Abstract
Idiopathic fetal growth restriction (FGR) is frequently associated with placental insufficiency. Previous reports have provided evidence that endocrine gland–derived vascular endothelial growth factor (EG-VEGF), a placental secreted protein, is expressed during the first trimester of pregnancy, controls both trophoblast proliferation and invasion, and its increased expression is associated with human FGR. In this study, we hypothesize that EG-VEGF-dependent changes in placental homeobox gene expressions contribute to trophoblast dysfunction in idiopathic FGR. The changes in EG-VEGF-dependent homeobox gene expressions were determined using a homeobox gene cDNA array on placental explants of 8–12 wks gestation after stimulation with EG-VEGF in vitro for 24 h. The homeobox gene array identified a greater-than-five-fold increase in HOXA9, HOXC8, HOXC10, HOXD1, HOXD8, HOXD9 and HOXD11, while NKX 3.1 showed a greater-than-two-fold decrease in mRNA expression compared with untreated controls. Homeobox gene NKX3.1 was selected as a candidate because it is a downstream target of EG-VEGF and its expression and functional roles are largely unknown in control and idiopathic FGR-affected placentae. Real-time PCR and immunoblotting showed a significant decrease in NKX3.1 mRNA and protein levels, respectively, in placentae from FGR compared with control pregnancies. Gene inactivation in vitro using short-interference RNA specific for NKX3.1 demonstrated an increase in BeWo cell differentiation and a decrease in HTR-8/SVneo proliferation. We conclude that the decreased expression of homeobox gene NKX3.1 downstream of EG-VEGF may contribute to the trophoblast dysfunction associated with idiopathic FGR pregnancies.
INTRODUCTION
Fetal growth restriction (FGR), also known as intrauterine growth restriction, is a pregnancy complication in which a fetus fails to reach its genetically determined growth potential in utero. Impairment in fetal development and growth in utero has been associated with lifelong adverse health sequelae, including diabetes and cardiovascular diseases (1,2), therefore, understanding the molecular mechanism of human FGR is of utmost importance. Obvious maternal, fetal and placental causes of FGR account only for one third of FGR cases (3), while the remaining two thirds are termed idiopathic. Idiopathic FGR is often associated with placental insufficiency (4) where the placentae are often smaller and show numerous morphological defects such as reduced trophoblast proliferation and abnormal villous vasculature with less branched terminal villi (5).
During the first trimester of human pregnancy, extravillous cytotrophoblasts (EVCT) grow from anchoring villi, invade the maternal decidua and remodel the uterine spiral arterioles (6). On the other hand, the mononuclear villous cytotrophoblasts (VCT) on the floating villi continue to proliferate and differentiate into the multinucleated syncytiotrophoblasts (ST). The ST layer plays a key role throughout pregnancy. It is the site for several important placental functions, including nutrient and ion exchange and the synthesis of hormones required for fetal growth and development (7,8). Thus, a syncytium that functions efficiently requires both fusion and differentiation of cytotrophoblasts (CTB). A dysfunction in CTB differentiation and proliferation leads to deficient EVCT invasion and ST apoptosis, both hallmarks of pregnancy pathologies, including human idiopathic FGR (6,9).
The proliferation, differentiation and invasion of the first-trimester human CTBs are influenced by multiple regulatory factors, including adhesion molecules, growth factors, cytokines, proteases, matrix-derived components, transcription factors and oxygen tension (10). Growth factors such as epidermal growth factor (EGF), hepatocyte growth factor (HGF) and transforming growth factor-β1 (TGF-β1), are widely studied as regulators of CTB proliferation, differentiation, migration and invasion (11–13).
In the last decade, we and others have been interested in the study of a new angiogenic factor, endocrine gland–derived vascular endothelial growth factor (EG-VEGF), that is characterized, sequenced and shown to be expressed in testis, adrenal gland, ovary and placenta (14). Human EG-VEGF exerts its effects via G protein–coupled receptors (PROKRs), termed PROKR1 and PROKR2 (15). We have recently shown that EG-VEGF and its receptors are highly expressed in the human placenta during the first-trimester of pregnancy, with the highest expression found in the ST; that its expression is up-regulated by hypoxia; and that its circulating levels are significantly higher during early pregnancy and in pregnancy pathologies including FGR and pulmonary embolism (PE) (16–19). In relation to placental development, we have demonstrated that EG-VEGF controls CTB proliferation, survival, differentiation and vascular development during the first-trimester of pregnancy, however, whether EG-VEGF control of these activities is a direct or an indirect process is far from being understood.
Growth factors including EG-VEGF and HGF have been shown to modulate the expression of the placental homeobox gene, HLX, and to alter CTB proliferation and migration (20,21). Homeobox genes, among other transcription factors, are known to play key roles in placental development and function (22–24). Also, it is proposed that differential responses to fluctuating hormone, or growth factor signals, such as those influenced by EG-VEGF, may constitute key controlling mechanisms for homeobox gene expressions in the human placenta (20).
In this study, we hypothesize that EG-VEGF-dependent change in placental homeobox gene expressions contribute to trophoblast dysfunction in idiopathic FGR. To investigate this, we employed first-trimester placental explant cultures and treated them with EG-VEGF to identify the candidate homeobox gene that showed a downregulated expression downstream of EG-VEGF in FGR placentae. The candidate gene was then validated in placentae obtained from third-trimester human idiopathic FGR and gestation-matched uncomplicated control pregnancies. Further, the candidate gene’s functional role on trophoblast proliferation and differentiation was determined using trophoblast-derived cell lines, in vitro.
MATERIALS AND METHODS
First-Trimester Placental Tissues
Human placentae from 8 to 14 wks gestation (n = 12) and second trimester pregnancies of 20 wks gestation (n = 3) were obtained from elective terminations. First trimester and second trimester placentae collection and processing were performed with the approval of the University Hospital ethics committees at the Broussais Hospital, Paris, France, and Grenoble University Hospital, Grenoble, France, and with the informed written consent of each patient.
Patient Details and Placental Tissue Sampling from Human Idiopathic FGR and Gestation-Matched Uncomplicated Control Pregnancies
Placentae from pregnancies complicated by idiopathic FGR (n = 25) and those from gestation-matched uncomplicated pregnancies as controls (n = 25) were collected following Cesarean section or vaginal delivery; gestation times ranged from 27 to 40 wks. Placenta collection was approved by the Research and Ethics Committee of The Royal Women’s Hospital, Melbourne. All patients were informed and consented. Table 1 reports the clinical characteristics of both the FGR and gestation-matched control groups included in this study (25,26). As illustrated in Table 2, the inclusion criteria for the FGR study group were a birth weight of less than the 10th percentile for gestation as determined by the Australian fetal growth charts (27), and at least two of the following diagnoses on antenatal ultrasound: abnormal umbilical artery Doppler flow velocimetry, oligohydramnios (Amniotic Fluid Index <7), or asymmetric growth of the fetus (head circumference-to-abdominal circumference ratio >95th percentile for gestation). Exclusion criteria for both cases and control groups were factors that are known causes of FGR and included maternal smoking and drug dependency during pregnancy, other underlying maternal diseases including gestational diabetes, preeclampsia, hypertension, placental abruption, prolonged rupture of membranes, fetal congenital anomalies and suspected intrauterine infection (25,26). Uncomplicated control pregnancies were selected to match FGR cases according to gestation times. All control patients gave birth to normally formed babies with birth weights appropriate for gestational age, and the placentae from these patients were also grossly normal (25,26).
Table 1.
Clinical characteristics of patient samples.
| Characteristics | FGR (n = 25) | GMC (n = 25) | Significance |
|---|---|---|---|
| Maternal characteristics | |||
| Gestational age (weeks)a | 34.4 ± 6.5 | 35.8 ± 6.6 | p = 0.25 |
| Maternal age (years)a | 33.2 ± 5.7 | 31.9 ± 6.5 | p = 0.4 |
| Placental weight (g)a | 409.3 ± 110.3 | 525.0 ± 148.2 | p < 0.005 |
| Parityb | |||
| Primiparous | 12 | 10 | p = 0.56 |
| Multiparous | 13 | 15 | |
| Mode of deliveryb | |||
| Vaginal | 10 | 6 | p = 0.35 |
| Cesarean section in labor | 3 | 2 | |
| Cesarean section not in labor | 12 | 17 | |
| New born characteristics | |||
| Genderb | |||
| Male | 8 | 12 | p = 0.25 |
| Female | 17 | 13 | |
| Birth weight | |||
| Weight (g)a | 2,051.4 ± 637.0 | 2,603.8 ± 857.0 | p < 0.05 |
| 10th–90th percentile | 0 | 25 | |
| 5th–10th percentile | 11 | ||
| <5th percentile | 14 | ||
Significance was determined by the Student t test. Results are expressed as mean ± standard error of the mean (SEM).
Significance was determined by the chi-square test. Results are expressed as mean ± standard error of the mean (SEM).
Table 2.
Clinical criteria for FGR-affected pregnancies.a
| Clinical characteristics | Number of samples |
|---|---|
| Birth weight <10th percentile | 25/25 |
| Abnormal umbilical artery Doppler velocimetry | |
| Elevated | 7/25 |
| Reversed | 6/25 |
| Absent | 8/25 |
| Not recorded | 4/25 |
| Head circumference to abdominal circumference ratio | |
| Asymmetry HC: AC >1.2 | 21/25 |
| Not recorded | 4/25 |
| Amniotic fluid index | |
| Oligohydramnios, AFI <7 | 18/25 |
| Normal, AFI ≥7 | 7/25 |
All FGR-affected pregnancies in this study met with the first selection criterion of birth weight being less than 10th percentile and at least two of the other ultrasound-determined selection criteria. Seventeen out of 25 FGR-affected pregnancies met all three ultrasound-determined selection criteria.
Placental bed samples (n = 10 each) from FGR and control pregnancies were collected with informed patient consent from the Department of Obstetrics and Gynecology at the hospital La Citadelle, Liège, Belgium, with the approval by the local ethics committee. Seventy percent of FGR cases were reported with abnormal umbilical Doppler velocimetry, while 20% FGR cases displayed reverse diastolic blood flow. FGR cases were associated with a significant decrease in both placental and infant birth weight (p < 0.05) (28).
All samples were processed within 20 min of placental delivery with tissues excised from randomly selected areas of central cotyledons following the removal by dissection of any attached decidua. Tissue pieces were thoroughly washed in 0.9% phosphate buffered saline (PBS) to minimize blood contamination, and then snap frozen and stored at −80°C for RNA and protein analysis or fixed in 10% formalin for immunolocalization studies.
Tissue and Cell Culture
Human villous explant cultures
First trimester human placentae were used to establish villous explant cultures (10–12 wks gestation, n = 6) as described previously (17). Fragments of placental villi (15–20 mg) were teased apart and seated on a transparent Biopore membrane (Millipore Corp.) of 12-mm diameter (inserts with a pore size of 0.4 μm). The inserts were precoated with 0.2 mL undiluted Matrigel (Becton Dickinson), polymerized at 37°C for 30 min and transferred to a 24-well culture dish. Explants were cultured in medium containing DMEM-Ham’s F-12 (DMEM/F12) (Invitrogen) supplemented with 0.25 μg/mL ascorbic acid, 100 μg/mL streptomycin and 100 IU/mL penicillin at pH 7.4. Villous explants were kept in culture for up to 72 h. The viability of the explants was assessed as described previously (17) by measuring human chorionic gonadotropin (hCG) production in the culture medium every 24 h. After 24 h culture, the medium was changed and explants incubated in the absence or presence of 50 ng/mL or 5 nmol/L recombinant human EG-VEGF (Tebu) (20).
Isolation and purification of villous (VCT) and extravillous (EVCT) trophoblasts
VCTs and EVCTs were prepared from first-trimester placental tissues (n = 12) as described previously (29). Briefly, trophoblasts were enriched by differential sequential trypsin digestion and the use of a Percoll gradient (20% to 60%). EVCTs were plated at a density of 5 × 104 cells/cm2 on Matrigel-coated (5 mg/mL) (BD Biosciences) 35-mm Falcon culture dishes and grown in HAM-F12/DME mol/L (v/v) supplemented with 10% fetal bovine serum (FBS), 100 IU/mL penicillin, 100 μg/mL streptomycin and 2 mmol/L glutamine. EVCT cell purity was assessed by positive staining for the specific markers of human invasive EVCTs, cytokeratin-7 (CK7) and human leukocyte antigen-G expression (29). VCTs were plated at a density of 1.5 × 104 cells/cm2 on 35 mm or 60 mm TPP (Techno Plastic Products) tissue culture dishes using DME mol/L supplemented with 10% FBS, 2 mmol/L glutamine, 100 μg/mL streptomycin and 100 IU/mL penicillin. The purity of VCT cultures was characterized by positive staining for CK7 (95% positive cells) and by microscopic observation of cell aggregates and syncytiotrophoblast formation at 48 to 72 h (29).
Human placental trophoblast-derived cell lines
The human placental trophoblast-derived choriocarcinoma BeWo cells (B30 clone) were grown in RPMI-1640 culture media supplemented with 10% FBS, 100 μg/mL streptomycin and 100 IU/mL penicillin (Invitrogen) and incubated at 37°C, 5% CO2 and 95% air. The HTR-8/SVneo cells were immortalized with the SV40 virus (30). In the present study, cells between 40–50 passages were used and grown in RPMI 1640 supplemented with 5% FBS, 100 μg/mL streptomycin, 100 IU/mL penicillin and amphotericin B (Invitrogen). Cells were maintained at 37°C in an atmosphere of 5% CO2 and 95% air.
Total RNA Extraction and cDNA Preparation
Total RNA from placental villous and placental bed tissues and cultured cells was extracted using the RNeasy Midi and Mini kits, respectively, as described previously (25). cDNA was prepared from 2 μg total RNA reverse-transcribed using Superscript III ribonuclease H-reverse transcriptase (Invitrogen) in a two-step reaction also as described previously (25,31).
Homeobox Gene cDNA Array
The Human HOX TaqMan PCR Array (Applied Biosystems) for gene profiling was used to screen for genes that showed differential expressions in EG-VEGF treated first-trimester placental explant cultures compared with untreated placental explant cultures. Briefly, 2 ng/well cDNA was used in a 20 μL reaction by the addition of TaqMan Master Mix (Applied Biosystems) and distributed into a TaqMan Array 96-well plate. The Human HOX Array plate contained 84 gene-specific primer sets with a panel of five housekeeping gene primers for normalization. The housekeeping genes consisted of 18S ribosomal RNA (18S), β-2-microglobulin (B2M), hypoxanthine phosphoribosyltransferase 1 (HPRT1), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin (ACTB). PCR was completed in an ABI Prism 7500 Sequencer according to the manufacturer’s instructions. The relative gene expression values, or fold changes, were analyzed using the DataAssist Software v3.0 (Applied Biosystems). Genes of interest were identified and prioritized based on the difference in gene mRNA expression in EG-VEGF treated placental tissues when compared with untreated control placental tissues.
Real-time PCR
Relative quantitation of mRNA was performed in an ABI Prism 7500 Sequencer using the Applied Biosystems prevalidated assay mix. Each assay mix consisted of unlabeled PCR primers and a TaqMan FAM-labeled MGB probe (NKX3.1 Hs00171834_m1; or gene that codes for chorionic gonadotropin-β [CGB] Hs00361224_g1, B-cell CLL/lymphoma 2 [BCL2] Hs00608023-m1 and BCL2 associated X-protein [BAX] Hs00180269_m1 [Applied Biosystems]). Gene expressions relative to the eukaryotic 18S rRNA endogenous control (#4319413E, VIC/MGB probe) (Applied Biosystems) were calculated according to the 2−ΔΔCT method (32).
Immunoblotting
Total cellular and tissue protein was extracted as described previously (25). Immunoblotting was performed with 25 μg of total protein using a 10% SDS/PAGE and electroblotting onto a nitrocellulose membrane (Pal Gelman) The membrane was blocked with 5% (w/v) skim milk for 1 h at room temperature, and followed by an overnight incubation in 0.02 μg/μL rabbit anti-human polyclonal NKX3.1 (H-50, SC25405, Santa Cruz Biotechnology) at 4°C. Antibody binding was visualized using horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (Invitrogen/Life Technologies), followed by autoradiography using the ECL-Western Chemiluminescence Detection Kit (GE Healthcare). Total protein load on the gel was determined by Coomassie staining, and the immunoreactive NKX3.1 protein was analyzed by scanning densitometry (ImageQuant, GE Healthcare) as described previously (25,26,33,34).
Immunofluorescence
Spatio-temporal distribution and localization of NKX3.1 protein in first- trimester and third-trimester FGR and control placental tissue sections was by immunofluorescence. Briefly, paraffin-embedded placental tissue sections, cut to 5-μm thickness, were dewaxed in xylene, hydrated in graded ethanol (100% to 50% ethanol) and blocked with 1% bovine serum albumin (BSA) prepared in PBS. Tissue sections were then incubated overnight at 4°C with the primary antibody, either rabbit anti-human NKX3.1 or mouse monoclonal anti-CK7 (RCK 105 clone, Abcam), at a concentration of 0.01 μg/μL in 1% BSA/PBS. Control sections were incubated with 0.02 μg/μL mouse IgG nonimmune rabbit serum prepared in 1% BSA/PBS (Dako). Fluorescence detection was by Alexa Fluor 488 and counterstaining with DAPI (Dako) according to the manufacturer’s recommendations.
Forskolin Mediated BeWo Cell Differentiation
BeWo cells were seeded at a density of 2 × 105 cells/well and serum starved in RPMI-1640 medium supplemented with 0.1% BSA (w/v). Forskolin, (FSK) (Invitrogen/Life Technologies) prepared in 1% dimethyl sulfoxide (DMSO), was added to a final concentration of 10 μmol/L (35) to induce differentiation over 72 h in culture. At the end of the incubation period, the medium was collected and stored at −20°C and the cells were processed for RNA and protein analysis. Untreated cells were maintained in RPMI 1640 medium supplemented with 1% DMSO and used as the control. Each experimental condition was implemented in duplicate, and all cell culture experiments repeated on three independent occasions.
β-hCG Protein Assay
Determination of β-hCG protein levels was by ELISA (α Diagnostic International) and carried out as described previously (36). The assay quantitation was performed on cell culture supernatant collected from n = 6 independent experiments. The minimum detection limit of this assay was 1.5 mIU/mL.
Gene Inactivation of NKX3.1 Expression in BeWo and HTR-8/SVneo Cells
BeWo cells were seeded at a density of 2 × 105 cells/well in 6-well plates and treated with FSK. They were then transfected with NKX3.1 siRNA (19 to 21 base pairs, Thermo Fisher Scientific Inc.) using the Hiperfect transfection reagent (Invitrogen). The optimum siRNA:RNAiFect ratio and incubation time for the culture was determined to be 1:6 and 72 h, respectively (data not shown).
HTR-8/SVneo cells were also seeded at a density of 2 × 105 cells/well in 6-well plates with supplemented culture medium and transfected as above for 48 h. Negative control (NC) siRNA used in these experiments consisted of an enzyme-generated pool of oligonucleotides (15–19 base pairs) that showed no DNA sequence similarity to any known human gene (AllStars Neg. siRNA AF 488, Qiagen).
Inhibition of EG-VEGF Receptors
To determine the role of EG-VEGF receptor-mediated signaling on NKX3.1 expression, BeWo cells grown in either normoxic or in hypoxia (2%) were treated with EG-VEGF receptor antagonists, PC-7 for PROKR1 (37) at 1 μmol/L and PKRA505 for PROKR2 (38) at 1 μmol/L, and incubated for 8 h. NKX3.1 mRNA was determined using real-time PCR as described above. Specificity of both antagonists was recently characterized (37,38).
Human Apoptosis cDNA Array
Human apoptosis cDNA arrays (Applied Biosystems) were used for gene profiling and to screen for genes that showed differential expression in BeWo and HTR-8/SVneo cells treated with NKX3.1 siRNA and NC siRNA. Briefly, cDNA was prepared and added to a TaqMan Master Mix (Applied Biosystems) and distributed in a TaqMan Array 96-well plate at approximately 2 ng/well in a 20 μL reaction. The array plate contained 84 gene-specific primer sets with a panel of five housekeeping gene primers for normalization. The housekeeping genes consisted of 18S ribosomal RNA (18S), β-2-microglobulin (B2M), hypoxanthine phosphoribosyltransferase 1 (HPRT1), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin (ACTB). PCR was performed in an ABI Prism 7500 Sequence Detector under the following cycling parameters: 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s and primer extension at 60°C for 1 min. Data (Ct values) were analyzed using the ABI Sequence Detector System software version 2.0 and the relative gene expression values, or fold changes, were analyzed using the DataAssist Software v3.0 (Applied Biosystems) and normalized to the average Ct value of the five housekeeping genes. Genes were prioritized based on the difference in gene mRNA expression in NKX3.1 siRNA treated cells when compared with NC.
The xCELLigence System
The functional consequence of NKX3.1 gene inactivation on HTR-8/SVneo cell proliferation was assessed by real-time monitoring of the xCELLigence RTCA DP system (ACEA Biosciences), as described previously (39). Briefly, the assay was performed in E-plates at 37°C, 5% CO2 and 95% humidified air. The xCELLigence system measures cell attachment expressed as an arbitrary parameter called the cell index (CI) and each experiment was monitored for at least 48 h.
Data Analysis
The significance of any differences between the clinical characteristics of the FGR-affected and the control pregnancies was by either the chi-square test or Student t test with the values described as mean ± SEM. The differences in mRNA and protein expressions of the FGR-affected placentae compared with gestation-matched controls were assessed by the Mann–Whitney U test. The value p <0.05 was considered significant and data analysis was performed using GraphPad Prism 5 (GraphPad Software Inc.).
RESULTS
Homeobox Gene Expression in Placental Explants
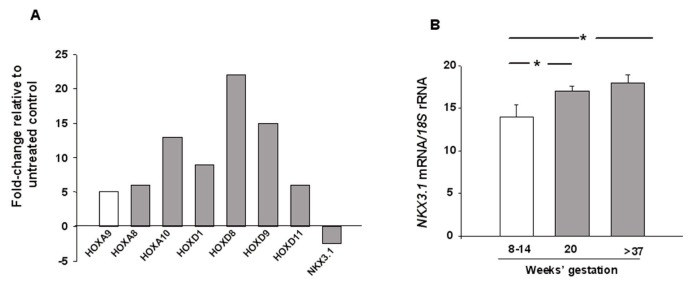
Figure 1A shows the homeobox gene expression profile identifying a greater-than-five-fold mRNA expression increase of HOXA9, HOXC8, HOXC10, HOXD1, HOXD8, HOXD9 and HOXD11, and a greater-than-two-fold decrease of NKX3.1 in placental explants stimulated with EG-VEGF compared with untreated controls. Homeobox gene NKX3.1 was the only gene that showed reduced expression in the presence of EG-VEGF, therefore, it was selected as the candidate gene for further gene expression validation and investigation of its functional role in trophoblastic proliferation and differentiation.
Figure 1.
Candidate gene identification and placental expression throughout pregnancy. (A) Effect of EG-VEGF on homeobox gene expression profile in first-trimester placental explants (n = 6). Real-time PCR was used to detect homeobox gene mRNA relative to 18S rRNA. Results are reported as fold changes relative to untreated controls. (B) Quantification of NKX3.1 mRNA expression levels in placental tissues at 8 to 14 wks gestation (n = 12), 20 wks gestation (n = 3) and at term (n = 6). 18S rRNA was used as the internal control. Results are reported as the mean ± SEM. Data were analyzed using the Mann–Whitney U test (significant differences when compared with the 8 to 14 wks gestation period, *p < 0.05).
Effect of Gestation on NKX3.1 mRNA Expression in Placental Tissues
NKX3.1 mRNA relative to 18S rRNA was determined in placental tissues at 8–14 wks gestation, 20 wks gestation and at term (>37 wks). As depicted in Figure 1B, a significant increase in NKX3.1 expression relative to 18S rRNA was observed in placental tissues with increasing gestation times.
FGR and Control Patient Characteristics
Table 1 describes the clinical features of FGR and the gestation-matched uncomplicated control pregnancies used in this study. Maternal age, parity, gestational age and mode of delivery were not significantly different between the two groups, conversely, mean placental weights and birth weights were significantly lower in FGR compared with gestation-matched control pregnancies (Table 1, p < 0.025). Using these placental samples, we have already demonstrated reliable gene expression differences for other homeobox genes (25,26,31). Table 2 describes the samples that were collected from a clinically well-defined cohort of FGR-affected pregnancies.
NKX3.1 Expression in FGR
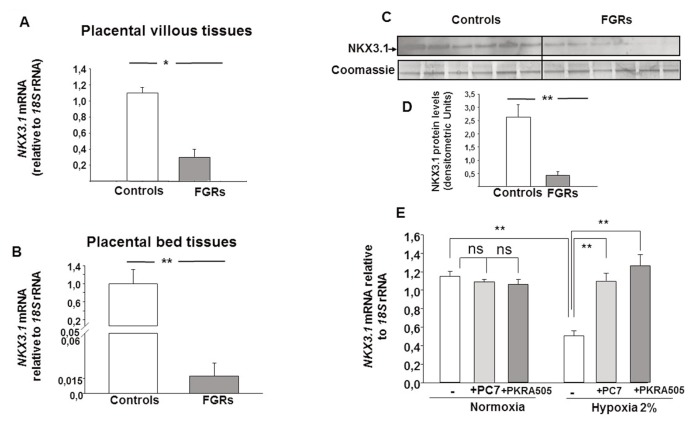
NKX3.1 mRNA levels and protein expressions were validated in FGR and control placentae by using real-time PCR and immunoblotting, respectively (Figure 2). The relative level of placental NKX3.1 mRNA was significantly decreased in placental villous tissues (Figure 2A) and in placental bed tissues (Figure 2B) in FGRs compared with controls. The decrease in NKX3.1 mRNA level was also further confirmed at the protein level. Figure 2C shows a representative immunoblot for the immunoreactive NKX3.1 protein (35 kDa) in FGR-affected placentae and in gestation-matched controls. A Coomassie-stained gel demonstrated equal loading of total placental protein, while densitometry showed that the immunoreactive placental NKX3.1 protein was decreased significantly in FGRs compared with controls (Figure 2D).
Figure 2.
Expression of NKX3.1 downregulated in FGR pregnancies. (A) Quantification of NKX3.1 mRNA expression levels in placental tissues from gestation-matched control (n = 25) and FGR pregnancies (n = 25). 18S rRNA was used as an internal control. (B) Quantification of NKX3.1 mRNA expression levels in placental bed tissues. 18S rRNA was used as an internal control. (C) Representative immunoblot for NKX3.1 that compares gestation-matched control (n = 6) versus FGR placentae (n = 6). Coomassie-stained gel depicting total protein load. Quantification of the intensity of the bands is illustrated in (D). Results are reported as the mean ± SE mol/L from n = 3 independent experiments. Data were analyzed using the Mann–Whitney U test (significant differences, *p < 0.05). Panel (E) reports NKX3.1 mRNA expression levels in BeWo cells that were incubated under normoxic or hypoxic conditions for 8 h, in the absence or presence of PROKR1 (PC7) or PROKR2 (PKRA505) antagonists. Both antagonists were used at 10 μmol/L. (significant differences, *p < 0.05; **p < 0.01).
Effect of Inhibition of EG-VEGF Receptors on NKX3.1 mRNA under Normoxic or Hypoxic Conditions
To determine the role of EG-VEGF receptor-mediated signaling on NKX3.1 expression, we performed an experiment using BeWo cells that were treated with EG-VEGF receptor inhibitors PC-7 for PROKR1 and PKRA505 for PROKR2 at 10 μmol/L concentrations each and incubated for 8 h under normoxic (20%) or hypoxic (2%) conditions (hypoxia being an environment that mimics the FGR pathology and is known to increase EG-VEGF expression and production) (17). Figure 2E shows that under normoxic conditions NKX3.1 mRNA was not significantly altered in the presence of inhibitors of EG-VEGF receptors in both BeWo. Interestingly, hypoxia induced a significant reduction in NKX3.1 mRNA expression that was completely abolished in the presence of PROKR1 and PROKR2 antagonists.
NKX3.1 Expression in Normal and FGR Human Placentae
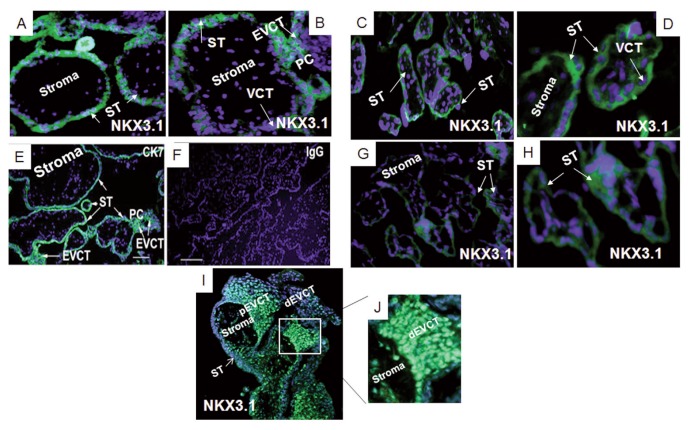
NKX3.1 protein localization in first-trimester placentae was detected in the nuclei of CTBs, the ST and in proliferating EVCTs of the proximal cell column (Figures 3A, B). In the third-trimester FGR (Figures 3G, H) and gestation-matched control placentae (Figures 3C, D) immunoreactive NKX3.1 protein was detected predominantly in the cytoplasm of the ST. Immunoreactivity to CK7 was used as a positive control in a term placental section (Figure 3E) while rabbit IgG was used as a negative control (Figure 3F). Figures 3I and 3J show nuclear immunostaining of NKX3.1 in placental columns from first trimester human placenta. NKX3.1 is mainly localized to the nuclei of villi cytotrophoblast cells and in the proximal extravillous trophoblast cells (pEVCT), that is, proliferative proximal cells within the column. NKX3.1 expression was decreased in distal EVCT (dEVCT). Photograph in Figure 3J is a higher magnification of the image reported in Figure 3I.
Figure 3.
NKX3.1 protein expression in first and third-trimester placental sections. Localization of NKX3.1 protein in a first-trimester placenta (A and B, n = 6) in a third-trimester FGR placenta (G and H, n = 6) and a gestation-matched control placenta (C and D, n = 6). NKX 3.1 staining (rabbit anti-human, green). Panels (E) and (F) show representative controls: cytokeratin7 (CK7) used as a positive control in a term placental section (E), rabbit IgG used as a negative control (F). Panels (I) and (J) show representative staining of the human placental column during the first trimester of pregnancy. Panel (J) is a higher magnification of the photograph in panel (I). Blue DAPI stain. Villous cytotrophoblast (VCT), extravillous cytotrophoblast (EVCT), proximal extravillous trophoblast cells (pEVCT) and distal EVCT (dEVCT), syncytiotrophoblast (ST), proximal cell column (PC). Panels (G) and (H), 200× magnification, and the scale bars are 100 μm.
In Vitro Differentiation and NKX3.1 Expression in Placental Cells
NKX3.1 mRNA levels were detected in cultured VCTs and EVCTs isolated from first-trimester placentae. In vitro differentiation of VCTs into ST occurred at 72 h and was associated with a significant increase in NKX3.1 mRNA, while NKX3.1 mRNA levels were detected in proliferating EVCTs at 24 h and decreased significantly in invasive EVCTs after 48 h (Figures 4A).
Figure 4.
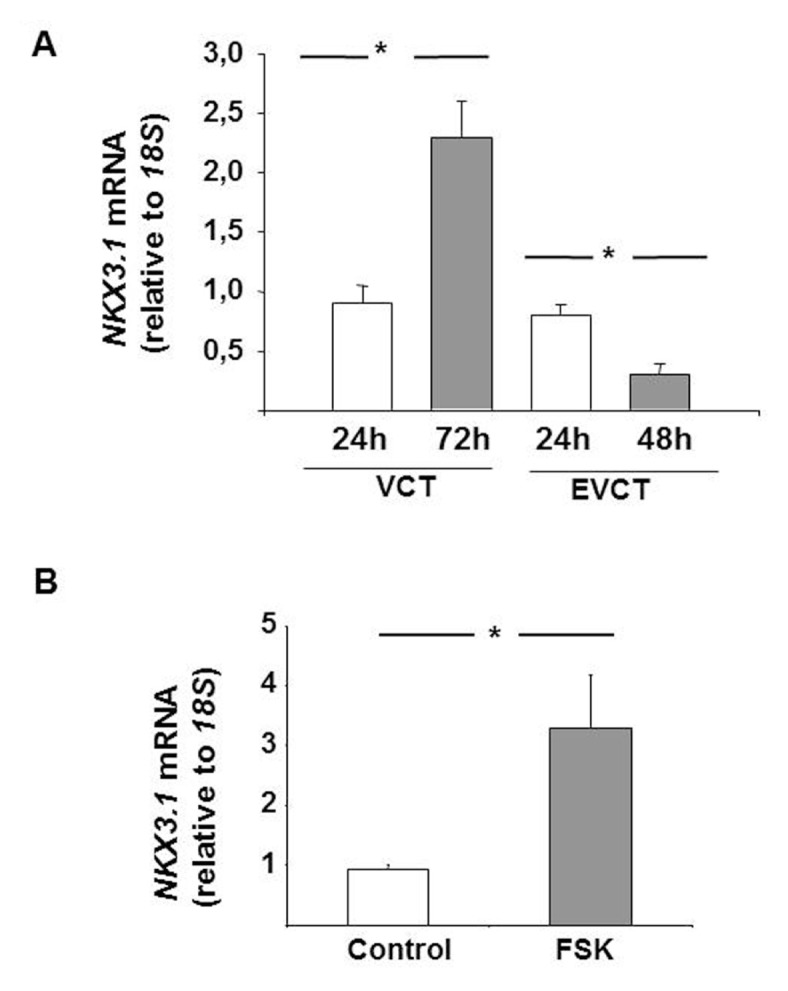
Effect of in vitro differentiation on NKX3.1 mRNA expression in placental cells. (A) Quantification of NKX3.1 mRNA expression levels in primary VCT and EVCT cultures from first-trimester placentae (n = 12). 18S rRNA was used as an internal control. (B) Effects of forskolin (10 μmol/L, 72 h) on NKX3.1 mRNA expression levels in BeWo cells. 18S rRNA was used as an internal control. Results are reported as the mean ± SE mol/L from n = 3 independent experiments. Data were analyzed using the Mann–Whitney U test (significant differences, *p < 0.05).
Effect of NKX3.1 Gene Inactivation on βhCG Production in BeWo Cells
To determine NKX3.1 expression during differentiation of BeWo cells, FSK was used. This induced a significant increase in NKX3.1 mRNA compared with the vehicle control (Figure 4B).
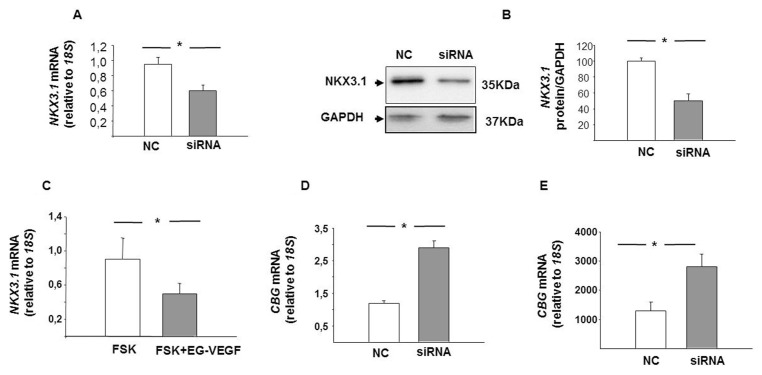
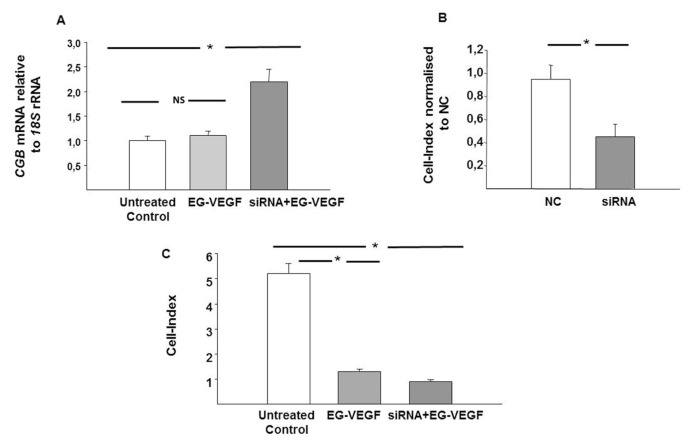
The functional consequence of a reduced placental NKX3.1 in FGR was modeled in cell culture by inactivating the NKX3.1 expression using NKX3.1 siRNA in FSK-induced BeWo cells. As shown in Figure 5, a significant decrease in NKX3.1 mRNA (Figure 5A) and protein (Figure 5B) was observed in the BeWo cells thus treated compared with the NC control. EG-VEGF treatment in FSK-induced BeWo cells also showed a significant decrease in NKX3.1 mRNA expression (Figure 5C). The effect of NKX3.1 inactivation on BeWo cell differentiation, as determined by real-time PCR and immunoassays, was an increased expression of CGB mRNA and β-hCG protein in NKX3.1 siRNA-treated cells compared with NC siRNA-treated cells (Figures 5D, E).
Figure 5.
Effect of NKX3.1 downregulation on cell differentiation in BeWo cells. (A) Quantification of the downregulation of NKX3.1 mRNA expression following treatment with NKX3.1 siRNA. (B) Shows a representative Western blot and quantification of the downregulation of NKX3.1 protein following the treatment with NKX3.1 siRNA. Cells were analyzed for the downregulation of NKX3.1 mRNA and protein expressions 72 h after transfection. Nonspecific siRNA served as control siRNA. (C) Expression of NKX3.1 mRNA in FSK ± EG-VEGF-treated BeWo cells. (D) Effect of NKX3.1 downregulation on CGB mRNA expression and β-hCG protein secretion in BeWo cells. Results are reported as the mean ± SE mol/L from n = 3 independent experiments. Data were analyzed using the Mann–Whitney U test (significant differences, *p < 0.05).
EG-VEGF Stimulation and NKX3.1 Inactivation Effect on Trophoblast Differentiation
The trophoblast differentiation marker CGB was measured following NKX3.1 siRNA transfection in the presence of EG-VEGF treatment (Figure 6A). Reduced NKX3.1 mRNA levels had significantly increased the CGB mRNA expression in BeWo cells, even in the presence of EG-VEGF stimulation.
Figure 6.
Effect of NKX3.1 inactivation on trophoblast differentiation and proliferation. (A) Expression of CGB mRNA in BeWo cells treated with EG-VEGF ± NKX3.1 siRNA. (B) Effect of NKX3.1 downregulation on HTR-8/SVneo proliferation. Results are reported as the mean ± SE mol/L from n = 3 independent experiments. Data were analyzed using the Mann–Whitney U test (significant differences, *p < 0.05).
Effect of NKX3.1 siRNA Knockdown on Cell Proliferation in HTR-8/SVneo Cells
The effect of NKX3.1 inactivation on HTR-8/SVneo proliferation was studied using the xCELLigence system in real-time monitoring over 48 h. As shown in Figure 6B, there was a significant decrease in the cell-index, suggesting decreased HTR-8/SVneo proliferation in NKX3.1 siRNA-treated cells compared with the NC control. In fact, proliferation was significantly decreased in siRNA-treated HTR-8/SVneo cells with and without EG-VEGF stimulation (p < 0.001) (Figure 6C).
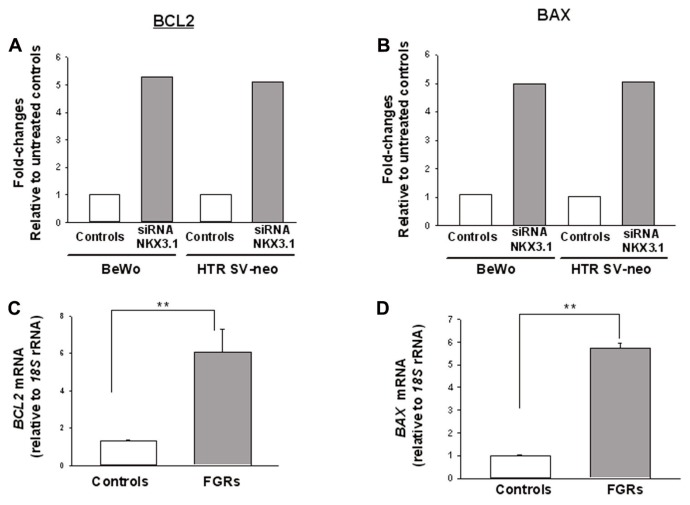
Figures 7A and 7B report the candidate gene BCL2 and BAX that showed differential expression (greater than five-fold) in human apoptosis array in both BeWo and HTR-8/SVneo cells when treated with NKX3.1 siRNA compared with the NC control, respectively. Further validation for mRNA expression for BCL2 and BAX showed a significant increase in FGR placentae compared with control (Figures 7C, D), respectively.
Figure 7.
Candidate gene identification using human apoptosis cDNA array and placental expression for candidate gene in FGR and control pregnancies. Effect of NKX3.1 gene inactivation on apoptosis gene expression profile identified BCL2 and BAX mRNA as candidate genes in BeWo and HTR-8/SVneo (A, B). Real-time PCR was used to detect apoptosis gene mRNA relative to 18S rRNA. Results are reported as fold changes relative to cells treated with NC. Quantification of candidate genes, BCL2 and BAX mRNA expression levels in FGR (n = 25) and control (n = 25) (C, D). 18S rRNA was used as the internal control. Results are reported as the mean ± SEM. Data were analyzed using the Mann–Whitney U test (significant differences when compared with control, *p < 0.05; **p < 0.01).
DISCUSSION
Placental NKX3.1 Expression in FGR
In this study, we report that EG-VEGF is an important regulator of homeobox genes in first trimester placental tissues. This was done using a clinically well-defined cohort of idiopathic FGR pregnancies that were carefully selected using strict clinical criteria indicative of placental insufficiency and underlying pathology (6,40–42). Thus, our study excluded pregnancies with delivery of constitutionally small-for-gestation-age babies, who were below the 10th percentile of birth weight but were otherwise healthy, and the clinical criteria for placental insufficiency was normal.
A recent study by Buckberry et al., reports that NKX3.1 is one of the sex-based transcription factors detectable in the human placenta (43). The human NKX3.1 gene, located on chromosome 8q21, is expressed at lower to moderate levels in tissues other than the placenta, including, heart, prostate and salivary glands (44). It regulates the proliferation of glandular epithelium and the formation of ducts in prostate and minor salivary glands (45). NKX3.1 is also known as a tumor suppressor. Its expression appears to be regulated by androgens while loss of its expression is linked to prostate cancer development (46). The data from this study show that growth factor-induced NKX3.1 regulation on cell proliferation and differentiation is not limited to prostate development but also occurs in placental trophoblast cells. Previous studies in the literature reported the expression of HOX genes in the normal and abnormal trophoblastic tissues and their role in the differentiation of trophoblasts (47,48). However until now, no data was reported on the expression and role of NKX3.1 in the control of placental developmental processes.
NKX3.1 and Trophoblast Differentiation in Primary Trophoblast Cells
This study has demonstrated the presence of NKX3.1 mRNA and protein in both primary EVCTs from first-trimester placentae and primary villous CTBs. The ST layer is a major site for numerous important placental functions, including nutrient and ion exchange and the synthesis of steroid and peptide hormones required for fetal development and growth (7,8). Here, we have used primary VCT and EVCT cells isolated from first-trimester placentae to determine the effect of NKX3.1 on in vitro differentiation as a fusion phenotype (ST) and as an invasive phenotype (EVCT), respectively, on NKX3.1 expression. We demonstrated that in vitro spontaneous differentiation of trophoblasts is associated with an increased NKX3.1 expression, suggesting NKX3.1 may have a significant role in villous trophoblast differentiation. A syncytium that functions normally requires both differentiation and fusion of trophoblasts. Thus, the role of NKX3.1 in the regulation of trophoblast fusion and differentiation will also be of interest in future studies. In this study we have used the independently derived, well-characterized human villous BeWo trophoblast cell line and HTR-8/SVneo extravillous trophoblast cell line to model for first-trimester VCT differentiation and EVCT proliferation, respectively.
NKX3.1 in the BeWo Cell Model for Differentiation of Trophoblasts
To study the effect NKX3.1 expression has on villous trophoblast differentiation, the human choriocarcinoma BeWo cell line was used as described previously (49). ST differentiation can be reproduced in vitro using different models (7,49–51), but BeWo cells exhibit a low spontaneous fusion rate that can be increased with FSK treatment (52). We demonstrated that FSK treatment increased NKX3.1 expression in BeWo cells, suggesting NKX3.1 may contribute to an optimum differentiation potential of trophoblasts.
Gene inactivation using NKX3.1 siRNA in FSK-treated BeWo cells demonstrated that NKX3.1 is involved in the regulation of BeWo cell differentiation, as demonstrated by a significant increase in CGB/βhCG gene expression and secretion. In our previous studies, we reported that FGR placental villi show an upregulated expression of CGB/βhCG (34,53). The decreased expression of NKX3.1 in FGR, therefore, may have clinical significance in contributing to the premature depletion of the pool of proliferating CTBs by favoring their differentiation. This may also be reflected by changes in the ST layer leading to trophoblast shedding and apoptosis in FGR (54).
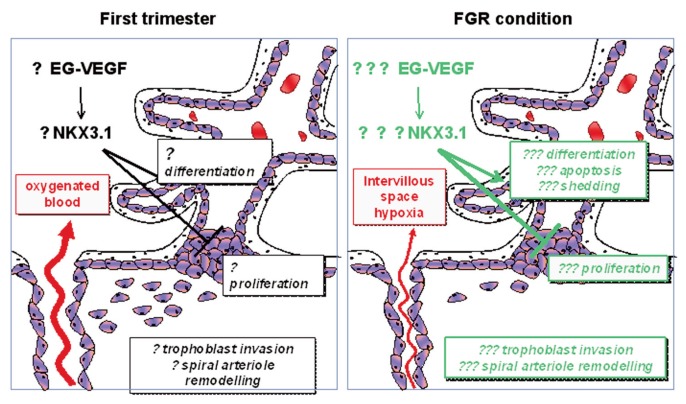
FGR also is associated with increased EG-VEGF levels (20). This elevation might, in turn, contribute to the decrease in NKX3.1 expression and to aberrant ST differentiation (Figure 8). Furthermore, in a recent pilot study, we have observed an increased expression of TP53 mRNA, a marker of apoptosis in BeWo cells, following siRNA NKX3.1 inactivation (data not shown). Altogether, our observations support a key role for NKX3.1 in the regulation of differentiation and its possible role in apoptosis of trophoblasts.
Figure 8.
Proposed model for the potential implication of NKX 3.1 in the pathogenesis of fetal growth restriction (FGR) by the end of the first trimester. Lack of maintenance of EG-VEGF has been associated with FGR (16–19). An EG-VEGF increase could lead to abnormal downregulation of NKX 3.1 expression leading to an increase in trophoblast differentiation, apoptosis and shedding as well as sustained inhibition of extravillous trophoblast (EVCT) invasion. The failure in spiral arteriole remodeling, known to be associated with FGR, would therefore contribute to abnormal persistence of placental hypoxia in the intervillous space, and could then act as a positive feedback loop to exacerbate EG- VEGF production.
Using BeWo cells that mimic syncytiotrophoblast cells, we demonstrated that EG-VEGF might have a direct effect on the regulation of NKX3.1 expression, since both PROKR1 and PROKR2 antagonists abolished the decrease in NKX3.1 expression observed under hypoxia, a condition that mimics FGR and increases EG-VEGF production. Further studies are needed to better dissect the signaling pathways involved in NKX3.1-EG-VEGF-dependent regulation.
NKX3.1 in the HTR-8/SVneo Model for Proliferation of Trophoblasts
In this study, immunohistochemical localization of NKX3.1 revealed the presence of the immunoreactive protein expressed primarily in the nuclei of proliferating CTB cell types in the anchoring villi of early pregnancy human placentae, but not in the nuclei of invading distal CTBs in the cell columns. Previous studies using HTR-8/SVneo cells as a model for proliferating trophoblasts confirmed that the HLX homeobox gene is an important regulator of EG-VEGF-mediated trophoblast cell proliferation (20). We also investigated whether NKX3.1 expression is a downstream effector gene in the EG-VEGF signaling pathway of trophoblast function. Upon stimulation of HTR-8/SVneo with EG-VEGF, in the presence of NKX3.1 siRNA, cell proliferation remained significantly decreased. This suggests that EG-VEGF is a negative regulator of NKX3.1-mediated trophoblast proliferation. Therefore, it is plausible that a decrease in NKX3.1 gene expression levels in cultured trophoblast cells may lead to substantially decreased trophoblast cell migration and invasion. Thus, the reduction in trophoblast proliferation would ultimately result in a decreased pool of cells acquiring the migratory and invasive phenotype. This may account for the characteristic shallow trophoblast invasion and reduced spiral arteriole remodeling observed in FGR.
Analyses on gene expression for apoptosis in BeWo and HTR-8/SVneo cells following gene inactivation using NKX3.1 identified proapoptotic genes BCL2 and BAX as direct or indirect downstream targets of NKX3.1. Further validation in placental tissues demonstrated a significant increase in both BCL2 and BAX in FGR compared with control, these observations are consistent with previous reports on increased indices of placental apoptosis in FGR (55,56).
CONCLUSION
In summary, EG-VEGF is a negative regulator of NKX3.1 expression in first-trimester placentae. NKX3.1 expression is decreased in placentae from pregnancies affected by idiopathic FGR when compared with gestation-matched controls. Furthermore, our functional studies suggest that decreased NKX3.1 expressions may lead to premature differentiation in the chorionic villous tissue of FGR-affected pregnancies. Our results also provide evidence for a reduced NKX3.1 expression leading to a decreased pool of extravillous trophoblasts leading, in turn, to the shallow invasion of trophoblasts seen in FGR.
Altogether these data suggest that maintenance of EG-VEGF expression over the first trimester of pregnancy is of vital importance and that abnormally increased levels during that trimester might affect the modulation of expression of homeobox genes such as NKX3.1, which in turn may contribute to the etiology of FGR. Nevertheless, it is still to be determined whether EG-VEGF increased levels in FGR are the cause or the consequence of the pathology. The causal hypothesis is currently under investigation by sustaining EG-VEGF production (osmotic pump delivering EG-VEGF) beyond early gestation in the gravid mouse and determining its direct effects on pregnancy outcome. The second hypothesis is also plausible as FGR condition is associated with the deregulation in multiple other predisposing factors such as hypoxia, a parameter known to increase EG-VEGF levels, which in turn will affect the expression of key homeobox genes. This hypothesis is supported by the in vitro experiments presented in this study.
ACKNOWLEDGMENTS
The authors wish to thank the consenting patients and the clinical and research midwives at the Pregnancy Research Centre, Department of Perinatal Medicine, The Royal Women’s Hospital for the supply of FGR and gestation-matched control placental tissues. INSERM (U1036), University Joseph Fourier, Commissariat à L’Energie Atomique (DSV/iRTSV/BCI). Funding support was provided from Groupement des Entreprises Françaises pour la Lutte contre le Cancer Comité Isère to N Alfaidy. Funding support for this work was provided from the Australian National Health and Medical Research Council (NHMRC New Investigator project grant #509140) award to P Murthi. We also thank F Balboni and QY Zhou for their collaboration. The human placental trophoblast-derived choriocarcinoma BeWo cell line (B30 clone) was a kind gift from Stephen Rogerson, The University of Melbourne, Department of Medicine, the Royal Melbourne Hospital, Victoria, Australia. The HTR-8/SVneo cells were a kind gift from Charles Graham, Queens University, Canada.
Footnotes
Online address: http://www.molmed.org
DISCLOSURE
The authors declare that they have no competing interests as defined by Molecular Medicine, or other interests that might be perceived to influence the results and discussion reported in this paper.
Cite this article as: Murthi P, et al. (2015) An EG-VEGF-dependent decrease in homeobox gene NKX3.1 contributes to cytotrophoblast dysfunction: a possible mechanism in human fetal growth restriction. Mol. Med. 21:645–56.
REFERENCES
- 1.Godfrey KM, Barker DJ. Fetal nutrition and adult disease. Am J Clin Nutr. 2000;71:1344S–52S. doi: 10.1093/ajcn/71.5.1344s. [DOI] [PubMed] [Google Scholar]
- 2.Illanes S, Soothill P. Management of fetal growth restriction. Semin Fetal Neonatal Med. 2004;9:395–401. doi: 10.1016/j.siny.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Brodsky D, Christou H. Current concepts in intrauterine growth restriction. J Intensive Care Med. 2004;19:307–19. doi: 10.1177/0885066604269663. [DOI] [PubMed] [Google Scholar]
- 4.Gagnon R. Placental insufficiency and its consequences. Eur J Obstet Gynecol Reprod Biol. 2003;110(Suppl 1):S99–107. doi: 10.1016/s0301-2115(03)00179-9. [DOI] [PubMed] [Google Scholar]
- 5.Kingdom J, Huppertz B, Seaward G, Kaufmann P. Development of the placental villous tree and its consequences for fetal growth. Eur J Obstet Gynecol Reprod Biol. 2000;92:35–43. doi: 10.1016/s0301-2115(00)00423-1. [DOI] [PubMed] [Google Scholar]
- 6.Chaddha V, Viero S, Huppertz B, Kingdom J. Developmental biology of the placenta and the origins of placental insufficiency. Semin Fetal Neonatal Med. 2004;9:357–69. doi: 10.1016/j.siny.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Pidoux G, et al. Review: Human trophoblast fusion and differentiation: lessons from trisomy 21 placenta. Placenta. 2012;33(Suppl):S81–6. doi: 10.1016/j.placenta.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Pidoux G, et al. A PKA-ezrin-Cx43 signaling complex controls gap junction communication and thereby trophoblast cell fusion. J Cell Sci. 2014;127:4172–85. doi: 10.1242/jcs.149609. [DOI] [PubMed] [Google Scholar]
- 9.Ishihara N, et al. Increased apoptosis in the syncytiotrophoblast in human term placentas complicated by either preeclampsia or intrauterine growth retardation. Am J Obstet Gynecol. 2002;186:158–66. doi: 10.1067/mob.2002.119176. [DOI] [PubMed] [Google Scholar]
- 10.Knofler M. Critical growth factors and signalling pathways controlling human trophoblast invasion. Int J Dev Biol. 2010;54:269–80. doi: 10.1387/ijdb.082769mk. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burton GJ, Jauniaux E, Charnock-Jones DS. Human early placental development: potential roles of the endometrial glands. Placenta. 2007;28(Suppl A):S64–9. doi: 10.1016/j.placenta.2007.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cartwright JE, et al. Trophoblast invasion of spiral arteries: a novel in vitro model. Placenta. 2002;23:232–5. doi: 10.1053/plac.2001.0760. [DOI] [PubMed] [Google Scholar]
- 13.Graham CH, Lala PK. Mechanisms of placental invasion of the uterus and their control. Biochem Cell Biol. 1992;70:867–74. doi: 10.1139/o92-135. [DOI] [PubMed] [Google Scholar]
- 14.LeCouter J, et al. Identification of an angiogenic mitogen selective for endocrine gland endothelium. Nature. 2001;412:877–84. doi: 10.1038/35091000. [DOI] [PubMed] [Google Scholar]
- 15.Lin DC, et al. Identification and molecular characterization of two closely related G protein-coupled receptors activated by prokineticins/endocrine gland vascular endothelial growth factor. J Biol Chem. 2002;277:19276–80. doi: 10.1074/jbc.M202139200. [DOI] [PubMed] [Google Scholar]
- 16.Brouillet S, et al. Molecular characterization of EG-VEGF-mediated angiogenesis: differential effects on microvascular and macrovascular endothelial cells. Mol Biol Cell. 2010;21:2832–43. doi: 10.1091/mbc.E10-01-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann P, Feige JJ, Alfaidy N. Expression and oxygen regulation of endocrine gland-derived vascular endothelial growth factor/prokineticin-1 and its receptors in human placenta during early pregnancy. Endocrinology. 2006;147:1675–84. doi: 10.1210/en.2005-0912. [DOI] [PubMed] [Google Scholar]
- 18.Hoffmann P, Feige JJ, Alfaidy N. Placental expression of EG-VEGF and its receptors PKR1 (prokineticin receptor-1) and PKR2 throughout mouse gestation. Placenta. 2007;28:1049–58. doi: 10.1016/j.placenta.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 19.Hoffmann P, et al. Role of EG-VEGF in human placentation: Physiological and pathological implications. J Cell Mol Med. 2009;13:2224–35. doi: 10.1111/j.1582-4934.2008.00554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brouillet S, et al. EG-VEGF controls placental growth and survival in normal and pathological pregnancies: case of fetal growth restriction (FGR) Cell Mol Life Sci. 2013;70:511–25. doi: 10.1007/s00018-012-1141-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rajaraman G, Murthi P, Brennecke SP, Kalionis B. Homeobox gene HLX is a regulator of HGF/c-met-mediated migration of human trophoblast-derived cell lines. Biol Reprod. 2010;83:676–83. doi: 10.1095/biolreprod.109.078634. [DOI] [PubMed] [Google Scholar]
- 22.Murthi P, et al. Homeobox genes and down-stream transcription factor PPARgamma in normal and pathological human placental development. Placenta. 2013;34:299–309. doi: 10.1016/j.placenta.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 23.Murthi P, Kalionis B, Rajaraman G, Keogh RJ, Da Silva Costa F. The role of homeobox genes in the development of placental insufficiency. Fetal Diagn Ther. 2012;32:225–30. doi: 10.1159/000339657. [DOI] [PubMed] [Google Scholar]
- 24.Quinn LM, Latham SE, Kalionis B. The homeobox genes MSX2 and MOX2 are candidates for regulating epithelial-mesenchymal cell interactions in the human placenta. Placenta. 2000;21(Suppl A):S50–4. doi: 10.1053/plac.1999.0514. [DOI] [PubMed] [Google Scholar]
- 25.Murthi P, et al. Homeobox gene HLX1 expression is decreased in idiopathic human fetal growth restriction. Am J Pathol. 2006;168:511–8. doi: 10.2353/ajpath.2006.050637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murthi P, et al. Homeobox gene ESX1L expression is decreased in human pre-term idiopathic fetal growth restriction. Mol Hum Reprod. 2006;12:335–40. doi: 10.1093/molehr/gal037. [DOI] [PubMed] [Google Scholar]
- 27.Guaran RL, Wein P, Sheedy M, Walstab J, Beischer NA. Update of growth percentiles for infants born in an Australian population. Aust N Z J Obstet Gynaecol. 1994;34:39–50. doi: 10.1111/j.1479-828x.1994.tb01037.x. [DOI] [PubMed] [Google Scholar]
- 28.Tsatsaris V, et al. Overexpression of the soluble vascular endothelial growth factor receptor in preeclamptic patients: pathophysiological consequences. J Clin Endocrinol Metab. 2003;88:5555–5563. doi: 10.1210/jc.2003-030528. [DOI] [PubMed] [Google Scholar]
- 29.Handschuh K, et al. Human chorionic gonadotropin produced by the invasive trophoblast but not the villous trophoblast promotes cell invasion and is down-regulated by peroxisome proliferator-activated receptor-gamma. Endocrinology. 2007;148:5011–9. doi: 10.1210/en.2007-0286. [DOI] [PubMed] [Google Scholar]
- 30.Graham CH, et al. Establishment and characterization of first trimester human trophoblast cells with extended lifespan. Exp Cell Res. 1993;206:204–11. doi: 10.1006/excr.1993.1139. [DOI] [PubMed] [Google Scholar]
- 31.Murthi P, et al. Homeobox gene DLX4 expression is increased in idiopathic human fetal growth restriction. Mol Hum Reprod. 2006;12:763–9. doi: 10.1093/molehr/gal087. [DOI] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Chui A, et al. Homeobox gene distal-less 3 is expressed in proliferating and differentiating cells of the human placenta. Placenta. 2010;31:691–7. doi: 10.1016/j.placenta.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 34.Pathirage NA, et al. Homeobox gene transforming growth factor beta-induced factor-1 (TGIF-1) is a regulator of villous trophoblast differentiation and its expression is increased in human idiopathic fetal growth restriction. Mol Hum Reprod. 2013;19:665–75. doi: 10.1093/molehr/gat042. [DOI] [PubMed] [Google Scholar]
- 35.Evseenko DA, Paxton JW, Keelan JA. Independent regulation of apical and basolateral drug transporter expression and function in placental trophoblasts by cytokines, steroids, and growth factors. Drug Metab Dispos. 2007;35:595–601. doi: 10.1124/dmd.106.011478. [DOI] [PubMed] [Google Scholar]
- 36.Chui A, et al. Homeobox gene Distal-less 3 (DLX3) is a regulator of villous cytotrophoblast differentiation. Placenta. 2011;32:745–51. doi: 10.1016/j.placenta.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 37.Balboni G, et al. Triazine compounds as antagonists at Bv8-prokineticin receptors. J Med Chem. 2008;51:7635–7639. doi: 10.1021/jm800854e. [DOI] [PubMed] [Google Scholar]
- 38.Curtis VF, et al. A PK2/Bv8/PROK2 antagonist suppresses tumorigenic processes by inhibiting angiogenesis in glioma and blocking myeloid cell infiltration in pancreatic cancer. PloS One. 2013;8:e54916. doi: 10.1371/journal.pone.0054916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keogh RJ. New technology for investigating trophoblast function. Placenta. 2010;31:347–50. doi: 10.1016/j.placenta.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 40.Garcia MG, et al. High expression of survivin and down-regulation of Stat-3 characterize the feto-maternal interface in failing murine pregnancies during the implantation period. Placenta. 2007;28:650–7. doi: 10.1016/j.placenta.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 41.Regnault TR, Galan HL, Parker TA, Anthony RV. Placental development in normal and compromised pregnancies— a review. Placenta. 2002;23(Suppl A):S119–29. doi: 10.1053/plac.2002.0792. [DOI] [PubMed] [Google Scholar]
- 42.Salafia CM. Placental pathology of fetal growth restriction. Clin Obstet Gynecol. 1997;40:740–9. doi: 10.1097/00003081-199712000-00008. [DOI] [PubMed] [Google Scholar]
- 43.Buckberry S, Bianco-Miotto T, Bent SJ, Dekker GA, Roberts CT. Integrative transcriptome meta-analysis reveals widespread sex-biased gene expression at the human fetal-maternal interface. Mol Hum Reprod. 2014;20:810–9. doi: 10.1093/molehr/gau035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhatia-Gaur R, et al. Roles for Nkx3.1 in prostate development and cancer. Genes Dev. 1999;13:966–77. doi: 10.1101/gad.13.8.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneider A, Brand T, Zweigerdt R, Arnold H. Targeted disruption of the Nkx3.1 gene in mice results in morphogenetic defects of minor salivary glands: parallels to glandular duct morphogenesis in prostate. Mech Dev. 2000;95:163–74. doi: 10.1016/s0925-4773(00)00355-5. [DOI] [PubMed] [Google Scholar]
- 46.Meeks JJ, Schaeffer EM. Genetic regulation of prostate development. J Androl. 2011;32:210–7. doi: 10.2164/jandrol.110.011577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amesse LS, Moulton R, Zhang YM, Pfaff-Amesse T. Expression of HOX gene products in normal and abnormal trophoblastic tissue. Gynecol Oncol. 2003;90:512–8. doi: 10.1016/s0090-8258(03)00357-3. [DOI] [PubMed] [Google Scholar]
- 48.Zhang YM, Xu B, Rote N, Peterson L, Amesse LS. Expression of homeobox gene transcripts in trophoblastic cells. Am J Obstet Gynecol. 2002;187:24–32. doi: 10.1067/mob.2002.122850. [DOI] [PubMed] [Google Scholar]
- 49.Orendi K, Gauster M, Moser G, Meiri H, Huppertz B. The choriocarcinoma cell line BeWo: syncytial fusion and expression of syncytium-specific proteins. Reproduction. 2010;140:759–66. doi: 10.1530/REP-10-0221. [DOI] [PubMed] [Google Scholar]
- 50.Fitzgerald JS, et al. Governing the invasive trophoblast: current aspects on intra- and extracellular regulation. Am J Reprod Immunol. 2010;63:492–505. doi: 10.1111/j.1600-0897.2010.00824.x. [DOI] [PubMed] [Google Scholar]
- 51.Gerbaud P, Pidoux G. An overview of molecular events occurring in human trophoblast fusion. Placenta. 2014;36(Suppl 1):S35–42. doi: 10.1016/j.placenta.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 52.Wice B, Menton D, Geuze H, Schwartz AL. Modulators of cyclic AMP metabolism induce syncytiotrophoblast formation in vitro. Exp Cell Res. 1990;186:306–16. doi: 10.1016/0014-4827(90)90310-7. [DOI] [PubMed] [Google Scholar]
- 53.Chui A, et al. Homeobox gene Distal-less 3 is a regulator of villous cytotrophoblast differentiation and its expression is increased in human idiopathic foetal growth restriction. J Mol Med (Berl) 2012;90:273–84. doi: 10.1007/s00109-011-0836-1. [DOI] [PubMed] [Google Scholar]
- 54.Huppertz B, Frank HG, Kingdom JC, Reister F, Kaufmann P. Villous cytotrophoblast regulation of the syncytial apoptotic cascade in the human placenta. Histochem Cell Biol. 1998;110:495–508. doi: 10.1007/s004180050311. [DOI] [PubMed] [Google Scholar]
- 55.Agata KB, Anita S, Urszula KK, Agnieszka NK, Grzegorz B. Expression of caspase-3, Bax nad Bcl-2 in placentas from pregnancies complicated by treated and non-treated fetal growth restriction. Ginekol Pol. 2009;80:652–6. [PubMed] [Google Scholar]
- 56.Karowicz-Bilinska A, Szczerba A, Kowalska-Koprek U, Nawrocka-Kunecka A. The evaluation of selected indices of apoptosis in placentas from pregnancies complicated by fetal growth restriction. Ginekol Pol. 2007;78:521–6. [PubMed] [Google Scholar]