Abstract
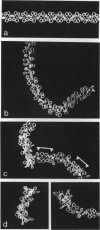
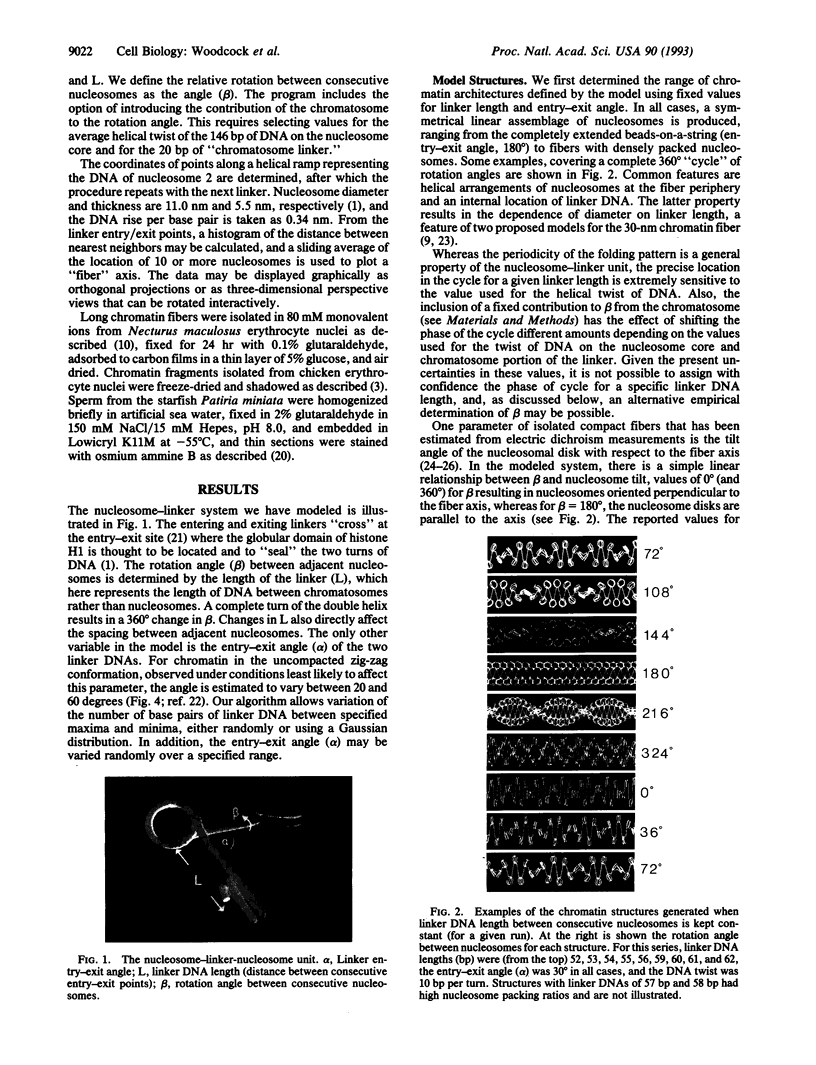
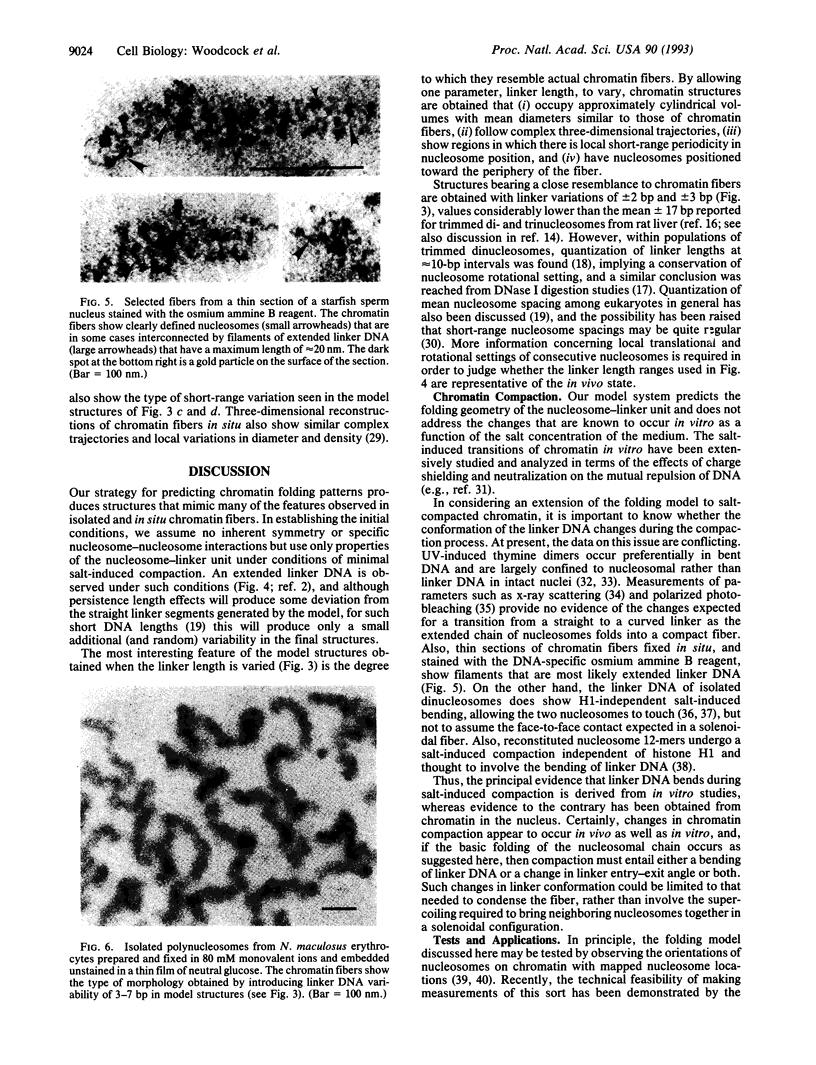
The "30-nm" chromatin fibers, as observed in eukaryotic nuclei, are considered a discrete level in a hierarchy of DNA folding. At present, there is considerable debate as to how the nucleosomes and linker DNA are organized within chromatin fibers, and a number of models have been proposed, many of which are based on helical symmetry and imply specific contacts between nucleosomes. However, when observed in nuclei or after isolation, chromatin fibers show considerable structural irregularity. In the present study, chromatin folding is considered solely in terms of the known properties of the nucleosome-linker unit, taking into account the relative rotation between consecutive nucleosomes that results from the helical twist of DNA. Model building based on this premise, and with a constant length of linker DNA between consecutive nucleosomes, results in a family of fiber- and ribbon-like structures. When the linker length between nucleosomes is allowed to vary, as occurs in nature, fibers showing the types of irregularity observed in nuclei and in isolated chromatin are created. The potential application of the model in determining the three-dimensional organization of chromatin in which nucleosome positions are known is discussed.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Athey B. D., Smith M. F., Rankert D. A., Williams S. P., Langmore J. P. The diameters of frozen-hydrated chromatin fibers increase with DNA linker length: evidence in support of variable diameter models for chromatin. J Cell Biol. 1990 Sep;111(3):795–806. doi: 10.1083/jcb.111.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordas J., Perez-Grau L., Koch M. H., Vega M. C., Nave C. The superstructure of chromatin and its condensation mechanism. II. Theoretical analysis of the X-ray scattering patterns and model calculations. Eur Biophys J. 1986;13(3):175–185. doi: 10.1007/BF00542561. [DOI] [PubMed] [Google Scholar]
- Clark D. J., Kimura T. Electrostatic mechanism of chromatin folding. J Mol Biol. 1990 Feb 20;211(4):883–896. doi: 10.1016/0022-2836(90)90081-V. [DOI] [PubMed] [Google Scholar]
- Davies H. G., Murray A. B., Walmsley M. E. Electron-microscope observations on the organization of the nucleus in chicken erythrocytes and a superunit thread hypothesis for chromosome structure. J Cell Sci. 1974 Nov;16(2):261–299. doi: 10.1242/jcs.16.2.261. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA bending and its relation to nucleosome positioning. J Mol Biol. 1985 Dec 20;186(4):773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- Dubochet J., Adrian M., Chang J. J., Homo J. C., Lepault J., McDowall A. W., Schultz P. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys. 1988 May;21(2):129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- Dustin I., Furrer P., Stasiak A., Dubochet J., Langowski J., Egelman E. Spatial visualization of DNA in solution. J Struct Biol. 1991 Aug;107(1):15–21. doi: 10.1016/1047-8477(91)90026-s. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G., McGhee J. D. Structure of the 30 nm chromatin fiber. Cell. 1986 Feb 14;44(3):375–377. doi: 10.1016/0092-8674(86)90456-3. [DOI] [PubMed] [Google Scholar]
- Garcia-Ramirez M., Dong F., Ausio J. Role of the histone "tails" in the folding of oligonucleosomes depleted of histone H1. J Biol Chem. 1992 Sep 25;267(27):19587–19595. [PubMed] [Google Scholar]
- Giannasca P. J., Horowitz R. A., Woodcock C. L. Transitions between in situ and isolated chromatin. J Cell Sci. 1993 Jun;105(Pt 2):551–561. doi: 10.1242/jcs.105.2.551. [DOI] [PubMed] [Google Scholar]
- Greulich K. O., Wachtel E., Ausio J., Seger D., Eisenberg H. Transition of chromatin from the "10 nm" lower order structure, to the "30 nm" higher order structure as followed by small angle X-ray scattering. J Mol Biol. 1987 Feb 20;193(4):709–721. doi: 10.1016/0022-2836(87)90353-6. [DOI] [PubMed] [Google Scholar]
- Horowitz R. A., Giannasca P. J., Woodcock C. L. Ultrastructural preservation of nuclei and chromatin: improvement with low-temperature methods. J Microsc. 1990 Feb;157(Pt 2):205–224. doi: 10.1111/j.1365-2818.1990.tb02959.x. [DOI] [PubMed] [Google Scholar]
- Horowitz R. A., Woodcock C. L. Alternative staining methods for Lowicryl sections. J Histochem Cytochem. 1992 Jan;40(1):123–133. doi: 10.1177/40.1.1370308. [DOI] [PubMed] [Google Scholar]
- Langmore J. P., Paulson J. R. Low angle x-ray diffraction studies of chromatin structure in vivo and in isolated nuclei and metaphase chromosomes. J Cell Biol. 1983 Apr;96(4):1120–1131. doi: 10.1083/jcb.96.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauderdale J. D., Stein A. Effects of plasmid length and positioned nucleosomes on chromatin assembly in vitro. Biochemistry. 1993 Jan 19;32(2):489–499. doi: 10.1021/bi00053a013. [DOI] [PubMed] [Google Scholar]
- Lohr D., Van Holde K. E. Organization of spacer DNA in chromatin. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6326–6330. doi: 10.1073/pnas.76.12.6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., Nickol J. M., Felsenfeld G., Rau D. C. Higher order structure of chromatin: orientation of nucleosomes within the 30 nm chromatin solenoid is independent of species and spacer length. Cell. 1983 Jul;33(3):831–841. doi: 10.1016/0092-8674(83)90025-9. [DOI] [PubMed] [Google Scholar]
- Mitra S., Sen D., Crothers D. M. Orientation of nucleosomes and linker DNA in calf thymus chromatin determined by photochemical dichroism. Nature. 1984 Mar 15;308(5956):247–250. doi: 10.1038/308247a0. [DOI] [PubMed] [Google Scholar]
- Pehrson J. R., Cohen L. H. Effects of DNA looping on pyrimidine dimer formation. Nucleic Acids Res. 1992 Mar 25;20(6):1321–1324. doi: 10.1093/nar/20.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehrson J. R. Thymine dimer formation as a probe of the path of DNA in and between nucleosomes in intact chromatin. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9149–9153. doi: 10.1073/pnas.86.23.9149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner J. B., Hamkalo B. A. Higher order structure in metaphase chromosomes. II. The relationship between the 250 A fiber, superbeads and beads-on-a-string. Chromosoma. 1978 Dec 6;69(3):373–379. doi: 10.1007/BF00332140. [DOI] [PubMed] [Google Scholar]
- Ris H., Kubai D. F. Chromosome structure. Annu Rev Genet. 1970;4:263–294. doi: 10.1146/annurev.ge.04.120170.001403. [DOI] [PubMed] [Google Scholar]
- Sen D., Mitra S., Crothers D. M. Higher order structure of chromatin: evidence from photochemically detected linear dichroism. Biochemistry. 1986 Jun 3;25(11):3441–3447. doi: 10.1021/bi00359a052. [DOI] [PubMed] [Google Scholar]
- Shimizu M., Roth S. Y., Szent-Gyorgyi C., Simpson R. T. Nucleosomes are positioned with base pair precision adjacent to the alpha 2 operator in Saccharomyces cerevisiae. EMBO J. 1991 Oct;10(10):3033–3041. doi: 10.1002/j.1460-2075.1991.tb07854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T. Nucleosome positioning: occurrence, mechanisms, and functional consequences. Prog Nucleic Acid Res Mol Biol. 1991;40:143–184. doi: 10.1016/s0079-6603(08)60841-7. [DOI] [PubMed] [Google Scholar]
- Strauss F., Prunell A. Nucleosome spacing in rat liver chromatin. A study with exonuclease III. Nucleic Acids Res. 1982 Apr 10;10(7):2275–2293. doi: 10.1093/nar/10.7.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss F., Prunell A. Organization of internucleosomal DNA in rat liver chromatin. EMBO J. 1983;2(1):51–56. doi: 10.1002/j.1460-2075.1983.tb01379.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subirana J. A. Order and disorder in 30 nm chromatin fibers. FEBS Lett. 1992 May 11;302(2):105–107. doi: 10.1016/0014-5793(92)80416-e. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979 Nov;83(2 Pt 1):403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F. Nucleosome positioning. Biochim Biophys Acta. 1992 Feb 28;1130(1):1–19. doi: 10.1016/0167-4781(92)90455-9. [DOI] [PubMed] [Google Scholar]
- Widom J. A relationship between the helical twist of DNA and the ordered positioning of nucleosomes in all eukaryotic cells. Proc Natl Acad Sci U S A. 1992 Feb 1;89(3):1095–1099. doi: 10.1073/pnas.89.3.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S. P., Athey B. D., Muglia L. J., Schappe R. S., Gough A. H., Langmore J. P. Chromatin fibers are left-handed double helices with diameter and mass per unit length that depend on linker length. Biophys J. 1986 Jan;49(1):233–248. doi: 10.1016/S0006-3495(86)83637-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock C. L., Frado L. L., Rattner J. B. The higher-order structure of chromatin: evidence for a helical ribbon arrangement. J Cell Biol. 1984 Jul;99(1 Pt 1):42–52. doi: 10.1083/jcb.99.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock C. L., McEwen B. F., Frank J. Ultrastructure of chromatin. II. Three-dimensional reconstruction of isolated fibers. J Cell Sci. 1991 May;99(Pt 1):107–114. doi: 10.1242/jcs.99.1.107. [DOI] [PubMed] [Google Scholar]
- Woodcock C. L., Woodcock H., Horowitz R. A. Ultrastructure of chromatin. I. Negative staining of isolated fibers. J Cell Sci. 1991 May;99(Pt 1):99–106. doi: 10.1242/jcs.99.1.99. [DOI] [PubMed] [Google Scholar]
- Yao J., Lowary P. T., Widom J. Direct detection of linker DNA bending in defined-length oligomers of chromatin. Proc Natl Acad Sci U S A. 1990 Oct;87(19):7603–7607. doi: 10.1073/pnas.87.19.7603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J., Lowary P. T., Widom J. Linker DNA bending induced by the core histones of chromatin. Biochemistry. 1991 Aug 27;30(34):8408–8414. doi: 10.1021/bi00098a019. [DOI] [PubMed] [Google Scholar]
- Zivanovic Y., Duband-Goulet I., Schultz P., Stofer E., Oudet P., Prunell A. Chromatin reconstitution on small DNA rings. III. Histone H5 dependence of DNA supercoiling in the nucleosome. J Mol Biol. 1990 Jul 20;214(2):479–495. doi: 10.1016/0022-2836(90)90195-R. [DOI] [PubMed] [Google Scholar]