Abstract
Background:
In order to select a better antibiotic choice for treatment of Pseudomonas aeruginosa infections, this study was conducted to determine the frequency of resistance to some antipseudomonal β-lactams in P. aeruginosa isolates from patients in Tehran, Iran. In addition, the relation between presence of genes known to be responsible for resistance to β-lactams (ampC, mexC1,2, and mexC3,4 genes) and resistance phenotype among P. aeroginosa isolates was evaluated.
Methods:
P. aeruginosa strains were isolated and identified by routine methods and PCR for oprL gene. Disk diffusion method was employed to determine the antimicrobial susceptibility pattern according to CLSI recommendations. PCR was used to detect the resistance genes.
Results:
Among 100 isolates of P. aeruginosa, 82% had ampC, 86% mexC1,2 and 89% mexC3,4 genes and combinations of these genes were seen in most of isolates and only 3% of isolates had none of these genes. Resistance to mezlocillin, cefepime, ceftazidime and piperacillin/ tazobactam was seen in 46%, 41%, 36% and 29% of isolates, respectively. Significant relation (P value ≤0.05 by Chi-square or Fisher Exact test) was observed between the presence of ampC gene and resistance to all the studied β-lactams in this study. No relation was observed for mexC genes, although many of isolates containing these two genes were phenotypically resistant.
Discussion:
This study had shown for the first time, the presence of ampC and mexC genes in significant percent of clinical isolates of P. aeruginosa in Tehran, Iran, and relation between presence of ampC gene and resistance to β-lactams.
Key Words: Pseudomonas aeruginosa, β-lactams, mexC, ampC
Introduction
Pseudomonas aeruginosa is a member of skin normal flora in humans but has emerged as a major nosocomial pathogen in immune-compromised patients as a result of burns or other severe trauma, and underlying diseases including cancer, diabetes and cystic fibrosis (1). This bacterium is associated with different kinds of infections such as otitis externa, burn wounds, urinary tract infections, ventilator associated pneumonia and septicemia (2). It is responsible for about 10% of nosocomial infections and is considered as a major cause of mortality and morbidity in these patients (3).
P. aeruginosa has different mechanisms of resistance against antimicrobial agents; therefore, it is an important problem in clinical centers (1). It uses special outer membrane porins to restrict the uptake of antibiotics and the secondary resistance mechanisms such as energy-dependent multidrug efflux and chromosomally producing β-lactamase (4). A major component of bacterial resistance to many classes of antibiotics is expelling them out of bacteria cells that occur due to the activity of membrane transporter proteins called drug efflux pumps (5). Extrusion of antibiotics and restricted uptake through porins of outer membrane can cause a decrease in intracellular concentration of antibiotics (6). There are many genes that encode putative efflux pumps. One of the important families of chromosomally encoded bacterial efflux pumps is the resistance nodulation division (RND) family. This kind of pumps has three components: a membrane fusion protein that is associated with the cytoplasmic membrane, a periplasmic accessory protein (such as MexA, MexC, MexE and MexX), and an outer membrane protein (OMP) (such as OprM, OprJ and OprN) (7-11). As a result of synergy between outer membrane impermeability and chromosomally encoded efflux pumps, P. aeruginosa shows a remarkable intrinsic resistance to various antibiotic families (9). This antibiotic resistance is mediated by several resistance genes using multiple mechanisms resulting in making the treatment of many Pseudomonal infections more complicated.
The existence of three resistance genes (ampC, mexC1,2 and mexC3,4) are related to resistance to antipseudomonal β-lactams in clinical isolates of P. aeruginosa (12). Due to the absence of information about distribution of these genes in Iran, this study was performed to determine the frequency of these genes among P. aeruginosa isolated from patients in Tehran, Iran. This study has shown for the first time, the presence of ampC and mexC genes in significant percent of clinical isolates of P. aeruginosa in Tehran, Iran, and relation between presence of ampC gene and resistance to β-lactams.
Materials and Methods
Bacterial isolates
P. aeruginosa isolated from patients in three hospital laboratories (Pars and Milad hospitals and Motahari Burn Center) in Tehran, Iran, in 2013 were collected and 100 of them selected randomly and used in this study. Identification of isolates as P. aeruginosa was done based on general phenotypic methods including colony pigmentation, Gram staining, oxidase test, oxidative/fermentative (OF) test for carbohydrate utilization, growth at 42˚C and growth on cetrimide agar (13).
PCR for detection of genes
Genomic DNA was extracted based on the Ozer et al. method (12) with some modifications. The isolates were screened for presence of resistance genes ampC, mexC1,2 and mexC3,4 genes according to PCR method of Ozer et al. (12). Molecular identification of P. aeruginosa was performed with PCR using oprL gene primers (14). Primers used in this study are shown in Table 1.
Table 1.
Primers used in this study
| Gene | Forward and Reverse Primers Sequences (5ʹ to 3ʹ) | Product length (bp) | Reference |
|---|---|---|---|
| ampC | CGGCTCGGTGAGCAAGACCTTC | 218 | (12) |
| AGTCGCGGATCTGTGCCTGGTC | |||
| mexC 1,2 | ATCCGGCACCGCTGAAGGCTGCG | 344 | (12) |
| CGGATCGAGCTGCTGGATGCGCG | |||
| mexC 3,4 | GTACCGGCGTCATGCAGGGTCC | 164 | (12) |
| TTACTGTTGCGGCGCAGGTGACT | |||
| oprL | ATGGAAATGCTGAAATTCGGC | 504 | (14) |
| CTTCTTCAGCTCGACGCGACG |
We performed Duplex PCR assay for the detection of studied genes in a thermal cycler (Techne, UK). This Duplex PCR reaction were carried out in a final volume of 25µl containing 12.5µl Master Mix (Amplicon Taq DNA Polymerase 2x Master Mix Red, ViraGene Company, Iran), 9.5µl DDW, 1µl of each primers (0.5 µl Forward primer and 0.5µl Reverse primer) and 1µl DNA template. Master Mix1 contained ampC and mexC1,2 primers, and Master Mix2 contained mexC3,4 and oprL primers. Program of amplification process was as follows: Initial denaturation at 93˚C for 5 min, 30 cycles of initiation at 93˚C, annealing at 55˚C and extension at 72˚C; each 1 min; and final extension at 72˚C for 5 min. The PCR products and 100bp DNA ladder were visualized under gel documentation system (UVItec, UK) after electrophoresis on a 1% agarose gel and staining by Ethidium Bromide.
Antimicrobial susceptibility testing
Disk diffusion method was used for detection of antimicrobial susceptibility pattern in clinical isolates of P. aeruginosa according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (15). The following antibiotic disks from MAST Group Ltd. (Merseyside, UK), were used: Mezlocillin (MEZ; 75µg), cefepime (CPM; 30µg), ceftazidime (CAZ; 30µg) and piperacillin/ tazobactam (PTZ; 100/10µg). Control strains used for piperacillin/ tazobactam was E. coli ATCC35218, and for other antibiotics was P. aeruginosa ATCC27853.
Data analysis
All collected data were analyzed and frequencies were computed by Statistical Package for Social Sciences version 20 (SSPS Inc, Chicago, IL, USA). The relation between antibiotic resistance and the presence of the resistance genes is determined by Chi-square or Fisher Exact test and P value ≤0.05 was considered statistically significant.
Results
From 100 collected P. aeruginosa isolates 67, 25 and 8 were isolated from patients in Pars Hospital, Milad Hospital, and Motahari Burn Center, respectively, which were isolated from sputum (50%), urine (35%), wound (13%), CSF (1%), and blood (1%). Characteristics of patients showed that 51 of them were male and 49 female, 62 were outpatient and 38 inpatients, and mean age was 52.57 ± 27.15 (12 cases had below 15, 20 cases 15-44, 23 cases 45-64 and 45 cases 65-94 years old.
In all isolates identified by phenotypically methods as P. aeruginosa, the oprL gene was also detected by PCR method. Altogether, ampC, mexC1,2 and mexC3,4 genes were detected in 82%, 86%, and 89% of isolates, respectively, and combination of genes were also seen in many of isolates and only three isolates had neither of these genes (table 2).
Table 2.
Frequency of studied genes in Iranian P. aeruginosa isolates
| Gene (s) | Percent |
|---|---|
| ampC | 82 |
| mexC 1,2 | 86 |
| mexC 3,4 | 89 |
| ampC & mexC 1,2 | 75 |
| ampC & mexC 3,4 | 75 |
| mexC 1,2 & mexC 3,4 | 80 |
| ampC & mexC 1,2 & mexC 3,4 | 70 |
| Without ampC & mexC1,2 & mexC3,4 | 3 |
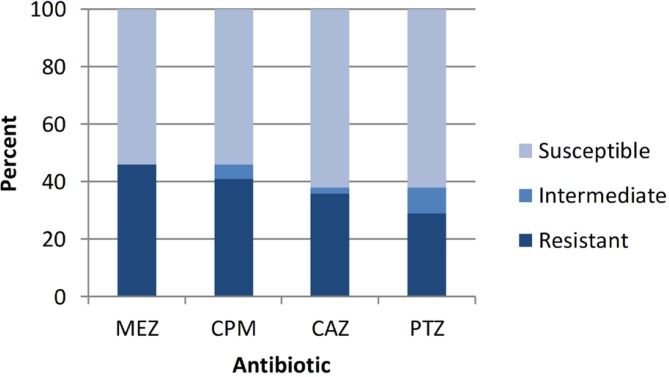
Antibiotic susceptibility of studied isolates had shown in Fig. 1. Resistance to mezlocillin, cefepime, ceftazidime and piperacillin/ tazobactam was seen in 46%, 41%, 36% and 29% of studied P. aeruginosa isolates, respectively. Relation between resistance to theses antipseudomonal β-lactams and the presence of ampC, mexC1,2 and mexC3,4 genes among studied P. aeruginosa isolates were also studied by statistical methods. Significant relation (P value ≤ 0.001) was shown between the resistance to each studied antibiotic and presence of ampC gene. This relation was not found for mexC genes, although a high number of resistant isolates had these genes.
Fig. 1.
Results of susceptibility test for Iranian P. aeruginosa isolates
Discussion
Because of the importance of P. aeruginosa in human infections, many studies are undertaken in the world about resistance to different antibiotics in clinical isolates of this bacterium. Many reports were published in Iran about frequency of resistance to different antibiotics in P. aeruginosa isolated from patients (16-23); although there is no report of resistance to mezlocillin. In this study we focused on determination of the frequency of resistance to four extended expectrum penicillins which are used in treatment of P. aeruginosa infections. Among clinical isolates of P. aeruginosa collected in three Tehran hospitals, resistance to ceftazidime was 36%, which was higher than Shahcheraghi et al. report (25%) and lower than other studies in Iran, which were 57.5% to 89.5% (17, 23-26). In our study, resistance to cefepime was 41% that was almost similar to a report with the result of 39%, but much less than other study which was 91.7% (17, 25). Furthermore we found that resistance to piperacillin/ tazobactam was 29% which showed similarity to study of Shahcheraghi et al. (28%), and a little higher than the result of Salimi et al. (19.1%), however, it was reported 87.2% resistance that was much higher than our study (17, 24, 26). The rates of resistance to ceftazidime, piperacillin/ tazobactam and mezlocillin in this study were 36%, 29%, and 46%, respectively, similar to a report (12) from Turkey, our neighbor country, which were reported respectively 30%, 24%, and 50%, but the resistance to cefepime in this study was higher than mentioned report (41% versus 18%). searching in studies of other countries, the rates of resistance to mezlocillin were reported 48% that shows higher rate in comparison with our findings (19, 20). Geographic differences in antimicrobial resistance were also shown in other studies, and some of the variables explaining these differences in population demographics, access to medical care and illicit drug use (27, 28).
P. aeruginosa use several genes to mediate resistance to β-lactam antibiotics including ampC, mexC1,2 and mexC3,4 genes (12, 29). The ampC gene encodes an inducible chromosomal β-lactamase. The mexC genes are related to MexCD-OprJ efflux system and are belonging to the RND family (30, 31). In the genetic map of this bacterium, ampC is located beside the genes of the MexCD-OprJ efflux system (30). Besides, it is shown that this efflux system is an inducible pump, which expression could be induced after more using of some inducer antibiotics, pressure of antibiotics and following mutation in mexR gene that control the expression of genes belonging to MexCD-OprJ efflux system (32, 33). Only a few reports were published about presence of ampC gene in clinical isolates of P. aeruginosa in Iran, such as study of Aghazadeh et al. (34), and presence of mexC genes have not be done before this study. The studied genes (ampC, mexC1,2 and mexC3,4) were detected in most of P. aeruginosa isolated from patients in Tehran, Iran. Unfortunately, we could not found any report about the rate of mexC genes in clinical isolates of P. aeruginosa in other countries for comparison, except the study of Ozer et al. in Turkey (12), which reported lower rates for these genes.
In this study, relation between the resistance to mezlocillin, cefepime, ceftazidime and piperacillin/ tazobactam and presence of ampC gene in P. aeruginosa isolated from patients in Iran were seen same to the Ozer et al. study in Turkey (12) but reported relation between the resistance to these antibiotics and presence of mexC genes was not shown in this study, although mexC genes were found in a high number of resistant isolates.
Conclusion
The study presented high frequency of resistance to mezlocillin, cefepime, ceftazidime, and piperacillin/ tazobactam in P. aeruginosa isolated from patient in Tehran, Iran, which could help in selection of the best antibiotic for empirical therapy in treatment of severe pseudomonal infections. Moreover, it was shown that most of these isolates had ampC and mexC genes and there was significant relation between resistant to used antipseudomonal β-lactams and presence of ampC gene.
Acknowledgment
Many thanks to Elham Faghihzadeh, Seyede Marzieh Mosavi, Rahim Nosrati and especially to microbiology laboratory staff of Pars, Milad, and Motahari hospitals for collaboration in this study. This survey was an MSc student thesis and financially supported by Research Council of Shahed University.
Conflict of interest
The authors declare that there is no conflict of interests.
References
- 1.Morales E, Cots F, Sala M, Comas M, Belvis F, Riu M, et al. Hospital costs of nosocomial multi-drug resistant Pseudomonas aeruginosa acquisition. BMC Health Serv Res. 2012;12(1):122. doi: 10.1186/1472-6963-12-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Satti L, Abbasi S, Qumar TA, Khan MS, Hashmi ZA. In Vitro Efficacy of Cefepime Against Multi-Drug Resistant Pseudomonas aeruginosa- An Alarming Situation in our Setup. Open Drug Resist J. 2011;1(1):12–6. [Google Scholar]
- 3.Tohidpour A, Najar Peerayeh S, Mehrabadi JF, Rezaei Yazdi H. Determination of the Efflux Pump-Mediated Resistance Prevalence in Pseudomonas aeruginosa, Using an Efflux Pump Inhibitor. Curr Microbiol. 2009;59(3):352–5. doi: 10.1007/s00284-009-9444-5. [DOI] [PubMed] [Google Scholar]
- 4.Hancock REW, Speert DP. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Resist Updates. 2000;3(4):247–55. doi: 10.1054/drup.2000.0152. [DOI] [PubMed] [Google Scholar]
- 5.Zaman GJR, Flens MJ, Van Leusden MR, De Haas M, Mulder HS, Lankelma J, et al. The human multidrug resistance-associated protein MRP is a plasma membrane drug-efflux pump. Proc Natl Acad Sci. 1994;91(19):8822–6. doi: 10.1073/pnas.91.19.8822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee A, Mao W, Warren MS, Mistry A, Hoshino K, Okumura R, et al. Interplay between Efflux Pumps May Provide Either Additive or Multiplicative Effects on Drug Resistance. J Bacteriol. 2000;182(11):3142–50. doi: 10.1128/jb.182.11.3142-3150.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piddock LJ. Clinically Relevant Chromosomally Encoded Multidrug Resistance Efflux Pumps in Bacteria. Clin Microbiol Rev. 2006;19(2):382–402. doi: 10.1128/CMR.19.2.382-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma D, Cook DN, Hearst JE, Nikaido H. Efflux pumps and drug resistance in gram-negative bacteria. Trends Microbiol. 1994;2(12):489–93. doi: 10.1016/0966-842x(94)90654-8. [DOI] [PubMed] [Google Scholar]
- 9.Poole K. Multidrug Efflux Pumps and Antimicrobial Resistance in Pseudomonas aeruginosa and related organisms. J Mol Microbiol Biotechnol. 2001;3(2):255–64. [PubMed] [Google Scholar]
- 10.Lomovskaya O, Warren MS, Lee A, Galazzo J, Fronko R, Lee M, et al. Identification and Characterization of Inhibitors of Multidrug Resistance Efflux Pumps in Pseudomonas aeruginosa: Novel Agents for Combination Therapy. Antimicrob Agents Chemother. 2001;45(1):105–16. doi: 10.1128/AAC.45.1.105-116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson JM, Church GM. Alignment and structure prediction of divergent protein families: periplasmic and outer membrane proteins of bacterial efflux pumps. J Mol Biol. 1999;287(3):695–715. doi: 10.1006/jmbi.1999.2630. [DOI] [PubMed] [Google Scholar]
- 12.Ozer B, Duran N, Onlen Y, Savas L. Efflux pump genes and antimicrobial resistance of Pseudomonas aeruginosa strains isolated from lower respiratory tract infections acquired in an intensive care unit. J Antibiot. 2012;65(1):9–13. doi: 10.1038/ja.2011.102. [DOI] [PubMed] [Google Scholar]
- 13.Phillips I. Identification Of Pseudomonas aeruginosa in the clinical laboratory. J Med Microbiol. 1969;2:9–16. doi: 10.1099/00222615-2-1-9. [DOI] [PubMed] [Google Scholar]
- 14.De Vos D, Lim Jr A, Pirnay JP, Struelens M, Vandenvelde C, Duinslaeger L, et al. Direct detection and identification of Pseudomonas aeruginosa in clinical samples such as skin biopsy specimens and expectorations by multiplex PCR based on two outer membrane lipoprotein genes, oprI and oprL. J Clin Microbiol. 1997;35(6):1295–9. doi: 10.1128/jcm.35.6.1295-1299.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. Twenty-third Informational Supplement. CLSI document M100-S23. 950 West Valley Road, Suite 2500, Wayne, Pennsylvania 19087 USA: Clinical and Laboratory Standards Institute; 2013. [Google Scholar]
- 16.Owlia P, Saderi H, Mansouri S, Salemi S, Ameli H. Drug resistance of isolated strains of Pseudomonas aeruginosa from burn wound infections to selected antibiotics and disinfectants. Iran J Pathol. 2006;1(2):64–9. [Google Scholar]
- 17.Moazami Goudarzi S, Eftekhar F. Assesment of carbapenem susceptibility and multidrug-resistance in Pseudomonas aeruginosa burn isolates in Tehran. Jondishapour J Microbiol. 2013;6(2):162–5. [Google Scholar]
- 18.Japoni A, Alborzi A, Kalani M, Nasiri J, Hayati M, Farshad S. Susceptibility patterns and cross-resistance of antibiotics against Pseudomonas aeruginosa isolated from burn patients in the South of Iran. Burns. 2006;32(3):343–7. doi: 10.1016/j.burns.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 19.Yetkin G, Otlu B, Cicek A, Kuzucu C, Duramaz R. Clinical, microbiologic, and epidemiologic characteristics of Pseudomonas aeruginosa infections in a Urinary Hospital, Malatya, Turkey. Am J Infect Control. 2006;34(4):188–92. doi: 10.1016/j.ajic.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 20.Ojeda Vargas MM, Alonso R, Vindel A, Monzon-Moreno C. Epidemiological considerations of infections caused by Pseudomonas aeruginosa in a Canary Islands hospitals. Med Mal Infect. 1993:434–7. [Google Scholar]
- 21.Alaghehbandan R, Azimi L, Rastegar Lari A. Nosocomial infections among burn patients in Tehran, Iran: a decade later. Ann Burns Fire Disasters. 2012;25(1):3–7. [PMC free article] [PubMed] [Google Scholar]
- 22.Saderi H, Karimi Z, Owlia P, Bahar MA, Akhavi Rad SMB. Phenotypic detection of Metalo-beta-Lactamase producing Pseudomonas aeruginosa strains isolated from burn patients. Iran J Pathol. 2008;3(1):20–4. [Google Scholar]
- 23.Ranjbar R, Owlia P, Saderi H, Mansouri S. Characterization of Pseudomonas aeruginosa strains isolated from burned patients hospitalized in a major burn center in Tehran, Iran. Acta Medica Iranica. 2011;49(10):675–9. [PubMed] [Google Scholar]
- 24.Shahcheraghi F, Badmasti F, Feizabadi MM. Molecular characterization of class 1 integrons in MDR Pseudomonas aeruginosa isolated from clinical settings in Iran, Tehran. FEMS Immunol Med Microb. 2010;58:421–5. doi: 10.1111/j.1574-695X.2009.00636.x. [DOI] [PubMed] [Google Scholar]
- 25.Hosseini Jazani N, Zahedi A, Garebagi N. Phenotipic detection of methalo-β-lactamase producing Pseudomonas aeruginosa isolated from Urmia hospitals. Afr J Microbiol Res. 2012;6(7):1387–92. [Google Scholar]
- 26.Salimi H, Yakhchali B, Owlia P, Lari AR. Molecular Epidemiology and Drug Susceptibility of Pseudomonas aeruginosa Strains Isolated From Burn Patients. Lab Medicine. 2010;41(9):540–4. [Google Scholar]
- 27.Hirsch EB, Tam VH. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev Pharmacoecon Outcomes Res. 2010;10(4):441–51. doi: 10.1586/erp.10.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Croft AC, D'Antoni AV, Terzulli SL. Update on the antibacterial resistance crisis. Med Sci Monit. 2007;13(6):RA103–18. [PubMed] [Google Scholar]
- 29.Conejo MC, Martinez-Martinez L, Garcia I, Picabea L, Pascual A. Effect of siliconized latex urinary catheters on the activity of carbapenems against Pseudomonas aeruginosa strains with defined mutations in ampC, oprD, and genes coding for efflux systems. Int J Antimicrob Agents. 2003;22(2):122–7. doi: 10.1016/s0924-8579(03)00119-5. [DOI] [PubMed] [Google Scholar]
- 30.Lambert PA. Mechanisms of antibiotic resistance in Pseudomonas aeruginosa. J R Soc Med. 2002;95(41):22–6. [PMC free article] [PubMed] [Google Scholar]
- 31.Strateva T, Yordanov D. Pseudomonas aeruginosa a phenomenon of bacteria resistance. J Med Microb. 2009;58:1133–48. doi: 10.1099/jmm.0.009142-0. [DOI] [PubMed] [Google Scholar]
- 32.Morita Y, Komori Y, Mima T, Kuroda T, Mizushima T, Tsuchiya T. Construction of a series of mutants lacking all of the four major mex operons for multidrug efflux pumps or possessing each one of the operons from Pseudomonas aeruginosa PAO1: MexCD-OprJ is an inducible pump. FEMS Microbiol Lett. 2001;202:139–43. doi: 10.1111/j.1574-6968.2001.tb10794.x. [DOI] [PubMed] [Google Scholar]
- 33.Narita ShI, Eda Sh, Yoshihara E, Nakae T. Linkage of the efflux-pump expression level with substrate extrusion rate in the MexAB-OprM efflux pump of Pseudomonas aeruginosa. Biochem Biophys Res Commun. 2003;308(4):922–6. doi: 10.1016/s0006-291x(03)01512-2. [DOI] [PubMed] [Google Scholar]
- 34.Aghazadeh M, Hojabri Z, Mahdian R, Nahaei MR, Rahmati M, Hojabri T, Pirzadeh T, Pajand O. Role of efflux pumps: MexAB-OprM and MexXY-OprA), AmpC cephalosporinase and OprD porin in non-metallo-β-lactamase producing Pseudomonas aeruginosa isolated from cystic fibrosis and burn patients. Infect Genet Evol. 2014;24:187–92. doi: 10.1016/j.meegid.2014.03.018. [DOI] [PubMed] [Google Scholar]