Abstract
Rust fungal pathogens of wheat (Triticum spp.) affect crop yields worldwide. The molecular mechanisms underlying the virulence of these pathogens remain elusive, due to the limited availability of suitable molecular genetic research tools. Notably, the inability to perform high-throughput analyses of candidate virulence proteins (also known as effectors) impairs progress. We previously established a pipeline for the fast-forward screens of rust fungal candidate effectors in the model plant Nicotiana benthamiana. This pipeline involves selecting candidate effectors in silico and performing cell biology and protein-protein interaction assays in planta to gain insight into the putative functions of candidate effectors. In this study, we used this pipeline to identify and characterize sixteen candidate effectors from the wheat yellow rust fungal pathogen Puccinia striiformis f sp tritici. Nine candidate effectors targeted a specific plant subcellular compartment or protein complex, providing valuable information on their putative functions in plant cells. One candidate effector, PST02549, accumulated in processing bodies (P-bodies), protein complexes involved in mRNA decapping, degradation, and storage. PST02549 also associates with the P-body-resident ENHANCER OF mRNA DECAPPING PROTEIN 4 (EDC4) from N. benthamiana and wheat. We propose that P-bodies are a novel plant cell compartment targeted by pathogen effectors.
Introduction
Plant pathogens colonize hosts by deploying virulence proteins known as effectors that manipulate plant cell structures and functions [1–2]. Once delivered into host tissues, effectors reside in the extracellular space (apoplastic effectors) or translocate into the plant cells (cytoplasmic effectors). Unravelling how effectors function in the host is key to understanding parasitism and to developing resistant plants [3]. Pathogen effectors are operationally plant proteins; they function in plant tissues, they associate with plant molecules, and their phenotypic expression in plants drives their evolution [4]. The field of effector biology has rapidly advanced in recent years due in large part to the availability of host plants that are amenable to molecular genetics and in which effectors can be heterologously expressed and studied [5]. However, due to the limited availability of functional genetic resources for crop species, characterising the effectors of crop pathogens remains challenging [6–7]. To study crop pathogen effectors, an alternative approach is the use a surrogate experimental plant system, such as Nicotiana benthamiana [8].
Nicotiana benthamiana (Solanaceae) is a well-established experimental system to study proteins in planta [9–10]. The agroinfiltration method allows transient expression of proteins in leaf cells, and a wide range of assays is available for functional investigations. Thus, N. benthamiana is extensively used in effector biology [11]. We recently used this plant to set up an effectoromics pipeline aimed at determining the plant cell compartments and protein complexes targeted by candidate effectors of rust fungi [8,12]. Such pipeline is a valuable tool for the rapid screening of candidate effectors.
Rust fungi (Pucciniales) are notorious for being destructive crop pathogens [13]. The species that infect wheat pose a constant threat to global food security [14]. These fungal pathogens include the yellow rust fungus Puccinia striiformis f sp tritici [15–16]. To date, effectors have not been functionally characterized for this species. However, genome and transcriptome analyses have predicted hundreds of candidate effectors, most of which are secreted proteins of unknown function [17–19]. Cantu and colleagues recently combined genome and in silico analyses to prioritize candidate effectors for further functional analyses [20].
Processing bodies (P-bodies) are protein/RNA complexes that reside in the cytosol of eukaryotic cells. They control the decapping, degradation, and storage of mRNA molecules [21]. In plants, P-bodies and P-body-resident proteins have important roles in post-embryonic development [22–23], salt stress tolerance [24], and immune responses [25]. In animals and yeast, limited evidence suggests that pathogenic bacteria and viruses target P-bodies [26–27]. To date, no connection has been made between P-bodies and filamentous pathogens.
In this study, we investigated sixteen candidate effectors of the wheat yellow rust fungus P. striiformis f sp tritici using the N. benthamiana effectoromics pipeline we previously developed [8]. We discovered that nine candidate effectors accumulate in distinct plant cell compartments and associate with specific protein complexes. Notably, the candidate effector PST02549 accumulates in P-bodies and associates with the wheat enhancer of mRNA decapping protein 4. Our findings point to P-bodies as a novel plant compartment targeted by pathogen effectors. We also propose that N. benthamiana can be used as an experimental system to screen candidate effectors of pathogens of monocot plants.
Results
Selection of 16 candidate effectors from Puccinia striiformis f sp tritici
The predicted effector complement of P. striiformis f sp tritici consists of hundreds of secreted proteins [20]. To select candidate effectors for functional investigations, we leveraged our recently developed pipeline [8] to select eleven proteins, using transcript enrichment in purified haustoria as the principal criterion for selection. We also included five proteins previously flagged as promising candidates [20] to obtain a final list of sixteen candidate effectors (Table 1, Sheet A in S1 Table). These sixteen candidates are Pucciniales-specific, and only seven show some sequence similarity to proteins of the wheat stem rust fungus Puccinia graminis f sp tritici (Table 1). This finding suggests that most of these candidate effectors recently emerged in the Pucciniaceae family.
Table 1. Puccinia striiformis f. sp. tritici candidate effectors analysed in this study.
| protein ID a | tribe b | protein length c | SP length c | cys d | gene expression be | similar proteins f |
|---|---|---|---|---|---|---|
| PST05258 | 54 | 256 | 21 | 0 | H | Pgt (3) |
| PST15391 | 54 | 256 | 19 | 0 | H5, I | Pgt (3) |
| PST11721 | 219 | 250 | 24 | 1 | H5, I | Pgt (1) |
| PST18220 | 238 | 110 | 19 | 0 | H5, I, TC | - |
| PST18221 | 238 | 112 | 20 | 0 | H1, I, TC | - |
| PST15642 | 308 | 102 | 21 | 3 | H5, I, TC | - |
| PST18447 | 318 | 146 | 24 | 4 | H5, I | Pgt (1), Mlp (1), Hv (1) |
| PST02549 | 320 | 297 | 21 | 1 | H5, I | - |
| PST05023 | 351 | 281 | 21 | 1 | H1, I10, TC | Pgt (1) |
| PST12160 | 354 | 168 | 19 | 1 | H5, I10 | - |
| PST10977 | 403 | 171 | 26 | 13 | H5, I | Pgt (1) |
| PST03196 | 478 | 206 | 25 | 2 | H5, I | Pgt (1) |
| PST05302 | 593 | 160 | 20 | 0 | H1, I, TC | - |
| PST15964 | 678 | 128 | 18 | 0 | H1, I | - |
| PST08468 | 1176 | 206 | 20 | 0 | H5, I | - |
| PST05006 | 1217 | 201 | 22 | 3 | H1, I | - |
a Protein IDs were adapted from [20] by removing the isolate ID for simplicity. Full-length protein IDs are indicated in Sheet A in S1 Table.
b Data mined from [20].
c Number of amino acids are indicated; SP: predicted signal peptide
d Number of cysteine residues in the mature form of the protein (i.e. without the signal peptide)
e H: transcripts were identified in isolated haustoria; H5: transcripts were in the top 5% of transcripts detected in isolated haustoria; H1: transcripts were in the top 1% transcripts detected in isolated haustoria; I: transcripts were identified in wheat tissues during infection; I10: transcripts were in the top 10% transcripts detected in wheat tissues during infection; TC: transcript accumulation was detected by RTq-PCR during a time-course infection of wheat.
f Pgt: Puccinia graminis f sp tritici; Mlp: Melampsora larici-populina; Hv: Hemileia vastatrix.
Candidate effector-fluorescent protein fusions accumulate in N. benthamiana leaf cells
To test whether the mature form (i.e. without the signal peptide) of the candidate effectors could be expressed in dicot cells, we generated candidate effector-green fluorescent protein (GFP) fusions and expressed them in N. benthamiana by agroinfiltration. Live-cell imaging and immunoblotting assays revealed that the sixteen fusion proteins accumulate in leaf cells at detectable levels, with no obvious sign of aggregation or degradation (Fig 1, S1 Fig). Some fusions showed a band signal at a lower molecular weight, in addition to the band signal at the theoretical size (PST18220, PST03196, and PST12160), or a band signal at a higher molecular weight than expected (PST02549 and PST05023), suggesting post-translational modifications (S1 Fig). As the proteins effectively accumulated in leaf cells, we inferred that transient assays in N. benthamiana are suitable for further in planta analyses.
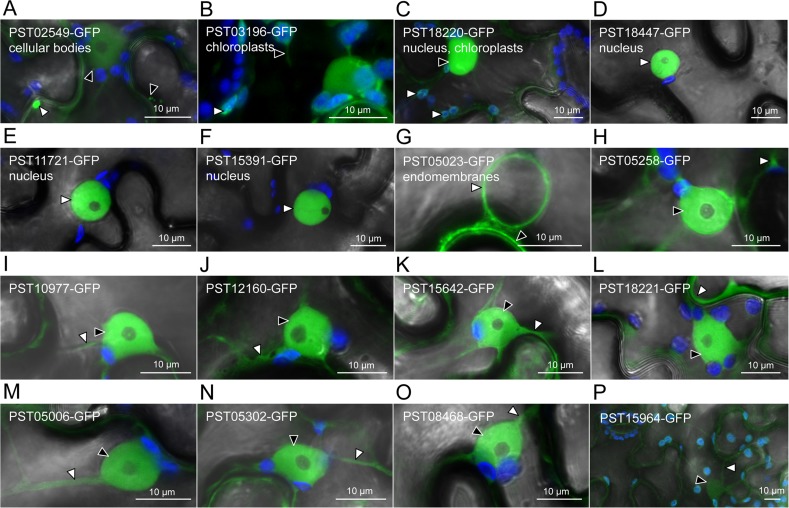
Fig 1. Seven candidate effectors show specific accumulation patterns in leaf cells.
Live-cell imaging of the 16 candidate effector-GFP fusion proteins accumulating in distinct subcellular compartments of N. benthamiana leaf cells. Proteins were transiently expressed in N. benthamiana leaf cells by agroinfiltration. Live-cell imaging was performed with a laser-scanning confocal microscope two days after infiltration. GFP and chlorophyll were excited at 488 nm. GFP (green) and chlorophyll (blue) fluorescence were collected at 505–525 nm and 680–700 nm, respectively. Images are single optical sections of 0.8 μm or a maximal projection of up to 47 optical sections (max. z-stack of 37.6 μm). Images displayed are overlays of the GFP signal, the chlorophyll signal, and bright field. For A-G, specific cellular compartments in which the GFP signal accumulates are indicated. White arrowheads indicate GFP-labelled cytosolic bodies (A), chloroplasts (B-C), nuclei (D-F), nuclear surrounding (G), or cytosolic fractions (H-P). Black arrowheads indicate GFP-labelled small cytosolic bodies (A), a stromule (B), a nucleus (C), the plasma membrane (G), or nuclei (H-P). In (P), the low level of accumulation of the fusion protein imposed higher laser power and gain, which resulted in non-specific signal for the GFP channel being visible in chloroplasts and ostiole edges.
Seven candidate effectors accumulate in specific plant cell compartments
To identify the plant cell compartments in which the candidate effectors accumulate, we performed live-cell imaging of cells expressing effector-GFP fusions. Seven out of the sixteen fusion proteins displayed an informative distribution in leaf cells (Fig 1). The fluorescence signal from PST02549-GFP and PST03196-GFP accumulated in small cytosolic bodies and chloroplasts, respectively, as well as in the nucleus and the cytosol. The fluorescence signal from PST18220-GFP labelled both chloroplasts and nuclei, suggesting a dual targeting to the two organelles. The fluorescence signal from PST18447-GFP, PST11721-GFP, and PST15391-GFP specifically accumulated in nuclei, and PST11721-GFP also labelled nuclear foci in some rare cases (S2 Fig). Finally, the fluorescence signal from PST05023-GFP labelled endomembrane compartments. The fluorescence signal from the remaining nine fusion proteins had a non-informative distribution in the nucleus and the cytosol, similar to the distribution of a free GFP control [8].
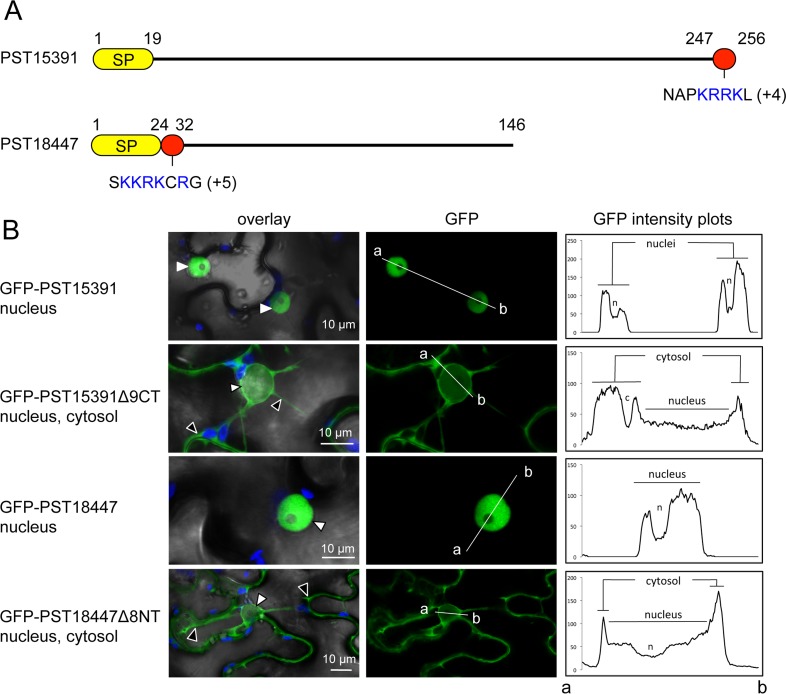
To explain the specific accumulation patterns observed, we examined the candidate effectors for subcellular targeting sequences. We focused on PST15391 and PST18447, because they showed specific, robust accumulation in nuclei (Figs 1 and 2). Previous analysis failed to identify a nuclear-localisation signal (NLS) for PST15391 and PST18447 [20]. However, we noted that both carry NLS-like stretches of amino acids enriched in positively charged residues at the C-terminus and N-terminus of their mature forms, respectively. Truncations lacking the NLS-like sequences were depleted from the nucleus and accumulated mainly in the cytosol, indicating that the NLS-like regions are necessary for specific nuclear accumulation. This set of experiments suggests that P. striiformis f sp tritici effectors use targeting sequences to traffic within plant cells.
Fig 2. PST15391 and PST18447 carry functional nuclear-localisation signals.
(A) Schematic representation of the protein primary structure of PST15391 and PST18447. Yellow: predicted signal peptide for secretion; red: amino acid sequence necessary for nuclear accumulation; blue: positively charged residues (net charge is indicated in parentheses). Numbers indicate amino acid positions. (B) Live-cell imaging of GFP-PST15391, GFP-PST15391Δ9CT, GFP-PST18447, and GFP-PST18447Δ8NT in N. benthamiana leaf cells. The cellular compartments in which the GFP signal accumulates are indicated. The left-hand side panel shows overlay images of bright field, chlorophyll, and GFP channels. The central panel shows GFP channel images. The right-hand side panel shows fluorescence intensity plots of the GFP along the line from a to b depicted in corresponding central panel images. Proteins were transiently expressed in N. benthamiana leaf cells by agroinfiltration. Live-cell imaging was performed with a laser-scanning confocal microscope two days after infiltration. GFP and chlorophyll were excited at 488 nm. GFP (green) and chlorophyll (blue) fluorescence were collected at 505–525 nm and 680–700 nm, respectively. Images are single optical sections of 0.8 μm or maximal projections of up to 3 optical sections (max. z-stack of 2.4 μm). White arrowheads: nuclei; black arrowheads: cytosol.
Six candidate effectors specifically and reliably associate with plant proteins
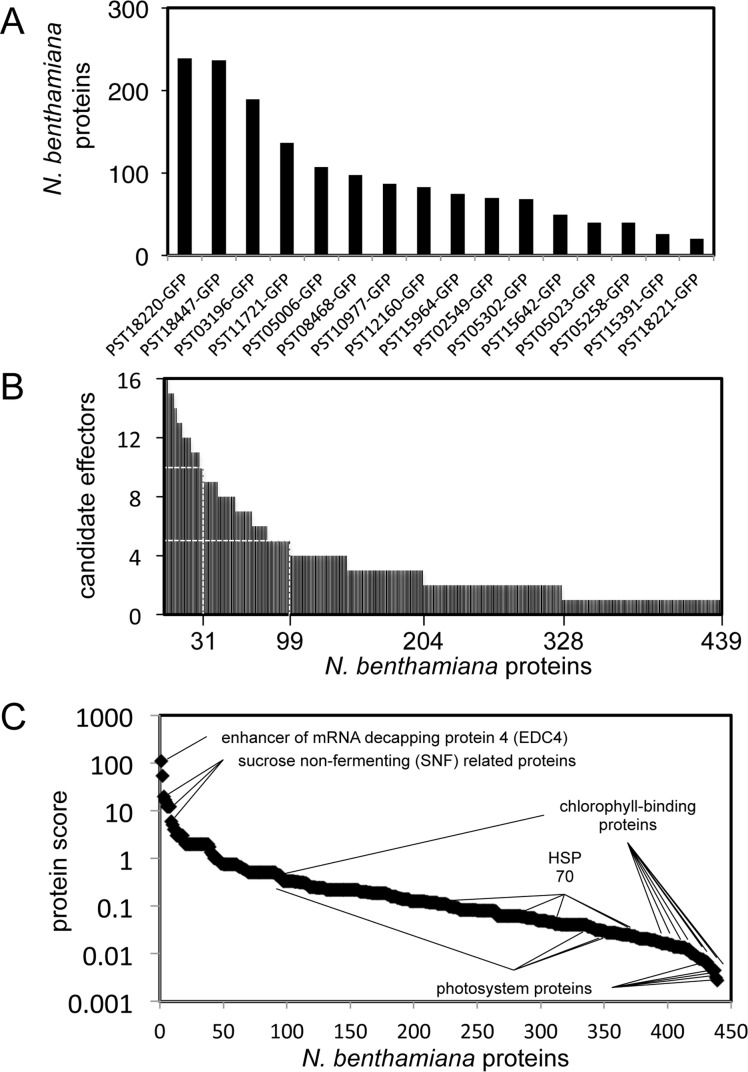
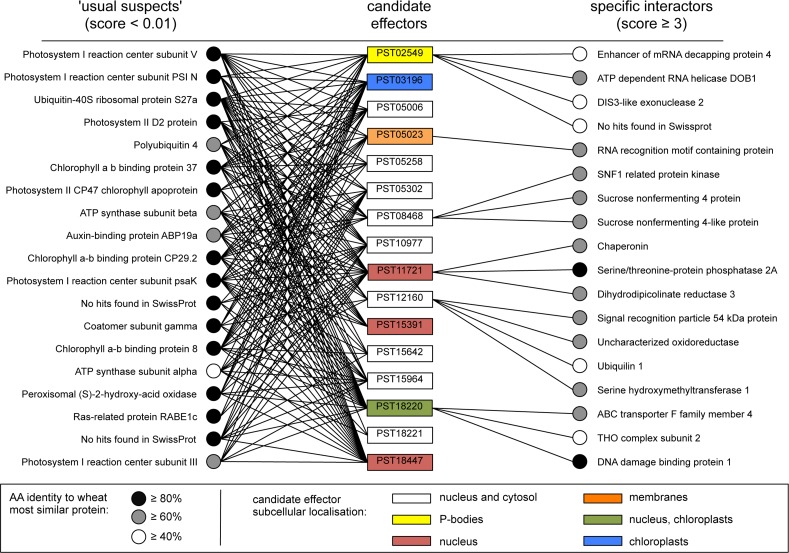
To gain further insight into the putative functions of the candidate effectors, we next aimed to identify the plant proteins they interact with in planta using anti-GFP coimmunoprecipitation/liquid chromatography-tandem mass spectrometry (coIP/MS) [8]. Using this approach, we identified 439 N. benthamiana proteins as potential interactors of the 16 candidate effectors (Sheet B and C in S1 Table, S3 Fig). A candidate effector associated with an average of 98 proteins, ranging from 20 to 236 (Fig 3). Conversely, a plant interactor associated with an average of 3.5 candidate effectors, ranging from 1 to 16. Given the high complexity of the dataset, we used a scoring method we previously developed [8] to discriminate reliable and specific interactors (high score) from redundant and non-specific ones (low score). Scores ranged from 0.003 to 108, with an average value of 0.91 (Fig 3). Eighteen proteins had a score of ≥ 3, and all specifically coimmunoprecipitated with a single candidate effector (Fig 4). For instance, the protein with the highest score (108) was an enhancer of mRNA decapping protein 4 (NbEDC4) that specifically and robustly immunoprecipitated with PST02549 (Sheet B in S1 Table).
Fig 3. Candidate effectors associate with distinct plant protein complexes.
(A) Number of N. benthamiana proteins associating with each candidate effector. Candidate effectors are arranged from left to right in descending order according to the number of interactors. (B) Number of candidate effectors associating with each N. benthamiana protein. The 439 interactors are arranged from left to right in descending order according to the number of associated candidate effectors. The X-axis legend indicates (from right to left) the number of N. benthamiana proteins that associated with at least one (439), two (328), three (204), five (99), and ten (31) candidate effectors. (C) For each N. benthamiana protein identified, we calculated a score following the formula "protein score = maximal peptide count/(redundancy)2”. Proteins are arranged from left to right in descending order based on their score. Selected proteins are indicated on the graph.
Fig 4. Nine candidate effectors have a specific subcellular localisation and/or a high-scoring plant protein interactor.
The 16 candidate effectors used in this study are shown in the middle column. Colours indicate specific subcellular localisation. The 16 plant proteins with the lowest scores (≤ 0.01; termed 'usual suspects') and the 18 plant proteins with the highest scores (≥ 3; termed 'specific interactors') are shown on the left- and right-hand sides, respectively. Black lines indicate the association between a candidate effector and a plant protein as detected by coIP/MS. For each N. benthamiana protein, the most similar wheat protein was identified by protein sequence similarity searches against the predicted proteome of the bread wheat Triticum aestivum L. using the BLASTp algorithm.
Proteins were transiently expressed in N. benthamiana leaf cells by agroinfiltration. Total proteins were isolated two days after infiltration. Plant protein complexes associated with the candidate effector-GFP fusions were purified by anti-GFP coimmunoprecipitation, separated with SDS-PAGE, and digested with trypsin. Trypsin-digested peptides were processed by LC-MS/MS and collected peaks were used to search a database containing the predicted proteome of N. benthamiana. After filtering out contaminants and proteins supported by a single peptide, and clustering similar proteins, a total of 439 non-redundant protein interactors were retained. The full dataset used to draw this figure is shown in Sheet B in S1 Table.
PST02549 associates with the wheat enhancer of mRNA decapping protein 4 (TaEDC4) in P-bodies
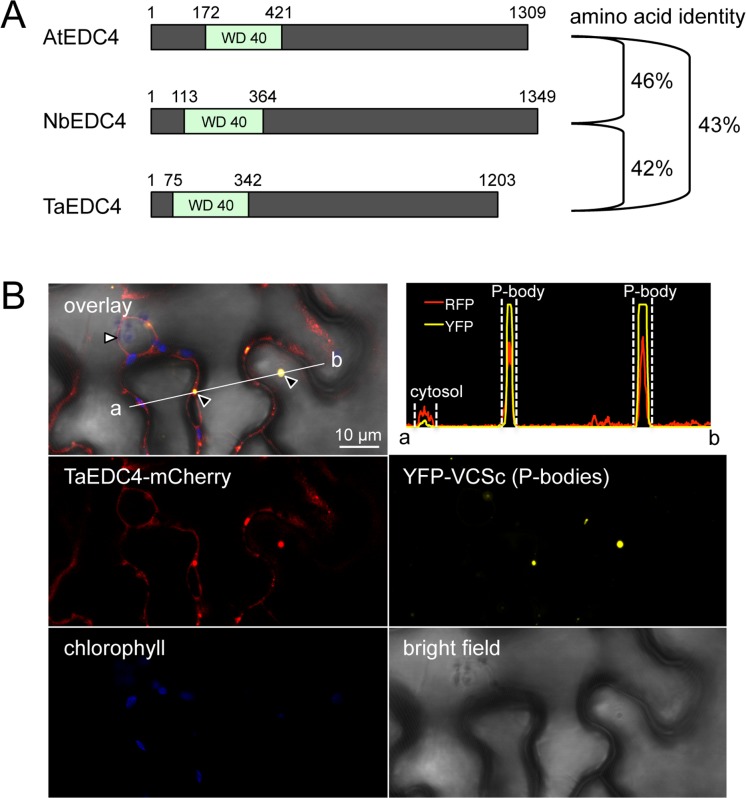
Our coIP/MS assays showed that PST02549 specifically associated with NbEDC4. To evaluate the biological significance of this association, we first identified and cloned the protein with the highest amino acid sequence similarity to NbEDC4 in bread wheat (Triticum aestivum), and named it TaEDC4. NbEDC4, TaEDC4, and Arabidopsis thaliana EDC4 (AtEDC4, also known as VARICOSE or VCS) are of a similar length (1203 to 1349 amino acids) and exhibit a pairwise amino acid sequence identity of between 42 and 46% (S4 Fig). The amino acid sequence identity between these proteins reaches 75% in the N-terminal WD40 domain (Fig 5). Next, we expressed a TaEDC4-mCherry fusion in N. benthamiana leaf cells. Confocal microscopy revealed that TaEDC4-mCherry accumulated in cytosolic foci in addition to the cytosol. Since EDC4 is a component of P-bodies, we hypothesized that the foci we observed were P-bodies. To test this hypothesis, we co-expressed TaEDC4-mCherry with YFP-VCSc, a marker of P-bodies [22]. Confocal microscopy revealed perfectly overlapping signals in cytosolic foci, confirming that TaEDC4 accumulates in P-bodies in N. benthamiana leaf cells (Fig 5).
Fig 5. TaEDC4 accumulates in P-bodies.
(A) Schematic representation of the protein primary structure of AtEDC4, NbEDC4, and TaEDC4. Numbers indicate amino acid positions. The percentage of pairwise amino acid sequence identity is indicated to the right of the diagram. (B) Live-cell imaging of TaEDC4-mCherry and YFP-VCSc in N. benthamiana leaf cells. Images show a single optical section of 0.8 μm. Proteins were transiently expressed in N. benthamiana leaf cells by agroinfiltration. Live-cell imaging was performed with a laser-scanning confocal microscope with a sequential scanning mode two days after infiltration. The YFP was excited at 514 nm; mCherry and chlorophyll were excited at 561 nm. YFP (yellow), mCherry (red), and chlorophyll (blue) fluorescence were collected at 525–550 nm, 580–620 nm, and 680–700 nm, respectively. White arrowhead: nuclei; black arrowhead: P-bodies. The intensity plot in the top right corner shows YFP and mCherry (RFP) relative fluorescence signal intensity along the white line connecting points a and b in the overlay image.
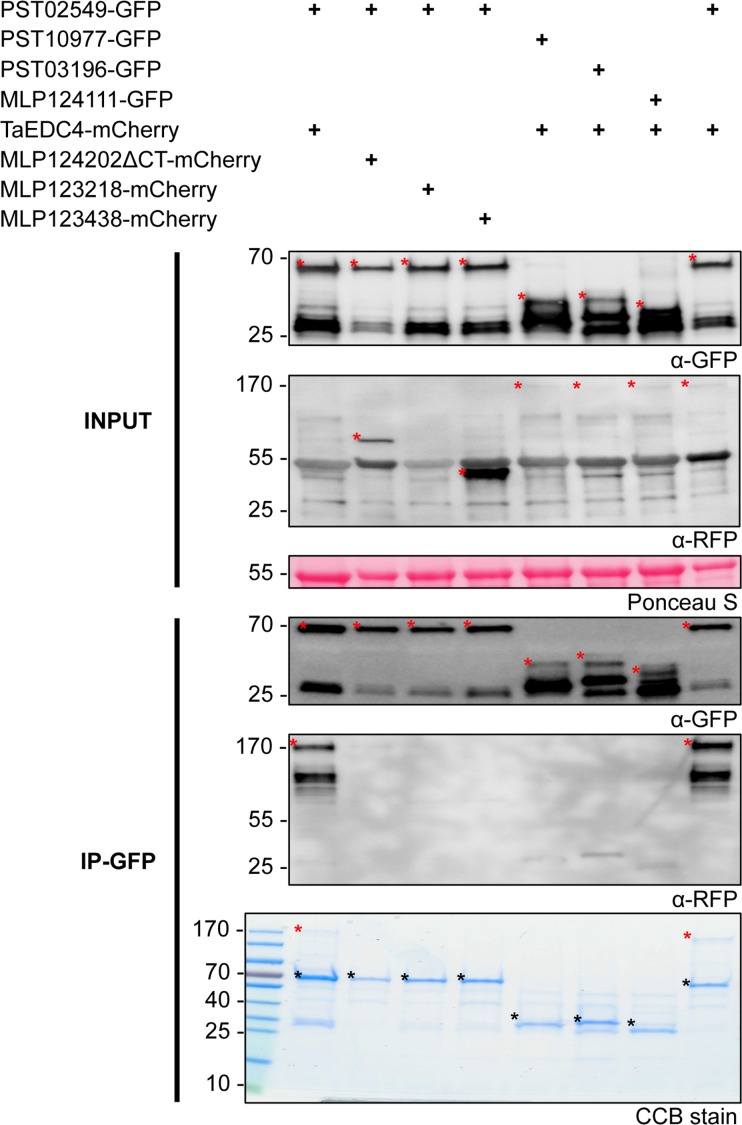
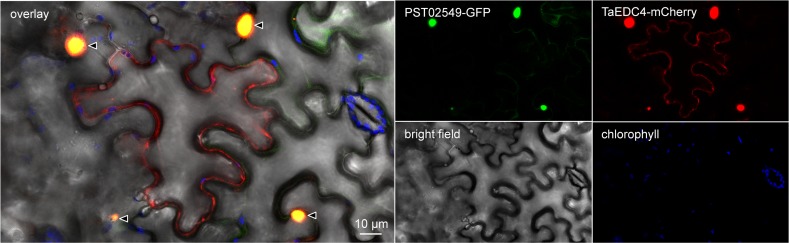
To determine whether PST02549 and TaEDC4 associate in planta, we co-expressed PST02549-GFP and TaEDC4-mCherry fusion proteins in N. benthamiana leaf cells and performed anti-GFP coimmunoprecipitation followed by immunoblotting or sodium dodecyl sulphate polyacrylamide gel electrophoresis/Coomassie Brilliant Blue (SDS-PAGE/CBB) staining. Both the anti-mCherry immunoblot and the SDS-PAGE/CBB assays revealed a specific band signal matching the predicted size of TaEDC4-mCherry in protein complexes immunoprecipitated with PST02549-GFP, indicating a strong and robust association between the two proteins (Fig 6). As negative controls, we used three GFP and three mCherry fusion proteins available in the lab (see Materials and Methods for details); none of these control proteins associated with either PST02549 or TaEDC4. Confocal microscopy revealed that the fluorescence signals from PST02549-GFP and TaEDC4-mCherry perfectly overlapped in cytosolic foci, indicating co-accumulation in P-bodies (Fig 7). Noteworthy, the intensity of PST02549-GFP and TaEDC4-mCherry band signals in the input blots of Fig 6 was similar across all conditions, indicating that co-expression of the two partners did not enhance their accumulation in leaf cells. From this set of experiments, we conclude that PST02549 and TaEDC4 specifically and robustly associate in P-bodies in N. benthamiana leaf cells.
Fig 6. PST02549 associates with TaEDC4 in planta.
Anti-green fluorescent protein (GFP) coimmunoprecipitation followed by immunoblot and sodium dodecyl sulphate-polyacrylamide gel electrophoresis/Coomassie Brilliant Blue (SDS-PAGE/CBB) analyses. Proteins were transiently expressed in N. benthamiana leaf cells by agroinfiltration. Total proteins were isolated two days after infiltration, and immediately used for anti-GFP immunoprecipitation. Immunoprecipitated protein mixtures were separated with SDS-PAGE. For direct protein visualization, the acrylamide gel was stained with CBB. For immunoblotting, proteins were electrotransferred onto polyvinylidene fluoride (PVDF) membranes. Immunodetection was performed with anti-GFP or anti-redu fluorescent protein (RFP) antibodies, and immunoblots were revealed with a chemiluminescent imager. Ponceau S staining of the PVDF membrane was used as a loading and transfer control. Theoretical protein size is indicated in parentheses in kilodalton (kDa) for each fusion protein. Numbers to the left of the blot and gel images indicate protein size in kDa. In the immunoblot images, red asterisks indicate specific protein bands. In the gel image, asterisks indicate specific protein bands (red: TaEDC4-mCherry; black: GFP fusions); the PageRuler ladder is shown to the left of the image. IP: immunoprecipitation. In the IP-GFP/ α-RFP blot, note that the weak band signals observed on the right side between 25 and 40 kDa are due to non-specific background detection of abundant GFP fusions by the anti-RFP antibodies.
Fig 7. PST02549 and TaEDC4 co-accumulate in P-bodies.
Live-cell imaging of PST02549-GFP and TaEDC4-mCherry in N. benthamiana leaf cells. Images show a single optical section of 0.8 μm. The white asterisk indicates a pavement cell expressing only the TaEDC4-mCherry fusion, in which no large P-body was detected. Proteins were transiently expressed in N. benthamiana leaf cells by agroinfiltration. Live-cell imaging was performed with a laser-scanning confocal microscope with a sequential scanning mode two days after infiltration. GFP and the chlorophyll were excited at 488 nm; the mCherry was excited at 561 nm. GFP (green), mCherry (red), and chlorophyll (blue) fluorescence were collected at 505–525 nm, 580–620 nm and 680–700 nm, respectively. Black arrowheads indicate P-bodies. White arrowheads: nuclei.
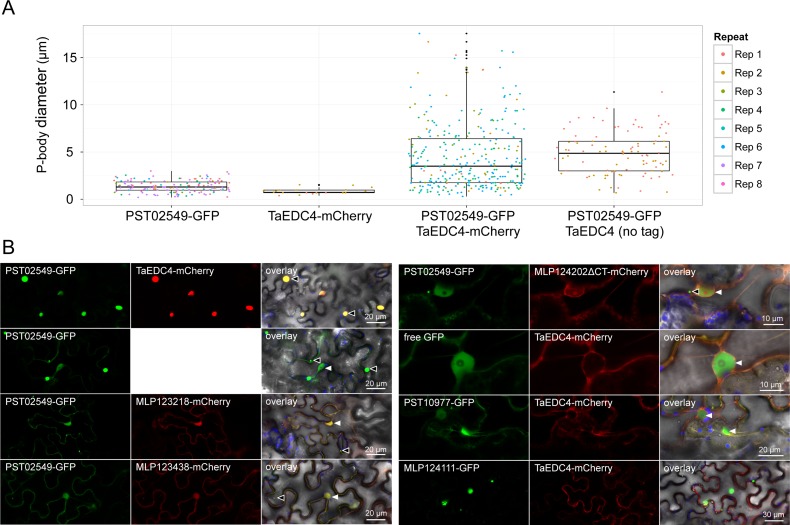
Co-expression of PST02549 and TaEDC4 increases the size of P-bodies
During confocal microscopy assays of N. benthamiana leaf cells co-expressing PST02549-GFP and TaEDC4-mCherry, we noted that the P-bodies appeared larger than usual. To quantify this phenomenon, we measured the diameter of P-bodies from confocal microscopy images. When PST02549-GFP was co-expressed with TaEDC4-mCherry or an untagged version of TaEDC4, the average diameters of the P-bodies were 4.5 ± 3.5 μm and 4.9 ± 2.2 μm, respectively (Fig 8, Sheet D in S1 Table). By contrast, when PST02549-GFP and TaEDC4-mCherry were expressed independently and/or with other control proteins, the average diameter of a P-body was 1.3 ± 0.6 μm. CoIP/MS assays confirmed the presence of the untagged TaEDC4, as well as the presence of the endogenous NbEDC4, in complex with PST02549 (Sheet E in S1 Table). None of the negative controls we tested co-localised with PST02549-GFP or TaEDC4-mCherry or triggered the formation of large P-bodies (Fig 8, Sheet D in S1 Table). We conclude that co-expression of PST02549 and TaEDC4 specifically increases the size of P-bodies.
Fig 8. Co-expression of PST02549 and TaEDC4 increases the size of P-bodies.
(A) Categorical scatterplots showing the diameter of P-bodies labelled by PST02549-GFP and/or TaEDC4-mCherry in leaf cells. Boxes depict the interquartile range and the median, vertical bars indicate the first and fourth quartile range, and outlier data points are depicted in black. P-body diameters were measured from laser scanning confocal microscope images acquired through two to eight independent agroinfiltration assays. The different colours correspond to independent observations (repeats). The following numbers of P-bodies were scored: PST02549-GFP (n = 150); TaEDC4-mCherry (n = 20), PST02549-GFP/TaEDC4-mCherry (n = 303), PST02549-GFP/TaEDC4 (n = 96). For treatments 'PST02549-GFP' and 'TaEDC4-mCherry', the fusion proteins were expressed alone or with additional control fusion proteins (see Sheet D in S1 Table for raw data). (B) Live-cell imaging of various GFP and mCherry fusion proteins in N. benthamiana leaf cells. Images present a single optical section of 0.8 μm of a maximal projection of up to 6 optical sections (max. z-stack of 4.8 μm). Overlay images merge GFP, mCherry, chlorophyll, and bright field signals. Note that for the PST02549-GFP/TaEDC4, TaEDC4 was untagged and the mCherry fluorescence signal was not recorded. Proteins were transiently expressed in N. benthamiana leaf cells by agroinfiltration. Live-cell imaging was performed with a laser-scanning confocal microscope with a sequential scanning mode two days after infiltration. GFP and the chlorophyll were excited at 488 nm; the mCherry was excited at 561 nm. GFP (green), mCherry (red), and chlorophyll (blue) fluorescence were collected at 505–525 nm, 580–620 nm and 680–700 nm, respectively. Black arrowheads indicate P-bodies. White arrowheads: nuclei. Note that the large protein aggregates formed by MLP124111-GFP do not show any TaEDC4-mCherry signal.
Discussion
In this study, we found that PST02549 accumulates in plant cell P-bodies and associates with a P-body-derived protein. This observation suggests that an effector that targets plant P-bodies has evolved in P. striiformis f sp tritici. To our knowledge, a connection between filamentous plant pathogens and P-bodies has not previously been established.
How would a pathogen benefit from manipulating host P-bodies? Some plant pathogen effectors target components of the host RNA silencing machinery [28–30]. In A. thaliana, two recent reports have connected P-bodies, or P-body-resident proteins, with plant immune responses [25, 31]. Therefore, pathogen effectors may target P-bodies to manipulate RNA metabolism and/or suppress immune responses. Interestingly, pathogenic bacteria and viruses that infect mammals are known to alter the structure and function of host P-bodies [32–33]. Therefore, diverse parasites of eukaryotes have evolved to target host P-bodies. Further mechanistic investigations of the pathogen effector/P-body interplay should reveal the biological significance of this phenomenon.
We observed an increase in P-body size upon co-expression of PST02549 and TaEDC4. The depletion or overexpression of P-body components is known to modify P-body integrity, which can lead to an increase in size [34]. It is therefore possible that the increase in P-body size observed in our study is due to over-accumulation of P-body-resident proteins such as PST02549 or TaEDC4. However, we observed this phenomenon only when the two proteins co-accumulated, indicating that both are required to increase P-body size. The biological significance of the association between PST02549 and TaEDC4 as well as the increase in P-body size remain to be further investigated in wheat.
The pipeline we used in this study allowed us to identify nine candidate effectors that target a given subcellular compartment and/or protein complex. We use this specific behaviour in leaf cells as a rationale to hypothesize that these proteins are bona fide cytoplasmic effectors. Thus, we flag them as high priority candidate effectors to further characterize in wheat. Noteworthy, we recently obtained similar informative data for 40% of a set of candidate effectors from another rust species, the poplar leaf rust fungus Melampsora larici-populina [8]. These findings establish the proof-of-concept that N. benthamiana is a valuable heterologous system for fast-forward effectoromic analysis of plant pathogens, regardless of their host plant.
In this study and in [8], we performed an in planta protein-protein interaction screen in the heterologous plant N. benthamiana. Such approach has advantages and disadvantages compared with traditional methods used to perform protein-protein interaction screens, the most common being yeast-two hybrid [35]. As a main advantage, our screen takes place in a leaf cell. Therefore, the prey proteins are in their natural context, i.e. in complex with their partners in the relevant cellular compartment. This argument holds true for the bait (i.e. the candidate effector), provided that it is indeed a bona fide cytoplasmic effector. The principal disadvantage of our approach is that it is prone to false negatives; we may miss interactors that are specific to the natural host species, or that are too divergent in N. benthamiana to associate with the bait. Importantly, P. striiformis f sp tritici does not infect solanaceous plants, and therefore its effectors do not associate with Solanaceous proteins in natura. Thus, follow up experiments are need to further validate the lead candidate interactors. The associations obtained via the screen help to rapidly identify interactors in the original host species, which are more likely to be biologically relevant.
The field of rust effector biology has progressed relatively slowly regarding the functional characterisation of effectors despite the availability of pathogen genome sequences [6]. In addition to PST02549, our study identified candidate effectors that should be treated as high priority proteins for further investigations. Notably, PST08468 specifically associated with members of the sucrose non-fermenting related protein kinases. These are key proteins that control the interplay between stress response and growth [36]. Studying PST08468 may help better understand how rust fungi manipulate this balance in their hosts.
We identified plant interactors of candidate effectors, some of which may represent bone fide effector targets. Growing evidence suggests that during evolution domains from effector targets have been incorporated into immune receptors such as nucleotide binding-leucine rich repeat (NB-LRR, also referred to as NLR) proteins to become ‘sensor domains’ that mediate recognition of specific effectors [37–39]. A recent genome-wide analysis predicted many NLR gene models in which protein domains that differ from typical NLR domains have been incorporated (Sarris et al., in review). Interestingly, six of the eighteen top scoring effector interactors identified in our study carry a protein domain that is predicted to be integrated into a plant NLR, including the WD40 protein domain of EDC4 (Sheet B in S1 Table). Therefore, our predicted host targets can be a valuable source of new ‘baits’ for engineering NLR genes with sensor domains.
Materials and Methods
In silico analyses
Predicted protein sequences were retrieved from the following sources: P. striiformis f sp tritici (http://yellowrust.com/) [20], N. benthamiana (http://solgenomics.net/) [10], and T. aestivum (http://www.plantgdb.org/TaGDB/). Protein sequence analysis was performed with ClustalX and Jalview programmes. Homology searches were performed with the BLAST+ programme. For the selection of the eleven candidate effectors, we applied an in silico pipeline previously developed [8]. We adapted it to fit the gene expression dataset we used [20]. Notably, we retained only candidate effectors whose transcripts were enriched in purified haustoria (top 5% category in [20]) compared to infected tissues. The set of candidate effectors selected via the pipeline was manually analysed to remove redundant family members. PST05258 and PST15391 from Tribe 54, as well as PST18220 and PST18221 from Tribe 238, were both retained due to high levels of polymorphism [20].
Cloning procedures and plasmids
The open reading frame (ORF) encoding the mature form (i.e. without the signal peptide) of P. striiformis f sp tritici small-secreted proteins or the full-length of T. aestivum EDC4 (Traes_6DL_3FBA5B70E.1) were amplified by polymerase chain reaction (PCR) using cDNA isolated from wheat leaves 14 days after inoculation with a virulent isolate of P. striiformis f sp tritici (isolate PST-08/21) [20], or were obtained through gene synthesis (Genewiz, London, UK), with codon optimization for plant expression and removal of internal BbsI and BsaI restriction sites. Primers and synthetic genes were designed to be compatible with the suite of Golden Gate vectors, as previously described [8] (Sheet A in S1 Table). Truncated versions of PST15391 and PST18447 were obtained by PCR cloning. All PCR-generated DNA fragments were verified by sequencing after cloning into level 0 Golden Gate vectors. Plasmids were multiplied and conserved in Escherichia coli (Subcloning Efficiency DH5α Competent Cells; Invitrogen, Carlsbad, California, USA) as previously described [8]. The fusion proteins built with candidate effectors from the poplar rust fungus Melampsora larici-populina (MLP124111, MLP123218, MLP123438 and MLP124202CT) were obtained in previous studies [8, 12] and were used as negative controls in coIP and confocal microscopy assays.
Transient protein expression in N. benthamiana leaf cells
Agrobacterium tumefaciens electrocompetent strain GV3101 (pMP90) was used to deliver T-DNA constructs in leaf cells of three- to four-week-old N. benthamiana plants, following the agroinfiltration method previously described [8]. The leaves were collected two days after infiltration for further protein isolation or microscopy.
Live-cell imaging by laser-scanning confocal microscopy
Confocal microscopy was performed as previously reported [8] with a Leica DM6000B/TCS SP5 laser-scanning confocal microscope (Leica Microsystems, Bucks, UK), using 10x (air) and 63x (water immersion) objectives. Each construct gave a similar localisation pattern in at least three independent observations. Image analysis was performed using the Fiji plugin of Image J 2.0.0 (http://fiji.sc/Fiji). To quantify the diameter of P-bodies, the ‘measure’ tool of Fiji was used to measure manually-drawn lines matching the apparent diameter of P-bodies in all the single optical section confocal images acquired in the course of this project. Categorical scatterplots were generated with R, using the ggplot2 package and an in-house developed script (S1 Text).
Protein isolation and immunoblot analyses
Frozen leaves were ground to a powder using a mortar and pestle. Total proteins were extracted as previously described [8]. Ten microliters of isolated protein was separated on a 15% SDS-PAGE gel, and the protein content was estimated by Coomassie Brilliant Blue (CBB) staining. Immunoblot analysis was performed as previously described [8], using GFP (B2):sc-9996 HRP-conjugated antibody (Santa-Cruz Biotechnology), rat anti-RFP 5F8 antibody (Chromotek, Munich, Germany) and a HRP-conjugated anti-rat antibody.
Coimmunoprecipitation and LC-MS/MS analyses
Coimmunoprecipitation procedures were performed as reported previously [8], using GFP_Trap_A beads (Chromotek, Munich, Germany). Various GFP and mCherry fusion proteins [8,12] were used as negative controls in confocal microscopy and coIP assays. Some proteins were specifically selected based on their ability to generate cytosolic aggregates (MLP124111), their similarity in size and accumulation pattern to tested proteins (MLP124202ΔCT), or their ‘stickyness in coIP/MS assays’; i.e. ability to associate with a high number of proteins, (PST10977 and PST03196). Sample preparation, liquid chromatography / tandem mass spectrometry (LC-MS/MS) and data analyses were performed as described in [8], using a hybrid mass spectrometer LTQ-Orbitrap XL (ThermoFisher Scientific, Carlsbad, California, USA) and a nanoflow-UHPLC system (NanoAcquity Waters Corp., Burnsville, Minnesota, USA). LC-MS/MS data were processed and plant interactors were scored as previously described [8] (Sheet B in S1 Table). Briefly, scores were calculated following the formula "protein score = maximal peptide count/(redundancy)2”. The redundancy value was calculated by integrating the coIP/MS data from [8].
Supporting Information
Proteins were transiently expressed in N. benthamiana leaf cells by agroinfiltration. Total proteins were extracted two days after infiltration by grinding leaves in liquid nitrogen and immediately extracting, reducing and denaturing proteins from the leaf powder. Proteins were separated on 15% SDS-PAGE gels and transferred onto a nitrocellulose membrane. Primary and secondary immune detection were performed with rabbit anti-GFP and goat anti-rabbit antibodies, respectively. Images originating from the same membrane and processed at the same time are grouped together. Blots were cropped to remove lanes previously published elsewhere (Petre et al., 2015a). Secondary antibodies and PageRuler signals were detected simultaneously using an infrared imager. The theoretical size of each fusion protein is indicated in kDa in parentheses. White asterisks indicate specific protein bands.
(TIF)
Live-cell imaging of PST11721-GFP in N. benthamiana leaf cells. Proteins were transiently expressed in N. benthamiana leaf cells by agroinfiltration. Live-cell imaging was performed with a laser-scanning confocal microscope two days after infiltration. The GFP was excited at 488 nm. GFP (green) fluorescence was collected at 505–550 nm. The image is a single optical section of 0.8 μm, showing an overlay of the GFP and bright field channels. The black arrowheads indicate GFP-labelled nuclear foci.
(TIF)
Protein mixtures isolated by anti-GFP immunoprecipitation were reduced and denatured in a Laemmli buffer. Proteins were separated with SDS-PAGE and stained with Coomassie Brilliant Blue. Trypsin-digested peptides were processed by LC-MS/MS and collected peaks were used to search a database containing the GFP sequence. The theoretical size of each fusion protein is indicated in parentheses in kilodalton (kDa). The number of peptides identified by LC-MS/MS and matching the GFP is indicated for each fusion protein between brackets. The size of the PageRuler ladder bands is indicated in kDa. Images originating from the same gel are grouped together. Black asterisks indicate detectable and specific protein bands.
(TIF)
Amino acid alignment of EDC4 of Arabidopsis thaliana (AtEDC4, AT3G13300.2), Nicotiana benthamiana (NbEDC4, NbS00023257g0003.1), and Triticum aestivum (TaEDC4, Traes_6DL_3FBA5B70E.1). Alignment was performed with ClustalX. Amino acid residues are colored according to the ClustalX scheme.
(TIF)
(XLSX)
(TXT)
Acknowledgments
We thank the Norwich Rust Group, members of the Kamoun Group, and LFCs for discussions, Dr. Vanessa Segovia for providing material, and support teams at The Sainsbury Laboratory and at The John Innes Centre.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
BP, CL, and SD were supported by the French National Research Agency through the Labex ARBRE (ANR-12-LABXARBRE-01) and the Young Scientist Grant POPRUST (ANR-2010-JCJC-1709-01). BP and CL are supported by an INRA Contrat Jeune Scientifique. BP was supported by the European Union, in the framework of the Marie-Curie FP7 COFUND People Programme, through the award of an AgreenSkills’ fellowship (under grant agreement n° 267196). BP, JS, JW, KVK, DGOS, and SK are supported by the Biotechnology and Biological Sciences Research Council and the Gatsby Charitable Foundation. DGOS is supported by a Leverhulme early career fellowship and a fellowship in computational biology at TGAC, in partnership with the John Innes Centre. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Dodds PN, Rathjen JP. 2010. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11:539–48 10.1038/nrg2812 [DOI] [PubMed] [Google Scholar]
- 2.Win J, Chaparro-Garcia A, Belhaj K, Saunders DG, Yoshida K, Dong S, et al. 2012. Effector biology of plant-associated organisms: concepts and perspectives. Cold Spring Harb Symp Quant Biol 77:235–47 10.1101/sqb.2012.77.015933 [DOI] [PubMed] [Google Scholar]
- 3.Dangl JL, Horvath DM, Staskawicz BJ. 2013. Pivoting the plant immune system from dissection to deployment. Science 341:746–51 10.1126/science.1236011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hogenhout SA, Van der Hoorn RA, Terauchi R, Kamoun S. 2009. Emerging concepts in effector biology of plant-associated organisms. Mol Plant Microbe Interact 22:115–22 10.1094/MPMI-22-2-0115 [DOI] [PubMed] [Google Scholar]
- 5.Martin F, Kamoun S. 2012. Effectors in Plant-Microbe Interactions. Wiley-Blackwell; Eds. ISBN: 978-0-470-95822-3 [Google Scholar]
- 6.Petre B, Joly DL, Duplessis S. 2014. Effector proteins of rust fungi. Front Plant Sci 5:416 10.3389/fpls.2014.00416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Upadhyaya NM, Mago R, Staskawicz BJ, Ayliffe MA, Ellis JG, Dodds PN. 2014. A Bacterial Type III Secretion Assay for Delivery of Fungal Effector Proteins into Wheat. Mol Plant Microbe Interact 27:255–264 10.1094/MPMI-07-13-0187-FI [DOI] [PubMed] [Google Scholar]
- 8.Petre B, Saunders DG, Sklenar J, Lorrain C, Win J, Duplessis S, et al. 2015a. Candidate Effector Proteins of the Rust Pathogen Melampsora larici-populina Target Diverse Plant Cell Compartments. Mol Plant Microbe Interact 28:689–700 [DOI] [PubMed] [Google Scholar]
- 9.Goodin MM, Zaitlin D, Naidu RA, Lommel SA. 2008. Nicotiana benthamiana: its history and future as a model for plant-pathogen interactions. Mol Plant Microbe Interact 21:1015–26 10.1094/MPMI-21-8-1015 [DOI] [PubMed] [Google Scholar]
- 10.Bombarely A, Rosli HG, Vrebalov J, Moffett P, Mueller LA, Martin GB. 2012. A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol Plant Microbe Interact 25:1523–30 10.1094/MPMI-06-12-0148-TA [DOI] [PubMed] [Google Scholar]
- 11.Pais M, Win J, Yoshida K, Etherington GJ, Cano LM, Raffaele S, et al. 2013. From pathogen genomes to host plant processes: the power of plant parasitic oomycetes. Genome Biol 14:211 10.1186/gb-2013-14-6-211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petre B, Lorrain C, Saunders DG, Win J, Sklenar J, Duplessis S, et al. 2015b. Rust fungal effectors mimic host transit peptides to translocate into chloroplasts. Cell Microbiol 10.1111/cmi.12530 [DOI] [PubMed] [Google Scholar]
- 13.Dean R, Van Kan JA, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, et al. 2012. The Top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol 13:414–30 10.1111/j.1364-3703.2011.00783.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beddow JM, Pardey PG, Chai Y, Hurley TM, Kriticos DJ, Braun H-J, et al. 2015. Research investment implications of shifts in the global geography of wheat stripe rust. Nat Plants 1:15132. [DOI] [PubMed] [Google Scholar]
- 15.Chen W, Wellings C, Chen X, Kang Z, Liu T. 2014. Wheat stripe (yellow) rust caused by Puccinia striiformis f. sp. tritici. Mol Plant Pathol 15:433–46 10.1111/mpp.12116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hubbard A, Lewis CM, Yoshida K, Ramirez-Gonzalez RH, de Vallavieille-Pope C, Thomas J, et al. 2015. Field pathogenomics reveals the emergence of a diverse wheat yellow rust population. Genome Biol 16:23 10.1186/s13059-015-0590-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cantu D, Govindarajulu M, Kozik A, Wang M, Chen X, Kojima KK, et al. 2011. Next generation sequencing provides rapid access to the genome of Puccinia striiformis f. sp. tritici, the causal agent of wheat stripe rust. PLoS One 6:e24230 10.1371/journal.pone.0024230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garnica DP, Upadhyaya NM, Dodds PN, Rathjen JP. 2013. Strategies for Wheat Stripe Rust Pathogenicity Identified by Transcriptome Sequencing. PLoS One 8:e67150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng W, Huang L, Huang J, Wang X, Chen X, Zhao J, et al. 2013. High genome heterozygosity and endemic genetic recombination in the wheat stripe rust fungus. Nat Commun 4:2673 10.1038/ncomms3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cantu D, Segovia V, MacLean D, Bayles R, Chen X, Kamoun S, et al. 2013. Genome analyses of the wheat yellow (stripe) rust pathogen Puccinia striiformis f. sp. tritici reveal polymorphic and haustorial expressed secreted proteins as candidate effectors. BMC Genomics 14:270 10.1186/1471-2164-14-270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan EKL, Fritzler MJ. 2012. Ten years of progress in GW/P body research Advances in Experimental Medicine and Biology. Springer Science and Business Media; Eds. [Google Scholar]
- 22.Xu J, Yang JY, Niu QW, Chua NH. 2006. Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell 18:3386–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu J, Chua NH. 2009. Arabidopsis decapping 5 is required for mRNA decapping, P-body formation, and translational repression during postembryonic development. Plant Cell 21:3270–9 10.1105/tpc.109.070078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steffens A, Bräutigam A, Jakoby M, Hülskamp M. 2015. The BEACH Domain Protein SPIRRIG Is Essential for Arabidopsis Salt Stress Tolerance and Functions as a Regulator of Transcript Stabilization and Localization. PLoS Biol 13:e1002188 10.1371/journal.pbio.1002188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maldonado-Bonilla LD, Eschen-Lippold L, Gago-Zachert S, Tabassum N, Bauer N, Scheel D, et al. 2014. The Arabidopsis tandem zinc finger 9 protein binds RNA and mediates pathogen-associated molecular pattern-triggered immune responses. Plant Cell Physiol 55:412–25 10.1093/pcp/pct175 [DOI] [PubMed] [Google Scholar]
- 26.Eulalio A, Fröhlich KS, Mano M, Giacca M, Vogel J. 2011. A candidate approach implicates the secreted Salmonella effector protein SpvB in P-body disassembly. PLoS One 6:e17296 10.1371/journal.pone.0017296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reineke LC, Lloyd RE. 2013. Diversion of stress granules and P-bodies during viral infection. Virology 436:255–67 10.1016/j.virol.2012.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weiberg A, Wang M, Bellinger M, Jin H. 2014. Small RNAs: a new paradigm in plant-microbe interactions. Annu Rev Phytopathol 52:495–516 10.1146/annurev-phyto-102313-045933 [DOI] [PubMed] [Google Scholar]
- 29.Qiao Y, Shi J, Zhai Y, Hou Y, Ma W. 2015. Phytophthora effector targets a novel component of small RNA pathway in plants to promote infection. Proc Natl Acad Sci U S A 112:5850–5 10.1073/pnas.1421475112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spanu PD. 2015. RNA-protein interactions in plant disease: hackers at the dinner table. New Phytol 207:991–5 10.1111/nph.13495 [DOI] [PubMed] [Google Scholar]
- 31.Roux ME, Rasmussen MW, Palma K, Lolle S, Regué AM, Bethke G, et al. 2015. The mRNA decay factor PAT1 functions in a pathway including MAP kinase 4 and immune receptor SUMM2. EMBO J 34:593–608 10.15252/embj.201488645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ariumi Y, Kuroki M, Kushima Y, Osugi K, Hijikata M, Maki M, et al. 2011. Hepatitis C virus hijacks P-body and stress granule components around lipid droplets. J Virol 85:6882–92 10.1128/JVI.02418-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pérez-Vilaró G, Fernández-Carrillo C, Mensa L, Miquel R, Sanjuan X, Forns X, et al. 2015. Hepatitis C virus infection inhibits P-body granule formation in human livers. J Hepatol 62:785–90 10.1016/j.jhep.2014.11.018 [DOI] [PubMed] [Google Scholar]
- 34.Eulalio A, Behm-Ansmant I, Izaurralde E. 2007. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol 8:9–22 [DOI] [PubMed] [Google Scholar]
- 35.Mukhtar MS, Carvunis AR, Dreze M, Epple P, Steinbrenner J, Moore J, et al. 2011. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science. 333:596–601 10.1126/science.1203659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emanuelle S, Doblin MS, Stapleton DI, Bacic A, Gooley PR. 2015. Molecular Insights into the Enigmatic Metabolic Regulator, SnRK1. Trends Plant Sci 10.1016/j.tplants.2015.11.001 [DOI] [PubMed] [Google Scholar]
- 37.Cesari S, Bernoux M, Moncuquet P, Kroj T, Dodds PN. 2014. A novel conserved mechanism for plant NLR protein pairs: the "integrated decoy" hypothesis. Front Plant Sci 5:606 10.3389/fpls.2014.00606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu CH, Krasileva KV, Banfield MJ, Terauchi R, Kamoun S. 2015. The "sensor domains" of plant NLR proteins: more than decoys? Front Plant Sci 6:134 10.3389/fpls.2015.00134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarris PF, Jones JD. 2015. Plant immune receptors mimic pathogen virulence targets. Oncotarget 6:16824–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Proteins were transiently expressed in N. benthamiana leaf cells by agroinfiltration. Total proteins were extracted two days after infiltration by grinding leaves in liquid nitrogen and immediately extracting, reducing and denaturing proteins from the leaf powder. Proteins were separated on 15% SDS-PAGE gels and transferred onto a nitrocellulose membrane. Primary and secondary immune detection were performed with rabbit anti-GFP and goat anti-rabbit antibodies, respectively. Images originating from the same membrane and processed at the same time are grouped together. Blots were cropped to remove lanes previously published elsewhere (Petre et al., 2015a). Secondary antibodies and PageRuler signals were detected simultaneously using an infrared imager. The theoretical size of each fusion protein is indicated in kDa in parentheses. White asterisks indicate specific protein bands.
(TIF)
Live-cell imaging of PST11721-GFP in N. benthamiana leaf cells. Proteins were transiently expressed in N. benthamiana leaf cells by agroinfiltration. Live-cell imaging was performed with a laser-scanning confocal microscope two days after infiltration. The GFP was excited at 488 nm. GFP (green) fluorescence was collected at 505–550 nm. The image is a single optical section of 0.8 μm, showing an overlay of the GFP and bright field channels. The black arrowheads indicate GFP-labelled nuclear foci.
(TIF)
Protein mixtures isolated by anti-GFP immunoprecipitation were reduced and denatured in a Laemmli buffer. Proteins were separated with SDS-PAGE and stained with Coomassie Brilliant Blue. Trypsin-digested peptides were processed by LC-MS/MS and collected peaks were used to search a database containing the GFP sequence. The theoretical size of each fusion protein is indicated in parentheses in kilodalton (kDa). The number of peptides identified by LC-MS/MS and matching the GFP is indicated for each fusion protein between brackets. The size of the PageRuler ladder bands is indicated in kDa. Images originating from the same gel are grouped together. Black asterisks indicate detectable and specific protein bands.
(TIF)
Amino acid alignment of EDC4 of Arabidopsis thaliana (AtEDC4, AT3G13300.2), Nicotiana benthamiana (NbEDC4, NbS00023257g0003.1), and Triticum aestivum (TaEDC4, Traes_6DL_3FBA5B70E.1). Alignment was performed with ClustalX. Amino acid residues are colored according to the ClustalX scheme.
(TIF)
(XLSX)
(TXT)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.