific cell lineages can be captured from flow cytometry studies, as well as changes in intracellular signaling and biochemistry in surface markers-defined cell populations. From the pluripotent hematopoietic stem cell (HSC) stage in the bone marrow, some cells respond to the granulocyte-monocyte colony stimulating factor (GM-CSF) present in the BM microenvironment and commit to the myeloid lineage giving rise to the common myeloid progenitors (CMP).
Fig1.
Bone marrow hematopoiesis.
Other cells respond to IL-7, which is also present in the BM microenvironment and commit to the lymphoid lineage, producing the common lymphoid progenitors (CLP). The two growth factors, GM-CSF and IL-7 are then required at various stages in the development and further differentiation of their respective lineages, thus demarcating the myeloid from the lymphoid cells by development and subsequently by function. From the myeloid lineage, some cells develop receptors and also respond to erythropoietin for a short time as they commit to and differentiate into erythroid cells (Molineux et al, 2009, Tong et al, 2005).
The early erythroid progenitors, BFU-E, are formed from CD34+ HSCs through the action of numerous growth factors including IL-3, IL-4, IL-6, eosinophil differentiation factor (EDF), and E-CSF found in BM niches. Erythropoietin (EPO) is required for optimal growth of BFU-E, but not as critical as for CFU-E, the next stage of erythroid progenitors following the BFU-E. EPO, which originates from fibroblasts in the renal cortex and regulated by GATA-2 at transcriptional level, is sufficient in supporting the commitment and proliferation of CFU-E (Harmening, 2009).
We focus on the stimulation of the individual lineages using the specific growth factors in order to monitor abnormal changes in development and function that result from xenobiotic exposures. This protocol summarizes the assays and techniques that are used to assess toxicity to the progenitor cells during development in the BM.
Basic Protocol 1
MOUSE FEMUR HARVEST AND BONE MARROW CELL ISOLATION
This protocol describes a procedure for harvesting mouse femurs and for performing subsequent BM cell isolation.
NOTE: All research involving animals must be pre-approved by the Institutional Animal Care and Use Committee at your institution.
Materials
C57BL/6J mice, 12 to 16 weeks old (The Jackson Laboratory)
CO2 Euthanasia Chamber
Dissecting board
Sterile surgical instruments (e.g. Scissors, Scalpels, and Forceps)
70% ethanol (v/v)
1X Hanks Balanced Salt Solution (HBSS)
Isocove’s Modified Dulbecco’s Medium (IMDM) with 2% Heat Inactivated FBS (see REAGENTS and SOLUTIONS for recipe)
Sterile 60 × 15 mm petri dishes
1 mL syringe with 25-G × 15.8 mm needle
9″ Pasteur Pipettes
Automated or manual cell counter
Mouse Femur Harvest
Place mouse in a CO2 euthanasia chamber and turn on the CO2 inflow to the chamber until the animal stops breathing (approximately 3-5 minutes). Once respiration has completely ceased, allow the mouse to remain in the chamber for at least an additional 1-2 minutes.
Obtain the weight of the mouse and transfer it to a dissection board so that the mouse is positioned with the ventral side facing up. Verify the mouse is dead by pinching the limbs and monitoring for reflex.
Spread the limbs apart, and secure each limb in position on the dissection board using push pins.
Wet the ventral fur with 70% ethanol to reduce the risk of contamination at site of incision.
Hold the abdominal skin with forceps and use sharp scalpel to make an incision from the top of the thigh to below each knee.
Dissect back of the fur and cut tissue to expose the femur bone, hip joint, and knee joint.
Separate the femur from the joints by cutting at the epiphyses and at the center of the hip joint. Collect each femur set from a mouse and place in 4 mL cold HBSS in a 15 mL tube. Place the femurs on ice until needed for BM cell isolation.
BM Cell Isolation
Place femurs in a sterile 60 × 15 mm dish containing cold IMDM with 2% FBS. Carefully, trim away excess tissue from the femurs so that the white of the bone is mostly visible. Cut both ends of each femur bone to expose the interior marrow shaft.
-
Flush cells from the interior marrow shaft of each femur bone into a sterile 60 × 15 mm dish containing cold IMDM with 2% FBS using a 1 mL syringe with a 25-G needle.
To maximize cell yield, this step usually requires passing 6 to 9 mL of media through each end of the femur.
Break up cell aggregates by passing the solution through a 9 in Pasteur pipette.
Transfer the medium containing cells from both femurs to a 15 mL centrifuge tube. Place on ice.
Centrifuge for 10 min at 400 × g, 4° C and resuspend in 5 mL IMDM with 2% FBS.
Make a 20μl sample suspension of 1:1 (v/v) with Acridine Orange/Propridium Iodide (or other appropriate stain, e.g. trypan blue.
-
Determine the cell concentration and viability using an automated or manual cell counting system.
It is expected that each femur set will yield approximately 30 × 106 cells.
Basic Protocol 2
ASSESSMENT OF BONE MARROW DERIVED LYMPHOID AND MYELOID PROGENITOR CELL ACTIVITY IN VITRO
Early hematopoietic cells, such as erythroid and lymphoid progenitors are dependent on various cytokines and growth factors in their microenvironment to stimulate proliferation, differentiation, and maturation. Colony forming cell (CFC) assays are based on this principle and utilize a semi-solid methylcellulose matrix supplemented with different cytokine milieus to drive the proliferation and differentiation of specific progenitor cell populations in vitro (Stem Cell Technical Manual version 3.2.0; R&D Systems). These colony forming units (CFU) are then identified and counted based on morphology. The following protocol describes the steps necessary for performing the CFU-E, CFU-B, and CFU-GM assays for assessing the toxicity of xenobiotics on the activity of erythroid, lymphoid and myeloid progenitor cells, respectively.
NOTE: Refer to the Stem Cell Technical Manual version 3.2.0 or R&D Systems for detailed instructions or specific product information.
Materials
3 mL syringes with 16-G × 3.8 cm Monoject aluminum hub blunt cannula needle
MethoCult GF M3534 (Stem Cell Technologies) for CFU-GM assay
Mouse methylcellulose complete media (R&D Systems) for CFU-B assay
MethoCult M3334 (Stem Cell Technologies) for CFU-E assay
Treated 35 mm cell culture dishes (Stem Cell Technologies)
IMDM with 2% Heat Inactivated FBS (see REAGENTS and SOLUTIONS for recipe)
100 mm petri dishes
35 mm petri dishes
Sterile water
LYMPHOID. MYELOID, ERUTHROID PROGENITOR CELL CFU ASSAYS
Prepare methylcellulose aliquots according to the manufacturer’s instructions.
Isolate bone marrow cells (as described in Basic Protocol 1) from each mouse following in vivo exposures, or pool together BM cells from two-three mice for in vitro exposures.
For CFU-E and CFU-B, suspend BM cells in IMDM with 2% FBS at 1 × 106 cells/, suspend BM cells in IMDM with 2% FBS at 2×105 cells/mL.
-
Transfer 400 μl (4 × 105 cells) of the 1 × 106 cells/mL cell suspension to a sterile tube containing 4 mL MethoCult M3334 for CFU-E or to 4 mL Mouse Methylcellulose Complete Media for Pre-B Cells . For CFU-GM, transfer 400 μl (8 × 104 cells) of the 2 × 105 cells/ml cell suspension to a sterile tube containing 4 mL MethoCult GF M3534.
For in vitro exposures, prepare treatments at the desired concentrations and add to each methylcellulose aliquot so that the combined cell and treatment volumes do not exceed 1:10 (v/v) of the methylcellulose volume. This will maintain the proper viscosity of the medium and ensure that cytokines and growth factors remain at the appropriate concentrations.
Vortex the tube contents thoroughly to ensure the cells are evenly mixed with the methylcellulose media. Allow the media to sit at room temperature for approximately 20-25 mins to allow air bubbles generated during mixing to escape.
-
Dispense 1 mL of the methylcellulose-cell mixture (1 × 105 cells for CFU-B and CFU-E or 2 × 104 cells for CFU-GM) using a 3 mL syringe with a 16-G × 3.8 cm Monoject Aluminum Hub, blunt cannula needle into three separate 35 mm culture dishes per mouse for triplicates.
Prior to dispensing the samples, draw approximately 0.5 mL of methylcellulose-cell mixture into the syringe and gently expel the mixture back into the tube several times to minimize the air gap between the sample and syringe plunger, which will improve seeding volume accuracy.
Disperse the methylcellulose-cell mixture evenly in each 35 mm culture dish by gently swirling and rocking the dish in a horizontal plane.
Place two culture dishes and one sterile 35 mm petri dish containing approximately 3 mL of water inside a covered 100 mm petri dish. Incubate the samples in a humidified incubator at 37°C, 5% CO2 for 48 h (CFU-E), 10 days (CFU-B), or 14 days (CFU-GM).
Count CFU-E, CFU-B, or CFU-GM colonies following the appropriate incubation period and record colony numbers for statistical analysis.
Basic Protocol 3
MOUSE BONE MARROW CELL SURFACE MARKERS AND INTRACELLULAR STAINING BY FLOW CYTOMETRY
It is important to select antibodies with appropriate fluorochromes. Using BD Fluorescence Spectrum Viewer (http://www.bdbiosciences.com/us/s/spectrumviewer) and BD FACSelect™ Multicolor Panel Designer (http://www.bdbiosciences.com/us/panelDesign) is an optional and easy way to design your own antibody panel for a specific experiment. Antibody titration and compensations are important steps to perform before acquiring the valuable data.
Materials
Sample cells
Flow tubes
Fluorochrome-conjugated antibodies
Ammonium chloride lysing solution (1.5 M ammonium chloride; 100 mM sodium bicarbonate; 10 mM disodium EDTA, pH 7.4)
Wash buffer (Dulbecco’s phosphate buffered saline (DPBS) w/o Ca+2 and Mg+2 containing 1% FBS and 0.9% sodium azide)
Compensation beads (optional)
90% methanol at −20 °C and 4% paraformaldehyde are needed for intracellular staining cytometer
Cell Surface Marker Staining
-
Make antibody cocktail and keep in dark.
Usually, for 1×106 bone marrow cells, 0.5 μg of each antibody should be enough.
Suspend 1 ×106 bone marrow cells in 100 μl wash buffer.
-
Combine appropriate amount of the cocktail with the cell suspension, mix well and incubate at room temperature in the dark for at least 15 minutes.
The incubation time may vary depending on your antibodies. Refer to the manual or instruction from the manufacturer for optimized staining results.
Lyse red blood cells by incubating with 2 ml of ammonium chloride lysing solution for 10 minutes.
Centrifuge cells at 275 × g for 10 minutes.
Aspirate the supernatant and wash cells with the wash buffer.
Re-suspended washed cells in 0.5 ml wash buffer.
-
Analyze at least 10,000 cells using a Flow cytometer.
Since flow cytometers differ in their settings, it is important to adjust the voltage and do compensation following the manual of your cytometer before running samples. Unstained samples can be used for gating.
If an isotope control is used, the fluorescence value can be subtracted from the samples.
Intracellular Marker Staining
-
Follow the surface marker staining procedure to stain the cell surface makers.
Cells should be stained with surface markers first before fixing, because fixed and permeabilized cells may have altered surface antigens, resulting in weakened or no signal.
Resuspend the cells with 100 μl wash buffer.
Add 100 μl of 4% paraformaldehyde (see REAGENTS AND SOLUTIONS for recipe) or cell fixative manufactured solutions such as IC Fixation Buffer from eBiosciences to cells and incubate at 4 °C for 15 minutes in the dark.
Wash fixed cells twice with wash buffer, resuspend cells in 200 μl of −20 °C methanol.
Incubate at −20 °C for 15 minutes.
Centrifuge cells at 275 × g for 10 minutes to pellet the cells.
Wash twice with wash buffer and resuspend the cells in 100 μl of wash buffer.
-
Add appropriate amount of intracellular staining antibody to cells and incubate at room temperature for 30 minutes in dark. Do not forget to add isotope control to the isotope control sample.
0.25 μg of antibody should be enough for 1×106 bone marrow cells, or follow the manual from the manufacturer.
-
Wash cells twice with wash buffer, resuspend in 500μl of wash buffer and analyze on flow cytometer.
Gate according to unstained samples and fluorescence minus one (FMO) (Tung et al, 2007). For intracellular staining, at least 20,000 cells should be analyzed.
Determine the amount of intracellular antigen from the mean channel fluorescence value. The isotope control is the background value which can be subtracted from the samples
REAGENTS AND SOLUTIONS
Recipe for Iscoves Modified Dulbecco’s Medium with 2% Heat Inactivated FBS:
500 mL Isocove’s Modified Dulbeccos’s Medium (Sigma Aldrich)
200 mM L-glutamine
10,000 U/mL penicillin with 10,000 μg/mL streptomycin sulfate (Gibco by Life Techonologies)
Fetal bovine Serum (e.g. HyClone, VWR), heated inactivated
Procedure
Add 10 mL heat inactivated HyClone fetal bovine serum Add 5 mL 200 mM L-glutamine
Add 5 mL 10,000 U/mL penicillin with 10,000 μg/ml streptomycin sulfate
Mix by inverting the tube. This medium can be stored at 4°C for approximately 1 month.
Recipe for 4% Paraformaldehyde solution:
Deionized H2O
HCl
NaOH solution (1N)
Paraformaldehyde powder
1X DPBS/PBS
Note: All steps should be performed with gloves and in ventilated chemical hood. Paraformaldehyde is a carcinogen, so the waste must be properly disposed following the institutional guidelines.
Procedure
Add 800 mL of 1X DPBS to a glass beaker on a stir plate in a ventilated chemicahood. Heat while stirring to 60 °C.
Add 40 g of paraformaldehyde powder, slowly raise the pH by adding NaOH dropwise from a pipette until the solution clears.
Filter the solution once it is cooled
Adjust the volume of the solution to 1 L with DPBS.
Use HCl to adjust the solution to pH 6.9. The solution can be stored at 4 °C for 60 days.
COMMENTARY
Background Information
Myeloid, lymphoid, and erythroid progenitor cells are derived from early hematopoietic stem cells (HSCs), that give rise to monocytes, neutrophils, erythrocytes, megakaryocytes, B, T, and NK cells. The earliest surface markers for mouse HSCs include lin(−), sca+ and c-kit+, and these cells are known as LSK stem cells. These cells are of interest to toxicologists because some endogenous agents, environmental chemicals and xenobiotics [e.g. dioxins, polychlorinated biphenyls (PCBs), and polycyclic aromatic hydrocarbons (PAHs)], activate aryl hydrocarbon receptors (AHR) that lead to immune modulation (Gasiewicz et al., 2010; Casado et al., 2011).
CFU assays are used to examine the ability of hematopoietic progenitor cells to proliferate into colonies in response to cytokine stimulation. The assays were originally developed in the 1960s (Till et al., 1961), and they are still widely used for assessing toxicity in BM progenitor cells. There are numerous types of commercially available methylcellulose-based media, such as complete medium with recombinant cytokines added or medium without cytokines, which allows users to customize cytokines based on the progenitor cell population of interest. The principle is based on clonal expansion, thus each colony on the semi solid medium originates from one progenitor. Progenitor cell populations can be collected from the culture and further characterized using methods such as, flow cytometry and quantitative real time PCR. The major limitation of the CFU assay is that it can only be used to detect committed and/or differentiated cells, but not HSC or immediate progenitors.
Below is a list of cytokines required by different cell colonies from mouse BM:
CFU-GM: GM-CSF.
CFU-E: EPO, Transferrin.
BFU-E: IL-3, IL-6, EPO, Transferrin.
CFU-pre-B: IL-7.
CFU-GEMM: GM-CSF.
For more detailed information of medium types and cytokines, refer to the CFU assay manual from STEMCELL™ Technologies (http://www.stemcell.com/~/media/Technical%20Resources/8/3/E/9/0/28405_methocult%20M.pdf).
Flow cytometry is a technique used for the measurement of multiple characteristics of suspended single cells in a flowing fluid stream. The flow cytometer was developed with the advances in optics and electronics during 1960s, as well as the widely-used fluorescein dyes in microscopy. Currently, there are more and more dyes coming to the market with detector for a wider range of light wavelength, making it possible to do immunophenotyping and intracellular staining in the same sample as a multiple antibody panel, as long as there are enough channels for different fluorochromes. The advantages of staining the surface and intracellular markers simultaneously is that it saves time, unlike sorting and then using traditional methods (such as Western Blot) for protein detection. Also, in the simultaneous staining we can look at expression/modification of certain targets at the same time in just one tube of sample. Flow cytometry has high accuracy because the results are based on data collected from thousands of single cells. Hematopoietic progenitor cells consist of both lymphoid and myeloid lineage cells. These cells can be identified by their surface makers. Listed in table 1 are some commonly-used marker panels for mouse bone marrow.
Table 1.
Markers for mouse BM cell populations
| Target mouse BM cell populations | Markers |
|---|---|
| Early B cells (including pro-B and pre-B cells) | CD45R (B220)+, CD127+ |
| Pro-B cells | CD45R (B220)+, CD127+, CD43+, CD19− |
| Pre-B cells | CD45R (B220)+, CD127+, CD43−, CD19+ |
| Erythroid progenitors (CFU-E, BFU-E) | CD71+, TER119− |
| Mesenchymal Stromal Cell | CD44+, CD29+, Sca-1+, CD90+, CD45− |
| Common myeloid progenitors | Sca-1−, CD34 +, CD16/32 − |
| Common lymphoid progenitors | c-Kit +, Sca-1 + |
| Granulocyte-macrophage progenitors | Lin−, IL-7R−, c-Kit+, Sca-1-CD34− |
| LSK cells | Lin−, Sca-1+, c-Kit+ |
Critical Parameters
CFU assays
Handle and prepare materials and reagents sterilely to avoid contamination. Most methylcellulose media do not contain antibiotics.
Seed cells at appropriate density for accuracy in colony identification and counting.
Ensure proper and even mixing of cells with methylcellulose medium to reduce variability in colony counts within each sample.
Properly identify and count appropriate colonies. Compare and confirm colony counts with a different individual.
Keep experiments small to moderate in size, and avoid prolonged exposure of cells to less than optimal conditions.
Reporting the results of the assay is a critical. Since the plated cell number is known, the number of the colonies per 1 × 106 cells is a common way to report results, especially for in vitro experiments. This indicator can also be easily transformed into a percentage of certain CFU in bone marrow cells, which contains the same information as colonies per 1 × 106 cells. However, for some in vivo experiments, colonies per 1 × 106 cells or cell percentage alone may not be appropriate, as the total cells recovered from each mouse may be altered due to toxicological effects on the cells. Therefore, the total cell recovery should also be taken into account, so that results can be reported as the number of colonies per femur or femur set. Proper analysis and reporting the data is very important and improper analysis may lead to erroneous or incomplete conclusions.
Flow cytometry assays
Before staining, cell viability must be measured to confirm that no significant apoptosis occurred. The surface markers and intracellular protein level may be altered due to stress and apoptosis.
As mentioned before, appropriate controls and compensation are critical for a successful flow cytometry experiment. If using antibody from BD Biosciences, compensation beads for different fluorescence can be purchased to do the compensation, which may be more convenient. FMO should be performed when using a panel with 4 or more fluorochromes. Isotope controls must be added for intracellular staining.
For cell populations, results can be reported as percentage in a certain amount of cells, or absolute cell number in an animal (i.e., total positive cells per femur). The reporting strategies depend on the kind of experiment and the targeted effects, which is the same as discussed in the CFU assay part.
The intracellular staining is measured by mean channel fluorescence. The value in isotope control is considered the background and may be subtracted from the values in the samples.
Troubleshooting
CFU assays
A common source of error in the CFU assay is the possible uneven dispersion of cells in the semi solid methylcellulose medium, which results in differences in cell numbers and colonies in inoculum of the same samples. This is very common and should be avoided by thoroughly mixing cells with the methylcellulose media and plating cells soon after air bubbles are released.
Flow cytometry assay
No anticipated cell population
Usually, the first gate to set is on the forward and size scatter graph for a single cell population. If the target population is not displayed, the axis can be changed from linear to log scale to expand the data points. Cell population may also be lost during the washing steps when aspirating.
Hard to gate out anticipated population
Appropriate fluorescence channel compensations must be performed before running the samples. If it is very hard to compensate, check the antibody concentration and do a titration. Also, if the samples have low viability, it is common for surface staining antibodies to go inside the cells and stick to the intracellular contents so that the background fluorescence will be high, and causing false positive to hinder the differentiation between positive and negative staining. On the other hand, apoptotic cells and dead cells may have reduced surface antigen to bind to the antibody. Therefore, make sure your sample of cells has acceptable viability before staining (> 80%). An alternative is to add a viability dye of appropriate color to gate out unwanted dead cells.
No significant difference between isotope control and samples
This is commonly seen when the cells are over-fixed and permeabilized. Samples cannot be left in fixing buffer or methanol for too long. 15 minutes is enough for most cells. On the other band, this could happen when the permeablizing step failed. Detergent such as 0.1% Triton X 100 can be added to cells for ~10 minutes if an additional permeablizing step is required, depending on the targeted cell type. Also, a negative result cannot be excluded, especially in chemical treated cells.
Anticipated Results
CFU assays
As a guide, for C57BL/6J mice, CFU-E assays usually yield 200-300 colonies/plate. CFU-B colonies are around 100/plate and CFU-GM colonies are approximately 50-60/plate. For more information, please refer to Stem Cell Technologies website and Technical Manual for Mouse Colony-Forming Unit Assays Using MethoCult™.
Cell surface and intracellular markers by flow cytometry
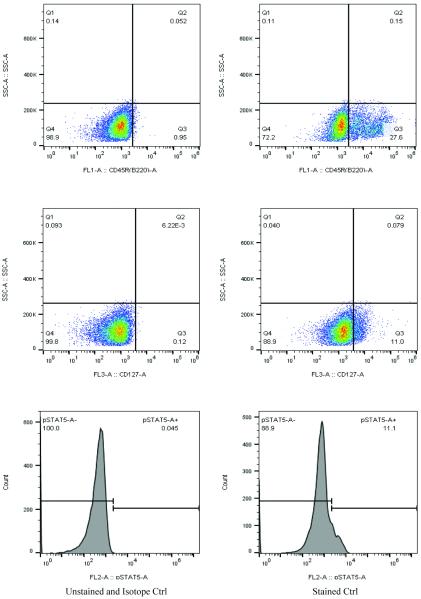
The bone marrow contains different cell populations. It is therefore necessary to use multiple gates to gate out the target cell population. Here is an example of a pre-B cell surface marker and intracellular phosphorylated STAT5 transcriptional factor staining in primary bone marrow cells (Fig. 2). CD45R and CD127 cell surface markers were used to separate the pre-B cell population (Table 1). The pSTAT5 intracellular staining showed a significant shift from the isotope control, indicating a successful staining of the cells.
Fig 2.
Primary bone marrow cells stained with CD45R, CD127 and intracellular phospho-STAT5 antibodies. The purpose of the staining is to see the pSTAT5 level in early B cells (Table 1). The left panel is showing an unstained sample for CD45R and CD127, as well as a sample stained with isotope control for phospho-STAT5. Right panel is showing a typical stained sample of CD45R, CD127 and phospho-STAT5.
Time Considerations
CFU assays
The time needed for femur harvest, BM cell isolation, and seeding of the cells for CFC assays takes approximately 1-2 hrs and may vary between labs, size of the experiment, expertise of the student or technician, and number of people assisting with the experiment. The incubation period is the major time consideration for CFC assays. The culture time for the CFU-E assay is 48 hours, CFU-pre-B is 10 days, and CFU-GM is 14 days.
Flow Cytometry
Flow cytometry based experiments have short turnaround time compared with several other techniques. Usually, the whole experiment including staining, setting up instrument, analyzing and interpreting data can be completed within 8 hours..
ACKNOWLEDGEMENT
This work was supported by a grant from the National Institute of Environmental Health Sciences (R01-ES019968 to SWB).We would also like to thank Fredine T. Lauer, Debra MacKenzie, and Shea McClain for their technical contributions.
Footnotes
INTERNET RESOURCES
STEMCELL™ Technologies, Mouse Colony-Forming Unit (CFU) Assays Using MethoCult™: (http://www.stemcell.com/~/media/Technical%20Resources/8/3/E/9/0/28405_methocult%20M.pdf).
R&D Systems, Mouse Methylcellulose Complete Media for Pre-B Cells: (https://www.rndsystems.com/products/mouse-methylcellulose-complete-media-for-pre-b-cells_hsc009#dsTab1).
BD Fluorescence Spectrum Viewer (http://www.bdbiosciences.com/us/s/spectrumviewer).
BD FACSelect™ Multicolor Panel Designer (http://www.bdbiosciences.com/us/panelDesign).
LITERATURE CITED
- Burchiel SW, Hadley WM, Barton SL, Fincher RH, Lauer LD, Dean JH. Persistent suppression of humoral immunity produced by 7, 12-dimethylbenz (a) anthracene (DMBA) in B6C3F1 mice: correlation with changes in spleen cell surface markers detected by flow cytometry. Int. J. Immunopharmacol. 1988;10:369–376. doi: 10.1016/0192-05619(88)90123-3. [DOI] [PubMed] [Google Scholar]
- Casado FL, Singh KP, Gasiewicz TA. Aryl hydrocarbon receptor activation in hematopoietic stem/progenitor cells alters cell function and pathway-specific gene modulation reflecting changes in cellular trafficking and migration. Mol Pharmacol. 2011;80:673–682. doi: 10.1124/mol.111.071381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha MCR, Lima FS, Vinolo MAR, Hastreiter A, Curi R, Borelli P, Fock RA. Protein malnutrition induces bone marrow mesenchymal stem cells commitment to adipogenic differentiation. PloS ONE. 2013;8:e58872. doi: 10.1371/journal.pone.0058872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezeh PC, Lauer FT, Mackenzie D, McClain S, Liu KJ, Hudson LG, Gandolfi AJ, Burchiel S. Arsenite selectively inhibits mouse bone marrow lymphoid progenitor cell development in vivo and in vitro and suppresses humoral immunity in vivo. PloS ONE. 2014;9:e93920. doi: 10.1371/journal.pone.0093920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezeh PC, Lauer FT, Liu KJ, Hudson LG, Burchiel SW. Arsenite interacts with DBC at low levels to suppress bone marrow lymphoid progenitors in mice. Biol Trace Elem Res. 2015;166:82–88. doi: 10.1007/s12011-015-0279-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantuzzi G, Faggioni R. Leptin in the regulation of immunity, inflammation and hematopoiesis. J. Leukoc. Biol. 2000;68:437–446. [PubMed] [Google Scholar]
- Gasiewicz TA, Singh KP, Casado FL. The aryl hydrocarbon receptor has an important role in the regulation of hematopoiesis: implications for benzene-induced hematopoietic toxicity. Chem Biol Interact. 2010;184:246–251. doi: 10.1016/j.cbi.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmening DM. Clinical hematology and fundamentals of hematostasis. 5th edition F. A. Davis Company; 2009. [Google Scholar]
- Iversen PO. Blood flow to the haematopoietic bone marrow. Acta Physiol Scand. 1997;159:269–276. doi: 10.1046/j.1365-201X.1997.00107.x. [DOI] [PubMed] [Google Scholar]
- Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- Koulnis M, Pop R, Porpiglia E, Shearstone JR, Hidalgo D, Socolovsky M. Identification and analysis of mouse erythroid progenitors using the CD71/TER119 flow-cytometric assay. J Vis Exp. 2011;54:e2809. doi: 10.3791/2809. PMCID: PMC3211121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molineux G, Foote M, Eliott S. Erythropoiesis and Erythropoietins. Second Edition Birkhauser; 2009. [Google Scholar]
- N’jai AU, Larsen M, Shi L, Jefcoate CR, Czuprynski JC. Bone marrow lymphoid and myeloid progenitor cells are suppressed in 7, 12 dimethylbenz(a)anthracene (DMBA) treated mice. Toxicology. 2010;271:27–35. doi: 10.1016/j.tox.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf RE, Aul C. Alcohol-induced disorders of the hematopoietic system. Z. Gastroenterol. 1988;26(Suppl 3):75–83. [PubMed] [Google Scholar]
- Sommadossi JP, Carlisle R. Toxicity of 3′-azido-3′-deoxythymidine and 9-(1,3-dihydroxy-2-propoxymethyl)guanine for normal human hematopoietic progenitor cells in vitro. Antimicrob. Agents. Chemother. 1987;31(3):452–4. doi: 10.1128/aac.31.3.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till JE, McCulloch EA. A direct measurement of the radiation sensitivity of normal mouse bone marrow cells. Radiat. Res. 1961;14:213–222. [PubMed] [Google Scholar]
- Tong W, Zang J, Lodish HF. Lnk inhibits erythropoiesis and Epo-dependent JAK2 activation and downstream signaling pathways. Blood. 2005;105(12):4604–4612. doi: 10.1182/blood-2004-10-4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung JW, Heydari K, Tirouvanziam R, Sahaf B, Parks DR, Herzenberg LA, Herzenberg LA. Modern flow cytometry: a practical approach. Clin. Lab. Med. 2007;27(3):453–68. doi: 10.1016/j.cll.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]