Summary
Repetitive DNA arrays are important structural features of eukaryotic genomes that are often heterochromatinized to suppress repeat instability. It is unclear, however, if all repeats within an array are equally subject to heterochromatin formation and gene silencing. Here, we show that in starving Saccharomyces cerevisiae, silencing of reporter genes within the ribosomal DNA (rDNA) array is less pronounced in outer repeats compared to inner repeats. This position effect is linked to the starvation-induced contraction of the nucleolus. We show that the chromatin regulators condensin and Hmo1 redistribute within the rDNA upon starvation; that Hmo1, like condensin, is required for nucleolar contraction; and that the position effect partially depends on both proteins. Starvation-induced nucleolar contraction and differential desilencing of the outer rDNA repeats may provide a mechanism to activate rDNA-encoded RNAPII transcription units without causing general rDNA instability.
Introduction
Repetitive DNA arrays are important constituents of eukaryotic genomes and affect many aspects of basic cell physiology (Richard et al., 2008). A prominent example is the ribosomal DNA (rDNA), which is found in large tandem-repeat arrays in most eukaryotes. In the yeast Saccharomyces cerevisiae, the rDNA locus consists of more than 100 copies of a 9.1-kb repeat unit and accounts for nearly 10% of the genome. The sheer size of this array raises intriguing questions about its spatial organization and whether all repeats are subject to the same regulation.
Gene expression in the yeast rDNA is tightly controlled. Each repeat encodes an RNAPI-transcribed 35S rRNA gene and an RNAPIII-transcribed 5S rRNA gene. In addition, RNAPII-transcribed sequences are encoded in the intergenic spacer sequences, IGS1 and IGS2, as well as on the anti-sense strand of the 35S rRNA gene (Coelho et al., 2002; Kobayashi and Ganley, 2005). A specialized chromatin state selectively silences RNAPII-dependent gene expression within the rDNA to protect repeats from non-allelic recombination, which is stimulated by intergenic transcription (Mekhail and Moazed, 2010; Ganley and Kobayashi, 2014). Silencing is controlled by several histone modifiers, including the sirtuin Sir2 (Huang, 2002; Kueng et al., 2013; Ryu and Ahn, 2014). In addition, the conserved condensin complex (comprising Smc2, Smc4, Brn1, Ycs4 and Ycg1) has been identified as a modulator of both Sir2-dependent and independent silencing pathways in the rDNA(Machin et al., 2004; Ryu and Ahn, 2014).
rRNA production in S. cerevisiae is down-regulated in response to gradual exhaustion of nutrients during saturating growth and upon acute shift to various low-nutrient conditions (Conrad et al., 2014). Although there are regulatory differences between the respective starvation responses (Klosinska et al., 2011), most involve signaling by TOR kinase, a conserved regulator of cell growth and ribosome biogenesis (Loewith and Hall, 2011). Inhibition of TOR kinase by rapamycin leads to reduced rRNA expression, increased rDNA stability and enhanced Sir2 binding in the rDNA (Ha and Huh, 2011). Altered transcriptional activity of the rDNA is also reflected in the size of the nucleolus, the subnuclear structure organized by the rDNA. The nucleolus occupies approximately one third of the nucleus in metabolically active yeast, but shrinks upon nutrient depletion or rapamycin treatment (Ashrafi et al., 1999; Sinclair and Guarente, 1997; Tsang et al., 2003). This nucleolar reorganization is condensin-dependent and involves a physical compaction of the rDNA array (Tsang et al., 2007).
Evidence suggests that there are physiological differences between individual rDNA repeats despite their identical DNA sequence. Repeats coexist in two chromatin states, harboring either dormant 35S rRNA genes with stable nucleosome occupancy (Dammann et al., 1993) or actively transcribed 35S genes enriched for the HMG protein Hmo1 (Merz et al., 2008). Furthermore, rDNA repeats either fire their replication origins or get passively replicated (Pasero et al., 2002). In both cases, this physiological heterogeneity appears independent of repeat position (Dammann et al., 1995; Pasero et al., 2002). By contrast, position effects are apparent in some meiotic mutants, in which rDNA recombination is specifically elevated in the outer repeats of the rDNA (Vader et al., 2011). Here, we investigate the possibility that RNAPII silencing may similarly depend on position within the rDNA.
Results
To test whether transcriptional silencing differs between individual repeats of the rDNA array, we took advantage of an existing collection of isogenic rDNA insertion lines in S. cerevisiae (Vader et al., 2011). These lines were created by random pop-in recombination of a URA3 reporter construct into the IGS1/2 region of the rDNA repeats (Figure S1A). Individual insertions were mapped to repeats 1, 3, 10, 12, 29 and 49, respectively, allowing us to assay gene expression at defined points within the left half of the approximately 100 repeats of the rDNA array (Figure 1A). As the reporters are inserted at homologous positions within the different repeats, any differential behavior of the reporters must be due to their relative position within the array.
Figure 1. Reporter gene expression in nutrient-depleted cells depends on position within the rDNA array.
(A) Cartoon depicting the 6 isogenic reporter lines used in this study. Each line carries a URA3 reporter insertion at a homologous position within a different rDNA repeat. (B) Northern blot detecting repeat-specific URA3 expression in nutrient-depleted cells (for the corresponding log-phase cultures, see Figure S1B). Asterisks indicate rDNA-derived read-through transcripts observed in nutrient-depleted cells. (C) Repeat-specific URA3 expression in nutrient-depleted cells measured by qRT-PCR and normalized within strains to UBC6 before normalization to repeat 49. (D) ChIP-qPCR detecting Pol II (8WG16) occupancy in 6 repeat-specific URA3 reporters. (E) ChIP-qPCR detecting histone H4 occupancy at the junctions of the repeat-specific reporter constructs. Primer pair A amplifies the junction between the reporter sequence and IGS1, B amplifies the junction with IGS2. Error bars in (C–E) show standard deviation (s.d.) of 3 independent experiments, each analyzed in triplicate qPCRs. Significance was determined by a Welch two-sample t-test. For all tests: * = p<0.05, ** = p<0.01, *** = p<0.001, n.s. = not significant (F) Plate growth assays of the repeat-specific reporter lines. 3.5-fold serial dilutions were plated onto synthetic glycerol (left) and glucose (right) media supplemented with either uracil (complete) or lacking uracil (-ura). The “Non-rDNA” control strain has URA3 integrated on Chr. XI between YKL075C and YKL077W. See also Figure S1.
To probe for position-dependent gene expression within the rDNA, the six reporter lines were grown in parallel and analyzed by Northern blotting. Reporter gene expression was measured for logarithmically proliferating (log-phase) and saturated cultures (hereafter referred to as nutrient-depleted). Log-phase cells produced mostly read-through transcripts (likely due to transcription initiation by RNAPI from the adjacent 35S rRNA promoter; Figure S1A, B). By contrast, nutrient-depleted cells expressed a distinct transcript of the expected length of the URA3 reporter construct (Figure 1B). The transcript first became unambiguously detectable upon exit from log phase (Figure S1C, D) and was produced at higher levels from the outer repeats of the rDNA (repeats 1 and 3) than from more central repeats (29 and 49). Thus, under conditions of nutrient depletion, reporter gene expression within the rDNA is influenced by the relative position of the reporter within the array.
Several findings indicate that this position effect is due to differential activity of RNAPII. First, the URA3 transcripts can be enriched using an oligo-dT primer and are thus polyadenylated. Quantification of these polyadenylated transcripts by qRT-PCR revealed at clear position dependency (Figure 1C). Second, the position effect is largely abolished upon disruption of Sir2 (Figure S1E, F), which specifically silences RNAPII-dependent transcription (Smith and Boeke, 1997). In this experiment, however, loss of position dependency may partly result from the increased reporter instability caused by loss of Sir2 (Gottlieb and Esposito, 1989). To further test the role of RNAPII, we performed repeat-specific ChIP-qPCR of RNAPII in nutrient-depleted wild-type cells. RNAPII occupancy was higher for reporters in the border repeats than in the central repeats (Figure 1D), indicating that RNAPII activity in the rDNA array is under positional control. By contrast, there was no overt positional bias for RNAPII occupancy in log-phase cells (Figure S1G), despite up to 8-fold higher enrichment (Figure S1H). Notably, in nutrient-depleted cells, histone occupancy next to the reporter constructs was lowest in the repeats with the highest reporter gene activity (Figure 1E). The functional significance of this anti-correlation is currently unclear, but these data indicate that chromatin states may differ between individual rDNA repeats in nutrient-depleted cells.
URA3 reporter gene activity in the rDNA is typically measured by growth assays on medium lacking uracil (–ura), although this assay is complicated by the fact that position-dependent URA3 expression is restricted to nutrient depletion. Accordingly, when colony growth was analyzed on –ura plates containing glucose as a favorable carbon source, we observed little difference in growth between repeat-specific reporter strains (Figure 1F). When cells were spotted on –ura plates containing a non-fermentable carbon source (glycerol), a position-dependent difference in the rate of colony growth became apparent that correlated with the differences in URA3 expression observed by Northern and qRT-PCR (Figure 1B, C). Thus, position-dependent expression of reporter genes in the rDNA is linked to unfavorable nutrient conditions as well as nutrient depletion.
As nutrient stress causes a contraction of the nucleolus, we tested whether the appearance of the transcriptional position effect was linked to nucleolar reorganization. To establish the timing of nucleolar contraction, we followed relative nucleolar volume in a time course as cells exhausted their nutrients, using immunofluorescence (IF) of Nop1 to mark the nucleolus (Sinclair and Guarente, 1997) and a nuclear GFP construct to control for the reduced average nuclear volume of starved cultures (Jorgensen et al., 2007). Relative nucleolar volume abruptly decreased at the earliest stages of exit from log phase (Figure 2A–C), whereas further nucleolar shrinkage was more gradual. URA3 transcription became detectable shortly after the initial volume decrease (Figure S1D). A correlation with nucleolar contraction was also apparent when cells were treated with rapamycin to induce nucleolar shrinkage in rich medium (Tsang et al., 2003). URA3 expression from repeat 1 was observed by Northern blotting within one hour of rapamycin addition (Figure S2A). qRT-PCR analysis (Figure 2D) revealed higher expression levels of URA3 at the border repeats relative to repeat 49 upon rapamycin treatment, but not in pretreatment or mock-treated control cells. A similar effect was also observed upon acute shift to low glucose (Figure S2B). We conclude that multiple conditions triggering nucleolar contraction render rDNA reporter gene expression dependent on repeat position.
Figure 2. Nucleolar contraction upon nutrient depletion requires condensin and Hmo1.
(A) Cell density of wild-type cells growing in rich medium (YPD) over time. Cells were seeded at low density (8.5 × 105 cells/ml), OD600 = 0.2. Arrow indicates exit from log phase. (B–C) Relative nucleolar volume from the same time course. Samples were taken at the indicated time points, fixed, and stained for Nop1 and nuclear GFP (tetR-NLS-GFP) to determine nucleolar and nuclear volume, respectively. (B) Representative images of Nop1 (red) and nuclear GFP (green) staining. (C) Relative nucleolar volume was determined for 25 cells for each time point and was normalized to the 4-hr time point. (D) Repeat-specific URA3 expression before and after treatment with rapamycin or ethanol (mock) was measured by qPCR. Values are normalized against UBC6 and repeat 49 per treatment. Error bars indicate s.d. of 3 independent experiments, each analyzed in triplicate. (E) Representative IF images of cells stained for Nop1 (red) and DAPI (blue) to label the nucleolus and non-nucleolar chromatin. Scale bar = 1μm. (F) Relative nucleolar volume compared to nuclear GFP normalized to wild type log phase at 23°C; n=25 for each condition. (G) Representative IF images of hmo1Δ mutants grown at 30°C in synthetic complete medium. Cells were stained for Nop1 (red) and DAPI (blue). Arrow indicates DAPI-negative gap in hmo1Δ mutants. (H) Relative nucleolar volume as in (F); n=25. Nucleolar volume was normalized to wild type log phase. (I) Distance between the centroids of the nucleolus and the non-nucleolar chromatin in wild type and hmo1Δ mutants as depicted in the schematic (top); n=35. Significance tests: (D) Welch two-sample t-test; (C, F, H, I) Wilcoxon signed-rank test. See also Figure S2.
To probe a possible mechanistic link between the transcriptional position effect and nucleolar remodeling, we investigated the genetic requirements for nucleolar contraction upon log-phase exit. Consistent with previous findings (Tsang et al., 2007), nucleolar contraction was largely abolished in temperature-sensitive mutants of the condensin subunit Ycs4 (ycs4-2) after 2 hours at the restrictive temperature (37°C; Figure 2E, F). Notably, ycs4-2 mutants also exhibited a defect in nucleolar contraction at the permissive temperature (23°C). Such temperature independence is in line with previous observations showing that some nucleolar phenotypes of condensin mutants can be uncoupled from the temperature-sensitive growth defect (Haeusler et al., 2008; Machin et al., 2004).
A search for additional factors involved in starvation-induced nucleolar contraction uncovered a role for the HMG protein Hmo1, which binds to open rDNA chromatin and responds to nutrient signaling by TOR kinase (Berger et al., 2007; Merz et al., 2008; Xiao and Grove, 2009). Analysis of Nop1 staining revealed that the relative nucleolar volume of hmo1Δ mutants did not significantly change upon nutrient depletion (Figure 2G, H) and remained at log-phase levels even after 48 hours of nutrient depletion to account for possible effects from the slower growth of hmo1Δ mutants. Thus, like condensin, Hmo1 promotes nucleolar contraction in response to starvation.
Loss of Hmo1 also led to changes in nucleolar morphology not observed in the condensin mutant. In contrast to the crescent-shaped nucleolus observed in wild type and condensin mutants, hmo1Δ mutants exhibited globular or ring-like Nop1 staining (Figure 2G). Moreover, the Nop1 domain was often separated from the nuclear chromatin by a DAPI-negative gap (arrow, Figure 2G), which increased the distance between the centroid of the nuclear chromatin and the centroid of the nucleolus by approximately 50% in log-phase hmo1Δ mutants compared to wild type (Figure 2I). However, because this gap was also observed in nutrient-depleted hmo1Δ mutants (Figure 2I) and was not apparent in condensin mutants, it is unlikely to be related to nucleolar contraction.
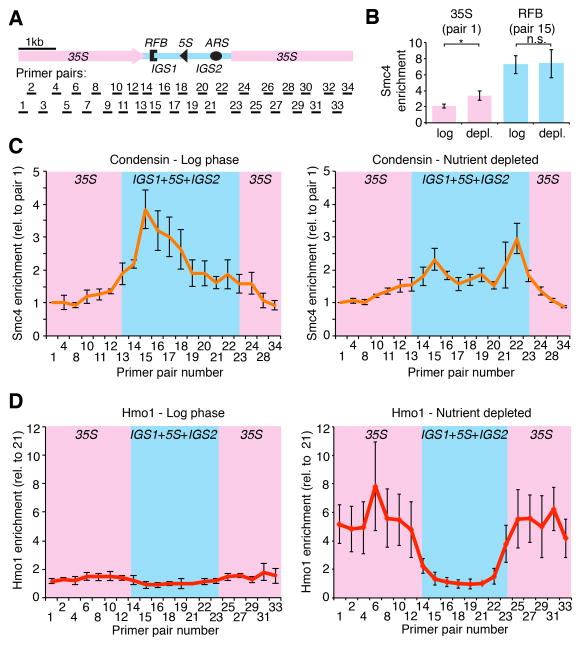
Given the requirement of condensin and Hmo1 for nucleolar contraction, we asked whether their binding patterns in the rDNA change following nutrient depletion. Condensin and Hmo1 are enriched on the rDNA, but localize to different parts within the repeats (Berger et al., 2007; Johzuka et al., 2006; Kasahara et al., 2007; Wang et al., 2004). We performed ChIP-qPCR for the condensin subunit Smc4 tagged with Pk9 in log-phase and nutrient-depleted cells using a previously published set of primers spanning an entire rDNA repeat (Figure 3A)(Huang and Moazed, 2003). Log-phase cells exhibited strong enrichment of Smc4 at the replication fork barrier (Figure 3B, C; RFB, primer pair 15), as observed previously for other condensin subunits (Johzuka et al., 2006). In nutrient-depleted cells, two changes were apparent. First, Smc4 binding was modestly increased over the 35S rRNA gene (Figure 3B), consistent with the increased binding of Ycs4-12xMYC observed upon TOR inhibition (Tsang et al., 2007). The degree of condensin enrichment seen here is substantially less dramatic, however, possibly due to experimental differences or as a result of the 12xMYC tag, which has been shown to alter Ycs4 functionality in starved cells (Yu and Koshland, 2003). Second, nutrient-depleted cells exhibited an additional strong peak of Smc4 enrichment near the origin of replication (Figure 3C; ARS, primer pair 22). These data indicate that nucleolar compaction is associated with a redistribution of condensin within the rDNA repeats.
Figure 3. Nutrient depletion causes a redistribution of condensin and Hmo1 within the rDNA.
(A) Schematic indicating the set of primers spanning an entire rDNA repeat (Huang and Moazed, 2003) used for the ChIP-qPCR experiments in (B–D). (B) Smc4-Pk9 enrichment in the 35S rDNA (primer pair 1) and near the RFB (primer pair 15) in log-phase and nutrient-depleted cells at 30°C. Samples were normalized to CUP1. Error bars indicate s.d. of 3 independent experiments, analyzed on the same qPCR plate. Significance: Welch two-sample t-test. (C) Smc4-Pk9 enrichment across an rDNA repeat in log-phase cells (left panel) and nutrient-depleted cells (right panel). Signals were normalized to a lowly enriched position (primer pair 1) for easier visualization of fold differences. (D) Hmo1 enrichment across an rDNA repeat in log-phase and nutrient-depleted cells. Signals are normalized to primer pair 21. Error bars in (C) and (D) indicate s.d. of 3 independent experiments, each analyzed in duplicate. See also Figure S3.
ChIP-qPCR analysis also revealed altered rDNA binding of Hmo1 in nutrient-depleted cells. Previous studies found Hmo1 enriched across the 35S rRNA gene (Berger et al., 2007; Kasahara et al., 2007). Under our conditions, this enrichment pattern was barely detectable in log-phase cells (Figures 3D and S3). However, in nutrient-depleted cells, Hmo1 binding across the 35S rRNA gene was strongly increased, leading to an overall 6-fold enrichment compared to other rDNA regions (IGS1+5S+IGS2; Figure 3D). Thus, the association patterns of both condensin and Hmo1 change upon nutrient depletion.
Despite their concurrent redistribution, neither factor influences the binding pattern of the other. Disruption of HMO1 does not alter Smc4 enrichment within IGS1 and IGS2 in log phase or nutrient-depleted cells (Figure S3A), and Hmo1 distribution is similarly unaffected in condensin (ycs4-2) mutants (Figure S3B–D). These data indicate that starvation-induced redistribution of condensin and Hmo1 occurs by independent mechanisms. Moreover, as loss of either factor prevents nucleolar contraction, nucleolar contraction occurs mechanistically downstream of the starvation-induced redistribution of condensin and Hmo1.
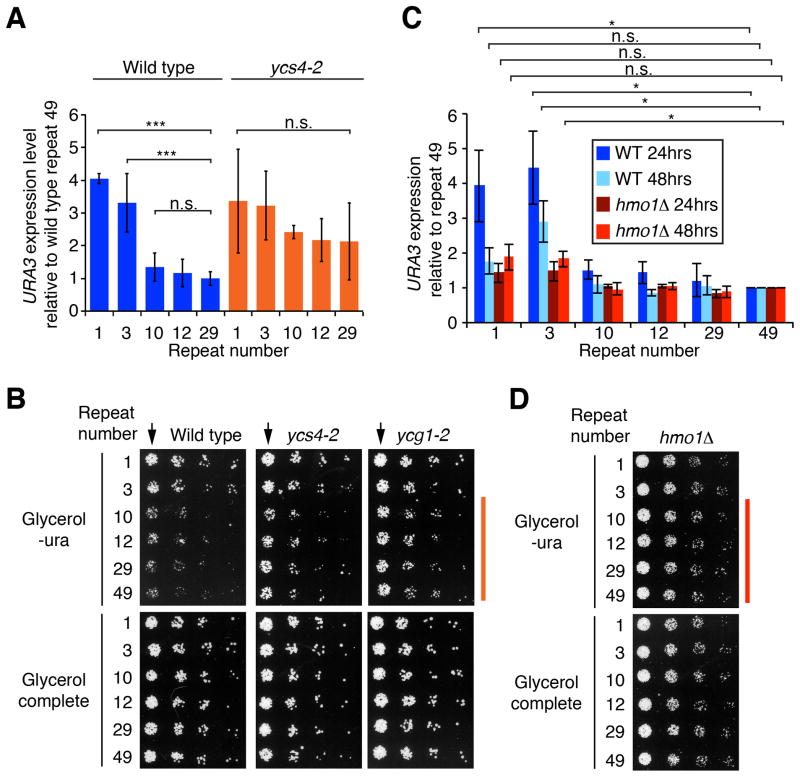
If nucleolar contraction is linked to position-dependent reporter gene expression in the rDNA, then this position effect should be reduced in the absence of condensin or Hmo1 function. Therefore, we introduced condensin and hmo1 mutations into the repeat-specific URA3 reporter lines. Reporter gene positions were in all cases confirmed by pulsed-field gel analysis and were found to be stable over the time frame of the experiment (e.g. Figure S4A). qRT-PCR analysis of nutrient-depleted cells revealed that the rDNA repeats in ycs4-2 mutants exhibited less profound positional differences in gene expression compared to wild type (Figure 4A), a trend that was also apparent from colony growth (Figure 4B). Similar results were obtained for another temperature-sensitive condensin mutant (ycg1-2, Figures 4B and S4B). Thus, condensin is at least partly required for position-dependent reporter gene regulation in the rDNA. In both cases, the position effect was lost at the permissive temperature (23°C). Therefore, this defect, like that of ycs4-2 mutants in nucleolar contraction, is largely temperature-independent.
Figure 4. Condensin and Hmo1 mediate the transcriptional position effect in the rDNA array.
(A) Repeat-specific URA3 expression was measured in nutrient-depleted ycs4-2 mutants at 23°C by qRT-PCR. Signals were normalized to UBC6 and repeat 29. Error bars indicate s.d. of 3 independent experiments, each analyzed in triplicate. (B) Plate growth assays of wild-type, ycs4-2 and ycg1-2 cells carrying repeat-specific reporter cassettes. 3.5-fold serial dilutions were plated onto synthetic glycerol medium supplemented with uracil (complete) or lacking uracil (-ura) and grown at 23°C. Arrows highlight the dilutions with the most obvious position-dependent differences in colony size. (C) Repeat-specific URA3 gene expression was determined in nutrient-depleted hmo1Δ cells by qRT-PCR. Values are normalized against UBC6 and repeat 49 for each condition. Because of the slow growth of hmo1Δ mutants, cells were harvested after 24 and 48 hrs of nutrient depletion. 48 hrs of nutrient depletion in hmo1Δ mutants roughly corresponds to 24 hrs of nutrient depletion in wild type, as determined by growth curves. Error bars as in (A). Significance in (A) and (C): Welch two-sample t-test. (D) Plate growth assays of hmo1Δ mutants carrying repeat-specific reporter cassettes. Analysis as in (B) but cells were grown at 30°C. See also Figure S4.
The position effect was also strongly reduced in the absence of Hmo1. qRT-PCR analysis revealed only small positional differences in repeat-specific reporter activity in hmo1Δ mutants after both 24 and 48 hours of nutrient depletion (Figure 4C). The same trend was apparent from colony growth (Figure 4D). Therefore, Hmo1 is required for efficient nucleolar contraction and establishment of the position effect. Unexpectedly, the position effect was also reduced in wild type after 48 hours of nutrient depletion (Figure 4C), despite a persistently contracted nucleolus (Figure 2E). Thus, although nucleolar contraction is linked to the establishment of the transcriptional position effect within the rDNA, nucleolar contraction alone is not sufficient to maintain the position effect over extended periods of nutrient depletion.
Discussion
Our results reveal unexpected positional differences in gene silencing within the rDNA of starved cells. Such position-dependent desilencing may result from a more permissive chromatin state spreading into the rDNA array upon nutrient depletion. It may also stem from increased exposure of the boundary-proximal repeats to a non-nucleolar environment that promotes gene activity or reduces silencing. Chromatin spreading may explain the repeat-dependent difference in histone occupancy (Figure 1E), although we note that we have so far failed to observe any difference in occupancy between the different reporter genes using ChIP analysis of Sir2, condensin or Hmo1 enrichment (Figure S1I and data not shown). Conversely, increased residence outside the nucleolus would be akin to the previously described exit of rDNA double-strand breaks from the nucleolus during DNA repair (Torres-Rosell et al., 2007). Indeed, the genome of starved yeast cells undergoes substantial reorganization (Rutledge et al., 2015), which may drive changes in gene activity. The possibility of spatial regulation is currently being tested using fluorescently labeled repeats as well as repeat-specific chromosome conformation capture.
We note that position-dependent desilencing may provide a solution for activating rDNA-encoded genes without causing the severe array instability associated with a complete loss of silencing (Mekhail and Moazed, 2010). The rDNA of many organisms, including yeast and humans, encodes RNAPII-expressed transcripts that must be expressed at certain stages without causing general instability (Coelho et al., 2002; Kobayashi and Ganley, 2005; McStay and Grummt, 2008). In yeast, the TAR1 gene, encoded in the anti-sense direction of the 35S gene, is of particular interest because it is specifically expressed upon nutrient depletion (Bonawitz et al., 2008; Galopier and Hermann-Le Denmat, 2011), coincident with positional desilencing. Strikingly, nutrient-dependent regulation of TAR1 is lost when the gene is analyzed on a plasmid (Galopier and Hermann-Le Denmat, 2011), suggesting that rDNA structure contributes regulatory control.
Our work adds to a growing body of evidence that position effects are inherent to the structural organization of repetitive DNA arrays. Previous analyses showed a position effect for meiotic recombination in the rDNA in specific mutants (Vader et al., 2011). We now demonstrate that position effects also affect reporter gene expression and occur in wild-type cells. An accompanying study by He et al. also uncovered transcriptional position effects in the repetitive pericentromeric regions of Schizosaccharomyces pombe. Thus, position effects are not limited to the rDNA and occur in evolutionarily highly diverged organisms. Tandem repetitive DNA arrays occupy a significant fraction of the most eukaryotic genomes and play crucial roles in regulating gene expression, chromosome organization and disease (Neguembor and Gabellini, 2010; Richard et al., 2008). We predict that higher-order organization and internal position effects represent an important mechanism of epigenetic regulation for the repetitive genome.
Experimental procedures
Strains and growth conditions
Strains used in this study are of the SK1 background and are listed in Table S1. Stability of the reporter positions was reconfirmed after all genetic manipulations. Details on strain construction and culture conditions are provided in the Supplemental Materials and Methods.
Estimation of relative nucleolar volume
Nucleolar and nuclear volumes were estimated from deconvolved z-stacks of fixed cells stained for Nop1 and nuclear GFP (tetR-NLS-GFP) using SoftWoRX2.50 software (Applied Precision).
Other procedures
Conditions for IF, ChIP, Northern blotting, reverse transcription and qPCR are outlined in the Supplemental Materials and Methods and Table S2.
Supplementary Material
Acknowledgments
We thank S. Ercan, F. Li, G. Vader, H. Blitzblau and H. Klein for discussions and critical reading of the manuscript, and S. Bell for the Sir2 antibody. This work was supported in part by NIH grant GM088248 to A.H. The authors declare no competing interests.
Footnotes
Author contributions
D.W., A.M., and A.H. designed the study and acquired, analyzed and interpreted the data. G.P. acquired initial data. All authors contributed to drafting and revision of the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashrafi K, Sinclair D, Gordon JI, Guarente L. Passage through stationary phase advances replicative aging in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1999;96:9100–9105. doi: 10.1073/pnas.96.16.9100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger AB, Decourty L, Badis G, Nehrbass U, Jacquier A, Gadal O. Hmo1 is required for TOR-dependent regulation of ribosomal protein gene transcription. Mol Cell Biol. 2007;27:8015–8026. doi: 10.1128/MCB.01102-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz ND, Chatenay-Lapointe M, Wearn CM, Shadel GS. Expression of the rDNA-encoded mitochondrial protein Tar1p is stringently controlled and responds differentially to mitochondrial respiratory demand and dysfunction. Curr Genet. 2008;54:83–94. doi: 10.1007/s00294-008-0203-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho PS, Bryan AC, Kumar A, Shadel GS, Snyder M. A novel mitochondrial protein, Tar1p, is encoded on the antisense strand of the nuclear 25S rDNA. Genes Dev. 2002;16:2755–2760. doi: 10.1101/gad.1035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M, Schothorst J, Kankipati HN, Van Zeebroeck G, Rubio-Texeira M, Thevelein JM. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev. 2014;38:254–299. doi: 10.1111/1574-6976.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann R, Lucchini R, Koller T, Sogo JM. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:2331–2338. doi: 10.1093/nar/21.10.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann R, Lucchini R, Koller T, Sogo JM. Transcription in the yeast rRNA gene locus: distribution of the active gene copies and chromatin structure of their flanking regulatory sequences. Mol Cell Biol. 1995;15:5294–5303. doi: 10.1128/mcb.15.10.5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galopier A, Hermann-Le Denmat S. Mitochondria of the yeasts Saccharomyces cerevisiae and Kluyveromyces lactis contain nuclear rDNA-encoded proteins. PLoS One. 2011;6:e16325. doi: 10.1371/journal.pone.0016325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb S, Esposito RE. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- Ha CW, Huh WK. Rapamycin increases rDNA stability by enhancing association of Sir2 with rDNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2011;39:1336–1350. doi: 10.1093/nar/gkq895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev. 2008;22:2204–2214. doi: 10.1101/gad.1675908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Moazed D. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev. 2003;17:2162–2176. doi: 10.1101/gad.1108403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. Transcriptional silencing in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Nucleic Acids Res. 2002;30:1465–1482. doi: 10.1093/nar/30.7.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johzuka K, Terasawa M, Ogawa H, Ogawa T, Horiuchi T. Condensin loaded onto the replication fork barrier site in the rRNA gene repeats during S phase in a FOB1-dependent fashion to prevent contraction of a long repetitive array in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:2226–2236. doi: 10.1128/MCB.26.6.2226-2236.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P, Edgington NP, Schneider BL, Rupes I, Tyers M, Futcher B. The size of the nucleus increases as yeast cells grow. Mol Biol Cell. 2007;18:3523–3532. doi: 10.1091/mbc.E06-10-0973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara K, Ohtsuki K, Ki S, Aoyama K, Takahashi H, Kobayashi T, Shirahige K, Kokubo T. Assembly of regulatory factors on rRNA and ribosomal protein genes in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:6686–6705. doi: 10.1128/MCB.00876-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klosinska MM, Crutchfield CA, Bradley PH, Rabinowitz JD, Broach JR. Yeast cells can access distinct quiescent states. Genes Dev. 2011;25:336–349. doi: 10.1101/gad.2011311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Ganley AR. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science. 2005;309:1581–1584. doi: 10.1126/science.1116102. [DOI] [PubMed] [Google Scholar]
- Kueng S, Oppikofer M, Gasser SM. SIR proteins and the assembly of silent chromatin in budding yeast. Annu Rev Genet. 2013;47:275–306. doi: 10.1146/annurev-genet-021313-173730. [DOI] [PubMed] [Google Scholar]
- Loewith R, Hall MN. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics. 2011;189:1177–1201. doi: 10.1534/genetics.111.133363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machin F, Paschos K, Jarmuz A, Torres-Rosell J, Pade C, Aragon L. Condensin regulates rDNA silencing by modulating nucleolar Sir2p. Curr Biol. 2004;14:125–130. [PubMed] [Google Scholar]
- McStay B, Grummt I. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu Rev Cell Dev Biol. 2008;24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- Mekhail K, Moazed D. The nuclear envelope in genome organization, expression and stability. Nat Rev Mol Cell Biol. 2010;11:317–328. doi: 10.1038/nrm2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz K, Hondele M, Goetze H, Gmelch K, Stoeckl U, Griesenbeck J. Actively transcribed rRNA genes in S. cerevisiae are organized in a specialized chromatin associated with the high-mobility group protein Hmo1 and are largely devoid of histone molecules. Genes Dev. 2008;22:1190–1204. doi: 10.1101/gad.466908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neguembor MV, Gabellini D. In junk we trust: repetitive DNA, epigenetics and facioscapulohumeral muscular dystrophy. Epigenomics. 2010;2:271–287. doi: 10.2217/epi.10.8. [DOI] [PubMed] [Google Scholar]
- Pasero P, Bensimon A, Schwob E. Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes Dev. 2002;16:2479–2484. doi: 10.1101/gad.232902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard GF, Kerrest A, Dujon B. Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol Mol Biol Rev. 2008;72:686–727. doi: 10.1128/MMBR.00011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge MT, Russo M, Belton JM, Dekker J, Broach JR. The yeast genome undergoes significant topological reorganization in quiescence. Nucleic Acids Res. 2015;43:8299–8313. doi: 10.1093/nar/gkv723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu HY, Ahn S. Yeast histone H3 lysine 4 demethylase Jhd2 regulates mitotic rDNA condensation. BMC Biol. 2014;12:75. doi: 10.1186/s12915-014-0075-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- Smith JS, Boeke JD. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- Torres-Rosell J, Sunjevaric I, De Piccoli G, Sacher M, Eckert-Boulet N, Reid R, Jentsch S, Rothstein R, Aragon L, Lisby M. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat Cell Biol. 2007;9:923–931. doi: 10.1038/ncb1619. [DOI] [PubMed] [Google Scholar]
- Tsang CK, Bertram PG, Ai W, Drenan R, Zheng XF. Chromatin-mediated regulation of nucleolar structure and RNA Pol I localization by TOR. EMBO J. 2003;22:6045–6056. doi: 10.1093/emboj/cdg578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang CK, Li H, Zheng XS. Nutrient starvation promotes condensin loading to maintain rDNA stability. EMBO J. 2007;26:448–458. doi: 10.1038/sj.emboj.7601488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vader G, Blitzblau HG, Tame MA, Falk JE, Curtin L, Hochwagen A. Protection of repetitive DNA borders from self-induced meiotic instability. Nature. 2011;477:115–119. doi: 10.1038/nature10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BD, Yong-Gonzalez V, Strunnikov AV. Cdc14p/FEAR pathway controls segregation of nucleolus in S. cerevisiae by facilitating condensin targeting to rDNA chromatin in anaphase. Cell Cycle. 2004;3:960–967. doi: 10.4161/cc.3.7.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Grove A. Coordination of Ribosomal Protein and Ribosomal RNA Gene Expression in Response to TOR Signaling. Curr Genomics. 2009;10:198–205. doi: 10.2174/138920209788185261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HG, Koshland DE. Meiotic condensin is required for proper chromosome compaction, SC assembly, and resolution of recombination-dependent chromosome linkages. J Cell Biol. 2003;163:937–947. doi: 10.1083/jcb.200308027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.