Figure 5. Identification of the Lamb1 mutation.
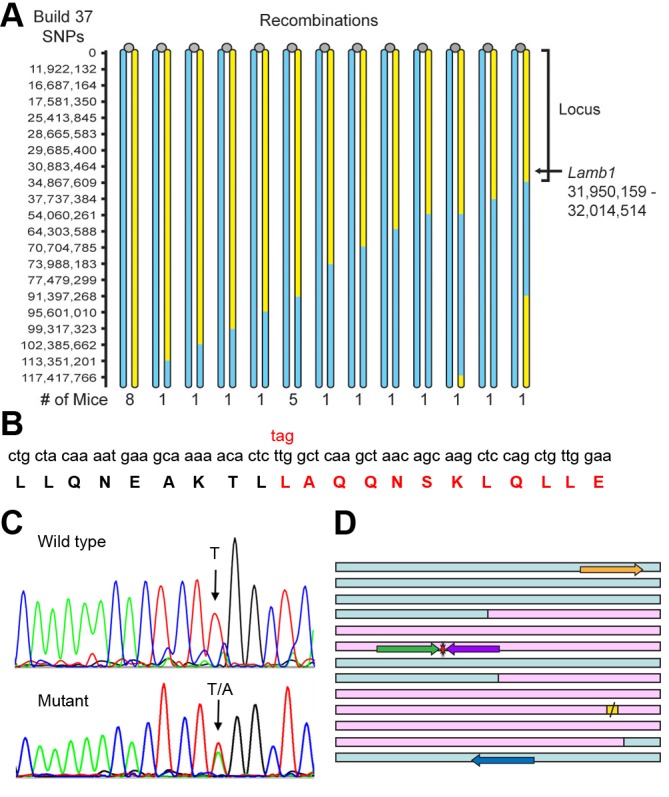
(A) SNP locus mapping summary for chromosome 12 in B6/FVB hybrids. The mutation is necessarily in B6 DNA (yellow); FVB DNA is blue. Mouse centromeres are at the top (gray ovals). There were no informative SNPs between base 0 and 11,922,132. (B) Exome sequencing result. Nucleotide and protein sequence for Lamb1 (laminin β1) amino acids 1721 to 1741 flanking the mutation. Mutation at a single nucleotide generated a stop codon, TAG, and the sequence in red and beyond (amino acids 1730 to 1786) was truncated. Eight other variants identified by exome sequencing in the locus were in exon-flanking intron sequence or 3'UTR and not predicted to be damaging. (C) We validated the mutation by Sanger sequencing. The identified causative Lamb1 mutation was not a reported variant in dbSNP, the Mouse Phenome Database, or the Sanger1 database. To date, the mutation has been verified in 33 symptomatic mice. (D) Allele-specific PCR design. Pink blocks are exons 32 and 33, the red symbol is the mutation, and the yellow square is the normal stop codon. The forward allele-specific primers (green) were longer than the reverse allele-specific primers (violet) because of high AT content. Each set of otherwise-identical internal primers ended with either T or A. If the mismatch is sufficiently destabilizing, priming will be absent or very low. If the mismatch is not sufficiently destabilizing, another base 5’ of the mutation can be changed to reduce stability and improve selectivity; the reverse WT allele-specific primer also had a substitution of G for C at –2. Forward and reverse outside primers (gold and blue) were predicted in the flanking DNA at convenient distances from the mutation based on melting temperatures matching the allele-specific primers. Diagnostic PCR was done with the gold/violet pair with the mutation, while the gold/blue pair served as a positive control.
