SUMMARY
Histone variants complement and integrate histone post-translational modifications in regulating transcription. The histone variant macroH2A1 (mH2A1) is almost three times the size of its canonical H2A counterpart due to the presence of a ~25kDa evolutionarily conserved non-histone macro domain. Strikingly, mH2A1 can mediate both gene repression and activation. However, the molecular determinants conferring these alternative functions remain elusive. Here, we report that mH2A1.2 is required for the activation of the myogenic gene regulatory network and muscle cell differentiation. H3K27 acetylation at prospective enhancers is exquisitely sensitive to mH2A1.2, indicating a role of mH2A1.2 in imparting enhancer activation. Both H3K27 acetylation and recruitment of the transcription factor Pbx1 at prospective enhancers are regulated by mH2A1.2. Overall, our findings indicate a role of mH2A1.2 in marking regulatory regions for activation.
INTRODUCTION
Histone post-translational modifications shape the epigenome and regulate transcription (Jenuwein and Allis, 2001) (Roadmap Epigenomics et al., 2015). The nucleosome incorporation of histone variants provides an additional regulatory layer which influences formation of chromatin states associated with either transcriptional repression or activation (Jin and Felsenfeld, 2007; Jin et al., 2009) (Barski et al., 2007; Maze et al., 2014). Localized replacement of canonical histones by histone variants modifies the chromatin structure to attract or repel transcription factors, chromatin writers, readers, and erasers (Skene and Henikoff, 2013). Among the different histone variants, the two isoforms macroH2A1.1 and 1.2 are characterized by the presence of an evolutionarily conserved, ~25kDa carboxyl-terminal globular region called the macro domain (Pehrson and Fried, 1992) serving as surface for interaction with metabolites and histone modifiers (Ladurner, 2003) (Kustatscher et al., 2005) (Chakravarthy et al., 2005) (Gamble and Kraus, 2010) (Hussey et al., 2014). A role for mH2A1 in mediating gene repression was initially suggested by observations linking it to female X-chromosome inactivation (Costanzi and Pehrson, 1998) (Csankovszki et al., 2001). More recently mH2A1 has been shown to contrast reprogrammed pluripotency (Gaspar-Maia et al., 2013) (Barrero et al., 2013) (Pasque et al., 2011), repress expression of the HoxA cluster (Buschbeck et al., 2009), of the α-globin locus in erythroleukemic cells (Ratnakumar et al., 2012), and suppress melanoma progression through regulation of cyclin-dependent protein kinase CDK8 (Kapoor et al., 2010). However, there is evidence to suggest that mH2A1 has a multifaceted function in controlling gene transcription (Gamble et al., 2010). Reducing mH2A1 levels not only does not result in generalized de-repression of mH2A1-bound genes but is in fact associated with failure to activate up to 75% of its targets (Gamble et al., 2010). Moreover, while inhibiting p300-dependent histone acetylation in vitro (Doyen et al., 2006), mH2A1 has been recently reported to cooperate with PARP-1 to regulate transcription by promoting CBP-mediated acetylation of histone H2B at lysines 12 and 120, with opposing effects on transcription (Chen et al., 2014). These and other observations (Creppe et al., 2012) (Podrini et al., 2014) indicate that mH2A1 may exert a dual function in regulating gene expression.
Here, we report that mH2A1.2 is involved in imparting enhancer competency in skeletal muscle cells. In agreement with previous findings, mH2A1.2 was localized to H3K27me3 promoter regions of repressed genes. However, mH2A1.2-occupied and repressed targets were not reactivated upon mH2A1.2 knock-down. Instead, activation of muscle enhancers was dependent on mH2A1.2, as its reduction brought about decreased H3K27 acetylation. Reducing mH2A1.2 impaired expression of the master developmental regulator Myogenin, resulting in defective activation of the myogenic gene regulatory network and muscle cell differentiation. Notably, mH2A1.2 mediated chromatin engagement of Pbx1, a homeodomain transcription factor priming MyoD gene targets for activation (Berkes et al., 2004) (Maves et al., 2007). In aggregate, these findings assign a role to mH2A1.2 in conferring enhancer marking and activation via regulation of transcription factors’ recruitment and H3K27 acetylation.
RESULTS
Genome-Wide Distribution of the Histone Variant MacroH2A1.2 Reveals Preferential Association with Regions of Active Transcription
To investigate the role of the histone variant mH2A1 in transcriptional regulation of cell differentiation, we employed the mouse skeletal muscle C2C12 cell line, as a model system. C2C12 cells recapitulate muscle differentiation in culture as they can be kept in an undifferentiated state as myoblasts (MB) and induced to differentiate to form multinucleated myotubes (MT) (Yaffe and Saxel, 1977). Both alternatively spliced mH2A1.1 and 1.2 isoforms (Rasmussen et al., 1999) (Costanzi and Pehrson, 2001) were expressed in C2C12 cells (Figure S1A). Since RNA-seq analysis indicated that the mH2A1.2 isoform was the most represented in MB and expressed at levels similar to those of mH2A1.1 in MT (Figure S1B), we chose to focus our study on the mH2A1.2 isoform. Analysis of ChIP-seq data generated from two experiments with two different mH2A1.2 antibodies (see Experimental Procedures), identified ~77,000 overlapping enriched genomic regions in MB and ~36,600 in MT, respectively (Figure S1C–D, Table S1). Peak calling with either MACS2 (Feng et al., 2012) or SICER (Zang et al., 2009) algorithm identified largely overlapping mH2A1.2-enriched regions (Figure S1E). Examples of mH2A1.2-occupied regions, as called by MACS2 algorithm, are illustrated in Figure S1F. A global reduction of the mH2A1.2 signal was observed after mH2A1.2 knock-down indicating that majority of peaks correspond to mH2A1.2 isoform (Figure S1G,H).
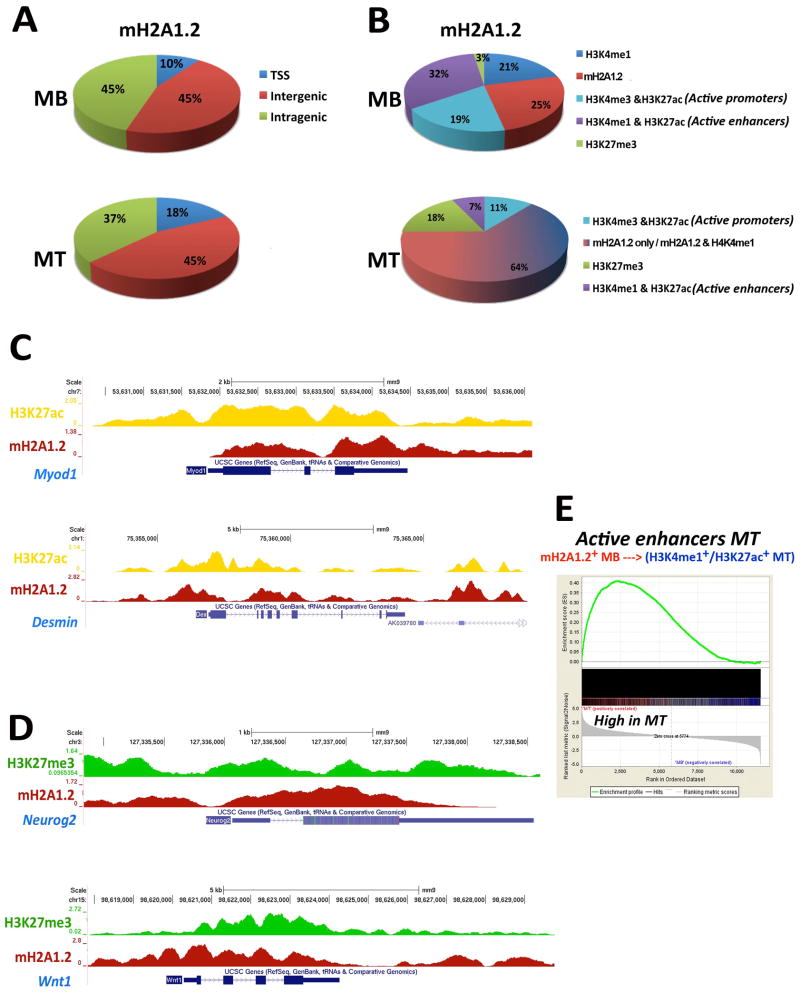
Genome-wide distribution of mH2A1.2 was similar in MB and MT (Figure 1A). Intersecting genome-wide maps of mH2A1.2 with those of active and repressive epigenetic marks in MB revealed that the majority of mH2A1.2 peaks was localized at active regions (Figure 1B). Specifically, 32% of mH2A1.2 peaks occurred at H3K4me1+/H3K27ac+ regions (active enhancers), 21% at H3K4me1+ regions, 19% overlapped with H3K4me3+/H3K27ac+ (active promoters), and 25% of mH2A1.2 peaks were located at regions not occupied by any of the epigenetic marks considered above (mH2A1.2 only). In contrast, only 3% of mH2A1.2 peaks co-localized with the repressive mark H3K27me3 (Figure 1B). Furthermore, among mH2A1.2-bound promoters, only 8% were H3K27me3+, while 67 % of these promoters were occupied by both H3K4me3 and H3K27ac (Figure S2A). In MT, the percentage of mH2A1.2+/H3K27me3+ regions increased to 18% (Figure 1B) and a GO analysis of the newly acquired mH2A1.2+/H3K27me3+ TSS identified terms related to, among others, “neuron differentiation”, “ pattern specification process” and “embryonic morphogenesis” (Table S1). Reduction of mH2A1.2 peaks at active enhancers (32% in MB vs. 7% in MT, Figure 1B) occurred at MT-specific enhancers (i.e., enhancers active in MT, see below) (Figure S2B) and coincided with increased mH2A1.2 occupancy at H3K4me1+ and otherwise non-epigenetically defined genomic regions (64%, Figure 1B). mH2A1.2 occupancy was also reduced, but more modestly, at constitutive enhancers (i.e., enhancers active in both MB and MT, see below) in MT (Figure S2C).
Figure 1. Genome-Wide Distribution of the Histone Variant MacroH2A1.2 and Associated Epigenetic Marks At Regulatory Regions of Skeletal Muscle Cells.
(A) Genome-wide distribution of mH2A1.2 in C2C12 MB and MT. (B) Co-localization of mH2A1.2 and epigenetic marks H3K4me3, H3K4me1, H3K27ac, and H3K27me3 in C2C12 MB and MT. (C) ChIP-seq profiles of mH2A1.2 and H3K27ac at Myod1 and Desmin loci. (D) ChIP-seq profiles of mH2A1.2 and H3K27me3 at Neurogenin2 and Wnt1 loci. Both H3K27ac and mH2A1.2 signals were corrected for input DNA. (E) GSEA of genes assigned to MT-active enhancers bound by mH2A1.2 in MB. Genes are ranked from left to right according to their Signal2Noise metric in MT. The enrichment score profile indicates that the gene set is enriched for upregulated genes in MT (p-value: <2.0e-4, FDR ~0).
Examples of expressed genes occupied by mH2A1.2 are shown in Figure 1C. Developmental regulators of other cell lineages, such as Neurog2 and Wnt1, which are transcriptionally silent in C2C12 cells (Mousavi et al., 2012), are among mH2A1.2-bound genes with H3K27me3 (Figure 1D).
We assigned MB-mH2A1.2+ active enhancers or MB-mH2A1.2+ regions acquiring either H3K4me1+ or H3K4me1+/H3K27ac+ in MT to genes by proximity (Whyte et al., 2013) (Mousavi et al., 2013) and queried gene expression changes occurring during the transition from MB to MT. Enhancers residing within 100Kb, 50Kb, or 20Kb from the closest promoter were considered. While the number of enhancer-assigned genes increased with increasing genomic intervals (Figure S2D), GO analyses for 100Kb and 50Kb intervals captured essentially all the terms returned by the analysis conducted for 20Kb interval, including “muscle cell differentiation” and “muscle and muscle system process” (Figure S2E, Table S1). Therefore, for further analysis, we considered a proximity measure of 20Kb to assign genes to identified enhancers. Genomic regions that became active enhancers in MT displayed a clear association with up-regulated genes (Figure 1E). Similarly, a smaller set comprising genes assigned to mH2A1.2+ regions and occupied by H3K4me1 and H3K27me3 marks in MB was also enriched for up-regulated genes in MT (Table S2). Overall, these results indicate that mH2A1.2 preferentially occupies transcriptionally active genomic regions in MB or regions programmed to be activated in MT.
MacroH2A1.2 Is Required for the Activation of the Myogenic Gene Regulatory Network and Differentiation of Skeletal Muscle Cells
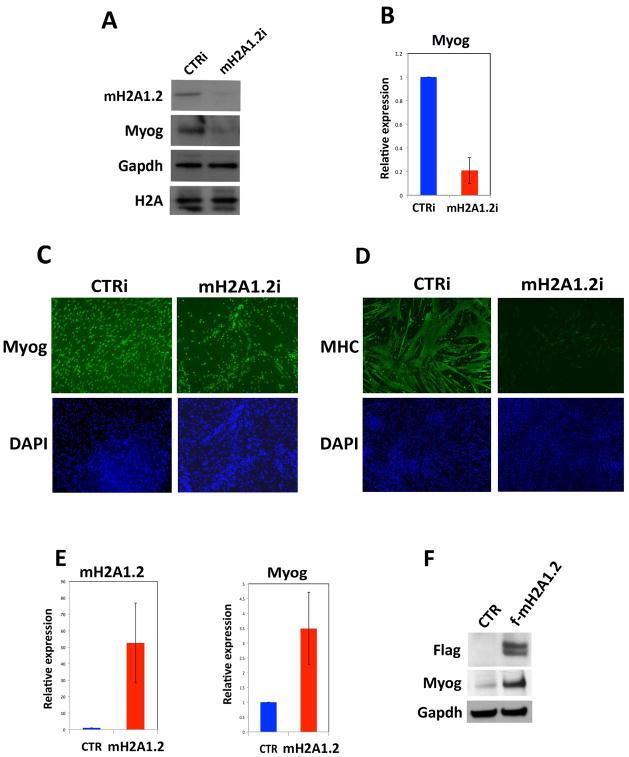
We addressed the function of mH2A1.2 during muscle cell differentiation by transfecting C2C12 cells with either control or two different mH2A1.2 siRNA (mH2A1.2 interference, mH2A1.2i) (Figure 2A and Figure S3A) and inducing them to differentiate to form MT. For further analysis, we chose to employ mH2A1.2i_2 siRNA, as they were the most effective (Figure S3A). mH2A1.2 siRNA specifically reduced mH2A1.2 and not the closely related mH2A1.1 isoform (Figure S3B). MB growth was not affected by mH2A1.2i (Figure S3C). However, Myogenin, a myogenic transcription factor required for muscle differentiation (Tapscott, 2005), was reduced (Figure 2A–C, Figure S3A,D) and formation of muscle-specific myosin heavy-chain (MHC)-positive, multinucleated MT compromised by mH2A1.2i (Figure 2D). The expression of the muscle-specific gene troponin T type 1 (Tnnt1) was also greatly reduced (Figure S3E). To complement knockdown experiments, exogenous Flag-tagged mH2A1.2 was expressed in C2C12 cells and found to increase Myogenin expression (Figure 2E,F).
Figure 2. Reducing MacroH2A1.2 Impairs Skeletal Muscle Cell Differentiation.
(A,B) Myogenin protein and mRNA evaluated after siRNA against mH2A1.2 in C2C12 cells. Gapdh and histone H2A were used as loading controls. Data are represented as mean ± SD. (C) Myogenin and (D) myosin heavy chain (MHC) immunofluorescence staining of control (CTRi) and mH2A1.2i C2C12 cells prompted to differentiate for 2 days. DAPI identifies nuclei. (E) mH2A1.2 and Myogenin mRNA expression in C2C12 cells transfected with Flag-empty (CTR) or Flag-mH2A1.2 (f-mH2A1.2) expression vector (0.8 g mH2A1.2 plasmid /1×105 cells). (F) Immunoblot for Flag, Myogenin and Gapdh in C2C12 transfected with Flag-empty (CTR) or Flag-mH2A1.2 vector. Data are represented as mean ± SD.
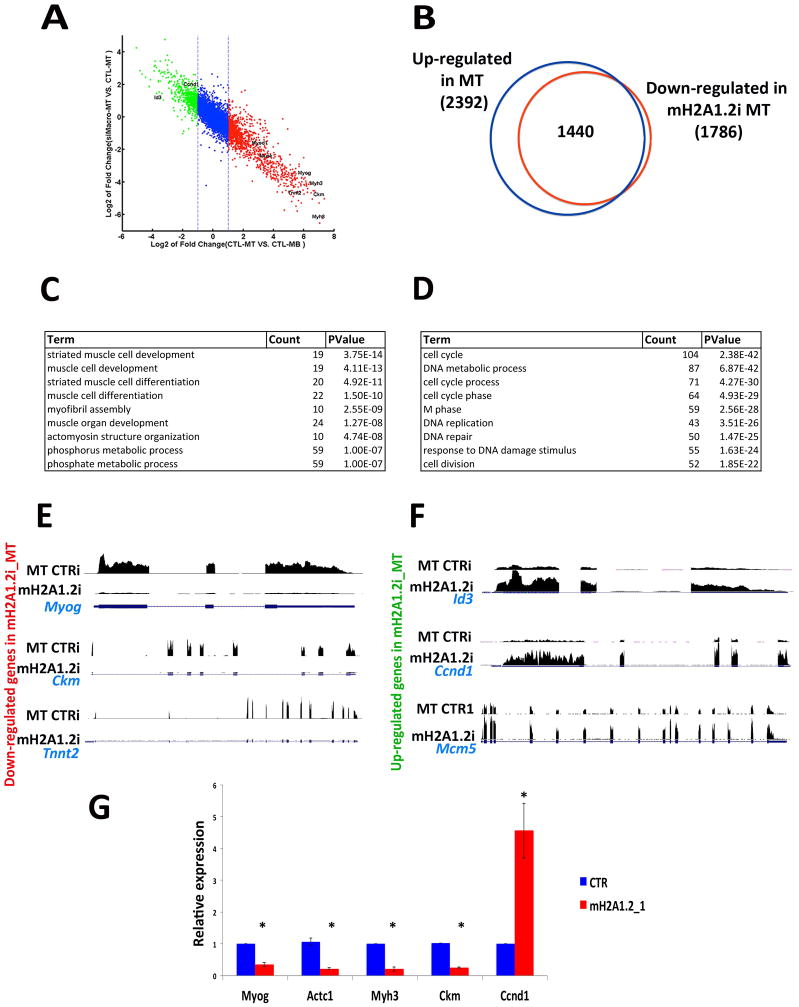
To define the global impact of reducing mH2A1.2 on the transcriptome, RNA-seq experiments were performed in control and mH2A1.2i C2C12 cells. When mH2A1.2i C2C12 MB were induced to differentiate, a profound effect on transcriptional dynamics was observed. As indicated in the scatter plot representing changes in gene expression (Figure 3A), genes physiologically up-regulated during cell differentiation failed to be properly activated in mH2A1.2i cells, while genes down-regulated during differentiation remained transcribed. In control cells, expression of 2,392 genes was increased during the transition from MB to MT (Figure 3B, Table S3). Compared to control MT, 1,786 gene transcripts were reduced by mH2A1.2i. Out of these 1,786 transcripts, 1,440 (80.5%) corresponded to transcripts increased during the differentiation of MB to MT (Figure 3B). Gene ontology (GO) analysis of the transcripts which failed to be appropriately up-regulated in mH2A1.2i cells returned terms related to “muscle cell development” and “muscle cell differentiation” (Figure 3C). GO terms for the transcripts which remained elevated in mH2A1.2i cells were related to “cell cycle”, “and “DNA replication” (Figure 3D). Myogenin and its downstream targets muscle creatine kinase (Ckm) and troponin T type 2 (Tnnt2) were not properly activated in mH2A1.2i cells (Figure 3E). Conversely, transcripts of the Inhibitor of DNA Binding 3 (Id3), a member of the Id family of helix-loop-helix proteins counteracting muscle differentiation (Benezra et al., 1990), cyclin D1 (Ccnd1), and the cell cycle regulator Mcm5, which are physiologically down-regulated upon C2C12 differentiation, remained abnormally elevated in mH2A1i cells (Figure 3F). To validate these findings, we employed a different mH2A1.2 siRNA (mH2A1.2i_1) (Figure S3A). In mH2A1.2i_1-transfected cells, transcripts of Myogenin, muscle-specific myosin heavy chain 3 (Myh3), cardiac actin (Actc1), creatine kinase (Ckm) were reduced while those of cyclin D (Ccnd1) remained elevated (Figure 3G). These findings indicate that mH2A1.2 is required to activate muscle gene expression during cell differentiation.
Figure 3. MacroH2A1.2 Regulates the Transcriptome of Differentiating Skeletal Muscle Cells.
(A) Scatter plot shows the inhibitory effect of mH2A1.2 knock down on transcriptome during differentiation. Each dot represents a gene, x-axis shows expression changes during differentiation in CTR, and y-axis shows the expression changes in mH2A1.2i versus CTR in MT. Genes marked red and green are up-regulated and down-regulated during differentiation, respectively. (B) Venn diagram illustrating number of genes up-regulated in control C2C12 MT and down-regulated in counterpart mH2A1.2i cells. (C) Gene ontology (GO) for genes down-regulated in differentiating mH2A1.2i C2C12 cells. (D) Gene ontology (GO) for genes whose transcription remains elevated in differentiating mH2A1.2i C2C12 cells. (E) RNA-seq profiles of down-regulated genes Myog, Ckm and Tnnt2 in differentiating CTRi and mH2A1.2i C2C12 cells. (F) RNA-seq profiles of up-regulated genes Id3, Ccnd1 and Mcm5 in differentiating CTLi and mH2A1.2i C2C12 cells. (G) Myogenin, Actc1, Myh3, Ckm and Ccnd1 mRNAs were evaluated after siRNA against mH2A1.2 in C2C12 cells. Data are represented as mean ± SD, *p<0.01.
MacroH2A1.2 Is Enriched at Prospective Enhancers and Is Necessary for Their Activation
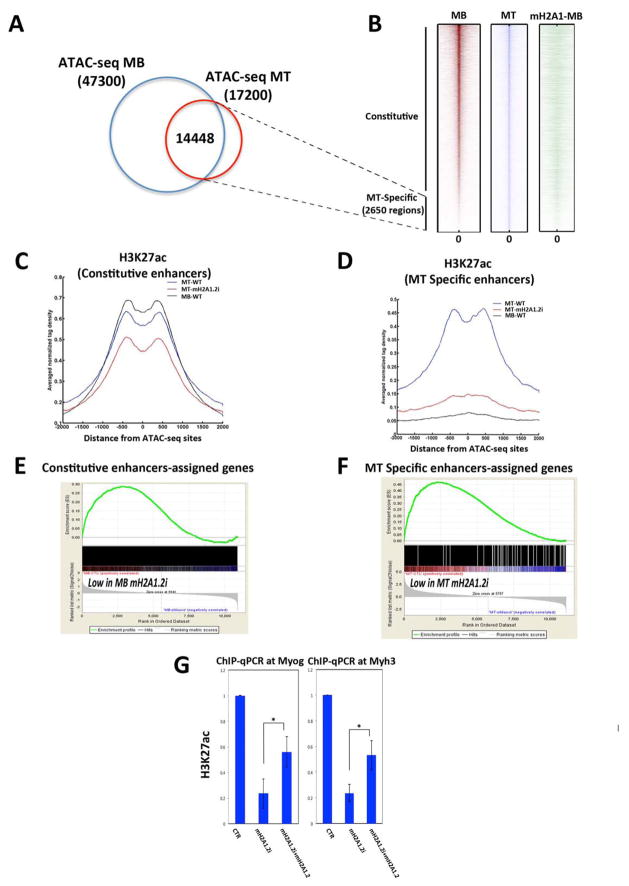
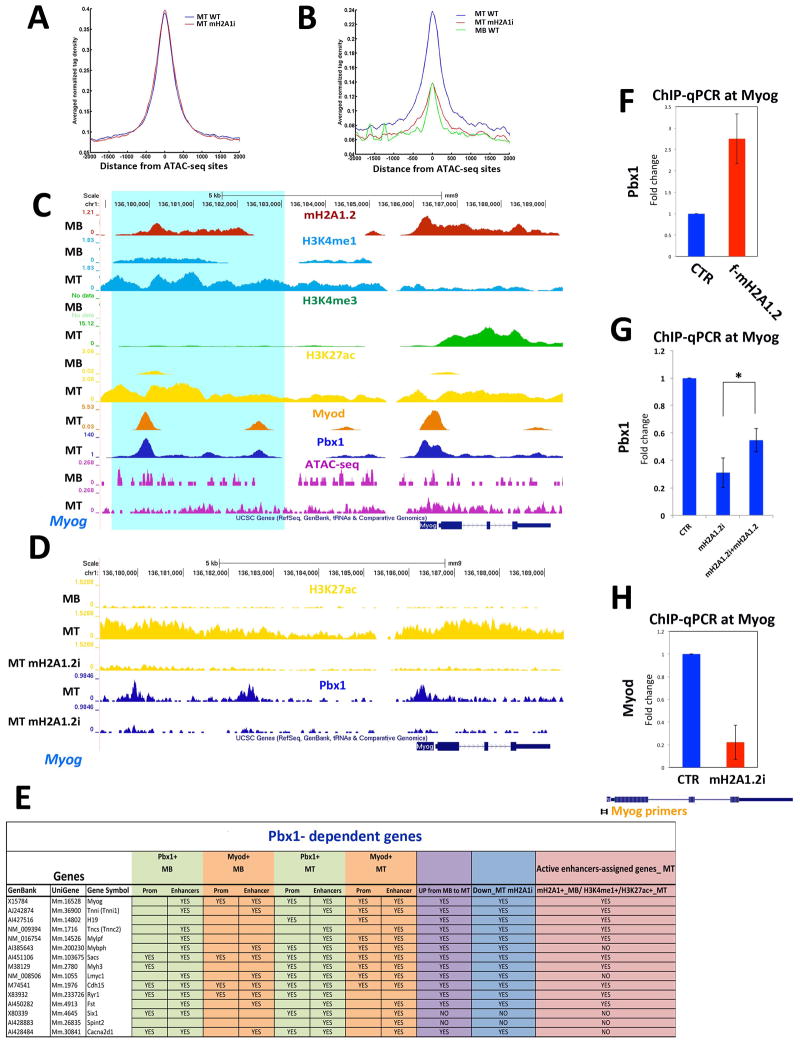
We employed Assay for Transposase-Accessible Chromatin with high-throughput sequencing (ATAC-seq) (Buenrostro et al., 2013) to define chromatin accessibility in C2C12 MB and MT. In ATAC-seq, tagging of nucleosome-free genomic regions is mediated by transposase-mediated delivery of sequencing adapters. Tagged regions correlate with DNase I hypersentitive sites (open chromatin), which are generally found within genomic regulatory functions. Using two independent replicates, ~47,300 and ~17,200 transposase-accessible or open chromatin regions were reproducibly identified in MB and MT, respectively (Figure 4A). More than 84% of these genomic regions (14,448/17,200) were open in both MB and MT (Figure 4B). The remaining ATAC-seq MT sites (~2,650) were closed in MB and open in MT, and almost all of them (~2,500) were located outside the promoter regions (Figure 4A,B). We refer to these two groups as constitutive (open in both MB and MT) and MT-specific enhancers (present only in MT), respectively. mH2A1.2 occupied both enhancer groups in MB (Figure 4B). While constitutive enhancers were similarly acetylated at H3K27 in both MB and MT (Figure 4C, compare MB-WT, black line, and MT-WT, blue line), MT-specific enhancers acquired H3K27ac only in MT (Figure 4D, compare MB-WT, black line, and MT-WT, blue line). To determine whether mH2A1.2 regulate the activity of constitutive and MT-specific enhancers, we assigned genes to these two groups of enhancers (based on proximity distance ±20kb) and evaluated how mH2A1.2i affected expression of the enhancer-assigned genes. Employing GSEA, we found that genes assigned to constitutive enhancers were positively correlated with genes whose expression was reduced by mH2A1.2i in MB (Figure 4E), whereas genes whose expression was diminished by mH2A1.2i in MT correlated with genes assigned to MT-specific enhancers (Figure 4F). Since H3K27 acetylation is a defining step associated with enhancer activation (Creyghton et al., 2010) (Heintzman et al., 2009) (Rada-Iglesias et al., 2011) (Zentner et al., 2011) (Bonn et al., 2012), we evaluated whether mH2A1.2 was involved in conferring H3K27 acetylation by performing H3K27ac ChIP-seq upon mH2A1.2i. H3K27 acetylation at constitutive enhancers was slightly reduced (Figure 4C, compare MT-WT, blue line with MT-mH2A1.2i, red line). A most profound effect of mH2A1.2i on H3K27 acetylation was observed at MT-specific enhancers. At these enhancers, mH2A1i.2 reduced H3K27ac to background levels observed in MB, where the chromatin of MT-specific enhancers is closed (Figure 4D, compare MT-WT, blue line with MT-mH2A1.2i, red line). Consistent with a more limited reduction of H3K27ac at constitutive enhancers (Figure 4C), transcription of genes assigned to constitutive enhancers was less affected than that of genes controlled by MT-specific enhancers (Figure 4E, ES<0.30, Figure 4F, ES>0.45) in mH2A1.2i cells. Next, we analyzed H3K27ac at promoter regions. mH2A1.2i did not modify H3K27ac at constitutive promoters but reduced it at MT-specific promoters (Figure S4A,B). These findings are consistent with impaired acquisition of MT-specific enhancer competency upon mH2A1.2i and consequent failure to induce promoter activation (H3K27ac) and gene transcription. To establish whether a direct link exists between reduced H3K27 acetylation and mH2A1.2i, we attempted rescue experiments by overexpressing mH2A1.2 in mH2A1.2i cells. mH2A1.2 overexpression partially restored H3K27ac at both the Myogenin and Myh3 loci in mH2A1.2i cells (Figure 4G). In summary, these results indicate that, during muscle cell differentiation, mH2A1.2 is involved in conferring enhancer activation by regulating H3K27 acetylation.
Figure 4. MacroH2A1.2 Influences H3K27 Acetylation at Enhancer Regions.
(A) Venn diagram representing ATAC-seq positive regions in C2C12 MB and MT. (B) Heat maps of tag densities representing distribution of ATAC-seq signals in C2C12 MB (red), MT (blue), and mH2A1.2 binding (green) in MB. (C) Average profile of H3K27ac signal in C2C12 MB (black line), MT (blue line), and MT-mH2A1.2i (red line) for constitutive enhancers. (D) Average profile of H3K27ac signal in MB (black line), MT (blue line) and MT-mH2A1.2i (red line) for MT-specific enhancers. (E) Gene set enrichment analysis (GSEA) of genes assigned to constitutive enhancers. Genes are ranked from left to right according to their Signal2Noise metric in MB-CTR vs. MB-mH2A1.2i. The enrichment score profile indicates that the gene set is enriched for downregulated genes in mH2A1.2i-MB (p-value: <2.0e-4, FDR <10%). (F) GSEA of genes assigned to MT-specific enhancers. Genes are ranked from left to right according to their Signal2Noise metric in MT-CTR vs. MT-mH2A1.2i. The enrichment score profile indicates that the gene set is enriched for genes strongly down-regulated in mH2A1.2i-MT (p-value: <2.0e-4, FDR <10%). (G) ChIP-qPCR for H3K27ac at the Myogenin and Myh3 loci in control (CTR), mH2A1.2i, and mH2A1.2i C2C12 cells transfected with mH2A1.2 expression vector. Data are represented as mean ± SD, *p<0.01.
Chromatin Engagement of the Transcription Factor Pbx1 At Muscle Regulatory Regions is Contingent on MacroH2A1.2
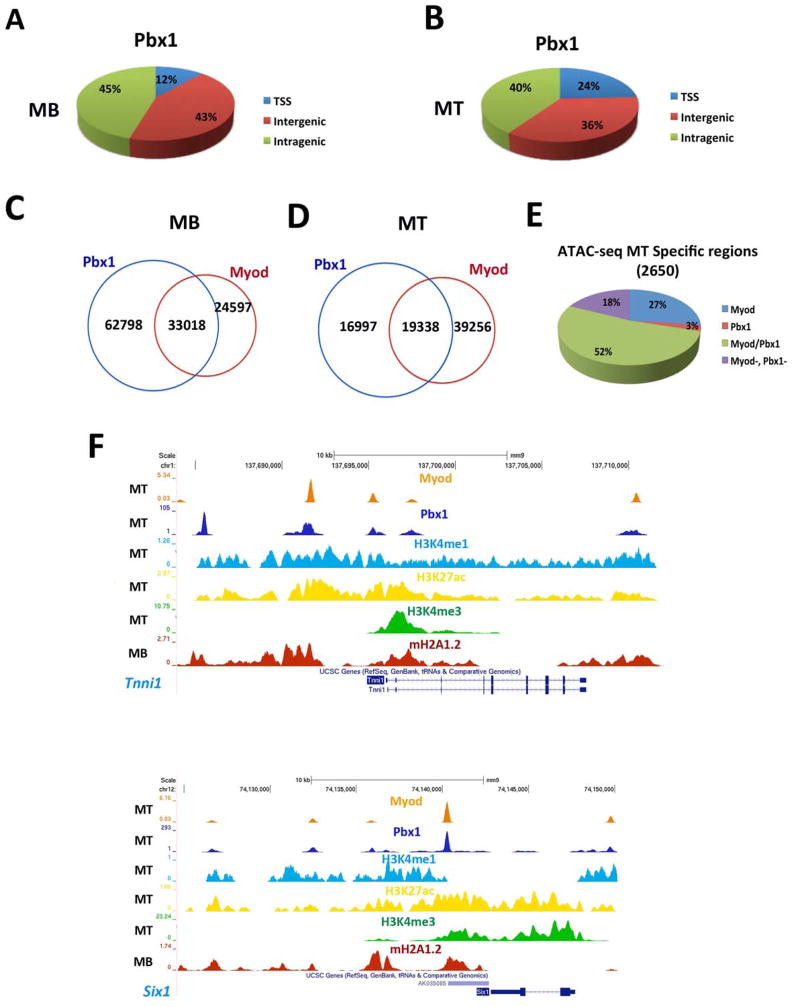
The presence of mH2A1.2 in MB at both TSS and enhancers destined to become activated in MT (MT-specific enhancers), as well as its requirement for their activation, prompted us to investigate a potential link between mH2A1.2 and the transcription factor Pbx1. The TALE (three- amino acids loop extension) homeodomain-containing transcription factor Pbx1 is required to assist MyoD-dependent activation of Myogenin (Berkes et al., 2004) (de la Serna et al., 2005). Pbx1 is constitutively bound to Myogenin gene in fibroblasts prior to MyoD-mediated conversion to muscle and, by directly interacting with two specific domains, ensures productive and stable MyoD recruitment at the Myogenin promoter (Berkes et al., 2004). More recently, Pbx1/MyoD interaction has been shown to regulate expression of a large cohort of MyoD-dependent genes (Fong et al., 2015). Suggesting a relationship between mH2A1.2 and Pbx1, analysis for DNA-binding motifs showed that, among others, the Pbx1 consensus binding motif was enriched within mH2A1.2-bound regions in MB (Figure S5A). We performed Pbx1 ChIP-seq and examined the overlap between Pbx1 and MyoD binding (Mousavi et al., 2013). Majority of Pbx1 peaks occurred at inter-and intragenic regions in both MB (88%) and MT (76%) (Figure 5A,B). Approximately 57% of the Pbx1 peaks overlapped with MyoD in MB and 33% in MT, respectively (Figure 5C,D). Moreover, in MT, MyoD and Pbx1 co-occupied 52% of the MT-specific ATAC-seq regions (Figure 5E). Examples of muscle genes co-occupied by MyoD and Pbx1 are shown in Figure 5F. In line with the above observations, the E-box (DNA recognition site for MyoD) emerged as one of the top enriched motifs within Pbx1-occupied regions (Figure S5B–C). Similarly, de novo motif analysis of common binding regions between MB and MT returned, among others, motifs with consensus matching MyoD/Myf5 and Pbx3 (Figure S5D). In MB, 5,902 Pbx1 peaks occurred at genomic regions acquiring epigenetic characteristics of active enhancers (H3K4me1+/H3K27ac+) in MT (Table S4). Of the genes assigned to MT-specific enhancers, 70% was also assigned to these Pbx1+ regulatory regions (Table S4). To investigate a potential dependency of Pbx1 binding on mH2A1.2, we conducted Pbx1 ChIP-seq in control and mH2A1.2i cells. While overall Pbx1 binding was not affected at constitutive enhancers (Figure 6A), it was markedly decreased at MT-specific enhancers, including the Myogenin locus (Figure 6B–D). Moreover, the promoters and/or enhancer regions of genes regulated by Pbx1 (Berkes et al., 2004) were co-occupied by Pbx1 and MyoD, and their transcription reduced by mH2A1.2i (Figure 6E). Next, we evaluated whether mH2A1.2 is sufficient to promote Pbx1 recruitment by expressing Flag-tagged mH2A1.2 and performing ChIP-qPCR for Pbx1 at Myogenin. Compared to control, Pbx1 recruitment at the Myogenin locus was increased in C2C12 mH2A1.2-transfected cells (Figure 6F). Importantly, Pbx1 transcripts were not affected by mH2A1.2 expression (Figure S6A). Thus, mH2A1.2 overexpression promotes Pbx1 engagement at Myogenin and activates its transcription (Figure 2E,F). Consistent with these findings, mH2A1.2 expression in mH2A1.2i cells partially restored Pbx1 binding at Myogenin (Figure 6G). In line with a role of Pbx1 in stabilizing MyoD binding (Berkes et al., 2004), MyoD engagement at Myogenin was also reduced by mH2A1.2i (Figure 6H). Overexpressed as well as endogenous and chromatin-bound mH2A1.2 interacted with Pbx1 (Figure S6B,C) and, employing bacterially produced and purified proteins, we detected an interaction of the macro domain – but not of the H2A-like region- of mH2A1.2 with Pbx1 (Figure S6D). Pbx1 also interacted with canonical H2A (data not shown). Altogether, the data reported in this paragraph indicate that mH2A1.2 regulates Pbx1 recruitment at developmental (MT-specific) enhancers and transcription of the associated genes.
Figure 5. Genome-Wide Analysis of Pbx1 and MyoD Binding in Skeletal Muscle Cells.
(A,B) Genome-wide distribution of Pbx1 binding in C2C12 MB and MT. (C,D) Venn diagrams representing Pbx1 and MyoD peaks in C2C12 MB and MT. (E) MyoD and Pbx1 distribution relative to MT-specific ATAC-seq regions. (F) ChIP-seq tracks at the Tnnt1 and Myh3 loci. Bottom to top: mH2A1.2 in MB (red track), H3K4me3 in MT (green track), H3K27ac in MT (yellow track), H3K4me1 in MT (blue track), Pbx1 in MT (blue tracks), MyoD in MT (orange track). The ChIP-seq signals were corrected for input DNA.
Figure 6. MacroH2A1.2 Regulates Recruitment of Pbx1 at Muscle Enhancer Regions.
(A) Average profile of Pbx1 signal in MT-WT (blue line) and differentiating mH2A1.2i C2C12 cells (red line) at constitutive enhancers; (B) Average profiles of Pbx1 signal in MT-WT (blue line), MT-mH2A1.2i (red line), and in MB-WT at MT-specific enhancers; (C) ChIP-seq tracks at the Myogenin locus. Top to bottom: mH2A1.2 in MT and MB (red tracks); H3K4me1 in MB and MT (light blue tracks); H3K4me3 in MB and MT (green tracks); H3K27ac in MB and MT (yellow tracks); MyoD in MT (orange track); Pbx1 in MT (blue tracks); ATACseq signal in MB and MT (purple tracks). Turquoise shading identifies a H3K27ac+/H3K4me1+/Pbx1+/MyoD+/H3K4me3− region. (D) ChIP-seq tracks at the Myogenin locus. Top to bottom: H3K27ac in MB, MT and MT_mH2A1.2i (yellow tracks); Pbx1 in MT CTR and mH2A1.2i (blue tracks). (E) Summary of Pbx1and MyoD occupancy in MB and MT, expression in MB and MT, expression in mH2A1.2i cells, and assignment to MT-specific enhancers of Pbx1-dependent genes reported in (Berkes et al., 2004). (F) Pbx1 ChIP-qPCR in CTR and mH2A1.2-overexpressing (2 g mH2A1.2 plasmid/1×105 cells). (G) ChIP-qPCR for Pbx1 at the Myogenin locus in control (CTR), mH2A1.2i, and mH2A1.2i C2C12 cells transfected with mH2A1.2 expression vector. Data are represented as mean ± SD, *p<0.01. (H) ChIP-qPCR for MyoD at the Myogenin locus in CTR and mH2A1.2i C2C12 cells. Data are represented as mean ± SD.
DISCUSSION
Here, we report that mH2A1.2 is a positive regulator of transcription and muscle cell differentiation. In agreement with previous studies, we have identified genomic regions co-occupied by mH2A1.2 and H3K27me3 (Buschbeck et al., 2009) (Ratnakumar et al., 2012) (Gaspar-Maia et al., 2013). However, mH2A1.2 knock-down neither modified H3K27me3 (data not shown) nor resulted in gene de-repression (Table S3), suggesting that, similarly to what observed at pluripotency genes (Gaspar-Maia et al., 2013), mH2A1.2 may play a redundant silencing role. Genome-wide distribution of mH2A1.2 localized at transcriptionally competent regulatory regions in undifferentiated C2C12 MB. However, competency of constitutive enhancers was only modestly affected by mH2A1.2i, indicating that, once enhancers are activated, mH2A1.2 may not be critical for their maintenance. Instead, mH2A1.2 exerted a critical function during the differentiation process. Reducing mH2A1.2 prevented activation of the myogenic gene regulatory network, with approximately 80% of the genes whose transcription is promoted during differentiation failing to be activated. This phenomenon coincided with the inability of muscle developmental enhancers to be appropriately H3K27 acetylated in mH2A1.2i cells.
The presence of mH2A1.2 at prospective enhancers and its requirement for their activation suggest that mH2A1.2 functions as a “marking” histone (Bell et al., 2011). Pioneer transcription factors can access silent chromatin by recognizing their complete or partial DNA motifs on nucleosomes followed by the subsequent binding of other transcription factors and chromatin remodelers (Zaret and Carroll, 2011) (Iwafuchi-Doi and Zaret, 2014) (Soufi et al., 2015). MyoD can convert non-myogenic cells to adopt the skeletal muscle phenotype (Davis et al., 1987). The ability of MyoD to initiate myogenesis in non-muscle cells is conferred by two independent domains, the cysteine-rich domain and the C-terminal helix III region (Gerber et al., 1997) (Bergstrom and Tapscott, 2001). These two domains ensure stable binding of MyoD to the Myogenin promoter via interaction with a protein complex containing Pbx1, a homeodomain transcription factor constitutively bound to the Myogenin promoter (Berkes et al., 2004) (de la Serna et al., 2005).Pbx1 has been proposed to act as a pioneer factor to guide chromatin recruitment of estrogen receptors in breast cancer (Magnani et al., 2011). Our findings indicate that mH2A1.2 exerts a licensing function for Pbx1 recruitment and H3K27 acetylation. The observed anti-correlation between mH2A1.2 occupancy and Pbx1 binding at MT-specific enhancers in MT (Figure S2B and Figure 6B) suggests that, once enhancers are bound by Pbx1 (and/or MyoD), the mH2A1.2-containing nucleosomes are disassembled and mH2A1.2 may dissociate from its target regions during chromatin remodeling events. It has been recently shown that pioneer activity can be achieved by different strategies. While the prototypic pioneer factor FoxA exploits the homology of its DNA binding domain with linker histone to interact with its DNA motif exposed on nucleosomes (Clark et al., 1993) (Ramakrishnan et al., 1993) (Cirillo and Zaret, 1999) (Cirillo et al., 2002), the reprogramming factor Oct4 can target partial sequences of its DNA binding motif using the two separate PouS and PouHD domains, and Sox2 may take advantage of the pre-bent conformation of its DNA binding motif as well as its nonspecific DNA binding properties (Soufi et al., 2015). To penetrate and remodel closed chromatin MyoD requires the two regions that interact with Pbx (Gerber et al., 1997) (Bergstrom and Tapscott, 2001) (Berkes et al., 2004) and point-mutations abolishing Pbx interaction redirect MyoD binding towards neuronal targets (Fong et al., 2015). Decreased Pbx1 recruitment at Myogenin after mH2A1.2 knock-down was partially rescued by mH2A1.2 overexpression. While the most parsimonious explanation of this phenomenon is that mH2A1.2 favors Pbx1 chromatin engagement, we cannot formally rule out that unidentified factor/s may directly or indirectly be involved. Our data suggest the possibility that by interacting with the macro domain of mH2A1.2, Pbx1 may gain access to repressed chromatin. However, canonical H2A also interacted with Pbx1. Despite the high homology between canonical H2A and the H2A-like domain of mH2A1.2 (Chakravarthy et al., 2005), the latter does not interact with Pbx1, indicating specificity of Pbx1 interaction within the mH2A1.2 moiety. As mH2A1.2 tends to form hybrid nucleosomes containing canonical H2A and H2B (Chakravarthy and Luger, 2006), Pbx1-binding specificity may arise from unique H2A-H2B-mH2A1.2 combinatorial composition of the nucleosomes. The mH2A1 macro domain interacts with histone deacetylases (Chakravarthy et al., 2005) and mH2A1 phosphorylation at serine 137 results in its exclusion from the heterochromatin of the inactive X chromosome (Bernstein et al., 2008). It is therefore possible that post-translational modifications may participate in imparting alternative functions to mH2A1 by modulating protein-protein interactions.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
All cells were cultured at 37°C with 5% CO2. Cell media were supplemented with 500 μg/ml Penicillin-streptomycin-glutamine (GIBCO). Both HEK293 and C2C12 cells (ATCC) were grown in 1x Dulbecco’s modified Eagle’s medium (DMEM) with 10% qualified fetal bovine serum (FBS) (GIBCO). For C2C12 cell differentiation, FBS was replaced with 2% horse serum and 1x insulin-transferrin-selenium (GIBCO). For siRNA experiments cells were transfected with Lipofectamine RNAiMax (Invitrogen) according to Manufacturer’s instructions. siRNA sequences are reported in Table S5. Plasmids were transfected in C2C12 cells using Lipofectamine 2000 (Invitrogen).
Antibodies
A list of the antibodies employed is reported in Extended Experimental Procedures.
Plasmid Construction
Plasmid construction is reported in Extended Experimental Procedures.
Protein expression and purification
GST fusion proteins were expressed in E. coli and purified using Glutathione Sepharose 4B (GE Healthcare Life Sciences) according to manufacturer’s protocol. His-Pbx1a was expressed in E. coli and purified using HisPur Cobalt Resin (Thermo Scientific) according to manufacturer’s protocol.
In vitro protein interaction
Purified His-Pbx1a and GST-macroH2A1.2 proteins were incubated with anti-Pbx1 antibody (Abnova, H00005087) in IP buffer (20 mM Tris-HCl [pH 8.0], 10% glycerol, 0.15 M KCl, 5 mM MgCl2, 0.1% NP40), the complexes bound to protein A agarose (Roche) were washed three times with IP buffer (with 0.5 M KCl) and once with IP buffer (with 0.15 M KCl). The interaction between Pbx1-a and GST proteins was detected by Western Blot with anti-Pbx1 and anti-GST (Santa Cruz, sc-459) antibodies.
Immunoprecipitations
For co-IP, HEK293T cells were co-transfected with plasmids expressing Pbx1a (Addgene, #21029) and Flag-tagged macroH2A1.2 and harvested with lysis buffer (20mM Tris-HCl [pH 8.0], 10% glycerol, 150 mM NaCl, 5 mM MgCl2, 0.1% NP-40, protease inhibitor cocktail). 1mg of whole cell lysate was incubated with anti-flag M2-agarose beads (Sigma). Protein interactions were detected by Western Blot with anti-Pbx1 (Abnova H00005087) and anti-flag (M2, Sigma).
Chromatin Fraction Isolation and Immunoprecipitation
Detailed protocols for chromatin isolation and immunoprecipitation are reported in Extended Experimental Procedures.
ChIP-qPCR and ChIP-Seq
Cells were cross-linked in 1% formaldehyde and processed according to published protocols (Metivier et al., 2003) (Mousavi et al., 2012). Briefly, cells were lysed in RIPA buffer (1x PBS, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) and centrifuged at 2000 rpm for 5 min. The chromatin fraction was sheared by sonication (4x30sec) in 1.5 ml siliconized Eppendorf tubes. The resulting sheared chromatin samples were cleared for 1 hour, immunoprecipitated overnight and washed in buffer I (20 mM TrisHCl pH 8.0, 150 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100), buffer II (20 mM TrisHCl pH 8.0, 500 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100), buffer III (10 mM TrisHCl pH 8.0, 250 mM LiCl, 1% NP- 40; 1% sodium deoxycholate, 1 mM EDTA) and Tris-EDTA (pH 8.0). All washes were performed at 4ºC for 5 min. Finally, cross-linking was reversed in elution buffer (100 mM sodium bicarbonate (NaHCO3), 1% SDS) at 65ºC overnight. Real Time qPCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystem) following standard procedure. List of primers used for qPCR is provided in Table S6. For ChIP-seq, 10ng of immuno-precipitated DNA fragments were used to prepare ChIP-seq libraries with the NEBNext RNA library prep kit (New England BioLabs) and Ovation SP Ultralow DR Multiplex system (Nugen) following the manufacturer’s protocol. The libraries were sequenced for 50 cycles on HiSeq 2000 or HiSeq2500 Illumina instrument.
RNA-Seq
mRNA-seq (poly(A)+ fraction) samples were prepared and processed according to the manufacturer's protocol (Illumina). Briefly, total RNA was extracted from approximately 1×106 C2C12 cells using the Trizol reagent. 500 ng of total RNA were retrotranscribed using High Capacity cDNA Reverse Transcription Kit (Applied Biosystem). qPCR was performed with Power SYBR Green PCR Master Mix (Applied Biosystem). All primers used for amplification are listed in Table S6. 1 μg to 3 μg of total RNA was employed to prepare RNA-seq libraries with the NEBNext RNA library prep kit (New England BioLabs) and Ovation SP Ultralow DR Multiplex system (Nugen) following the manufacturer’s protocol. The libraries were sequenced for 50 cycles (single-end reads) on HiSeq 2000 or HiSeq2500 Illumina instrument,
ATAC-Seq
ATAC-seq was performed according to a published protocol (Buenrostro et al., 2013) with minor modification. Briefly, 5×104 C2C12 cells were pelleted, washed with 50ul of 1xPBS and lysed in 50ul of Lysis Buffer (10mM Tris-HCl, pH7.4, 10mM NaCl, 3mM MgCl2, 0.1% of IGEPAL CA-630). To tag and fragment accessible chromatin, nuclei were centrifuged at 500x g for 10min and re-suspended in 40ul of transposition reaction mix with 2ul Tn5 transposase (Illumina Cat# FC-121-1030). The reaction was incubated at 37ºC with shaking at 300rpm for 30min. DNA-fragments were then purified, and amplified by PCR (12–15 cycles based on the amplification curve). C2C12 MB and MT samples were multiplexed using primers Ad2.1-4 paired with Ad1 for final library amplification as described previously (Buenrostro et al., 2013). Purified libraries were then sequenced on HiSeq2500 Illumina instrument.
Venn diagrams
The area-proportional Venn diagrams were drawn based on images generated using a free online software (http://bioinforx.com/free/bxarrays/venndiagram.php).
Bioinformatic Analysis
RNA-Seq Analysis
Whole transcriptome sequencing (RNA-seq) of C2C12 MB and MT for control and mH2A1.2i in three biological replicates were completed on HiSeq2000/2500 Illumina instruments, using cDNA libraries generated from poly(A)+ purified mRNA samples. 50bp single-end reads were mapped to mouse genome (mm9 assembly) using TopHat (Trapnell et al., 2009) and gene transcript levels were determined via Cuffdiff in the form of FPKM (RPKM) values by correcting for multi reads and using geometric normalization (Trapnell et al., 2013). Up- and down-regulated genes were selected using 1.5-fold change cutoff, and only genes with mean RPKM value of greater than one in at least one condition were included. Gene ontology analyses for selected list of genes were performed by the online bioinformatics resource DAVID (National Institute of Allergy and Infectious Diseases, NIH) (Huang da et al., 2009a, b).
ChIP-seq and ATAC-seq Analyses
ChIP-seq data from two biological replicates for each sample were obtained using HiSeq 2000/2500 Illumina instruments, de-multiplexed through Illumina pipeline, and mapped to the mouse genome (mm9 assembly) using Bowtie algorithm (Langmead et al., 2009) with default parameters except for seed length set to 32 and suppressing all alignments for reads if more than 20 were presented. ChIP-seq data generated from genomic DNA (Input DNA) or IgG were used as a control for calling enriched regions. Peaks for macroH2A1 and Pbx1 were called using MACS version 2 (Zhang et al., 2008) with q-value set at 0.05. Previously published ChIP-seq data for MyoD in MB and MT (Mousavi et al., 2013) were re-analyzed using similar parameters. Regions of open chromatin were identified using MACS from ATAC-Seq data obtained from two biological replicates in C2C12 MB and MT. Only regions called in both replicates were used in downstream analysis. In all cases, redundant reads were removed and only one mapped read to each unique regions of the genome was kept and used in peak calling. Peaks were assigned to promoters if they were located in ±1000bp vicinity of TSS, assigned to intragenic if they were located in gene body excluding +1000bp of TSS, and assigned to intergenic otherwise. For generating the profile of different marks across TSS or ATAC-Seq sites, aligned reads, after removing redundant reads, were directly mapped to sliding windows of 100bp in 25bp steps, at ±2000bp around the center of ATAC-Seq peaks or ±5000bp around the TSS. Signals were averaged across all sites and normalized to the total number of reads for each sample. Profiles and HeatMap and other downstream analysis were done using custom programing in MATLAB. Gene set enrichment analysis were done using GSEA tools (Subramanian et al., 2005) (Mootha et al., 2003) with number of permutation set to 5000, and permutation was applied to gene set. Gene lists were generated by assigning genes to the genomic regions of interest (e.g enhancers) using proximity distance of ±20kbp of gene body (region of interest lies within the interval of [TSS- 20kbp, TES+20kbp]), increasing the proximity distance to ±50kbp or ±100 kbp, whiles increased the number of total and false positive assigned genes, did not returned any new enriched GO terms. Bedtools package (Quinlan and Hall, 2010) was handy for several applications including intersecting regions, generating bedgraph files converted to bigwig files presented in genome browser tracks, filtering reads, etc. Motif enrichment and de-novo motif analysis was carried out using Homer package (Heinz et al., 2010) for the regions of 200bp-long around peaks summit.
Supplementary Material
Acknowledgments
We thank members of the Sartorelli’s laboratory for helpful discussion and technical advice. This work was supported in part by the Intramural Research Program of NIAMS.
Footnotes
ACCESSION NUMBERS
The accession number for the RNA-seq and ChIP-seq datasets reported in this paper is GEO: GSE76010
Supplemental Information includes Supplemental Experimental Procedures, six figures, and five tables and can be found with this article online at
AUTHOR CONTRIBUTIONS
S.D.O., H.Z., and V.S. designed the experiments. S.D.O., A.H.W., H-Y.S. and K.S. conducted the experiments and analyzed the data. H.Z. designed and conducted computational analysis. L.B. and A.G.L. contributed reagents. G.G-C supervised sequencing experiments. J.J.O’S supervised H-Y.S. and analyzed the data. S.D.O., H.Z., and V.S. wrote the manuscript with input from the other authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barrero MJ, Sese B, Kuebler B, Bilic J, Boue S, Marti M, Izpisua Belmonte JC. Macrohistone variants preserve cell identity by preventing the gain of H3K4me2 during reprogramming to pluripotency. Cell reports. 2013;3:1005–1011. doi: 10.1016/j.celrep.2013.02.029. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bell O, Tiwari VK, Thoma NH, Schubeler D. Determinants and dynamics of genome accessibility. Nature reviews Genetics. 2011;12:554–564. doi: 10.1038/nrg3017. [DOI] [PubMed] [Google Scholar]
- Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- Bergstrom DA, Tapscott SJ. Molecular distinction between specification and differentiation in the myogenic basic helix-loop-helix transcription factor family. Mol Cell Biol. 2001;21:2404–2412. doi: 10.1128/MCB.21.7.2404-2412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkes CA, Bergstrom DA, Penn BH, Seaver KJ, Knoepfler PS, Tapscott SJ. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Molecular cell. 2004;14:465–477. doi: 10.1016/s1097-2765(04)00260-6. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Muratore-Schroeder TL, Diaz RL, Chow JC, Changolkar LN, Shabanowitz J, Heard E, Pehrson JR, Hunt DF, Allis CD. A phosphorylated subpopulation of the histone variant macroH2A1 is excluded from the inactive X chromosome and enriched during mitosis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:1533–1538. doi: 10.1073/pnas.0711632105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczynski B, Riddell A, Furlong EE. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nature genetics. 2012;44:148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschbeck M, Uribesalgo I, Wibowo I, Rue P, Martin D, Gutierrez A, Morey L, Guigo R, Lopez-Schier H, Di Croce L. The histone variant macroH2A is an epigenetic regulator of key developmental genes. Nature structural & molecular biology. 2009;16:1074–1079. doi: 10.1038/nsmb.1665. [DOI] [PubMed] [Google Scholar]
- Chakravarthy S, Gundimella SK, Caron C, Perche PY, Pehrson JR, Khochbin S, Luger K. Structural characterization of the histone variant macroH2A. Mol Cell Biol. 2005;25:7616–7624. doi: 10.1128/MCB.25.17.7616-7624.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy S, Luger K. The histone variant macro-H2A preferentially forms “hybrid nucleosomes”. The Journal of biological chemistry. 2006;281:25522–25531. doi: 10.1074/jbc.M602258200. [DOI] [PubMed] [Google Scholar]
- Chen H, Ruiz PD, Novikov L, Casill AD, Park JW, Gamble MJ. MacroH2A1.1 and PARP-1 cooperate to regulate transcription by promoting CBP-mediated H2B acetylation. Nature structural & molecular biology. 2014;21:981–989. doi: 10.1038/nsmb.2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Molecular cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- Cirillo LA, Zaret KS. An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Molecular cell. 1999;4:961–969. doi: 10.1016/s1097-2765(00)80225-7. [DOI] [PubMed] [Google Scholar]
- Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- Costanzi C, Pehrson JR. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature. 1998;393:599–601. doi: 10.1038/31275. [DOI] [PubMed] [Google Scholar]
- Costanzi C, Pehrson JR. MACROH2A2, a new member of the MARCOH2A core histone family. J Biol Chem. 2001;276:21776–21784. doi: 10.1074/jbc.M010919200. [DOI] [PubMed] [Google Scholar]
- Creppe C, Janich P, Cantarino N, Noguera M, Valero V, Musulen E, Douet J, Posavec M, Martin-Caballero J, Sumoy L, et al. MacroH2A1 regulates the balance between self-renewal and differentiation commitment in embryonic and adult stem cells. Mol Cell Biol. 2012;32:1442–1452. doi: 10.1128/MCB.06323-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. From the Cover: Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csankovszki G, Nagy A, Jaenisch R. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. The Journal of cell biology. 2001;153:773–784. doi: 10.1083/jcb.153.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Berkes CA, Bergstrom DA, Dacwag CS, Tapscott SJ, Imbalzano AN. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol Cell Biol. 2005;25:3997–4009. doi: 10.1128/MCB.25.10.3997-4009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyen CM, An W, Angelov D, Bondarenko V, Mietton F, Studitsky VM, Hamiche A, Roeder RG, Bouvet P, Dimitrov S. Mechanism of polymerase II transcription repression by the histone variant macroH2A. Mol Cell Biol. 2006;26:1156–1164. doi: 10.1128/MCB.26.3.1156-1164.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nature protocols. 2012;7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong AP, Yao Z, Zhong JW, Johnson NM, Farr GH, 3rd, Maves L, Tapscott SJ. Conversion of MyoD to a Neurogenic Factor: Binding Site Specificity Determines Lineage. Cell reports. 2015;10:1937–1946. doi: 10.1016/j.celrep.2015.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble MJ, Frizzell KM, Yang C, Krishnakumar R, Kraus WL. The histone variant macroH2A1 marks repressed autosomal chromatin, but protects a subset of its target genes from silencing. Genes & development. 2010;24:21–32. doi: 10.1101/gad.1876110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble MJ, Kraus WL. Multiple facets of the unique histone variant macroH2A: from genomics to cell biology. Cell cycle. 2010;9:2568–2574. doi: 10.4161/cc.9.13.12144. [DOI] [PubMed] [Google Scholar]
- Gaspar-Maia A, Qadeer ZA, Hasson D, Ratnakumar K, Leu NA, Leroy G, Liu S, Costanzi C, Valle-Garcia D, Schaniel C, et al. MacroH2A histone variants act as a barrier upon reprogramming towards pluripotency. Nature communications. 2013;4:1565. doi: 10.1038/ncomms2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AN, Klesert TR, Bergstrom DA, Tapscott SJ. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes & development. 1997;11:436–450. doi: 10.1101/gad.11.4.436. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hussey KM, Chen H, Yang C, Park E, Hah N, Erdjument-Bromage H, Tempst P, Gamble MJ, Kraus WL. The histone variant MacroH2A1 regulates target gene expression in part by recruiting the transcriptional coregulator PELP1. Mol Cell Biol. 2014;34:2437–2449. doi: 10.1128/MCB.01315-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, Zaret KS. Pioneer transcription factors in cell reprogramming. Genes & development. 2014;28:2679–2692. doi: 10.1101/gad.253443.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes & development. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G. H3.3/H2A.Z double variant-containing nucleosomes mark 'nucleosome-free regions' of active promoters and other regulatory regions. Nature genetics. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A, Goldberg MS, Cumberland LK, Ratnakumar K, Segura MF, Emanuel PO, Menendez S, Vardabasso C, Leroy G, Vidal CI, et al. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature. 2010;468:1105–1109. doi: 10.1038/nature09590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustatscher G, Hothorn M, Pugieux C, Scheffzek K, Ladurner AG. Splicing regulates NAD metabolite binding to histone macroH2A. Nature structural & molecular biology. 2005;12:624–625. doi: 10.1038/nsmb956. [DOI] [PubMed] [Google Scholar]
- Ladurner AG. Inactivating chromosomes: a macro domain that minimizes transcription. Molecular cell. 2003;12:1–3. doi: 10.1016/s1097-2765(03)00284-3. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani L, Ballantyne EB, Zhang X, Lupien M. PBX1 genomic pioneer function drives ERalpha signaling underlying progression in breast cancer. PLoS genetics. 2011;7:e1002368. doi: 10.1371/journal.pgen.1002368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maves L, Waskiewicz AJ, Paul B, Cao Y, Tyler A, Moens CB, Tapscott SJ. Pbx homeodomain proteins direct Myod activity to promote fast-muscle differentiation. Development. 2007;134:3371–3382. doi: 10.1242/dev.003905. [DOI] [PubMed] [Google Scholar]
- Maze I, Noh KM, Soshnev AA, Allis CD. Every amino acid matters: essential contributions of histone variants to mammalian development and disease. Nature reviews Genetics. 2014;15:259–271. doi: 10.1038/nrg3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature genetics. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Mousavi K, Zare H, Dell'orso S, Grontved L, Gutierrez-Cruz G, Derfoul A, Hager GL, Sartorelli V. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Molecular cell. 2013;51:606–617. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi K, Zare H, Wang AH, Sartorelli V. Polycomb protein Ezh1 promotes RNA polymerase II elongation. Molecular cell. 2012;45:255–262. doi: 10.1016/j.molcel.2011.11.019. [DOI] [PubMed] [Google Scholar]
- Pasque V, Gillich A, Garrett N, Gurdon JB. Histone variant macroH2A confers resistance to nuclear reprogramming. The EMBO journal. 2011;30:2373–2387. doi: 10.1038/emboj.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehrson JR, Fried VA. MacroH2A, a core histone containing a large nonhistone region. Science. 1992;257:1398–1400. doi: 10.1126/science.1529340. [DOI] [PubMed] [Google Scholar]
- Podrini C, Koffas A, Chokshi S, Lelliott CJ, White JK, Adissu HA, Williams R, Greco A. MacroH2A1 isoforms are associated with epigenetic markers for activation of lipogenic genes in fat-induced steatosis. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2014 doi: 10.1096/fj.14-262717. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan V, Finch JT, Graziano V, Lee PL, Sweet RM. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature. 1993;362:219–223. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- Rasmussen TP, Huang T, Mastrangelo MA, Loring J, Panning B, Jaenisch R. Messenger RNAs encoding mouse histone macroH2A1 isoforms are expressed at similar levels in male and female cells and result from alternative splicing. Nucleic acids research. 1999;27:3685–3689. doi: 10.1093/nar/27.18.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnakumar K, Duarte LF, LeRoy G, Hasson D, Smeets D, Vardabasso C, Bonisch C, Zeng T, Xiang B, Zhang DY, et al. ATRX-mediated chromatin association of histone variant macroH2A1 regulates alpha-globin expression. Genes & development. 2012;26:433–438. doi: 10.1101/gad.179416.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics C. Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Henikoff S. Histone variants in pluripotency and disease. Development. 2013;140:2513–2524. doi: 10.1242/dev.091439. [DOI] [PubMed] [Google Scholar]
- Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, Zaret KS. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell. 2015;161:555–568. doi: 10.1016/j.cell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe D, Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977;270:725–727. doi: 10.1038/270725a0. [DOI] [PubMed] [Google Scholar]
- Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes & development. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome research. 2011;21:1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.