Abstract
Diverse changes in coloration across distant taxa are mediated through alterations in certain highly conserved pigmentation genes. Among these genes, Mc1r is a frequent target for mutation, and many documented alterations involve coding sequence changes. We investigated whether regulatory mutations in Mc1r may also contribute to pigmentation loss in the blind Mexican cavefish, Astyanax mexicanus. This species comprises multiple independent cave populations that have evolved reduced (or absent) melanic pigmentation as a consequence of living in darkness for millions of generations. Among the most salient cave-associated traits, complete absence (albinism) or reduced levels of pigmentation (brown), have long been the focus of degenerative pigmentation research in Astyanax. These two Mendelian traits have been linked to specific coding mutations in Oca2 (albinism) and Mc1r (brown). However, four of the seven caves harboring the brown phenotype exhibit unaffected coding sequences compared to surface fish. Thus, diverse genetic changes involving the same genes likely impact reduced pigmentation among cavefish populations. Using both sequence and expression analyses, we show that certain cave-dwelling populations harboring the brown mutation have substantial alterations to the putative Mc1r cis-regulatory region. Several of these sequence mutations in the Mc1r 5′ region were present across multiple, independent cave populations. This study suggests that pigmentation reduction in Astyanax cavefish evolves through a combination of both coding and cis-regulatory mutations. Moreover, this study represents one of the first attempts to identify regulatory alterations linked to regressive changes in cave-dwelling populations of Astyanax mexicanus.
Keywords: Mc1r, regressive evolution, Astyanax, cavefish, pigmentation, cis-regulatory
Introduction
Natural variants of the gene Mc1r explain several variable pigmentation phenotypes in natural populations. These phenotypes are often caused by coding sequence alterations, such as the extension (e) locus in mouse models. Recessive Mc1r extension (e) alleles produce a yellow phaeomelanin phenotype that impacts plumage in birds, and hair color variation in humans. At present, more than 60 coding sequence mutations have been identified in Mc1r [reviewed in García-Borrón et al. 2005].
Mc1r sequence alterations with functional impact often result from base pair deletions in the coding sequence, or amino acid substitutions [reviewed in Hoekstra 2006]. Fewer studies have identified a role for regulatory mutations affecting pigmentation through altered Mc1r expression. However, cis-regulatory alterations affecting pigmentation gene expression patterns have been identified in other systems. For instance, Drosophila wing spot coloration is influenced by cis-regulatory mutations impacting the yellow gene [Jeong et al. 2006]. Peromyscus deer mice demonstrating melanism (dark pigmentation) harbor a 125-kb deletion in the Agouti gene (antagonist of Mc1r) inclusive of a putative regulatory region, which reduces Agouti expression [Kingsley 2009]. In stickleback fish, lighter pigmentation in the gills and ventrum of freshwater populations are caused by cis-regulatory changes affecting Kitl expression [Miller et al. 2007].
Cave animals are excellent models for investigating changes in coloration due to the recurrent loss of pigmentation in organisms living amidst the dark, subterranean environment. Among cave-dwelling models, the Mexican tetra Astyanax mexicanus is particularly powerful since closely related surface-dwelling fish are available, enabling direct comparisons between a surrogate ‘ancestral’ form and the 29 derived cave-dwelling populations. The genetic basis for two key pigmentation phenotypes have been described in multiple independent populations, including absence of melanin (albinism; Oca2) [Protas et al. 2006] and reduction in melanin content (brown; Mc1r) [Gross et al. 2009].
For both albinism and brown, diverse loss-of-function or hypomorphic mutations have been characterized. For example, a 2-bp deletion (Δ23, 24) in the Mc1r open reading frame of Pachón cavefish causes a frame-shift leading to a premature stop codon. A second (hypomorphic) mutation (C490T), identified in individuals from the Yerbaniz and Japonés cavefish populations, causes a cysteine substitution at position R164C homologous to the R160W mutant in humans [Gross et al. 2009]. Although only three caves demonstrate coding sequence alterations in Mc1r, complementation tests suggested that the same gene might be responsible for brown in seven distinct cave populations (Table 1). However, in four of the populations harboring brown, the Mc1r coding sequence is identical to surface fish.
Table 1.
Distribution of the brown phenotype among Astyanax cave populations
| Cave locality | Method of identification | Genetic alteration | References |
|---|---|---|---|
|
Brown affected populations
| |||
| Chica | Observed in the wild | Unknown | Şadoğlu and McKee 1969 |
| Sabinos | Observed in the wild | Unknown |
Şadoğlu and McKee 1969 Şadoğlu and McKee 1969 |
| Observed in the wild | Wilkens and Strecker 2003 | ||
| Pachón | Complementation tests | Δ23/24 | Gross et al. 2009 |
| Curva | Complementation tests | Unknown | Wilkens and Strecker 2003 |
| Piedras | Complementation tests |
Unknown C490T |
Wilkens and Strecker 2003 Wilkens and Strecker 2003 |
| Yerbaníz (Japonés) | Complementation tests | Δ23/24 | Gross et al. 2009 |
|
| |||
|
Unaffected populations (confirmed absent)
| |||
| Molino | Complementation tests | None | Wilkens and Strecker 2003 |
|
| |||
|
Presence of brown unknown
| |||
| Tinaja | Unknown | Unknown | -- |
In this study, we investigated possible involvement of cis-regulatory mutations leading to the brown phenotype in three cavefish populations compared to the (normally-pigmented) surface-dwelling form. We evaluated the upstream genomic sequence of Mc1r in representative individuals drawn from ten Astyanax populations. Indeed, when characterizing sequence variation in the 5′ Mc1r region, we found a surprisingly high level of sequence diversity across cave-dwelling populations, with several instances of sequence variation within highly conserved non-coding elements. Quantitative gene expression analyses in adult fish revealed substantial changes in Mc1r expression differing between surface-dwelling fish and three cave populations – Pachón, Tinaja and Chica. There are no sequence differences in the Mc1r coding region of Tinaja or Chica compared to surface-dwelling fish based on a prior analysis in which the Mc1r open reading frame was sequenced and compared across thirteen populations [Gross et al. 2009]. Previous functional analyses of Mc1r, in which mRNA transcripts were abrogated using morpholino knock-down approaches, demonstrated that reduced levels of Mc1r recapitulate the brownish eyes and reduced melanic content found in the classic brown phenotype [Gross et al. 2009]. Thus, the functional role of Mc1r, combined with the divergent expression patterns we report here, may explain the parallel evolution of reduced pigmentation in wild cavefish. Further, this study indicates that pigmentation reduction in geographically diverse cavefish populations may occur through both coding and regulatory mutations impacting the same gene.
Materials and Methods
Sequence analysis of the 5′ Mc1r region
Genomic DNA was isolated from fin clips derived from adult surface and nine cave populations including: Pachón, Molino, Japonés, Yerbaníz, Sabinos, Piedras, Tinaja, Curva, and Chica (Qiagen; Valencia, CA; n= 1 fin clip per locality). All cavefish used in this study were derived from wild-caught specimens collected and generously provided by Dr. Richard Borowsky (New York University). Lab-reared surface fish used in this study were derived from specimens collected in the wild at Arroyo Sarco (at the Río Sabinos drainage, Mexico) and the Río Valles drainage. Chica cavefish used in qPCR studies were obtained from a commercial supplier (Quality Marine; Los Angeles, CA). Roughly 2500 bp of the upstream genomic regions of Mc1r were isolated using the Universal GenomeWalker Kit (Clontech; Mountain View, CA) and amplified using overlapping primer sets in ~500 bp segments (forward 1: TAGAATACTCAAGCTATGCATCCAA, reverse 1: ACGCATTTAGTTTTATATTAAGTTCCA; forward 2: TGTAAACCTTAAATAAACCATCACAC, reverse 2: GTCAACCAGCACAGTCAAGC; forward 3: CATTTTTGTTAAATTAAGCAGTTTGA, reverse 3: AAGTGGCTTAGATTTAGGTTTACATTT; forward 4: TGCATTGATCTTTTTGAGTTGC, reverse 4: CAGACAAAGGTTCTACACACCAA; forward 5: AAAAATAGCACCACTGTTTAGTCAAA, reverse 5: ACTGGAACCTCTTTGGCTCT; forward 6: GTGCGGAACCATGATCAGTA, reverse 6: CCCACTGCTCCTGGACTTT) with Taq DNA polymerase (0.125 μl per 25 μl reaction, Roche; Indianapolis, IN) using PCR cycling parameters as follows: 94 °C for 2:00, then cycled to 94 °C for 0:30 seconds, 56 °C for 1:00, 72 °C for 2:30, and repeated for 34 additional cycles; then 72 °C for 10:00 and then 4 °C). PCR amplicons were subcloned into the pGEM-T Easy vector (Promega; Madison, WI) and sequenced (Operon; Huntsville, AL). Sequences were aligned using SeqMan Pro (DNASTAR.v.11.0; Madison, WI) and analyzed with MegAlign (DNASTAR.v.11.0; Madison, WI) using the Martinez-Needleman-Wunsch method for closely-related sequences.
Prediction of conserved non-coding elements
Consensus sequences were evaluated for conserved non-coding elements using the mVISTA online tool for comparative genomics (http://genome.lbl.gov/vista) [Frazer et al. 2004] using the MLAGAN algorithm. Reference genomic sequence and annotation data from Danio rerio for Mc1r was retrieved using Ensembl (Zv9; www.ensembl.org). Program parameters were adapted from similar VISTA studies [Frazer et al. 2004]. The regulatory VISTA (rVISTA) software program predicted regulatory element motifs for TFE, TFEB, Mitf and E-box transcription factors, based on TRANSFAC Professional (v.9.2; 2005). We selected these specific transcription factors for our screen because they have previously been shown to interact with the putative promoter region of Mc1r [Moro et al. 1999; Miccadei et al. 2008].
RNA-seq analyses
RNA was isolated from pools of surface or Pachón cave embryos (n=50 for each morphotype) at 10 hours post-fertilization (hpf), 24 hpf, 36 hpf, and 72 hpf with the RNeasy Kit (Qiagen; Valencia, CA). Total RNA from whole juveniles was extracted and pooled from three individuals at four months of age. DNase treatment was performed using genomic DNA ‘eliminator’ filter columns provided in the RNeasy kit (Qiagen; Valencia, CA). RNA quality was assessed via spectrophotometric analysis using a Nanovue Plus instrument (GE Healthcare Life Sciences; Pittsburgh, PA). A260/A280 absorbance ratio measurements yielded values of ~2.0 – 2.1 for all RNA pools, indicating the RNA extracts were pure (uncontaminated) samples. RNA-sequencing was performed in triplicate for 10 hpf – 72 hpf embryonic stages and in duplicate for juveniles using Illumina HiSeq 2500 Technology (TruSeq v.2 kit) at the Cincinnati Children’s Hospital Core Sequencing Facility (Cincinnati, OH). Sequencing reads (from fastq-formatted files) were aligned to a previously published comprehensive transcriptome template [Gross et al. 2013] inclusive of the Mc1r sequence. Gene expression levels were calculated using the QSeq module of the ArrayStar software program (DNAStar v.11.0; Madison, WI) using an RPKM normalization strategy [Mortazavi et al. 2008; Gross et al. 2013]. Our transcriptome template was generated using SeqMan NGen software (DNAStar v.11.0; Madison, WI) optimized for use with our 50-bp, unpaired read sets. We utilized the RPKM method to evaluate differential expression, which has been previously performed using the same software programs employed in this report [Leyva-Pérez et al. 2014; Guaiquil et al. 2014; Youngblood et al. 2014]. Expression differences were subsequently evaluated using quantitative PCR (see below). We tested for significant differences between surface and cavefish samples using a Student’s t-test with FDR (Benjamini Hochberg; DNASTAR.v.11.0). Sequencing reads are accessioned to the NCBI SRA (BioProject ID: PRJNA258661).
Quantitative PCR expression analyses
RNA pools were generated from four populations (surface fish, Pachón cavefish, Tinaja cavefish, and Chica cavefish) from adult fin clips (n=1 per locality) using the RNeasy Kit (includes the genomic column eliminator DNase treatment; described above). Cavefish from Pachón and Chica harbor the brown mutation based on direct observations, classical genetic crosses, and complementation studies [Şadoğlu and McKee 1969; Wilkens and Strecker, 2003]. We obtained Chica cavefish from a commercial supplier (Quality Marine; Los Angeles, CA), similarly described in prior studies [Espinasa and Jeffery 2006]. Although Tinaja cavefish have not been formally reported to harbor brown in the literature, they display the phenotypic characteristics of brown based on direct observation (reduced eumelanin content). Further, the Tinaja cave entrance is ~2.5 km from the Sabinos cave which does harbor brown, and prior reports suggest migration has occurred between these populations [Bradic et al. 2012]. Thus, Tinaja was used for this study because lab-reared specimens are available, this population breeds consistently, and is derived from the older “El Abra” lineage which is closely related to several reported brown populations [Şadoğlu and McKee 1969].
cDNA pools were synthesized using the Transcriptor RT kit (Invitrogen; Waltham, MA; Roche; Indianapolis, IN). Briefly, 1 μg of total RNA was hybridized to Oligo dT primers (Invitrogen) at 65°C for 10 min, then cooled on ice for ~5 min. We then added 4.0 μl of 5x RT buffer, 0.5 μl of Protector RNAse Inhibitor, 2.0 μl dNTP mixture, 0.5 μl of Transcriptor RT, and incubated for 50°C for 1 hr. Following incubation, the cDNA pool was inactivated via incubation at 85°C for 5 min, and used immediately for qPCR assays or stored at −20°C. qPCR primers were designed to amplify ~100 bp fragment for detecting Mc1r (forward: CACGTCCAGCTCACTCTTCA; reverse: TAGAGCCCGGCAGTGAATAC). Fragments were amplified using EvaGreen super mix (Bio-Rad; Hercules, CA) with the following cycling parameters: step 1—95°C for 30 s, step 2—95°C for 5 s, step 3—55.1°C for 10 s, plate read, repeated for 39 additional cycles. Standard curves were generated to check amplification efficiency, and qPCR efficiency was calculated to be 100% for all samples analyzed in this study (reported by CFX Manager Software v.3.1). A melt curve analysis was performed on all qPCR amplicons (60°C to 95°C, in 0.5°C increments). All melt curve analyses yielded a single peak for both genes analyzed in this study, Mc1r and Glyceraldehyde 3-phosphate dehydrogenase (GapDH). Normalized expression (ΔΔCq) was calculated from samples in sextuplet with CFX Manager Software (BioRad.v.3.1) using reference gene GAPDH (forward: TGTGTCCGTGGTGGATCTTA; reverse: TGTCGCCAATGAAGTCAGAG-3′), and surface fish samples served as an inter-run calibration across replicate experiments. Significant differences in Mc1r gene expression were calculated for each cavefish population compared to surface fish using a two-tailed Student’s t-test (CFX Manager Software, BioRad.v.3.1). All qPCR experiments were performed in accordance with MIQE guidelines [Bustin et al. 2009; Taylor et al. 2010].
Results and Discussion
5′ Mc1r sequence analyses in cavefish identify multiple genetic alterations that co-localize with highly conserved non-coding regions
Previous studies in Astyanax have identified specific coding sequence alterations leading to regressive pigmentation traits, but this study is among the first to describe the involvement of prospective cis-regulatory alterations in degenerative, cave-associated phenotypes. Here, we characterized the putative regulatory region of Mc1r (2510 bp) – a gene with well-documented physiological roles governing melanin dynamics in vertebrates. Mc1r is a single-exon gene; hence it does not harbor introns that could otherwise include regulatory elements. In other systems, the minimal Mc1r promoter lies immediately upstream of the translational start codon [Moro et al. 1999; Miccadei et al. 2008]. Moreover, the 5′-genomic region of Mc1r in humans harbors characteristics of G protein-coupled receptor (GPCR) promoters, including several GC-rich sequences, lack of a TATA box, and presence of numerous transcriptional start sites and E-box motifs [Moro et al. 1999]. Though the gene Mc1r mediates reduced pigmentation in brown individuals derived from the Pachón, Yerbaníz and Japonés Astyanax cave populations, little is known about how Mc1r governs reduced pigmentation the other cave lineages harboring the brown phenotype (Chica, Sabinos, Curva and Piedras caves). To explore this, we evaluated the 5′ region of Mc1r in nine cavefish populations – the seven caves known to express brown, one lineage that likely harbors brown (Tinaja) and one additional cave where brown is confirmed to be absent (Molino) [Wilkens and Strecker 2003].
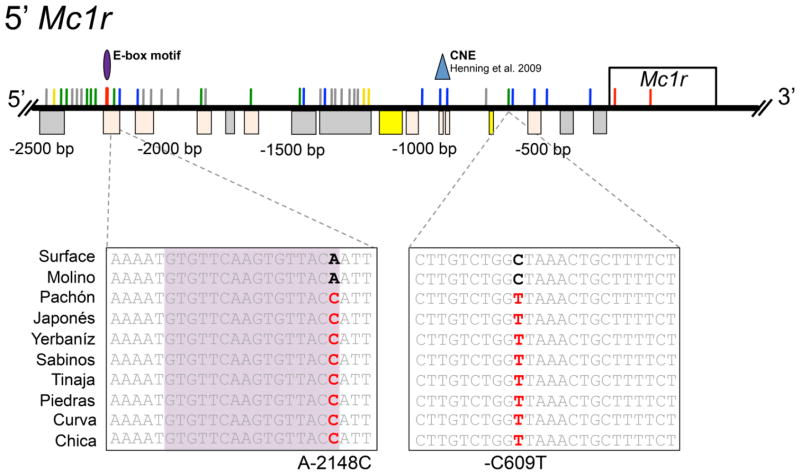
Comparisons of cavefish and epigean forms identified 42 unique mutations in the 5′ region upstream of Mc1r (Fig. 1; Table S1). Three of the genetic alterations we discovered were present in all eight caves including Molino, therefore these likely do not contribute to brown. Additionally, 22 mutations were only present in a single cavefish population and 6 mutations were present in two or more caves, but not across all of the “brown” caves. However, 11 mutations were fixed in all seven cavefish lineages harboring brown, excluding the Molino cave, rendering these alterations as intriguing candidates for future functional promoter studies (e.g., Fig. 1 A-2147C, C-609T).
Fig. 1. Multiple 5′ Mc1r genetic mutations in cavefish co-localize to highly conserved non-coding elements and one transcription factor binding motif.
Sequence analyses reveal the presence of numerous mutations throughout the 5′ region of Mc1r. Any sequence alterations shared with Molino cave likely serve no functional significance (gray). Some mutations identified are only present in a single cavefish lineage (blue) and others are randomly present in two or more of the caves assayed (yellow). However, some alterations are present in all eight caves (excludes Molino; green). Boxes below the line indicate the highly conserved CNEs between Danio and Astyanax sequences detected with the VISTA alignments (Gray- <55% conserved; Peach- >55%; Yellow- >75%). The highly conserved fish-specific CNE described in Henning et al. (2010) is marked with a blue triangle. Additionally, the E-box binding motif detected with rVISTA is represented by the purple oval, which co-localizes with the A-2148C mutation (red colored mutation) that is fixed in all caves except Molino.
Additional sequence analyses, including a pairwise alignment of sequences across representatives from nine caves revealed that the mean sequence identity shared between cave and surface morphs was 99.18% and the average sequence index, which includes a penalty for gaps/deletions, was 98.4% for the putative promoter region of Mc1r. Furthermore, distance comparisons indicated a similar level of sequence divergence across the nine cave lineages compared to surface in 5′Mc1r (mean= 0.49). These analyses indicated that, despite a moderate level of sequence variation across cave populations, the overall similarity of sequences between cave and surface fish was rather modest.
We then explored whether any of the mutations we identified may potentially impact the expression of Mc1r by altering highly conserved regions of non-coding DNA that harbor regulatory binding sites. To identify these putative regulatory regions, we first compared the upstream Mc1r promoter region derived from surface and nine cave-dwelling Astyanax populations to the same genomic interval in zebrafish (Danio rerio) using VISTA alignments. Recent studies document significant genomic similarity shared between Danio and Astyanax (both are members of the Ostariophysian superorder) despite ~150 My of divergence [Gross 2012]. We identified 16 highly conserved non-coding elements (CNEs) based on our comparisons between Danio and Astyanax for Mc1r (Fig. 1; gray, peach and yellow boxes). Interestingly, many of the 5′ Mc1r mutations not shared with Molino co-localized to these CNEs. For instance, six alterations were found uniquely in a single cavefish population. Three sequence mutations were present in two or more cave populations. Interestingly, five mutations were present in all eight cavefish lineages, except Molino (Table S1). Considering that some of these mutations reside in highly conserved non-coding regions, it is possible that distinct cavefish lineages are converging on the same brown phenotype via Mc1r, but as a consequence of mutations that are specific to each cave. This would be similar to albinism, wherein Pachón and Molino caves demonstrate different coding sequence mutations (exon deletions) in the gene Oca2 [Protas et al. 2006].
Next, we evaluated these conserved non-coding regions by identifying specific transcription factor binding sites within these regions. Cavefish-specific mutations that co-localize to these motifs could potentially hinder the binding efficiency, and ultimately, affect the expression of Mc1r. We focused our search for binding motifs based on prior characterizations of transcription factors found to interact with the regulatory region of Mc1r. In humans, the 5′ region upstream of Mc1r harbors several E-box motifs, and a band-shift assay demonstrated the binding of micropthalmia transcription factor (MITF), a melanocyte-specific regulator, to a 150 bp region upstream of the Mc1r start codon in pigment cells [Moro et al. 1999; Miccadei et al. 2008]. MITF is a basic helix-loop-helix leucine zipper (bHLH-zip) transcription factor with highly conserved N-terminal domains and common putative phosphorylation sites. Therefore, we searched for other MITF family members including TFE and TFEB. In sum, we focused our search for binding motifs to the E47 (binds E-boxes), MITF (binds M-boxes), TFE, and TFEB transcription factors.
An rVISTA analysis identified 8 putative binding motifs in the 5′Mc1r region, all of which were conserved across both surface and hypogean populations with the exception of one motif present in a region 5′ of Mc1r. Among the putative transcription factor sites discovered, one particular motif was mutated (nucleotide position -2148) in all brown cave populations, but this mutation was absent from both surface fish and the Molino cave population (which does not harbor brown) [Strecker and Wilkens 2003]. This polymorphism resides in a prospective E-box binding motif for the E47 transcription factor (Fig. 1; purple oval). The A-2148C mutation is not present in the core E47 motif (CAAGTG). However, recent evidence shows that the genomic sequence neighboring an E-box transcription factor motif can influence binding specificity of bHLH factors by impacting DNA molecular conformation [Yasumoto et al. 1994; Gordan et al. 2013]. It is possible that these binding motifs may have accumulated mutations in cavefish lineages, leading to a change in gene expression (and therefore the brown phenotype) in distinct subterranean populations. Sequence alignments between Astyanax and Danio demonstrate that the E-box site at position -2148 is present within a highly conserved non-coding element. This level of sequence similarity may indicate that this binding site is functional and serves a critical role in the regulation of Mc1r. To date, the Mc1r regulatory regions tested with gene deletion studies are limited to human-specific sequences which only tested functional binding of one prospective transcription factor (Mitf) [Moro et al. 1999; Miccadei et al. 2008]. Therefore, future functional analyses (e.g., promoter deletion constructs) will both identify the specific Mc1r minimal promoter region required to drive gene expression in teleosts, and determine whether the A-2148C is a causative regulatory lesion shared among brown cave populations.
Quantitative analyses reveal reduced Mc1r expression in later stages of development across independent populations cavefish
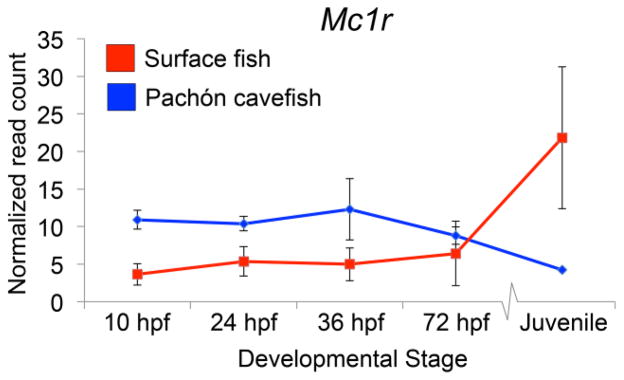
If sequence mutations in the putative Mc1r promoter confer reduced expression leading to loss of pigmentation in cave-dwelling forms of Astyanax, we would predict a reduction in transcriptional abundance of this gene. To examine this possibility, we first evaluated Mc1r gene expression during early development (pigmentation appears at ~24 hpf) using a next-generation sequencing approach comparing surface morphs and Pachón cavefish. Differential gene expression of Mc1r was evaluated in triplicate across four early developmental stages (10 hpf, 24 hpf, 36 hpf, 72 hpf) and juvenile (~4 months old; duplicates), encompassing both early and juvenile stages of pigmentation development in Astyanax.
Interestingly, during early stages of development (10 hpf to 72 hpf), Mc1r is expressed slightly higher in Pachón cavefish compared to surface fish. However, by the juvenile stage, Mc1r demonstrates a moderate increase (~5.155 fold) in surface fish, compared to cavefish (Fig. 2). Although we detected subtle differences in Mc1r gene expression, none of the early time points we tested showed statistically significant differences between surface and Pachón cavefish. This may be due to the fact that many pigmentation genes are expressed at extremely low levels in vivo.
Fig. 2. RNA-seq analyses demonstrate subtle differences in pigmentation gene expression between morphotypes in early development.
Surprisingly, degrees of differential expression observed were relatively subtle between surface and Pachón cave-dwelling forms over early development. Mc1r appears slightly higher in cavefish than surface until juvenile hood. Points represent the RPKM normalized mean, and bars indicate the standard error derived from RNA-sequencing replicates.
Since the Pachón Mc1r transcript harbors a destructive 2-bp deletion, an alternative explanation for its reduced transcriptional abundance may be through degradation via nonsense-mediated decay. This scenario could mean that upstream changes to the 5′-UTR may not have functional consequences in this cave population, but still be consistent with the qPCR results we report here. However, this mRNA deprivation could coincide with or – occur independently from – the upstream sequence alterations we discovered. Distinguishing between these two (not mutually exclusive) scenarios in Pachón cavefish cannot be easily answered with the approaches we employed in this study. However, it is interesting that two other (putatively) brown cave populations demonstrate reduced Mc1r expression despite having an intact Mc1r coding sequence.
Although RNA-seq studies indicated subtle differences between surface and Pachón cavefish, the polarity of Mc1r expression difference was consistent with our predictions at the juvenile stage. Therefore, we sought to further characterize Mc1r expression in later development (i.e., adulthood) in multiple brown cavefish lineages using qPCR. We quantified mRNA transcriptional abundance in surface fish and cavefish derived from three depigmented populations: Pachón, Tinaja, and Chica using quantitative PCR. Pachón and Chica cavefish exhibit brown, however this phenotype has not been formally evaluated in Tinaja cavefish [Gross et al. 2009]. Tinaja fish, however, exhibit dramatically reduced levels of melanic-based pigment [Protas et al. 2006] (Fig. 3A–D). Our results demonstrated that Mc1r expression was significantly reduced in all three cavefish populations compared to surface fish (Fig. 3E; p< 0.0001). Interestingly, Mc1r expression patterns were not identical across these independent troglomorphic localities. For example, the normalized expression in Tinaja and Chica cavefish was appreciably lower than the Mc1r levels detected in Pachón cavefish (Fig. 3E).
Fig. 3. qPCR analyses reveal reduced Mc1r expression present in independent populations of adult cavefish.
Varying levels of coloration are observed in Astyanax mexicanus. The surface-dwelling form is highly pigmented (black, A), compared to albino Pachón cavefish (blue, B). A severe reduction in pigmentation is also observed in both Tinaja (orange, C) and Chica cavefish (yellow, D). Normalized Mc1r gene expression levels for Pachón, Tinaja and Chica cave populations were significantly lower compared to the (ancestral) surface morphotype (p< 0.0001; E).
Shedding light on the evolutionary and genetic mechanisms leading to pigmentation loss in the subterranean environment
Degenerative loss of pigmentation consistently recurs in animals that invade the cave environment. However, the evolutionary mechanism(s) leading to regressive loss has long perplexed cave biologists, including Charles Darwin, who attributed eye loss in cavefish to “disuse”. The regression of characters in cave animals may evolve through selection (direct or indirect) or neutral mutation (through genetic drift). The precise benefit(s) of pigmentation loss in subterranean animals is poorly understood, however some other cave-related traits, namely eye loss, may be evolving through indirect selection through the expansion of non-visual sensory systems [Reviewed in Gross 2012]. This concept is highly dependent upon whether the character is ecologically relevant. In cave-dwelling Astyanax populations, melanic-based pigment translocation (for background adaptation) is useless in the dark, subterranean environment. Thus, the absence of light may permit genes related to pigmentation to accumulate mutations following colonization of the cave habitat. A combination of previous QTL studies, and the presence of the Oca2 loss-of-function allele in a non-albino cave population indicate that pigmentation loss in cave morphs may be evolving as a consequence of neutral forces [Gross and Wilkens 2013].
Alternatively, it may be that genes contributing to pigmentation may have pleiotropic consequences that are adaptive in cavefish. For example, a recent study showed experimentally reduced levels of Oca2 in cavefish (using morpholino knock-down) influences levels of catecholamines, which in turn controls other behaviors in cavefish such as sleep and feeding [Bilandžija et al. 2013]. These behavioral alterations may provide a benefit to cavefish in the nutrient-poor cave environment, however the precise mechanism through which this has evolved remains unclear. Forthcoming genomic scans will clarify if certain genetic mutations, such as those discovered for Mc1r and Oca2, have risen to high frequencies in independent cavefish populations as a consequence of strong natural selection.
Conclusions
Little is known about the regulatory structure controlling Mc1r expression. The regulatory structure of Mc1r is best characterized in human and mouse. A luciferase promoter assay in two independent experiments suggested that the Mc1r minimal promoter lies immediately 5′ of the translational start and is approximately 500 bp in length [Moro et al. 1999; Miccadei et al. 2008]. Interestingly, a VISTA alignment comparing putative regulatory regions of Mc1r in several fish species including pufferfish, medaka, stickleback, and Midas cichlids identified a CNE present ~818 bp – 855 bp upstream of the translational start [Henning et al. 2010] (Fig. 1; blue triangle). Together, these results suggest that sequence mutations present in the upstream Mc1r cis-regulatory region may be associated with reduced pigmentation in Astyanax cavefish. Although seven caves harbor the brown mutation, four cave populations demonstrated no coding sequence alterations compared to surface fish [Gross et al. 2009]. However, complementation crosses indicated that the same genomic region underlies the brown phenotype [Wilkens and Strecker 2003]. Previous studies of albinism in cavefish similarly showed that independent cavefish populations likely acquired albinism through the same (or closely linked) locus. A subsequent study supported this notion – demonstrating that different coding sequence mutations in the same gene (Oca2) govern albinism in distinct populations [Protas et al. 2006]. In this study, we identified substantial sequence variation – some shared mutations and others unique – in the 5′ Mc1r region in independent cavefish lineages. Since experimentally reduced Mc1r expression recapitulates the brown phenotype [Gross et al. 2009], this study suggests that regulatory changes may also contribute to the brown phenotype. The work presented here represents one of the first examples of a potential role for regulatory alterations mediating a classic regressive phenotype in Astyanax cavefish.
Alterations in key regulatory regions underlie morphological and physiological variation across broad taxa, including amphibians, birds, mammals, and insects [reviewed in Hoekstra 2006]. For instance, pelvic spine reduction has evolved repeatedly in sticklebacks through deletion of a Pel enhancer, located directly upstream of the Pitx1 promoter [Chan et al. 2010]. The pattern of pigmentation gene expression demonstrated by Mc1r varied dynamically across development, but only demonstrated significantly different morphotype-specific expression at the adult stage. Forthcoming larger-scale expression analyses, evaluating more pigmentation-related genes across early development and juvenilehood, will better inform how particular genes impact complex pigmentation phenotypes in Astyanax.
In conclusion, brown cave populations that demonstrate no coding sequence errors in Mc1r still demonstrate reduced expression of this gene. This degenerative pigmentation phenotype may have evolved through the accumulation of sequence mutations affecting the 5′ regulatory region. Thus, regressive pigmentation in Astyanax cavefish may have evolved through a combination of both coding and cis-regulatory alterations. The contribution of both forms of sequence variation implies a role for regulatory alterations, alongside coding sequence variation, in the evolution of cave-associated traits.
Supplementary Material
Table S1. Detailed sequence analysis of mutations and conserved non-coding regions in the 5′ Mc1r region
Acknowledgments
Funding
This study was funded by the National Science Foundation, Washington D.C., U.S.A. (grant number DEB-1457630 to JBG).
The authors wish to thank Dr. Richard Borowsky (New York University) for generously providing fin clips used in this report. In addition, we thank the members of the Gross laboratory for helpful discussions. Two anonymous reviewers provided helpful criticisms on an earlier draft of this manuscript.
Footnotes
Conflict of Interest Statement
The authors declare that they have no conflict of interest.
Ethical Statement
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. All procedures performed in studies involving animals were in accordance with the ethical standards of the institution or practice at which the studies were conducted. The protocol was approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Cincinnati (Protocol Number 10-01-21-01).
Informed Consent
This article does not contain any studies involving human participants performed by any of the authors. Therefore, for this type of study, formal consent is not applicable.
Literature Cited
- Bilandžija H, Ma L, Parkhurst A, Jeffrey WR. A potential benefit of albinism in Astyanax cavefish: downregulation of the Oca2 gene increases tyrosine and catecholamine levels as an alternative to melanin synthesis. PLOS ONE. 2013;8:e80823. doi: 10.1371/journal.pone.0080823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradic M, Beerli P, García-de León F, Esquivel-Bobadilla S, Borowsky RL. Gene flow and population structure in the Mexican blind cavefish complex (Astyanax mexincanus) BMC Evol Biol. 2012;12:9. doi: 10.1186/1471-2148-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–22. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Chan YF, Marks ME, Jones FC, Villarreal G, Jr, Shapiro MD, Brady SD, Southwick AM, Absher DM, Grimwood J, Schmutz J, Meyers RM, Petrov D, Jónsson B, Schluter D, Bell MA, Kingsley DM. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327:302–305. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyva-Pérez M, de la O, Valverde-Corredor A, Valderrama R, Jiménez-Ruiz J, Muñoz-Merida A, Trelles O, Barroso JB, Luque F. Early and delayed long-term transcriptional changes and short-term transient responses during cold acclimation in olive leaves. DNA Res. 2014;22:1–11. doi: 10.1093/dnares/dsu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinasa L, Jeffery WR. Conservation of retinal circadian rhythms during cavefish eye degeneration. Evol Dev. 2006;8(1):16–22. doi: 10.1111/j.1525-142X.2006.05071.x. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;2:W273–9. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Borrón JC, Sánchez-Laorden BL, Jiménez-Cervantes C. Melanocortin-1 receptor structure and functional regulation. Pig Cell Res. 2005;18:393–410. doi: 10.1111/j.1600-0749.2005.00278.x. [DOI] [PubMed] [Google Scholar]
- Gordan R, Shen N, Dror I, Zhou T, Horton J, Rohs R, Bulyk ML. Genomic regions flanking E-Box binding sites influence DNA binding specificity of bHLH transcription factors through DNA shape. Cell Reports. 2013;3:1093–1104. doi: 10.1016/j.celrep.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JB, Borowsky R, Tabin CJ. A novel role for Mc1r in the parallel evolution of depigmentation in independent populations of the cavefish Astyanax mexicanus. PLoS Genet. 2009;5:e1000326. doi: 10.1371/journal.pgen.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JB. Cave evolution. Encyclopedia of Life Sciences (eLS; Wiley Press) John Wiley & Sons Ltd; Chichester, UK: 2012. [DOI] [Google Scholar]
- Gross JB, Furterer A, Carlson BM, Stahl BA. An integrated transcriptome-wide analysis of cave and surface dwelling Astyanax mexicanus. PLOS ONE. 2013;8(2):e55659. doi: 10.1371/journal.pone.0055659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JB, Wilkens H. Albinism in phylogenetically and geographically distinct populations of Astyanax cavefish arises through the same loss-of-function Oca2 allele. Heredity. 2013;111:122–130. doi: 10.1038/hdy.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guaiquil VH, Pan Z, Karagianni N, Fukuoka S, Alegre G, Rosenblatt MI. VEGF-B selectively regenerates injured peripheral neurons and restores sensory and trophic functions. Proc Natl Acad Sci U S A. 2014;111:17272–17277. doi: 10.1073/pnas.1407227111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henning F, Renz AJ, Fukamachi S, Meyer A. Genetic, comparative genomic, and expression analyses of the Mc1r locus in the polychromatic Midas cichlid fish (Teleostei, Cichlidae Amphilophus sp.) species group. Jour Mol Evol. 2010;70:405–412. doi: 10.1007/s00239-010-9340-4. [DOI] [PubMed] [Google Scholar]
- Hoekstra HE. Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity. 2006;97:222–234. doi: 10.1038/sj.hdy.6800861. [DOI] [PubMed] [Google Scholar]
- Jeong S, Rokas A, Carroll SB. Regulation of body pigmentation by the abdominal-B hox protein and its gain and loss in Drosophila evolution. Cell. 2006;125:1387–1399. doi: 10.1016/j.cell.2006.04.043. [DOI] [PubMed] [Google Scholar]
- Kingsley EP, Manceau M, Wiley CD, Hoekstra HE. Melanism in Peromyscus Is caused by independent mutations in Agouti. PLOS ONE. 2009;4:e6435. doi: 10.1371/journal.pone.0006435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miccadei S, Pascucci B, Picardo M, Natali PG, Civitareale D. Identification of the minimal melanocyte-specific promoter in the Melanocortin receptor 1 gene. Jour Exp Clin Can Res. 2008;27:71. doi: 10.1186/1756-9966-27-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CT, Beleza S, Pollen AA, Schluter D, Kittles RA, Shriver MD, Kingsley DM. cis-regulatory changes in Kit Ligand expression and parallel evolution of pigmentation in sticklebacks and humans. Cell. 2007;131:1179–1189. doi: 10.1016/j.cell.2007.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro O, Ideta R, Ifuku O. Characterization of the promoter region of the human Melanocortin-1 Receptor (MC1R) gene. Biochem Biophys Res Comm. 1999;262:452–460. doi: 10.1006/bbrc.1999.1228. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Protas ME, Hersey C, Kochanek D, Zhou Y, Wilkens H, Jeffrey WR, Zon LI, Bowowsky R, Tabin CJ. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat Genet. 2006;38:107–111. doi: 10.1038/ng1700. [DOI] [PubMed] [Google Scholar]
- Şadoğlu P, McKee A. A second gene that affects eye and body color in Mexican blind cave fish. J Hered. 1969;60:10–14. doi: 10.1093/oxfordjournals.jhered.a107917. [DOI] [PubMed] [Google Scholar]
- Taylor S, Wakem M, Dijkman G, Alsarraj M, Nguyen M. A practical approach to RT-qPCR–Publishing data that conform to the MIQE guidelines. Methods. 2010;50:S1–5. doi: 10.1016/j.ymeth.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Wilkens H, Strecker U. Convergent evolution of the cavefish Astyanax (Characidae, Teleostei): genetic evidence from reduced eye-size and pigmentation. Biol J Linn Soc. 2003;80:545–554. [Google Scholar]
- Yasumoto K, Yokoyama K, Shibata K, Tomita Y, Shibahara S. Microphthalmia-associated transcription factor as a regulator for melanocyte-specific transcription of the human tyrosinase gene. Mol Cell Biol. 1994;14:8058–8070. doi: 10.1128/mcb.14.12.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngblood BA, Grozdanov PN, MacDonald CC. CstF-64 supports pluripotency and regulates cell cycle progression in embryonic stem cells through histone 3′ end processing. Nucleic Acids Res. 2014;42:8330–8342. doi: 10.1093/nar/gku551. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Detailed sequence analysis of mutations and conserved non-coding regions in the 5′ Mc1r region