Abstract
In this article, we study the causal inference problem with a continuous treatment variable using propensity score-based methods. For a continuous treatment, the generalized propensity score is defined as the conditional density of the treatment-level given covariates (confounders). The dose–response function is then estimated by inverse probability weighting, where the weights are calculated from the estimated propensity scores. When the dimension of the covariates is large, the traditional nonparametric density estimation suffers from the curse of dimensionality. Some researchers have suggested a two-step estimation procedure by first modeling the mean function. In this study, we suggest a boosting algorithm to estimate the mean function of the treatment given covariates. In boosting, an important tuning parameter is the number of trees to be generated, which essentially determines the trade-off between bias and variance of the causal estimator. We propose a criterion called average absolute correlation coefficient (AACC) to determine the optimal number of trees. Simulation results show that the proposed approach performs better than a simple linear approximation or L2 boosting. The proposed methodology is also illustrated through the Early Dieting in Girls study, which examines the influence of mothers’ overall weight concern on daughters’ dieting behavior.
Keywords: boosting, distance correlation, dose–response function, generalized propensity scores, high dimensional
1 Introduction
Much of the literature on propensity scores in causal inference has focused on binary treatments. In the past decade, a few studies (e.g., Lechner (2002), Imai and Van Dyk (2004), Tchernis, Horvitz-Lennon, and Normand (2005), Karwa, Slavković, and Donnell (2011) and McCaffrey, Griffin, Almirall, Slaughter, Ramchand, and Burgette (2013)) have extended propensity score-based approaches to categorical treatments with more than two levels.
In this article, we consider the problem of causal inference when the treatment is quantitative. Quantitative treatments are very common in practice, such as dosage in biomedical studies (Imbens, 2000), number of cigarettes in prevention studies (Imai and Van Dyk, 2004), and duration of training in labor studies (Kluve, Schneider, Uhlendorff, and Zhao, 2012). In the special case of continuous treatments, a main objective is to estimate the dose-response function. Hirano and Imbens (2004) propose a two-step procedure for estimating the dose-response function and suggest a technique called “blocking” to evaluate the balance in the covariates after adjusting for the propensity scores. An alternative approach (Robins, 1999) is based on marginal structural models (MSMs). In MSMs, we specify a response function and employ IPW to consistently estimate the parameters of the function.
A key step in both approaches is to estimate the generalized propensity score, which is defined as the conditional density of the treatment level given the covariates. Conditional density is usually estimated nonparametrically, such as kernel estimation or local polynomial estimation (e.g., Hall, Wolff, and Yao (1999), Fan, Yao, and Tong (1996)). When there are a large number of covariates in the study, the nonparametric estimation of the conditional density suffers from the curse of dimensionality. Given the limited literature on this topic, we propose a boosting algorithm to estimate the generalized propensity score. In boosting, the number of trees to be generated is an important tuning parameter, which essentially determines the trade-off between bias and variance of the targeted causal estimator. In the binary treatment case, it has been suggested that the optimal number of trees be determined by minimizing the average standardized absolute mean (ASAM) difference between the treatment group and the control group (McCaffrey, Ridgeway, and Morral, 2004). The standardized mean difference is also a well-established criterion to assess balance in the potential confounders after weighting. This idea can easily be extended to the categorical treatment case. Similarly, for a continuous treatment, we could divide the treatment into several categories and draw causal inference based on the categorical treatment. However, doing so may introduce subjective bias and information loss. Instead, we aim to develop an innovative criterion that minimizes the correlation between the continuous treatment variable and the covariates after weighting.
This article proceeds as follows. In Section 2, we review the concepts of dose-response function, generalized propensity scores and the ignorability assumption. In Section 3, we propose a boosting method to estimate the generalized propensity scores and propose an innovative criterion to determine the optimal number of trees in boosting. A detailed algorithm is described and the corresponding R code is provided in the Appendix. In Section 4, we compare the proposed methods through simulation studies, and a data analysis application to the Early Dieting in Girls study is presented in Section 5. Some discussion concludes Section 6.
2 Dose-Response Function
2.1 Definition and Assumptions
Let Y denote the response of interest, T be the treatment level, and X be a p-dimensional vector of baseline covariates. The observed data can be represented as (Yi, Ti, Xi), i = 1,…,n, a random sample from (Y, T, X). In addition to the observed quantities, we further define Yi(t) as the potential outcome if subject i were assigned to treatment level t. Here, T is a random variable and t is a specific level of T. The dose-response function we are interested in estimating is μ(t) = E[Yi(t)], and we assume Yi(t) is well defined for t ∈ τ, where τ is a continuous domain.
Similar to the binary case, the ignorability assumption is as follows:
where f (t|·) refers to the conditional density. That is, the treatment assignment is conditionally independent of the potential outcomes given the covariates. In other words, we assume there are no unmeasured covariates that may jointly influence the treatment assignment and potential outcomes.
Denote the generalized propensity score as r(t, X) ≡ fT|X(t|X), which is the conditional density of observing the treatment level t given the covariates (Imbens, 2000). The ignorability assumption implies
That is, to adjust for confounding, it is sufficient to condition on the generalized propensity scores instead of conditioning on the vector of covariates, which might be high-dimensional.
2.2 Estimation based on Marginal Structural Models
Under the ignorability assumption, we focus on the marginal structural model approach to estimate the dose-response function proposed by Robins (1999) and Robins, Hernán, and Brumback (2000). The method works by building a marginal structural model for the potential outcomes. For example, we may assume a linear model:
| (1) |
Model (1) is marginal because it is defined for the expected value of potential outcomes without conditioning on any covariates (which is different from regression models). Based on the observed data (Yi, Ti, Xi), i = 1,…,n, the parameters in (1) can be consistently estimated by IPW. For the ith subject, the weight is
| (2) |
There are two important issues related to this approach: (i) the estimation of the inverse probability weights; (ii) the functional form of the outcome model in (1). The first issue is the main topic of this article and will be explored in the next section. Here we briefly discuss the second issue. The consistency result of MSM estimators relies on the correct specification of the outcome model. However, the true form of E[Y(t)] is unknown in reality and a flexible model is always preferred. In the data application, we assume a regression spline function (Eubank, 1988) for the outcome model:
| (3) |
where is a truncated power function and u+ = max(0, u). p is the order of the polynomial function and K is the total number of inner knots. The inner knots are either distributed evenly on τ or defined as the equally spaced sample quantiles of Ti, i = 1,…,n. That is,
where T(i) is the ith quantile of T1, T2,…, Tn.
To determine the regression spline function, we need to find the optimal p and K. The traditional model selection criteria, such as AIC and BIC, are based on a simple random sample. These criteria can be extended to determine the form of the marginal structural model for a non-randomized sample (Hens, Aerts, and Molenberghs, 2006, Platt, Brookhart, Cole, Westreich, and Schisterman, 2012). Under the assumption that Y is normally distributed, the weighted AIC can be defined as
| (4) |
where l is the total number of parameters. In (3), l = K + p + 1. Notice that in this stage, we treat as fixed. Similarly, we define the weighted BIC as
| (5) |
We will illustrate the specification of the outcome model through the data application in Section 5.3. In the next section, we will focus on the first issue and propose a boosting algorithm for estimating the generalized propensity scores.
3 The Proposed Method
3.1 Modeling the generalized propensity scores
In the MSM approach, the estimation of in (2) is essential. For simplicity, we assume T follows a normal distribution so that r(Ti) can be easily estimated by normal density. To be noticed, if the normality assumption does not hold for T, we can always employ a nonparametric method, such as Kernel density estimation, to estimate r(Ti). To estimate r(Ti, Xi), a traditional way is to assume
| (6) |
Then, the estimation of r(Ti, Xi) follows two steps (Robins et al., 2000):
Run a multiple regression of Ti on Xi, i = 1,…,n, and get T̂i and σ̂;
- Calculate the residuals ε̂i = Ti − T̂i; r(Ti, Xi) can be approximated by
(7)
Because the ignorability assumption is untestable, researchers usually collect a large number of covariates, which means X is high-dimensional. In this case, equation (6) may not hold. A more general approach is to assume
| (8) |
where m(X) is the mean function of T given X. We advocate a machine learning algorithm, boosting, to estimate m(X). The advantage of boosting is that it is a nonparametric algorithm that can automatically pick important covariates, nonlinear terms and interaction terms among covariates (McCaffrey et al., 2004). It fits an additive model and each component (base learner) is a regression tree. Mathematically, it can be written as:
| (9) |
where M is the total number of trees, Km is the number of terminal nodes for the m-th tree, Rmj is the indicator of rectangular region in the feature space spanned by X and cmj is the predicted constant in region Rmj. Km and Rmj are determined by optimizing some nonparametric information criterion, such as Entropy, misclassification rate or Gini Index. cmj is simply the average value of Ti in the training data that falls in the region Rmj. Details about how to construct a tree classifier can be found in Breiman, Friedman, Stone, and Olshen (1984).
In boosting, M is an important tuning parameter. It determines the trade-off between bias and variance of the causal estimator. In inverse weighted methods, if subject i receives a weight wi, it means the subject will be replicated wi − 1 times; that is, there will be wi − 1 replications in the weighted pseudo sample. In the weighted sample, if the propensity scores are correctly estimated, the treatment assignment and the covariates are supposed to be unconfounded under the ignorability assumption (Robins et al., 2000). Consequently, the causal effect can be estimated as in a simple randomized study without confounding. Therefore, a reasonable criterion is to stop the algorithm at the number of trees such that the treatment assignment and the covariates are independent (unconfounded) in the weighted sample. Next, we propose stopping criteria for boosting based on this idea.
3.2 Algorithm
We propose four different criteria (summarized in Table 1) for how to measure the degree of independence/correlation between the treatment and each covariate. These criteria are named as “Pearson/polyserial”, ”Spearman”, “Kendall” and “distance”. Pearson/polyserial, Spearman and Kendall correlations are commonly used correlation matrices; distance correlation (Székely, Rizzo, and Bakirov, 2007, Székely and Rizzo, 2009) is the most recently proposed and is gaining popularity due to its nice property: it can be defined for two variables of arbitrary dimensions and arbitrary types. Next, we will briefly describe these four correlations.
Table 1.
Stopping criteria based on different correlation coefficients
| Criterion | Continuous Xj | Categorical Xj |
|---|---|---|
| Pearson/polyserial | Pearson ρ | polyserial ρ |
| Spearman | Spearman ρ | Spearman ρ |
| Kendall | Kendall τ | Kendall τ |
| distance | distance r | distance r |
We denote one of the covariates as Xj, for j = 1,2,…,J, where J is the total number of covariates. If both T and Xj are normally distributed, the Pearson correlation coefficient will be zero given that T and Xj are independent. When Xj is categorical, the Pearson correlation coefficient could be biased (Olsson, 1979). Instead, we should use the polyserial correlation coefficient (Olsson, Drasgow, and Dorans, 1982), which essentially assumes that the categorical variable is obtained by classifying an underlying continuous variable . The unknown parameters of can be estimated by maximum likelihood. Then, the polyserial correlation is calculated as the Pearson correlation between T and . Spearman and Kendall correlation coefficients are rank-based correlations, that can be applied to both continuous and categorical variables. If T and Xj are independent, we would expect the Spearman and Kendall correlation coefficients to be close to zero. A more flexible measurement of correlation/independence is distance correlation. The distance correlation takes values between zero and one and it equals zero if and only if T and Xj are independent, regardless of the type of Xj.
In the binary treatment case, to check whether the propensity scores adequately balance the covariates, we calculate the standardized difference in the weighted mean between the treatment group and the control group. If balance is achieved, the difference should be small. In the continuous case, we propose a general algorithm that uses bootstrapping to calculate the weighted correlation coefficient. The procedure requires the following steps for each value of M (number of trees).
- Calculate r̂(Ti, Xi) using boosting with M trees. Then, calculate
Sample n observations from the original dataset with replacement. Each data point is sampled with the inverse probability weight obtained from the first step. Calculate the corresponding coefficient between T and Xj on the weighted sample and denote it as dji;
Repeat Step (2) k times and get dj1, dj2,…,djk. Calculate the average correlation coefficient between T and Xj, denoted as d̄j;
- Perform a Fisher transformation on d̄j, i.e.,
(10) Average the absolute value of zj over all the covariates and get the average absolute correlation coefficient (AACC).
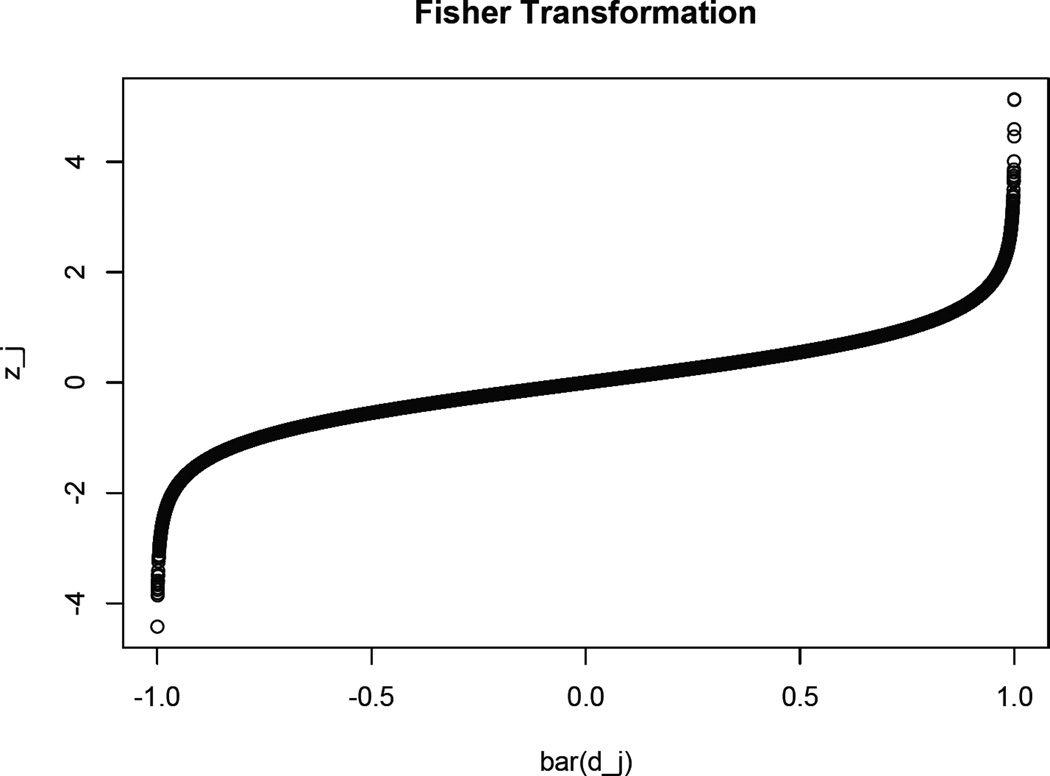
For each value of M = 1,2,…,20000, calculate AACC and find the optimal number of trees that lead to the smallest AACC value. The R code for calculating AACC is displayed in Appendix A. An alternative suggestion is to replace Step 5 by calculating the maximum value of the absolute correlation coefficient (MACC) over all the covariates and find the optimal number of trees that lead to the smallest MACC value. After the value of M is determined, the generalized propensity score is estimated by (9) and (7). The Fisher transformation in Step 4 is mainly for the determination of the cut-off value for AACC (MACC). We know that, in the binary treatment case, a well-accepted cut-off value for ASAM is 0.2 (McCaffrey, Ridgeway, and Morral, 2004). In the continuous treatment case, we set the cut-off value for AACC (MACC) to be 0.1. That is, when AACC < 0.1 (MACC < 0.1), we claim that the confounding effect between the treatment and the outcome is small. Appendix B shows a heuristic proof for the claimed cut-off value. Figure 1 is an illustration of the Fisher transformation. As can be seen, when |d̄j| is small, the transformation is almost the identity; when |d̄j| is large, |zj| increases faster than |d̄j|. This is another advantage of using Fisher transformation: when we try to minimize AACC, the larger absolute values of correlation coefficients will get more penalty compared to the original scale.
Figure 1.
An illustration of Fisher Transformation
4 Simulation
4.1 Simulation Setup
In this section, we conduct simulation studies to compare the performance of the proposed methods to the existing methods. The generation of observed (Y, T, X) are as follows. First, the vector of baseline covariates (potential confounders), denoted as X = (X1, X2,…,X10), are generated from the following distributions: X1, X2 ~ N(0,1), X3 ~ Bernoulli(0.5), X4 ~ Bernoulli(0.3), X5,…,X7 ~ N(0,1) and X8,…,X10 ~ Bernoulli(0.5). Among the ten covariates, X1 − X4 are real confounders related to both the treatment and the outcome.
The continuous treatment is generated from N(m(X),1), where the mean function is defined for different scenarios. In Scenario (A), m(X) is a linear combination of the real confounders. In Scenario (B), we consider a nonparametric model that is similar to a tree structure with main effects and one quadratic term while in Scenario (C), we add two interaction terms. The true forms of m(X) in different scenarios are displayed as follows:
Scenario (A): m(X) = 6 + 0.3X1 + 0.65X2 − 0.35X3 − 0.4X4;
Scenario (B): m(X) = 6 + 0.3I{X1 > 0.5} + 0.65I{X2 < 0} − 0.35I{X3 = 1} − 0.4I{X4 = 0} + 0.65I{X1 > 0}I{X1 > 1};
Scenario (C): m(X) = 6 + 0.3I{X1 > 0.5} + 0.65I{X2 < 0} − 0.35I{X3 = 1} − 0.4I{X4 = 0} + 0.65I{X1 > 0}I{X1 > 1} + 0.3I{X1 > 0}I{X4 = 1} − 0.65I{X2 > 0.3}I{X3 = 0}.
The potential outcome function for a subject with covariates X is generated from
where ε ~ N(0,1). Based on the data generation process, the true dose-response function is
| (11) |
4.2 Results
To compare different methods, we set the value of the parameter of interest to be 0.4, which is the coefficient of T in the dose-response function (11). We apply IPW to estimate the coefficient and employ four different methods to estimate m(X) in the generalized propensity scores: (1) linear approximation (Equation 6) using all the covariates; (2) linear approximation with variable selection; (3) L2 boosting by minimizing the empirical quadratic loss, called mboost by Bühlmann and Yu (2003); (4) boosting with the proposed stopping criteria: Pearson/polyserial, Spearman, Kendall and distance. In Method (2), we employ a variable selection technique that is similar to the idea suggested by Hirano and Imbens (2004) to select covariates in the generalized propensity score model. First, we divide the treatment variable into three groups with equal sizes. Then, we test if each covariate is distributed the same in different treatment “groups” using ANOVA at the significance level of 0.05. We only include those covariates that are significantly different among treatment “groups”. In Table 2, we denote Method (1) as Linear1 and Method (2) as Linear2. We generate 1000 data sets with a sample size of 500. The simulation results are shown in Table 2.
Table 2.
Simulation results for Scenarios (A), (B) and (C)
| True Causal Effect: 0.4 | |||||
|---|---|---|---|---|---|
| Method | Mean | Bias | SD | MSE | CI Cov (%) |
| Scenario (A) | |||||
| Linear1 | 0.417 | 0.017 | 0.097 | 0.0097 | 87.7 |
| Linear2 | 0.409 | 0.009 | 0.096 | 0.0093 | 87.9 |
| Mboost | 0.431 | 0.031 | 0.079 | 0.0072 | 87.3 |
| Proposed (Peason/polyserial) | 0.423 | 0.023 | 0.076 | 0.0064 | 91.9 |
| Proposed (Spearman) | 0.421 | 0.021 | 0.077 | 0.0064 | 92.2 |
| Proposed (Kendall) | 0.422 | 0.022 | 0.077 | 0.0064 | 91.6 |
| Proposed (distance) | 0.427 | 0.027 | 0.075 | 0.0064 | 89.8 |
| Scenario (B) | |||||
| Linear1 | 0.445 | 0.045 | 0.071 | 0.0070 | 90.6 |
| Linear2 | 0.434 | 0.034 | 0.069 | 0.0060 | 91.9 |
| Mboost | 0.376 | −0.024 | 0.065 | 0.0048 | 93.7 |
| Proposed (Peason/polyserial) | 0.383 | −0.017 | 0.066 | 0.0046 | 94.5 |
| Proposed (Spearman) | 0.383 | −0.017 | 0.067 | 0.0048 | 94.2 |
| Proposed (Kendall) | 0.384 | −0.016 | 0.067 | 0.0048 | 94.3 |
| Proposed (distance) | 0.380 | −0.020 | 0.068 | 0.0051 | 93.5 |
| Scenario (C) | |||||
| Linear1 | 0.437 | 0.037 | 0.078 | 0.0075 | 92.8 |
| Linear2 | 0.426 | 0.026 | 0.076 | 0.0064 | 93.3 |
| Mboost | 0.385 | −0.015 | 0.067 | 0.0047 | 94.5 |
| Proposed (Peason/polyserial) | 0.390 | −0.010 | 0.068 | 0.0047 | 95.7 |
| Proposed (Spearman) | 0.390 | −0.010 | 0.069 | 0.0048 | 95.6 |
| Proposed (Kendall) | 0.390 | −0.010 | 0.068 | 0.0047 | 95.2 |
| Proposed (distance) | 0.389 | −0.011 | 0.068 | 0.0047 | 95.7 |
In Scenario (A), the true mean function m(X) is linear in the covariates, and hence the linear approximation proposed by Robins et al. (2000) leads to the smallest bias. In addition, Linear2 has smaller bias and MSE than Linear1, which indicates that variable selection in the propensity score model does improve the performance. Compared to Linear1 and Linear2, the proposed methods yield much smaller variances and MSEs, as well as better confidence interval coverages. Compared to the proposed methods, mboost based on L2 boosting yields larger bias and MSE. In Scenarios (B) and (C) where m(X) follows a tree structure, Linear2 performs better than Linear1. In addition, the proposed methods are superior in terms of the bias, MSE and 95% confidence interval coverage. Our simulation results are not very sensitive to the choice of the correlation matrices among Pearson/polyserial, Spearman and Kendall correlations. Distance leads to slightly more biased estimates compared to the other proposed criteria.
To further explore the proposed algorithm, we randomly select 100 datasets from the simulated datasets in each scenario. We then compare the number of trees selected by each criterion with the optimal number of trees that leads to the smallest absolute bias with respect to the true causal effect (0.4). Table 3 shows the average number of trees based on the 100 datasets. Compared to the “best” model with the optimal number of trees, “distance” tend to select smaller models, which explains that “distance” yields relatively larger bias than the other three criteria in the simulation. The differences in the number of trees between the “best” model and the models selected by “Pearson/polyserial”, “Spearman” and “Kendall” are relatively small. In Scenario (A) and (C), they yield slightly more complex models than the “best” models and in Scenario (B), they yield slightly smaller models.
Table 3.
Average number of trees in boosting selected by each criterion
| Pearson/polyserial | Spearman | Kendall | distance | best | |
|---|---|---|---|---|---|
| Scenario (A) | 11782.05 | 12053.14 | 11189.09 | 9953.57 | 11011.24 |
| Scenario (B) | 11695.03 | 12160.33 | 11502.69 | 10437.27 | 12282.18 |
| Scenario (C) | 10155.62 | 10232.12 | 10962.81 | 7981.67 | 9703.48 |
5 Data Analysis Example
5.1 Early Dieting in Girls Study
It is reported that dieting increases the likelihood of overeating, weight gain, and chronic health problems (Neumark-Sztainer, Wall, Story, and Standish, 2012). We analyze the Early Dieting in Girls study, which is a longitudinal study that aims to examine parental influences on daughters’ growth and development from ages 5 to 15 (Fisher and Birch, 2002). The study involves 197 daughters and their mothers, who are from non-Hispanic, White families living in central Pennsylvania. The participants were assessed at five different waves. At each wave, daughters and their mothers were interviewed during a scheduled visit to the laboratory.
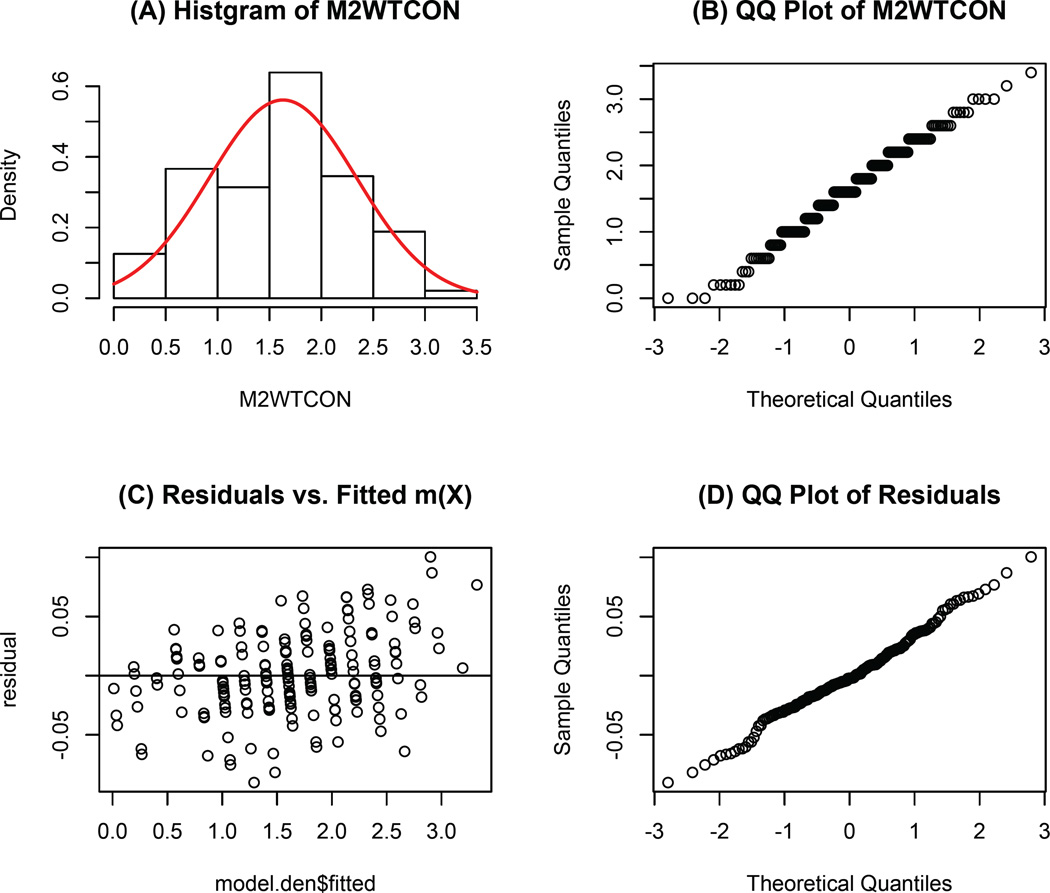
In this analysis, we study the influence of mothers’ weight concern on girls’ early dieting behaviour. The treatment variable is mother’s overall weight concern (M2WTCON), which is measured at daughter’s age 7. It is the average score of five questions in the questionnaire. A higher value implies the mother is more concerned about gaining weight. In the dataset, its values range from 0 to 3.4. The histogram in Figure 2 (A) and the QQ plot in Figure 2 (B) show that the treatment is approximately normally distributed. The outcome is whether the daughter diets between ages 7 and 11. We exclude those daughters who reported dieting before age 7, which results in 158 subjects, of which 45 daughters reported dieting. There are 50 potential baseline confounders in this study regarding participants’ characteristics, such as: family history of diabetes and obesity, family income, daughter’s disinhibition, daughter’s body esteem, mother’s perception of mother’s current size and mother’s satisfaction with daughter’s current body (Birch and Fisher, 2000).
Figure 2.
Model diagnostics for the fitted boosting model.
5.2 Estimation of the Generalized Propensity Scores
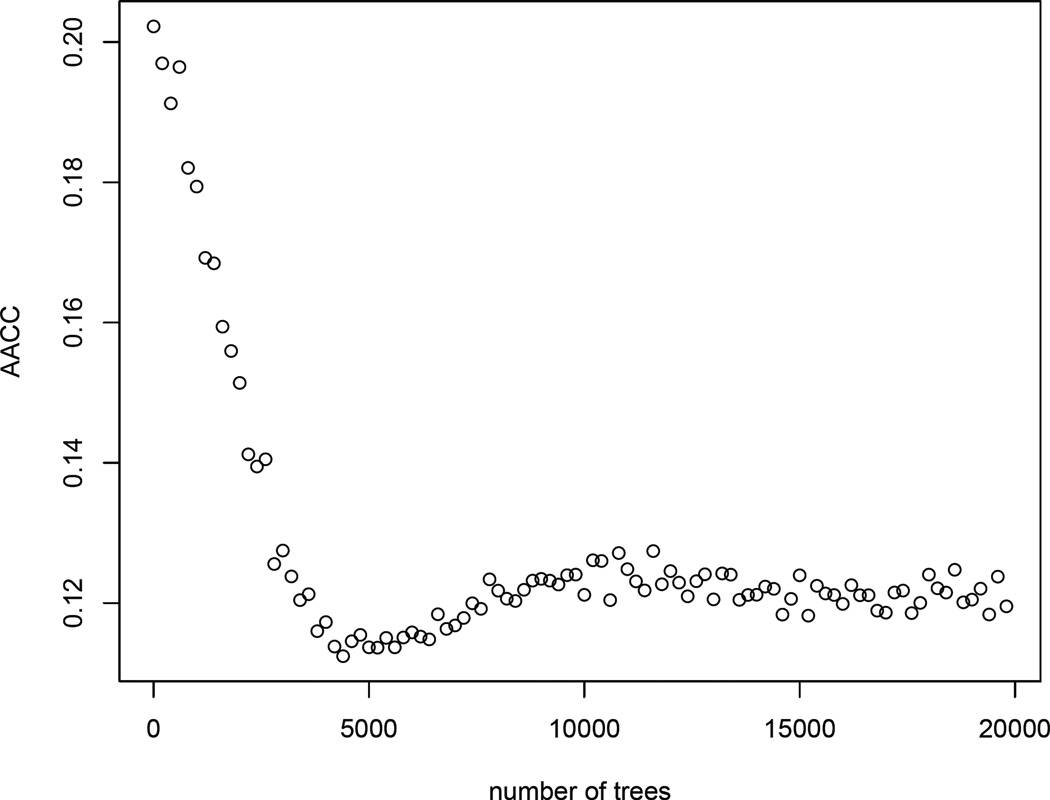
Since the treatment is self-selected, to draw causal inferences, we need to adjust for the confounders in the study. Given a large number of potential confounders, we employ a boosting algorithm to estimate the generalized propensity scores. The simulation studies in Section 4 shows that the estimation results are not sensitive to the choice of the correlation matrices among Pearson/polyserial, Kendall and Spearman. Therefore, we use “Pearson/polyserial” as the main criterion to select the optimal number of trees in boosting. Figure 3 displays the AACC value versus the number of trees from 1 to 20000. Based on the data, the optimal number of trees M = 4846 and AACC = 0.11. Figure 2 (C) & (D) show that the residuals from the boosting model (Ti − m̂ (Xi)) have approximately constant variance and they are normally distributed. Based on the residual plots, we conclude that the boosting model sufficiently estimates the treatment level given covariates.
Figure 3.
AACC value for M = 1,…, 20000.
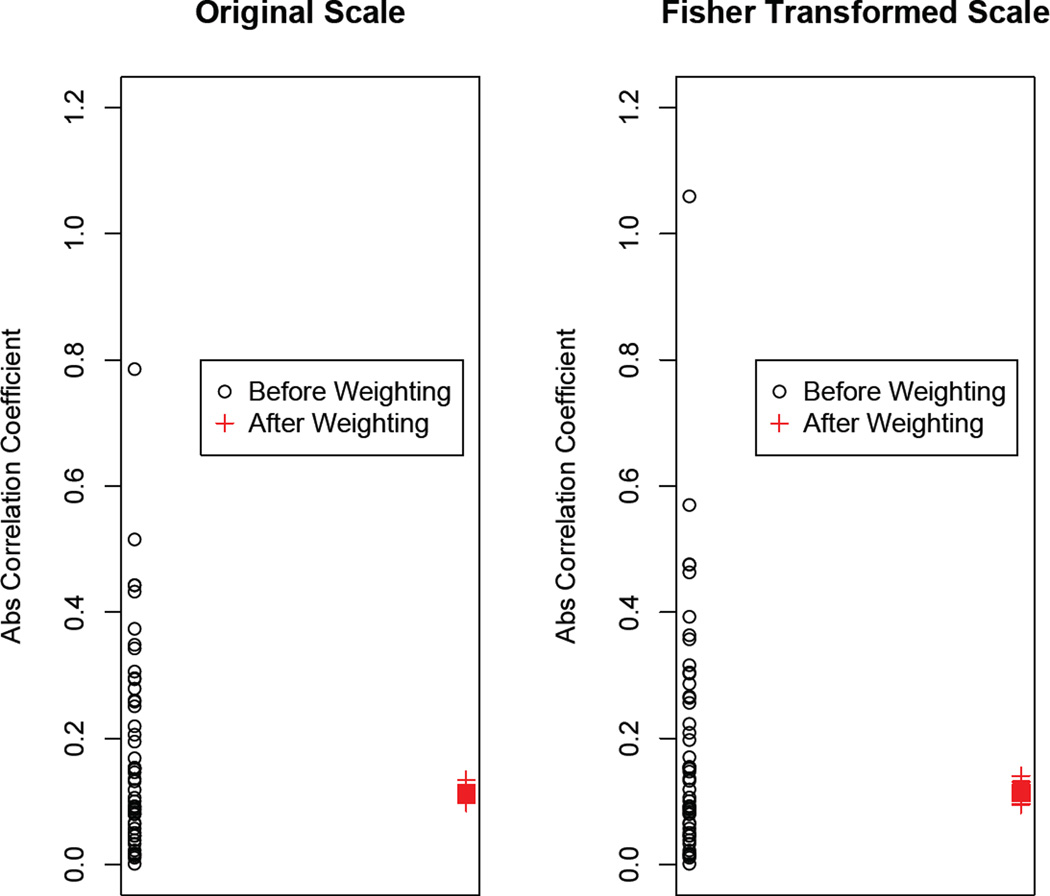
Now we will focus on assessing the balance in the covariates. Notice that in the original data, AACC = 0.177, which is much larger than 0.1, the cut-off value. In addition, if we look at each covariate separately, there are many covariates whose absolute correlation coefficients with T are larger than 0.2. As shown in Figure 4, after applying the weights, most of the absolute correlations among the treatment and each covariate in the weighted sample are below 0.1 on both the original scale and the Fisher transformed scale. This indicates that the confounding effect of the covariates between the treatment and the outcome is greatly reduced after weighting.
Figure 4.
Absolute value of correlation coefficients between T and each covariate before weighting (black dots) and after weighting (red plus signs).The left panel shows the original scale and the right panel shows the Fisher transformed scale.
5.3 Modeling the Mean Outcome
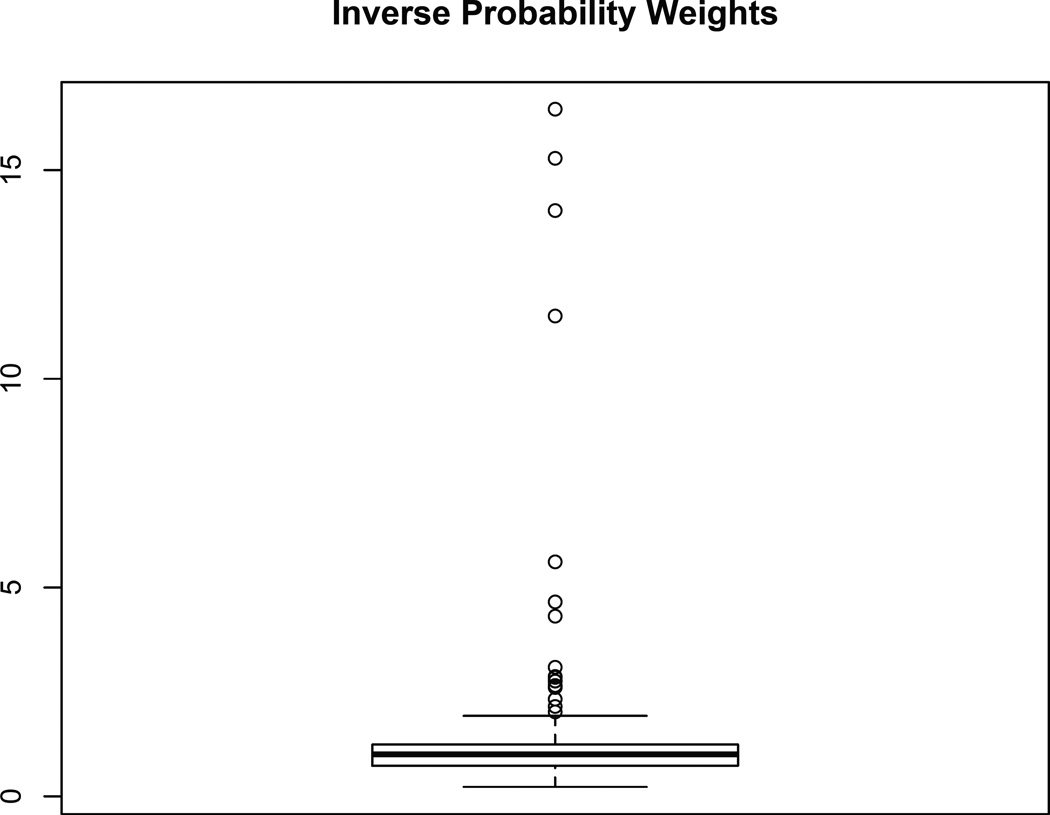
To draw causal inferences, the next step is to determine the functional form of the outcome model. The boxplot of the estimated inverse weights from our proposed method is displayed in Figure 5. As shown, most weights are distributed around the value of 1. However, there are some extreme weights with values larger than 10. Extreme weights are harmful to the analysis because they increase the variance of the causal estimates (Kang and Schafer, 2007). When we estimate the dose-response function, we shrink the top five percent of the weights to the 95th quantile.
Figure 5.
Boxplot of the inverse probability weights using Pearson/polyserial correlation.
Since all the potential confounders are well-adjusted by the propensity model, we can model the outcome as a function of the treatment. Otherwise, we may also include covariates that are related to the treatment in the outcome model. We then assume a regression spline function as in (3). For binary outcomes, the regression spline function is
The weighted AIC and BIC displayed in (4) and (5) are employed to determine the optimal p and K. For a binary outcome, the first part of (4) and (5) should be replaced by
where p̂i is the estimated probability of early dieting for subject i. We consider three different values for p: p = 1,2, 3, which corresponds to piecewise linear, quadratic and cubic models. We consider K = 0,1,2,…, 9. We select the optimal number of p and K based on the values of BICw, which are displayed in Table 4. As shown, the best model is when p = 1 and K = 0. Therefore, the model we fit is
The causal log odds ratio (β) is estimated as 0.1782 with a standard error 0.2865 (p-value=0.5349). The standard error is obtained using sandwich formula by the survey package in R. We may also employ bootstrapping to estimate the standard error by repeatedly taking a bootstrap sample with replacement from the original dataset and applying the same estimating procedure. Based on 1000 replications, the bootstrapping estimate of the standard error is 0.2374, which is slightly smaller than the sandwich formula. It indicates that the probability of daughter’s early dieting increases when the mother’s weight concern increases. However, the causal effect is not significant at the significance level of 0.1
Table 4.
BICw for different K and p
| K | p = 1 | p = 2 | p = 3 |
|---|---|---|---|
| 0 | 215.20 | 219.43 | 221.42 |
| 1 | 220.20 | 220.33 | 225.84 |
| 2 | 219.88 | 227.68 | 228.32 |
| 3 | 225.69 | 226.35 | 234.54 |
| 4 | 225.24 | 230.33 | 235.60 |
| 5 | 231.95 | 233.94 | 239.36 |
| 6 | 231.51 | 235.47 | 242.31 |
| 7 | 236.21 | 238.53 | 248.72 |
| 8 | 240.47 | 243.61 | 249.15 |
| 9 | 242.38 | 249.14 | 251.93 |
6 Discussion
In this article, we focused on the causal inference problem with a continuous treatment variable. We are mainly interested in estimating the dose-response function. IPW based on marginal structural models is a useful tool to estimate the causal effect. When the treatment variable is continuous, the generalized propensity score is defined as the conditional density of the treatment given covariates. Because the dimension of covariates is usually large, it is suggested that the conditional density can be estimated in two steps. First, a mean function of the treatment given covariates is estimated; secondly, the conditional density is normally approximated using residuals from the first step or nonparametrically estimated by a kernel method. We suggest using a boosting algorithm to estimate the mean function and propose an innovative stopping criterion based on the correlation metrics. The proposed stopping criterion is similar to generalized boosted model proposed by McCaffrey et al. (2004) for the binary treatment. Simulation results show that the proposed method performs better than the existing methods, especially when the function of the treatment given covariates is not linear.
It is known that, in causal inference problems, propensity scores are nuisance parameters and the parameter of interest is the causal treatment effect. It has been shown that a propensity score model with a better predictive performance may not lead to better causal treatment effect estimates (Drake, 1993, Lunceford and Davidian, 2004, Schafer and Kang, 2008). Therefore, while modeling propensity scores, we should really focus on the property of the causal estimates (Brookhart and van der Laan, 2006, Galdo, Smith, and Black, 2008). However, the true causal treatment effect is unknown in practice. For example, in Brookhart and van der Laan (2006), an over-fitted parametric propensity score model is used as the reference model; Galdo et al. (2008) proposed a weighted cross-validation technique to approximate mean integrated squared error of the counterfactual mean function. From another perspective, some recent literature has focused on the estimation of propensity scores by achieving balance in the covariates (e.g., McCaffrey et al. (2004), Hainmueller (2012), Imai and Ratkovic (2014)). The underlying idea is that by achieving balance, the bias in the treatment effect estimate due to measured covariates can be reduced (Harder, Stuart, and Anthony, 2010). The stopping rules proposed for boosting in this study also falls into this realm: we select the optimal number of trees in boosting by achieving balance in the covariates. The balance is measured through correlation between the treatment variable and the covariates in the weighted pseudo sample.
There are several potential areas for future research. For example, in the proposed algorithm described in Section 3.2, the correlations in the weighted pseudo sample are estimated using bootstrapping with unequal probabilities. However, bootstrapping in this case is computationally intensive, especially when the stopping criterion is based on distance correlation. A more straightforward approach is to develop the weighted Pearson or distance correlation for nonrandom samples using estimating equations. It may greatly improve the computation time.
In the data application, we use a cut-off value of 0.1 for AACC. In other words, if AACC < 0.1, we claim that the confounding effect of the covariates (potential confounders) is small; Based on Cohen’s effect sizes for the Pearson correlation coefficient (Cohen, 1988), we may also claim that, when 0.1 < AACC < 0.3, the confounding effect is medium; and when AACC > 0.55, the confounding effect is large. However, more theoretical and empirical justification is needed for the choice of the cut-off value, which can be explored in future work.
Finally, we should point out that the estimation of the generalized propensity scores is a much more challenging task than the case of a binary treatment. The reason is that we are concerned about all moments of the conditional distribution of the treatment given covariates, while in the binary case, we are only interested in the conditional mean. In the proposed method, we follow a two-step procedure by first modeling the mean function of the treatment given covariates. As shown in (8), we assume that the random errors are normally distributed and have constant variance. If the model diagnostics (e.g., Figure 2) show that either of the two assumptions is invalid, we may transform the treatment variable (see the lottery example in Hirano and Imbens (2004)) or use nonparametric methods to estimate the density. For example, replace (7) by a kernel density estimator. Future research may explore the application of other machine learning algorithms or a mixture of normal distributions to estimate the conditional density.
Acknowledgments
The project described was supported by Award Number P50DA010075 from the National Institute on Drug Abuse and 5R21 DK082858-02 from the National Institute on Diabetes and Digestive and Kidney Disorders. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute on Drug Abuse or the National Institutes of Health. We would also like to thank Jennifer Savage Williams and Leann Birch for permission to use the Early Dieting in Girls Study data, which was funded by National Institute of Child Health and Human Development R01 HD32973. The content is solely the responsibility of the authors and does not necessarily represent the official views of NICHD.
Appendix A: R codes
The following R function calculates the average absolute correlation coefficient (AACC) among a continuous treatment and covariates after applying the inverse probability weights. The subsequent R codes demonstrate how to estimate the dose-response function using a real dataset.
F.aac.iter=function(i,data,ps.model,ps.num,rep,criterion) {
# i: number of iterations (trees)
# data: dataset containing the treatment and the covariates
# ps.model: the boosting model to estimate p(T_i|X_i)
# ps.num: the estimated p(T_i)
# rep: number of replications in bootstrap
# criterion: the correlation metric used as the stopping criterion
GBM.fitted=predict(ps.model,newdata=data,n.trees=floor(i),
type="response")
ps.den=dnorm((data$T-GBM.fitted)/sd(data$T-GBM.fitted),0,1)
wt=ps.num/ps.den
aac_iter=rep(NA,rep)
for (i in 1:rep){
bo=sample(1:dim(data)[1],replace=TRUE,prob=wt)
newsample=data[bo,]
j.drop=match(c("T"),names(data))
j.drop=j.drop[!is.na(j.drop)]
x=newsample[,-j.drop]
if(criterion=="spearman"|criterion=="kendall"){
ac=apply(x, MARGIN=2, FUN=cor, y=newsample$T,
method=criterion)
} else if (criterion=="distance"){
ac=apply(x, MARGIN=2, FUN=dcor, y=newsample$T)
} else if (criterion=="pearson"){
ac=matrix(NA,dim(x)[2],1)
for (j in 1:dim(x)[2]){
ac[j]=ifelse (!is.factor(x[,j]), cor(newsample$T, x[,j],
method=criterion),polyserial(newsample$T, x[,j]))
}
} else print("The criterion is not correctly specified")
aac_iter[i]=mean(abs(1/2*log((1+ac)/(1-ac))),na.rm=TRUE)
}
aac=mean(aac_iter)
return(aac)
}
# Create the data frame for the covariates
x = data.frame(BMIZ, factor(DIABETZ1), G1BDESTM, G1WTCON,
factor(INCOME1), M1AGE1, M1BMI, factor(M1CURLS), factor(M1CURMT),
M1DEPRS, M1ESTEEM, M1GFATCN, M1GNOW, factor(M1GSATN),
M1MFATCN, M1MNOW, factor(M1MSAT), factor(M1NOEX), M1OGIBOD,
M1PCEAFF, M1PCEEFF, M1PCEEXT, M1PCEIMP, M1PCEPER, M1PDSTOT,
M1RLOAD, factor(M1SMOKE), M1WGTTES, M1WTCON, M1YRED, factor(OBESE1),
g1discal, g1obcdc, g1ovrcdc, g1pFM, g1wgttes, m1cfqcwt, m1cfqenc,
m1cfqmon, m1cfqpwt, m1cfqrsp, m1cfqrst, m1cfqwtc, m1dis, m1hung,
m1lim, m1picky, m1rest, m1zsav, m1zsweet)
# Find the optimal number of trees using Pearson/polyserial correlation
library(gbm)
library(polycor)
mydata=data.frame(T=M2WTCON,X=x)
model.num=lm(T˜1,data=mydata)
ps.num=dnorm((mydata$T-model.num$fitted)/(summary(model.num))$sigma,0,1)
model.den=gbm(T˜.,data=mydata, shrinkage = 0.0005,
interaction.depth=4, distribution="gaussian",n.trees=20000)
opt=optimize(F.aac.iter,interval=c(1,20000), data=mydata, ps.model=model.den,
ps.num=ps.num,rep=50,criterion="pearson")
best.aac.iter=opt$minimum
best.aac=opt$objective
# Calculate the inverse probability weights
model.den$fitted=predict(model.den,newdata=mydata,
n.trees=floor(best.aac.iter), type="response")
ps.den=dnorm((mydata$T-model.den$fitted)/sd(mydata$T-model.den$fitted),0,1)
weight.gbm=ps.num/ps.den
# Outcome analysis using survey package
library(survey)
dataset=data.frame(earlydiet,M2WTCON, weight.gbm)
design.b=svydesign(ids= ˜1, weights=˜weight.gbm, data=dataset)
fit=svyglm(earlydiet˜M2WTCON, family=quasibinomial(),design=design.b)
summary(fit)
Appendix B: Cut-off value for AACC
In this section, we provide a heuristic proof for the cut-off value for AACC. In the continuous treatment case, denote a covariate as Xj and the treatment variable as T. If (Xj, T) has a bivariate normal distribution and Xj, T are independent, the Fisher transformed Pearson’s correlation coefficient, zj, has the following asymptotic distribution:
On the other hand, if we dichotomize the continuous treatment to a binary treatment with a sample size of n1 for the treatment group and n0 for the control group (n1 + n0 = n), we know the Cohen’s effect size is defined as:
where s is the pooled standard deviation. If there is no difference between the two groups, asymptotically,
Therefore, if the cut-off value for the standardized mean difference is 0.2 in the binary treatment case, the cut-off value for AACC in the continuous treatment case should be , which has a maximum value of 0.1 when . In fact, this cut-off value is consistent with what has been claimed regarding the effect size of Pearson correlation coefficient r (Cohen, 1988). That is, when |r| < 0.1, the effect size is small; when 0.1 < |r| < 0.3, the effect size is medium; when |r| > 0.5, the effect size is large.
Contributor Information
Yeying Zhu, Email: yeying.zhu@uwaterloo.ca, Department of Statistics and Actuarial Science, University of Waterloo, 200 University Ave W, Waterloo, ON N2L 3G1, Canada.
Donna L. Coffman, Email: dcoffman@psu.edu, The Methodology Center, The Pennsylvania State University, University Park, PA, USA.
Debashis Ghosh, Email: ghoshd@psu.edu, Department of Statistics and Public Health Sciences, The Pennsylvania State University, University Park, PA, USA.
References
- Bia M, Mattei A. A stata package for the estimation of the dose-response function through adjustment for the generalized propensity score. The Stata Journal. 2008;8:354–373. [Google Scholar]
- Birch LL, Fisher JO. Mothers’ child-feeding practices influence daughters’ eating and weight. The American Journal of Clinical Nutrition. 2000;71:1054–1061. doi: 10.1093/ajcn/71.5.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L, Friedman J, Stone C, Olshen R. Classification and regression trees. Chapman & Hall/CRC; 1984. [Google Scholar]
- Brookhart MA, van der Laan MJ. A semiparametric model selection criterion with applications to the marginal structural model. Computational Statistics & Data Analysis. 2006;50:475–498. [Google Scholar]
- Bühlmann P, Yu B. Boosting with the l2 loss: regression and classification. Journal of the American Statistical Association. 2003;98:324–339. [Google Scholar]
- Cohen J. Statistical power analysis for the behavioral sciences. Psychology Press; 1988. [Google Scholar]
- Drake C. Effects of misspecification of the propensity score on estimators of treatment effect. Biometrics. 1993;49:1231–1236. [Google Scholar]
- Eubank RL. Spline smoothing and nonparametric regression. Marcel Dekker; 1988. [Google Scholar]
- Fan J, Yao Q, Tong H. Estimation of conditional densities and sensitivity measures in nonlinear dynamical systems. Biometrika. 1996;83:189–206. [Google Scholar]
- Fisher JO, Birch LL. Eating in the absence of hunger and overweight in girls from 5 to 7 y of age. The American journal of clinical nutrition. 2002;76:226–231. doi: 10.1093/ajcn/76.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdo JC, Smith J, Black D. Bandwidth selection and the estimation of treatment effects with unbalanced data. Annales d’Economie et de Statistique. 2008:189–216. [Google Scholar]
- Hainmueller J. Entropy balancing for causal effects: A multivariate reweighting method to produce balanced samples in observational studies. Political Analysis. 2012;20:25–46. [Google Scholar]
- Hall P, Wolff RC, Yao Q. Methods for estimating a conditional distribution function. Journal of the American Statistical Association. 1999;94:154–163. [Google Scholar]
- Harder VS, Stuart EA, Anthony JC. Propensity score techniques and the assessment of measured covariate balance to test causal associations in psychological research. Psychological methods. 2010;15:234. doi: 10.1037/a0019623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hens N, Aerts M, Molenberghs G. Model selection for incomplete and design-based samples. Statistics in Medicine. 2006;25:2502–2520. doi: 10.1002/sim.2559. [DOI] [PubMed] [Google Scholar]
- Hirano K, Imbens GW. The propensity score with continuous treatments. Applied Bayesian modeling and causal inference from incomplete-data perspectives. 2004:73–84. [Google Scholar]
- Imai K, Ratkovic M. Covariate balancing propensity score. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2014;76:243–263. [Google Scholar]
- Imai K, Van Dyk D. Causal inference with general treatment regimes. Journal of the American Statistical Association. 2004;99:854–866. [Google Scholar]
- Imbens GW. The role of the propensity score in estimating dose-response functions. Biometrika. 2000;87:706–710. [Google Scholar]
- Kang JDY, Schafer JL. Demystifying double robustness: a comparison of alternative strategies for estimating a population mean from incomplete data. Statistical Science. 2007;22:523–539. doi: 10.1214/07-STS227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karwa V, Slavković AB, Donnell ET. Causal inference in transportation safety studies: Comparison of potential outcomes and causal diagrams. The Annals of Applied Statistics. 2011;5:1428–1455. [Google Scholar]
- Kluve J, Schneider H, Uhlendorff A, Zhao Z. Evaluating continuous training programmes by using the generalized propensity score. Journal of the Royal Statistical Society: Series A (Statistics in Society) 2012;175:587–617. [Google Scholar]
- Lechner M. Program heterogeneity and propensity score matching: an application to the evaluation of active labor market policies. Review of Economics and Statistics. 2002;84:205–220. [Google Scholar]
- Lunceford JK, Davidian M. Stratification and weighting via the propensity score in estimation of causal treatment effects: a comparative study. Statistics in Medicine. 2004;23:2937–2960. doi: 10.1002/sim.1903. [DOI] [PubMed] [Google Scholar]
- McCaffrey DF, Griffin BA, Almirall D, Slaughter ME, Ramchand R, Burgette LF. A tutorial on propensity score estimation for multiple treatments using generalized boosted models. Statistics in medicine. 2013;32:3388–3414. doi: 10.1002/sim.5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey DF, Ridgeway G, Morral AR. Propensity score estimation with boosted regression for evaluating causal effects in observational studies. Psychological Methods. 2004;9:403–425. doi: 10.1037/1082-989X.9.4.403. [DOI] [PubMed] [Google Scholar]
- Neumark-Sztainer D, Wall M, Story M, Standish AR. Dieting and unhealthy weight control behaviors during adolescence: associations with 10-year changes in body mass index. Journal of Adolescent Health. 2012;50:80–86. doi: 10.1016/j.jadohealth.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson U. Maximum likelihood estimation of the polychoric correlation coefficient. Psychometrika. 1979;44:443–460. [Google Scholar]
- Olsson U, Drasgow F, Dorans NJ. The polyserial correlation coefficient. Psychometrika. 1982;47:337–347. [Google Scholar]
- Platt RW, Brookhart AM, Cole SR, Westreich D, Schisterman EF. An information criterion for marginal structural models. Statistics in Medicine. 2012;32:1383–1393. doi: 10.1002/sim.5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins J. Association, causation, and marginal structural models. Synthese. 1999;121:151–179. [Google Scholar]
- Robins J, Hernán M, Brumback B. Marginal structural models and causal inference in epidemiology. Epidemiology. 2000;11:550–560. doi: 10.1097/00001648-200009000-00011. [DOI] [PubMed] [Google Scholar]
- Schafer JL, Kang J. Average causal effects from nonrandomized studies: A practical guide and simulated example. Psychological Methods. 2008;13:279–313. doi: 10.1037/a0014268. [DOI] [PubMed] [Google Scholar]
- Székely GJ, Rizzo ML. Brownian distance covariance. The Annals of Applied Statistics. 2009;32:1236–1265. doi: 10.1214/09-AOAS312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Székely GJ, Rizzo ML, Bakirov NK. Measuring and testing dependence by correlation of distances. The Annals of Statistics. 2007;35:2769–2794. [Google Scholar]
- Tchernis R, Horvitz-Lennon M, Normand SLT. On the use of discrete choice models for causal inference. Statistics in Medicine. 2005;24:2197–2212. doi: 10.1002/sim.2095. [DOI] [PubMed] [Google Scholar]