Abstract
In higher eukaryotic genomes, Long Interspersed Nuclear Element 1 (LINE-1) retrotransposons represent a large family of repeated genomic elements. They transpose using a reverse transcriptase (RT), which they encode as part of the ORF2p product. RT inhibition in cancer cells, either via RNA interference-dependent silencing of active LINE-1 elements, or using RT inhibitory drugs, reduces cancer cell proliferation, promotes their differentiation and antagonizes tumor progression in animal models. Indeed, the non-nucleoside RT inhibitor efavirenz has recently been tested in a phase II clinical trial with metastatic prostate cancer patients. An in-depth analysis of ORF2p in a mouse model of breast cancer showed ORF2p to be precociously expressed in precancerous lesions and highly abundant in advanced cancer stages, while being barely detectable in normal breast tissue, providing a rationale for the finding that RT-expressing tumors are therapeutically sensitive to RT inhibitors. We summarize mechanistic and gene profiling studies indicating that abundant LINE-1-derived RT can “sequester” RNA substrates for reverse transcription in tumor cells, entailing the formation of RNA:DNA hybrid molecules and impairing the overall production of regulatory miRNAs, with a global impact on the cell transcriptome. Based on these data, LINE-1-ORF2 encoded RT has a tumor-promoting potential that is exerted at an epigenetic level. We propose a model whereby LINE1-RT drives a previously unrecognized global regulatory process, the deregulation of which drives cell transformation and tumorigenesis with possible implications for cancer cell heterogeneity.
Keywords: LINE-1 retrotransposons, reverse transcriptase, tumorigenesis, differentiation therapy, cancer heterogeneity, epigenetics
Introduction: The retrotransposition machinery in the genesis of genomic and epigenomic landscapes
The complete sequencing of the human genome has disclosed the unexpected finding that coding genes account for a mere 1.2% of the total genome, while the remaining portion is constituted by non-coding DNA (International Human Genome Sequencing Consortium, 2001). Branded by a historically “bad reputation”, non-coding sequences have been defined as “junk” (Ohno, 1972) or “selfish” (Orgel and Crick, 1980) DNA, a view further strengthened by the evidence that nearly 50% of the human genome is constituted by apparently functionless transposable “genetic parasites” thought to increasingly litter all chromosomes during evolution.
Two main families of transposable elements characterize eukaryotic genomes: DNA transposons, which mobilize through a “cut and paste” mechanism (Muñoz-López and García-Pérez, 2010), and retrotransposons, which mobilize instead through “copy and paste,” a process that requires the reverse transcription of RNA intermediates into cDNA copies as a preliminary step in retrotransposition (Levin and Moran, 2011), promoting the broad expansion of retroelements in eukaryotic genomes.
A key player in this mechanism is the enzyme reverse transcriptase (RT) encoded by LINE-1 retrotransposons themselves. The latter are a source of the RT activity required to promote retrotransposition in human cells (Brouha et al., 2003). LINE-1 elements actually harbor two open reading frames, ORF1 and ORF2, which respectively encode ORF1p, an RNA-binding protein, and ORF2p, with reverse transcriptase (RT) and endonuclease (EN) activities (reviewed in Babushok and Kazazian, 2007). The LINE-1-derived retrotransposition machinery, constituted by ORF1 and ORF2 proteins, has cis-preference for its own LINE-1 RNA (Esnault et al., 2000; Wei et al., 2001; Kulpa and Moran, 2006). LINE-1-derived RT is also used for retrotranscription /retrotransposition of other RNAs, including Alu elements (Dewannieux et al., 2003), SVA (SINE-R/VNTR/ALU) elements (Ostertag et al., 2003) and mRNAs that give rise to a large population of processed pseudogenes nearly as numerous as the original coding genes (Pink et al., 2011). It is now well established that RT-originating sequences contribute to shape genomes and constitute a large proportion of evolutionarily conserved chromosomal DNA, accounting altogether for nearly 50% of the human genome (International Human Genome Sequencing Consortium, 2001). Such extensive preservation suggests a functional importance of retrotransposons. Not surprisingly, retrotransposons are increasingly being implicated in fundamental genomic functions, in both normal and pathological contexts (Rebollo et al., 2012). Indeed, as highly dynamic components of genomes, they contribute a relentless source of genetic and epigenetic variations and novelty (Feschotte, 2008; Bourque, 2009; Beck et al., 2011) and, on the long run, a major driving force in genome evolution (Oliver and Greene, 2011). A detailed description of all functional implications of retrotransposition in genome biology and evolution would be out of scope in this article, but extensive information is discussed in excellent reviews (Feschotte, 2008; Goodier and Kazazian, 2008; Bourque, 2009; Beck et al., 2011; Oliver and Greene, 2011; Rebollo et al., 2012).
The advent of high-throughput technologies in recent years has provided an accurate localization of new genomic insertions, shifting the focus from a gene-centric to a genome-wide view. This has revolutionized the traditional paradigms of genome organization by disclosing novel and unexpectedly complex genomic landscapes. Studies now show that genomes are crowded with sequences of reverse-transcribed origin, many of which are correlated with the insurgence of a variety of pathologies (for a review Hancks and Kazazian, 2012), in particular cancer (Belancio et al., 2010).
The ENCODE Project Consortium (The ENCODE Project Consortium, 2012) showed that approximately 80% of the human genome is pervasively transcribed; actually, a relevant proportion of small and long non-coding transcripts are functional components of genome-wide regulatory networks (Djebali et al., 2012). The groundbreaking finding of an astounding landscape of small RNAs—classified as microRNAs (miRNAs), endogenous small interfering RNAs (endo-siRNAs or siRNAs) (Piatek and Werner, 2014) and Piwi-interacting RNAs (piRNAs) (Kim et al., 2009), depending on their origin and the proteins they interact with - has unveiled an RNA-mediated regulatory network that controls the genome architecture and transcriptomic profile (Aalto and Pasquinelli, 2012; Li, 2014), influencing a multitude of biological processes. Growing data show a dual relationship between small RNAs and retroelements: on the one hand, small RNAs act as “guardians of the genome” in transposon-defense pathways aimed at repressing retroelement mobility (Yang and Kazazian, 2006; reviewed in Malone and Hannon, 2009); on the other hand, retroelements are intimately involved in their biogenesis, because a growing number of small RNAs in all three classes have a recognized retrotransposon-derived origin (Borchert et al., 2011; Watanabe et al., 2011).
Long non-coding RNA (lncRNAs) are components of the mammalian transcriptome and constitute a heterogeneous class of thousands of polymerase II-transcribed RNA species, polyadenylated, spliced, mostly localized in the nucleus (reviewed by Zhang et al., 2013). A large proportion of lncRNAs, with either oncogenic or tumor suppressor roles, are constituted by antisense RNAs; the latter, together with sense transcripts, are being identified in genome-wide regulatory networks that epigenetically fine-tune genome expression, with implications in tumorigenesis, differentiation and development (reviewed in Pelechano and Steinmetz, 2013, Fatica and Bozzoni, 2014). lncRNAs also have tight connections with transposable elements of both the DNA-based and the retroelements families, which occur within nearly 80% of mature lncRNA transcripts and account for about 30–40% of total human lncRNA sequences (Kelley and Rinn, 2012; Kapusta et al., 2013). Also of RT-derived origin are a large proportion of genomic sequences highly preserved throughout evolution and classified as conserved, highly-conserved and ultra-conserved elements (UCRs), according to the level of conservation throughout species (Bejerano et al., 2004; Woolfe et al., 2005, for a recent review see Nelson and Wardle, 2013).
From an ample survey encompassing the genomes of 29 mammalian species (Lowe and Haussler, 2012), a vision of genomes emerges as complex integrated functional systems, in which a considerable proportion of non-exonic sequences were exapted from mobile element insertions (Nishihara et al., 2006; Lowe and Haussler, 2012) to assemble large-scale regulatory circuits. Deregulation of these circuits is implicated in a variety of diseases, including cancer (Esteller, 2011).
LINE-1-encoded RT as a new underestimated player in cancer
While retrotransposable elements are extensively studied and characterized, somewhat surprisingly the retrotransposon-encoded RT activity has long failed to attract an equivalent attention. The RT encoded by infective retroviruses has actually been intensively studied since the time of its discovery in 1970 by Baltimore (1970) and Temin and Mizutani (1970), due to its clinical implications (Herschhorn and Hizi, 2010). In contrast, the endogenous cellular RT has long been overlooked, despite many clues implicating it in as relevant processes as embryogenesis and tumorigenesis. Decades after the discoveries of Baltimore and Temin, a body of evidence has shown that endogenous RT expression is developmentally modulated: low levels of RT, if any, are expressed in differentiated non pathological tissues; increased expression is instead typical of cells characterized by low differentiation and high proliferation, e.g., early embryos (for a review see Sciamanna et al., 2011) and transformed cells (for a review see Sinibaldi-Vallebona et al., 2011). Overall, that is consistent with the notion that LINE-1 increased expression (Chen et al., 2012a; Rodić et al., 2014) and retroelement mobilization are implicated in tumorigenesis (Hancks and Kazazian, 2012; Kaer and Speek, 2013).
In contrast to differentiated quiescent cells, tissues and cells with low differentiation and high proliferation states are sites of high RT expression and provide permissive contexts for retrotransposition. Following up on that line, several studies have pursued RT inhibition in cancer cells, either using non-nucleoside RT inhibitors (nevirapine and efavirenz; Mangiacasale et al., 2003; Landriscina et al., 2005; Sciamanna et al., 2005, 2013), or RNA interference (RNAi)-mediated downregulation of RT-encoding LINE-1 elements (Sciamanna et al., 2005; Oricchio et al., 2007). In the latter case, the RNAi assays were carried out using double-stranded siRNA oligonucleotide targeted against the ORF-1 encoding domain of human full-length, highly expressed LINE-1s (Brouha et al., 2003). Both the drug-mediated and the RNAi-mediated approaches to reduce LINE-1-derived RT were found to reduce proliferation, promote differentiation and reprogram the global transcription profiles of coding and non-coding sequences in several cancer cell lines (human melanoma, glioblastoma, osteosarcoma and prostate, colon and small cell lung carcinomas). This provided early evidence for the implication of the LINE-1-encoded RT in tumorigenesis. The inhibitory effects of efavirenz on LINE-1 reverse transcription and retrotransposition were further tested in in vitro assays (Dai et al., 2011), and its antiproliferative and differentiating potential have been recently confirmed in breast (Patnala et al., 2013) and pancreatic (Hecht et al., 2015) cancer cell lines. Moreover, efavirenz treatment of mice xenografted with human tumorigenic cells caused the arrest, or a significant slow down, of progression of several tumor types in vivo (Sciamanna et al., 2005). Importantly, RNAi-mediated LINE-1 downregulation drastically reduced the tumorigenic potential of human cancer cells in nude mice (Oricchio et al., 2007). These effects are reversible and, upon discontinuation of RT inhibitory treatments, tumor cells return to their original de-differentiated phenotype and unrestrained proliferation capacity (Sciamanna et al., 2005); these obervations provided initial hints to an epigenetic role of RT.
The high levels of RT activity found in tumor cells and tissues, reported by our (Mangiacasale et al., 2003; Gualtieri et al., 2013) and other laboratories (Patnala et al., 2013), correlate well with the enhanced rate of retrotransposition observed in many human tumors, a phenomenon that dramatically contributes to shape cancer genomes (Iskow et al., 2010; Lee et al., 2012; Solyom et al., 2012; Shukla et al., 2013; Ewing et al., 2015). In a MMTV-PyVT transgenic mouse strain (Guy et al., 1992), whose females spontaneously develop breast carcinoma, a burst in the copy number of both LINE-1 and SINE B1 elements was depicted very early at tumor onset; their copy number further increases along with tumor progression (Gualtieri et al., 2013). These data converge to indicate that tumors constitute a highly permissive environment for retrotranscription, yet do not answer the question of whether overexpression and amplification of LINE-1 elements act as oncological “drivers” or as mere “passengers” (Rodic and Burns, 2013). The findings that pharmacological inhibition of RT is sufficient to reduce cancer cell proliferation, promote differentiation and antagonize tumor progression in animal models, similar to the effects obtained by RNAi-specific downregulation of LINE-1 expression, strongly support a causative role of LINE-1-encoded RT in tumorigenesis. In an applied clinical perspective, therefore, RT can be regarded as a target and RT inhibitors as potential therapeutic agents in a novel cancer differentiation therapy. Efavirenz has recently been tested in a phase II trial with metastatic prostate cancer patients, suggesting that relatively high dosage (over 600 mg per day) may be beneficial as a novel anticancer treatment (Houédé et al., 2014).
The role of RT encoded by LINE-1 in tumorigenesis is distinct from that of RT activities produced from the other two potential sources, i.e., endogenous retroviruses (HERVs) and telomerase-associated RT (TERT). First, RNAi-mediated downregulation of HERV-K expression showed negligible effects on the rate of proliferation and differentiation of cancer cells, in contrast with the dramatic effects observed after LINE-1-specific RNAi (Oricchio et al., 2007). Second, inhibitors of LINE-1 derived RT elicit rapid changes in treated cells in our experiments (Mangiacasale et al., 2003; Sciamanna et al., 2005), differently from drugs targeting telomerase, which reduce cancer cell proliferation after a long tratement (about 120 days; Damm et al., 2001); these data therefore rule out the possibility that TERT contributes to the rapid response of cells to RT inhibitors. It should be noted, however, that LINE-1 RT is critical for telomere maintenance, given that LINE-1 knockdown in cancer cells correlates with: (i) reduced length of telomeres, (ii) decreased telomerase activity, and (iii) decreased telomerase mRNA level (Aschacher et al., 2016). Together these results reveal that LINE-1 RT has a functional impact on TERT. Thus, while TERT is not involved in the changes elicited by inhibitors targeting the retrotransposon-derived genuine RT, the level of activity of LINE-1 elements may impact on TERT. These findings again strengthen the view that LINE-1 RT is a major player in tumorigenesis.
LINE-1 ORF2-encoded RT activity in cancer progression
The ORF2-encoded RT has been recently assessed for its suitability as a tumor marker (Gualtieri et al., 2013) in females of the cancer-prone MMTV-PyVT described above (Guy et al., 1992). In these females, breast cancer tissues withdrawn at different times after birth are representative of progressive cancer stages. ORF2p cannot be detected in normal breast tissue by immunohistochemistry (IHC), but increased expression is triggered very early in tumorigenesis, preceding the appearance of typical histological alterations and accepted cancer markers (e.g., Ki67 and epidermal growth factor receptor ERB2); further upregulation takes place during tumor growth. These findings correlate well with the notion that hypomethylated LINE-1 sequences, from which ORF2p is produced, are typical of cancer genomes and precancerous lesions compared to their normal tissues counterpart (Miousse and Koturbash, 2015).
The abundant expression of LINE-1 products in preneoplastic mammary tissues suggests an exploitable tool as a potential diagnostic biomarker for early cancer detection: the identification of cancer-prone foci marked by increased RT before the appearance of recognizable histological alterations, can expand the window of opportunities for therapeutical intervention, which can possibly be most effective if associated with the development of RT inhibitory treatment. Interestingly, the abundance and subcellular localization of LINE-1 products are also proposed to have prognostic value in human metastatic breast cancer (Chen et al., 2012a).
Compelling objectives of “the war on cancer” currently include the definition of novel early markers identifying cancer-prone lesions before their spreading, as well as the development of novel therapeutic approaches in possible replacement of conventional cytotoxic chemotherapy. In a recent critical reappraisal, Hanahan (2014) has pointed out that the war on cancer, if not lost, is certainly not won and has suggested that therapeutic strategies should avoid fragmenting along multiple, highly diversified narrow paths targeting many substrates, each of which is highly selective for a specific cancer. Rather, the therapeutic “bullets” ought to hit fewer targets shared by a large spectrum of cancers (Hanahan, 2014). LINE-1 ORF2-encoded RT would fulfill these criteria, representing, at the same time, an early diagnostic cancer marker, a worth pursuing therapeutic target and the driving component of a newly emerging cancer-promoting mechanism.
The molecular bases of the RT-dependent cancer-promoting mechanism
As briefly recalled above, retrotransposition events have had a fundamental role not only in shaping the genomic landscape, but also in directing regulatory networks aimed to fine-tune a variety of genomic functions. Data obtained in the last few years growingly indicate that the retrotransposon machinery, besides being a well-known source of genomic variations caused by new insertions (Böehne et al., 2008; Bourque, 2009), also exerts a global epigenetic regulatory role on the cellular transcriptome. LINE-1 ORF2-encoded RT is a new player in this mechanism.
Prompted by the finding that tumor cell lines are endowed with abundant LINE-1-encoded RT, Sciamanna et al. (2013) began to address the mechanism through which RT might act by comparing the global transcription profile of melanoma cells before and after RT inhibition by microarray analysis. The results showed that RT inhibition modulates the expression of a broad range of coding genes, but also long and small non-coding sequences, including UCRs and miRNAs. miRNAs actually emerged as crucial components of the RT-depending mechanism; indeed, a subpopulation, known to be involved in cell differentiation, cell growth, tumorigenesis and metastatic progression proved highly responsive to RT inhibition. Many miRNA-encoding genes are significantly associated with genomic regions enriched in closely spaced Alu repeats, further strengthening the link between miRNAs and retrotransposons. The physical association of pre-miRNA genomic loci with high density retroelements actually suggests that the latter can exert a regulatory “position effect” on miRNA expression (Slotkin and Martienssen, 2007). Experimental evidence supporting an orchestrating role of the RT enzyme emerged from cesium chloride density centrifugation analysis of nucleic acids extracted from melanoma and prostate carcinoma cell lines, harboring either “native” or efavirenz-inhibited RT: by buoyant density analysis, LINE-1- and Alu-containing molecules with the density of DNA:RNA hybrids were selectively identified in tumor cells, which disappeared upon treatment with efavirenz and were absent in non-transformed human fibroblasts (Sciamanna et al., 2013). Thus, the DNA:RNA hybrids are an especially abundant, if not exclusive, component of cancer cells, generated by reverse transcription of RNA templates, largely—albeit not exclusively—provided by LINE-1 and Alu transcripts. These data suggest that a cancer-promoting RT-dependent mechanism is active in tumor cells and can be blocked by inhibiting the LINE-1 RT. Based on these data, Sciamanna et al. proposed a model (Sciamanna et al., 2014) whereby the highly expressed LINE-1 RT in cancer cells can intercept RNAs and convert them in RNA:DNA hybrids via reverse transcription. Central to the model is the RT-dependent production of RNA:DNA hybrids, associated with altered functional miRNA profiles, observed under conditions of high LINE-1-derived RT in cancer cells and modulatable by RT inhibitors. A wealth of data show that miRNA expression is indeed downregulated in cancer cells, with profound implications for cell fates (Lu et al., 2005; Gaur et al., 2007; Jansson and Lund, 2012). A variety of small RNAs, including 7SL RNA (Ullu and Weiner, 1984), tRNAs (Kaçar et al., 1992), small nuclear RNAs (Doucet et al., 2015), and YRNAs (Perreault et al., 2005), are known to act as templates for reverse transcription in intermediate steps of the genesis of pseudogenes. It is not unreasonable to hypothesize that miRNA precursors may also be retrotranscribed. The observation that the production of hybrid RNA-DNA molecules is associated with aberrant miRNA profiles in cancer cells actually suggests that RT can “subtract” RNA precursors, thus preventing or impairing the formation of double stranded (ds) RNA dicer substrates for the biogenesis of mature miRNAs: this would ultimately contribute to establish favorable conditions for the onset of cancer phenotypes.
RT inhibition results in restored miRNA biogenesis, likely re-establishing their regulatory networks, consistent with its empirically established capacity to revert the cancer phenotype (Sciamanna et al., 2013).
In agreement with this idea, a subset of LINE-1-specific siRNAs, targeting LINE-1 expression and capable to induce methylation of their promoters, are found to be down-modulated in breast cancer compared to normal cells (Chen et al., 2012b). Conversely, LINE-1 inhibition by siRNAs up-modulate the expression of miRNAs involved in tumor suppression (Ohms et al., 2014). Taken as a whole, these findings indicate an orchestrating role of LINE-1-encoded RT in setting a cancer-permissive cellular state.
Although, the LINE-1 enzymatic machinery preferentially reverse transcribes its own RNA (Esnault et al., 2000; Wei et al., 2001; Kulpa and Moran, 2006), the presence of intronless pseudogenes scattered throughout mammalian genomes points out that mRNAs transcribed from protein-coding genes are also substrates for reverse transcription by the endogenous RT (Pink et al., 2011). This suggests that the RT-depending mechanism, in addition to targeting miRNAs, can also target several more RNA classes, coding and non-coding, small- and long-RNAs, though with a possible preferential bias for those associated with, or derived from, retroelement sequences. Consistent with this view, Sciamanna et al. (2013) found that about one third of the efavirenz-downmodulated miRNAs in melanoma cells are clustered on chromosome 19 (C19MC) in a locus characterized by a high density of primate-specific Alu repeats, which were shown to have co-evolved with miRNAs coding genes (Lehnert et al., 2009). An independent study also reported that LINE-1 silencing caused a deregulated profile of miRNA expression in breast cancer cells (Ohms and Rangasamy, 2014).
In summary, LINE-1 expression and small RNA networks emerge as the balanced components of a RT-depending regulatory mechanism placed at the intersection between normally differentiated and transformed non-differentiated cellular states: when one component raises the other one decays.
It is worth stressing that the partial inactivation of miRNA function is not an exclusive feature of cancer, but is a physiological phenomenon, shared with early preimplantation embryos, a context where again miRNA pathways become transiently suppressed (Suh et al., 2010). Moreover, miRNA inactivation is concomitant with a burst of LINE-1 activity in both tumorigenesis and embryogenesis. In the next paragraph we discuss this striking analogy and suggest that physiological and pathological processes have in common the same RT-dependent mechanism.
The RT-based mechanism as global regulator of differentiation in tumorigenesis and embryogenesis
In prior developmental studies, the presence of LINE-1-encoded RT activity and protein was assessed in gametes and early embryos to address the potential role of this enzyme in embryogenesis. Unexpectedly, Giordano et al. (2000) found an RT activity in mature murine spermatozoa, providing the first hint that RT might somehow be involved in early embryogenesis. The sperm endogenous RT, far from being a nonfunctional remnant encoded by “genomic parasites,” has a full enzymatic activity able to reverse transcribe exogenous RNA molecules, taken up and internalized by spermatozoa, in cDNA copies that could then be delivered to embryos at fertilization (Giordano et al., 2000; reviewed in Spadafora, 2008). Pittoggi et al. (2003) further found that RT is also present in early embryos and is strictly required for preimplantation development: indeed, exposing zygotes to RT inhibitor, or antisense-mediated downregulation of LINE-1 (Beraldi et al., 2006), both caused a drastic arrest of development at the two- and four-cell embryo stages with globally altered gene expression profiles. Interestingly, fertilization activates a reverse transcription wave in zygotes within a few hours, which then propagates throughout the first cell division; that is concomitant with the production of new LINE-1 copies that mostly remain as non integrated extrachromosomal structures (Vitullo et al., 2012). Indirect evidence for an embryonic RT activity also emerge from reports that somatic LINE-1 retrotranspositions occur in human stem cells (Garcia-Perez et al., 2007; Coufal et al., 2009) and in very early stages of development in humans (van den Hurk et al., 2007) and rodents using transgenic murine and rat models (Kano et al., 2009). These findings indicate that the endogenous RT is active in early stages of embryogenesis, where it appears to have implications for epigenetic regulation of gene expression and to be necessary for the unfolding of the developmental program.
Cancer and embryo developmental studies convergingly point to the conclusion that an RT-based mechanism is physiologically activated in early embryogenesis and repressed in differentiated tissues; its unscheduled re-activation in somatic cells has cancer-promoting effects, yielding increased cell proliferation and loss of differentiation, in analogy with embryonic growth. It is a well-established notion that tumors and embryos share a variety of cellular, biochemical and molecular features and that genes typically expressed in embryogenesis, yet silenced in normal differentiated tissues, are re-expressed in tumors (Ma et al., 2010). These circumstances support the conclusion that tumorigenesis often recapitulates developmental patterns (Kaiser et al., 2007). In this conceptual framework, Spadafora (Spadafora, 2015) proposed that the RT-dependent mechanism is a source of the functional analogies shared by the physiological and pathological processes connecting embryogenesis and tumorigenesis.
The genesis of cancer heterogeneity
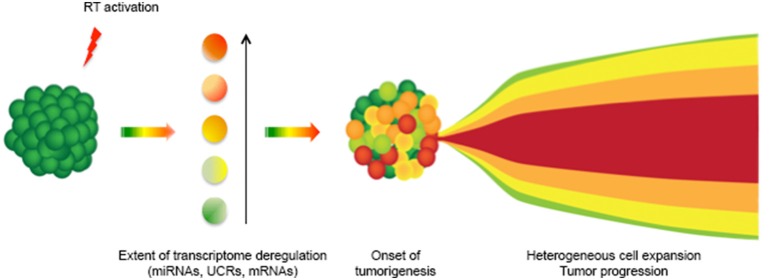
The retrotransposon machinery is highly sensitive to stressing stimuli (Hagan and Rudin, 2002; Terasaki et al., 2013). In response, LINE-1 expression can be activated at differential levels in different cells, depending on the nature and the intensity of endogenous or exogenous stressors. We propose that the differential activation of RT, including by stress, can generate the heterogeneously differentiated cell populations that typically characterize human cancers (Meachem and Morrison, 2013). It is currently unclear whether the cellular heterogeneity observed in cancer reflects the existence of cell populations undergoing a progressive transformation “trajectory,” initiating as a primary cancer state and sequentially evolving into metastatic cells, or whether a broad array of cellular variations simultaneously arise in a single stress-responding event. Based on the data discussed above, it is tempting to speculate that the latter is the case; cells with varying degrees of malignancy—some of which may confer metastatic capacity—may concomitantly originate in a single genome-wide burst of stress-activated LINE-1-RT expression. In this hypothetical model, schematized in Figure 1, burst(s) of LINE-1 expression, triggered by exogenous and/or endogenous stimuli in normal cells (in green), would generate an array of cell populations endowed with various levels of LINE-1-dependent RT activity (indicated by different shades of colored cells), coinciding with the emergence of preneoplastic lesions. We propose that the different levels of LINE-1 activation correspond to different degrees of cell de-differentiation; in the process, embryonic regulatory patterns can be reactivated and induce somatic cells to revert back to embryo-like states (Kaiser et al., 2007). LINE-1 activation at low levels would exert modest de-defferentiation effects, while higher levels would determine a more extensive reactivation of embryonic patterns, with the ensuing production of more aggressive “embryo-like” transformed cells. The cell populations concomitantly originating from the activation of RT expression would then differentially propagate throughout cancer progression, thus contributing to cancer heterogeneity. The model represented in Figure 1 was inspired by the recently proposed “Big Bang” hypothesis for the genesis of human cancer, in which a single ancestral event is thought to originate the heterogeneity of cancer cell populations (Sottoriva et al., 2015), which would then progress and expand in parallel (Klein, 2009). The simultaneous genesis of cells with heterogeneous invasive potential would also offer a possible explanation for the genesis and spreading of metastatic tumors of unknown primary origin: these are a relatively rare class of metastatic tumors detected in patients in which the primary tumor cannot be identified, and account for 3–5% of all cancer diagnoses (Natoli et al., 2011; Stella et al., 2012).
Figure 1.
A model for RT-dependent induction of cancer cell heterogeneity. The deregulated expression of LINE-1 elements in somatic cells (green) causes a burst of RT activity (red flash), which deregulates the transcriptome of individual cells at various levels (represented by different color shades): this originates heterogeneous cancer cell populations. In the model, cancer cell heterogeneity would therefore set in following the early burst of differentially expressed RT activity in different cells. Cancer would then progress with the expansion of various cell populations (on the right).
In more general terms, the model predicts a relatively minor role for DNA mutations in cancer progression, as cell transformation is rather viewed as originating from an RT-mediated reactivation of “embryonic” regulatory circuits mostly acting at the epigenetic level in differentiated cells (Spadafora, 2015). Although needing further experimental testing, the model builds on emerging evidence indicating the global reach of RT onto several RNA classes (Sciamanna et al., 2013; Ohms et al., 2014; Ohms and Rangasamy, 2014) and is compatible with its reversible character by modulating RT levels (Sciamanna et al., 2005).
Retrotransposable elements also clearly impinge on genome function by generating extensive variations via insertional mutagenesis. Although large numbers of mutations are identified by high-throughput sequencing data in cancer contexts, their role(s) in tumorigenesis is often undefined (Kandoth et al., 2013): predisposing gene mutations in fact play a documented causative role only in 5–10% of human cancers (Nagy et al., 2004).
Recent excellent works have reported that the genomes of different tumor types harbor hundreds of de novo somatic insertions, selectively found in cancer genomes (Iskow et al., 2010; Lee et al., 2012; Solyom et al., 2012; Shukla et al., 2013; Doucet-O'Hare et al., 2015; Ewing et al., 2015; Rodić et al., 2015). Despite of these reports, however, the general implication of L1 retrotransposition events as either “driver” mutations (i.e., with a causative role in tumorigenesis), or as “passengers” (i.e., manifesting a consequence of the loss of genome regulation associated with cell transformation), remains an open question (Rodic and Burns, 2013). Insertions were documented and were attributed causative trigger in specific cases; among others, LINE-1 insertion were found within the c-myc gene (Morse et al., 1988), or in the tumor-suppressing gene apc (Miki et al., 1992) in breast and colon carcinoma, respectively; in those instances, LINE-1 insertions should have an activating (c-myc) or inhibitory (apc) role, respectively. In a different context, Alu insertions also result in neurofibromatosis type 1 (Wallace et al., 1991). It is worth recalling, however, that together LINE-1, Alu and SVA insertions account only for a marginal contribution (<0.5%) to the genesis of cancer (Callinan and Batzer, 2006). This leaves ample room for non-inserational mechanisms of tumorigenesis that implicate retrotransposons. In addition, the concept that new insertions might cause tumorigenesis would be hard to reconcile with the full reversibility of the “therapeutic” effect associated with LINE-1 RT inhibition in various cancer cells, observed in our and other laboratories (Sciamanna et al., 2005; Oricchio et al., 2007; Patnala et al., 2013). In our model, therefore, insertional mutagenesis, though not being totally ruled out, plays a minor role. We believe that most retrotranspositional insertions observed in many tumors reflect a failure to repress the activity of retroelements (a frequent failure in cancer), rather than being a cause of tumorigenesis.
In an extreme view, mutations may often represent a tolerated consequence of the tumor-associated global deregulation rather than the cause. The evidence summarized so far suggest that deregulated RT activity, likely acting in combination with other key epigenetic processes such as global DNA hypomethylation and chromatin remodeling, contributes to shape pro-tumorigenic expression profiles, and thus favors the phenotypic plasticity and diversity of cancer cells.
Cancer as a reversible “developmental” disorder and differentiation therapy
As discussed above, the non-coding RNA profiles modulated by RT can globally regulate cell differentiation. Evidence is emerging that unscheduled reactivation of RT, as occurring in cancer cells, or its developmentally regulated repression, as in normal cells, are sufficient to promote cell de-differentiation or, on the contrary, stabilize the differentiated state, respectively. Tumorigenesis can be viewed as the erroneous resumption of genome-wide networks active in embryogenesis and silenced in adult life, and the differentiation process can be regarded as a sequence of transient and reversible cellular states in which RT activity is variably activated. According to this view, cancer would also be a reversible phenomenon and, as such, potentially modulatable by RT-inhibitory differentiation-inducing agents. The idea that the “normal” differentiation program can be restored to cancer cells, with the loss or attenuation of tumorigenic phenotype, has inspired much research and clinical work in the last decades. Perhaps the best known example is the development of retinoic acid-based differentiation therapy, successfully applied to treat acute promyelocytic leukemia (APML). Retinoic acid is a powerful morphogen and differentiating agent and has been the object of intense studies in the last decades, the outcome of which cannot be exhaustively discussed here (reviewed by Tang and Gudas, 2011). Other attempts to apply the same principle to solid tumors, however, have had more limited results so far (reviewed by Leszczyniecka et al., 2001; Cruz and Matushansky, 2012). The data obtained from in vitro assays, preclinical tests on animal models and a recent human trial, converge in viewing the LINE-1-encoded RT as an effective target for a non-cytotoxic, differentiation-inducing cancer therapy; RT inihibition appears to be the common condition sufficent to reverse tumorigenicity and restore differentiation to a wide variety of cancer cells.
Conclusions
Growing data undermine the concept of terminal differentiation as a stably acquired condition, revealing that: (i) differentiation states should rather be viewed as transient conditions, and (ii) even in the presence of genomic alterations, epigenetics often wins over genetics (Lotem and Sachs, 2002). Epigenetic changes can effectively bypass the genetic alterations associated with, or caused by, tumorigenesis and reprogram gene expression profiles, reverting, or mitigating, the malignant phenotypes of cells. LINE-1-encoded RT is emerging as a key epigenetic regulator at the intersection between normal and pathological development. As such, the level of RT activity has the potential to shift the biological balance of cells in one or the other direction. In our view, these findings and emerging concepts, besides their clinical implications, fulfill the early prediction by Temin that endogenous RT activity plays roles both in normal development, as in embryogenesis, and in pathologies as such as cancer (Temin, 1971).
Funding
Work in our laboratory was supported by the Italian Ministry of Health grant n. 15ONC/8 “Endogenous Reverse Transcriptase as a tumor marker and causative agent of tumor onset and progression,” to CS.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the skillful assistance of Cosimo Curianò with drawing preparation.
References
- Aalto A. P., Pasquinelli A. E. (2012). Small non-coding RNAs mount a silent revolution in gene expression. Curr. Opin. Cell Biol. 24, 333–340. 10.1016/j.ceb.2012.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschacher T., Wolf B., Enzmann F., Kienzl P., Messner B., Sampl S., et al. (2016). LINE-1 induces hTERT and ensures telomere maintenance in tumour cell lines. Oncogene 35, 94–104. 10.1038/onc.2015.65 [DOI] [PubMed] [Google Scholar]
- Babushok D. V., Kazazian H. H., Jr. (2007). Progress in understanding the biology of the human mutagen LINE-1. Hum. Mutat. 28, 527–539. 10.1002/humu.20486 [DOI] [PubMed] [Google Scholar]
- Baltimore D. (1970). RNA-dependent DNA polymerase in virions of RNA tumor viruses. Nature 226, 1209–1211. 10.1038/2261209a0 [DOI] [PubMed] [Google Scholar]
- Beck C. R., Garcia-Perez J. L., Badge R. M., Moran J. V. (2011). LINE-1 elements in structural variation and disease. Annu. Rev. Genomics Hum. Genet. 12, 187–215. 10.1146/annurev-genom-082509-141802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano G., Pheasant M., Makunin I., Stephen S., Kent W. J., Mattick J. S., et al. (2004). Ultraconserved elements in the human genome. Science 304, 1321–1325. 10.1126/science.1098119 [DOI] [PubMed] [Google Scholar]
- Belancio V. P., Roy-Engel A. M., Deininger P. L. (2010). All y'all need to know'bout retroelements in cancer. Semin. Cancer Biol. 20, 200–210. 10.1016/j.semcancer.2010.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beraldi R., Pittoggi C., Sciamanna I., Mattei E., Spadafora C. (2006). Expression of LINE-1 retroposons is essential for murine preimplantation development. Mol. Reprod. Dev. 73, 279–287. 10.1002/mrd.20423 [DOI] [PubMed] [Google Scholar]
- Böehne A., Brunet F., Galiana-Arnoux D., Schultheis C., Volff J. N. (2008). Transposable elements as drivers of genomic and biological diversity in vertebrates. Chromosome Res. 16, 203–215. 10.1007/s10577-007-1202-6 [DOI] [PubMed] [Google Scholar]
- Borchert G. M., Holton N. W., Williams J. D., Hernan W. L., Bishop I. P., Dembosky J. A., et al. (2011). Comprehensive analysis of microRNA genomic loci identifies pervasive repetitive-element origins. Mob. Genet. Elements 1, 8–17. 10.4161/mge.1.1.15766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G. (2009). Transposable elements in gene regulation and in the evolution of vertebrate genomes. Curr. Opin. Genet. Dev. 19, 607–612. 10.1016/j.gde.2009.10.013 [DOI] [PubMed] [Google Scholar]
- Brouha B., Schustak J., Badge R. M., Lutz-Prigge S., Farley A. H., Moran J. V., et al. (2003). Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. U.S.A. 100, 5280–5285. 10.1073/pnas.0831042100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callinan P. A., Batzer M. A. (2006). Retrotransposable elements and human disease. Genome Dyn. 1, 104–115. 10.1159/000092503 [DOI] [PubMed] [Google Scholar]
- Chen L., Dahlstrom J. E., Chandra A., Board P., Rangasamy D. (2012a). Prognostic value of LINE-1 retrotransposon expression and its subcellular localization in breast cancer. Breast Cancer Res. Treat. 136, 129–142. 10.1007/s10549-012-2246-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Dahlstrom J. E., Lee S. H., Rangasamy D. (2012b). Naturally occurring endo-siRNA silences LINE-1 retrotransposons in human cells through DNA methylation. Epigenetics 7, 758–771. 10.4161/epi.20706 [DOI] [PubMed] [Google Scholar]
- Coufal N. G., Garcia-Perez J. L., Peng G. E., Yeo G. W., Mu Y., Lovci M. T., et al. (2009). L1 retrotransposition in human neural progenitor cells. Nature 460, 1127–1131. 10.1038/nature08248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz F. D., Matushansky I. (2012). Solid tumor differentiation therapy - is it possible? Oncotarget 3, 559–567. 10.18632/oncotarget.512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L., Huang Q., Boeke J. D. (2011). Effect of reverse transcriptase inhibitors on LINE-1 and Ty1 reverse transcriptase activities and on LINE-1 retrotransposition. BMC Biochem. 2:18. 10.1186/1471-2091-12-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm K., Hemmann U., Garin-Chesa P., Hauel N., Kauffmann I., Priepke H., et al. (2001). A highly selective telomerase inhibitor limiting human cancer cell proliferation. EMBO J. 20, 6958–6968. 10.1093/emboj/20.24.6958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewannieux M., Esnault C., Heidmann T. (2003). LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 35, 41–48. 10.1038/ng1223 [DOI] [PubMed] [Google Scholar]
- Djebali S., Davis C. A., Merkel A., Dobin A., Lassmann T., Mortazavi A., et al. (2012). Landscape of transcription in human cells. Nature 489, 101–108. 10.1038/nature11233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet A. J., Droc G., Siol O., Audoux J., Gilbert N. (2015). U6 snRNA Pseudogenes: markers of retrotransposition dynamics in mammals. Mol. Biol. Evol. 32, 1815–1832. 10.1093/molbev/msv062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet-O'Hare T. T., Rodic N., Sharma R., Darbari I., Abril G., Choi J. A., et al. (2015). LINE-1 expression and retrotransposition in Barrett's esophagus and esophageal carcinoma. Proc. Natl. Acad. Sci. U.S.A. 112, E4894–E900. 10.1073/pnas.1502474112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C., Maestre J., Heidmann T. (2000). Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 24, 363–367. 10.1038/74184 [DOI] [PubMed] [Google Scholar]
- Esteller M. (2011). Non-coding RNAs in human disease. Nat. Rev. Genet. 12, 861–874. 10.1038/nrg3074 [DOI] [PubMed] [Google Scholar]
- Ewing A. D., Gacita A., Wood L. D., Ma F., Xing D., Kim M. S., et al. (2015). Widespread somatic L1 retrotransposition occurs early during gastrointestinal cancer evolution. Genome Res. 25, 1536–1545. 10.1101/gr.196238.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A., Bozzoni I. (2014). Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. 15, 7–21. 10.1038/nrg3606 [DOI] [PubMed] [Google Scholar]
- Feschotte C. (2008). Transposable elements and the evolution of regulatory networks. Nat Rev. Genet. 9, 397–405. 10.1038/nrg2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Perez J. L., Marchetto M. C. N., Muotri A. R., Coufal N. C., Gage F. H., O'shea K. S., et al. (2007). LINE-1 retrotransposition in human embryonic stem cells. Hum. Mol. Genet. 16, 1569–1577. 10.1093/hmg/ddm105 [DOI] [PubMed] [Google Scholar]
- Gaur A., Jewell D. A., Liang Y., Ridzon D., Moore J. H., Chen C., et al. (2007). Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 67, 2456–2468. 10.1158/0008-5472.CAN-06-2698 [DOI] [PubMed] [Google Scholar]
- Giordano R., Magnano A. R., Zaccagnini G., Pittoggi C., Moscufo N., Lorenzini R., et al. (2000). Reverse transcriptase activity in mature spermatozoa of mouse. J. Cell Biol. 148, 1107–1113. 10.1083/jcb.148.6.1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier J. L., Kazazian H. H., Jr. (2008). Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 135, 23–35. 10.1016/j.cell.2008.09.022 [DOI] [PubMed] [Google Scholar]
- Gualtieri A., Andreola F., Sciamanna I., Sinibaldi- Vallebona P., Serafino A., Spadafora C. (2013). Increased expression and copy number amplification of LINE-1 and SINE B1 retrotransposable elements in murine mammary carcinoma progression. Oncotarget 4, 1882–1893. 10.18632/oncotarget.1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy C. T., Cardiff R. D., Muller W. J. (1992). Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol. Cell. Biol. 12, 954–961. 10.1128/MCB.12.3.954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan C. R., Rudin C. M. (2002). Mobile genetic element activation and genotoxic cancer therapy potential clinical implications. Am. J. Pharmacogenomics 2, 25–35. 10.2165/00129785-200202010-00003 [DOI] [PubMed] [Google Scholar]
- Hanahan D. (2014). Rethinking the war on cancer. Lancet 383, 558–563. 10.1016/S0140-6736(13)62226-6 [DOI] [PubMed] [Google Scholar]
- Hancks D. C., Kazazian H. H., Jr. (2012). Active human retrotransposons: variation and disease. Curr. Opin. Genet. Dev. 22, 191–203. 10.1016/j.gde.2012.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht M., Erber S., Harrer T., Klinker H., Roth T., Parsch H., et al. (2015). Efavirenz has the highest anti-proliferative effect of non-nucleoside reverse transcriptase pnhibitors against pancreatic cancer cells. PLoS ONE 10:e0130277. 10.1371/journal.pone.0130277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschhorn A., Hizi A. (2010). Retroviral reverse transcriptases. Cell. Mol. Life Sci. 67, 2717–2747. 10.1007/s00018-010-0346-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houédé N., Pulido M., Mourey L., Joly F., Ferrero J. M., Bellera C., et al. (2014). A phase II trial evaluating the efficacy and safety of efavirenz in metastatic castration-resistant prostate cancer. Oncologist 19, 1227–1228. 10.1634/theoncologist.2014-0345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–921. 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Iskow R. C., McCabe M. T., Mills R. E., Torene S., Pittard W. S., Neuwald A. F., et al. (2010). Natural mutagenesis of human genomes by endogenous retrotransposons. Cell 141, 1253–1261. 10.1016/j.cell.2010.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson M. D., Lund A. H. (2012). MicroRNA and cancer. Mol. Oncol. 6, 590–610. 10.1016/j.molonc.2012.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaçar Y., Thomann H. U., Gross H. J. (1992). The first human genes for tRNA(ArgICG), tRNA(GlyUCC), and tRNA(ThrIGU) and more tRNA(Val) pseudogenes: expression and pre-tRNA maturation in HeLa cell-free extracts. DNA Cell Biol. 11, 781–790. 10.1089/dna.1992.11.781 [DOI] [PubMed] [Google Scholar]
- Kaer K., Speek M. (2013). Retroelements in human disease. Gene 518, 231–241. 10.1016/j.gene.2013.01.008 [DOI] [PubMed] [Google Scholar]
- Kaiser S., Park Y. K., Franklin J. L., Halberg R. B., Yu M., Jessen W. J., et al. (2007). Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol. 8:R131. 10.1186/gb-2007-8-7-r131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C., McLellan M. D., Vandin F., Ye K., Niu B., Lu C., et al. (2013). Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339. 10.1038/nature12634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano H., Godoy I., Courtney C., Vetter M. R., Gerton G. L., Ostertag E. M., et al. (2009). L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 23, 1303–1312. 10.1101/gad.1803909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapusta A., Kronenberg Z., Lynch V. J., Zhuo X., Ramsay L. A., Bourque G., et al. (2013). Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet. 9:e1003470. 10.1371/journal.pgen.1003470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D., Rinn J. (2012). Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol. 13:R107. 10.1186/gb-2012-13-11-r107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V. N., Han J., Siomi M. C. (2009). Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10, 126–139. 10.1038/nrm2632 [DOI] [PubMed] [Google Scholar]
- Klein C. A. (2009). Parallel progression of primary tumours and metastases. Nat. Rev. Cancer 9, 302–312. 10.1038/nrc2627 [DOI] [PubMed] [Google Scholar]
- Kulpa D. A., Moran J. V. (2006). Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat. Struc. Mol. Biol. 13, 655–660. 10.1038/nsmb1107 [DOI] [PubMed] [Google Scholar]
- Landriscina M., Fabiano A., Altamura S., Bagala C., Piscazzi A., Cassano A., et al. (2005). Reverse transcriptase inhibitors downregulate cell proliferation in vitro and in vivo and restore TSH signaling and iodine uptake in human thyroid anaplastic carcinoma. J. Clin. Endocrinol. Metab. 90, 5663–5671. 10.1210/jc.2005-0367 [DOI] [PubMed] [Google Scholar]
- Lee E., Iskow R., Yang L., Gokcumen O., Haseley P., Luquette L. J., III., et al. (2012). Landscape of somatic retrotransposition in human cancers. Science 337, 967–971. 10.1126/science.1222077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnert S., Van Loo P., Thilakarathne P. J., Marynen P., Verbeke G., Schuit F. (2009). Evidence for co-evolution between human microRNAs and Alu-repeats. PLoS ONE 4:e4456. 10.1371/journal.pone.0004456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leszczyniecka M., Roberts T., Dent P., Grant S., Fisher P. B. (2001). Differentiation therapy of human cancer: basic science and clinical applications. Pharmacol. Ther. 90, 105–196. 10.1016/S0163-7258(01)00132-2 [DOI] [PubMed] [Google Scholar]
- Levin H. L., Moran J. V. (2011). Dynamic interactions between transposable elements and their hosts. Nat. Rev. Genet. 12, 615–627. 10.1038/nrg3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. C. (2014). Chromatin remodeling by the small RNA machinery in mammalian cells. Epigenetics 9, 1–8. 10.4161/epi.26830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotem J., Sachs L. (2002). Epigenetics wins over genetics: induction of differentiation in tumor cells. Semin. Cancer Biol. 12, 339–346. 10.1016/S1044-579X(02)00054-8 [DOI] [PubMed] [Google Scholar]
- Lowe C. B., Haussler D. (2012). 29 Mammalian genomes reveal novel exaptations of mobile elements for likely regulatory functions in the human genome. PLoS ONE 7:e43128. 10.1371/journal.pone.0043128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Getz G., Miska E. A., Alvarez-Saavedra E., Lamb J., Peck D., et al. (2005). MicroRNA expression profiles classify human cancers. Nature 435, 834–838. 10.1038/nature03702 [DOI] [PubMed] [Google Scholar]
- Ma Y., Zhang P., Wang F., Yang J., Yang Z., Qin H. (2010). The relationship between early embryo development and tumourigenesis. J. Cell. Mol. Med. 14, 2697–2701. 10.1111/j.1582-4934.2010.01191.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone C. D., Hannon G. J. (2009). Small RNAs as guardians of the genome. Cell 136, 656–668. 10.1016/j.cell.2009.01.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiacasale R., Pittoggi C., Sciamanna I., Careddu A., Mattei E., Lorenzini R., et al. (2003). Exposure of normal and transformed cells to nevirapine, a reverse transcriptase inhibitor, reduces cell growth and promotes differentiation. Oncogene 22, 2750–2761. 10.1038/sj.onc.1206354 [DOI] [PubMed] [Google Scholar]
- Meachem C. E., Morrison S. J. (2013). Tumour heterogeneity and cancer cell plasticity. Nature 501, 328–337. 10.1038/nature12624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki Y., Nishisho I., Horii A., Miyoshi Y., Utsunomiya J., Kinzler K. W., et al. (1992). Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 52, 643–645. [PubMed] [Google Scholar]
- Miousse I. R., Koturbash I. (2015). The Fine LINE: methylation drawing the cancer landscape. Biomed Res. Int. 2015:131547. 10.1155/2015/131547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse B., Rotherg P. G., South V. J., Spandorfer J. M., Astrin S. M. (1988). Insertional mutagenesis of the myc locus by a LINE-1 sequence in a hu- man breast carcinoma. Nature 333, 87–90. 10.1038/333087a0 [DOI] [PubMed] [Google Scholar]
- Muñoz-López M., García-Pérez J. L. (2010). DNA Transposons: Nature and application in Genomics. Curr. Biol. 11, 115–128. 10.2174/138920210790886871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy R., Sweet K., Eng C. (2004). Highly penetrant hereditary cancer syndromes. Oncogene 23, 6445–6470. 10.1038/sj.onc.1207714 [DOI] [PubMed] [Google Scholar]
- Natoli C., Ramazzotti V., Nappi O., Giacomini P., Palmeri S., Salvatore M., et al. (2011). Unknown primary tumors. Biochim. Biophys. Acta 1816, 13–24. 10.1016/j.bbcan.2011.02.002 [DOI] [PubMed] [Google Scholar]
- Nelson A. C., Wardle F. C. (2013). Conserved non-coding elements and cis regulation: actions speak louder than words. Development 140, 1385–1395. 10.1242/dev.084459 [DOI] [PubMed] [Google Scholar]
- Nishihara H., Smit A. F. A., Okada N. (2006). Functional noncoding sequences derived from SINEs in the mammalian genome. Genome Res. 16, 864–874. 10.1101/gr.5255506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohms S., Lee S. H., Rangasamy D. (2014). LINE-1 retrotransposons and let-7 miRNA: partners in the pathogenesis of cancer? Front Genet. 5:338 10.3389/fgene.2014.00338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohms S., Rangasamy D. (2014). Silencing of LINE-1 retrotransposons contributes to variation in small noncoding RNA expression in human cancer cells. Oncotarget 5, 4103–4117. 10.18632/oncotarget.1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. (1972). So much “junk” DNA in our genome. Brookhaven Symp. Biol. 23, 366–370. [PubMed] [Google Scholar]
- Oliver K. R., Greene W. K. (2011). Mobile DNA and the TE-Thrust hypothesis: supporting evidence from the primates. Mob. DNA 2:8. 10.1186/1759-8753-2-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgel L. E., Crick F. H. (1980). Selfish DNA: the ultimate parasite. Nature 284, 604–607. 10.1038/284604a0 [DOI] [PubMed] [Google Scholar]
- Oricchio E., Beraldi R., Sciamanna I., Tolstonog G. V., Schumann G. G., Spadafora C. (2007). Distinct roles for LINE-1 and Herv-K retroelements in cell proliferation, differentiation and tumor progression. Oncogene 26, 4226–4233. 10.1038/sj.onc.1210214 [DOI] [PubMed] [Google Scholar]
- Ostertag E. M., Goodier J. L., Zhang Y., Kazazain H. H., Jr. (2003). SVA elements are nonautonomous retrotransposons that cause disease in humans. Am. J. Hum. Genet. 73, 1444–1451. 10.1086/380207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnala R., Lee S. H., Dahlstrom J. E., Ohms S., Chen L., Dheen S. T., et al. (2013). Inhibition of LINE-1 retrotransposon-encoded reverse transcriptase modulates the expression of cell differentiation genes in breast cancer cells. Breast. Cancer Res. Treat 143, 239–253. 10.1007/s10549-013-2812-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V., Steinmetz L. M. (2013). Gene regulation by antisense transcription. Nat Rev. 14, 880–893. 10.1038/nrg3594 [DOI] [PubMed] [Google Scholar]
- Perreault J., Noël J. F., Brière F., Cousineau B., Lucier J. F., et al. (2005). Retropseudogenes derived from the human Ro/SS-A autoantigen-associated hY RNAs. Nucleic Acids Res. 33, 2032–2041. 10.1093/nar/gki504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatek M. J., Werner A. (2014). Endogenous siRNAs: regulators of internal affairs. Biochem. Soc. Trans. 42, 1174–1179. 10.1042/BST20140068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pink R. C., Wicks K., Caley D. P., Punch E. K., Jacobs L., Carter D. R. F. (2011). Pseudogenes: Pseudo-functional or key regulators in health and disease? RNA 17, 792–798. 10.1261/rna.2658311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittoggi C., Sciamanna I., Mattei E., Beraldi R., Lobascio A. M., Mai A., et al. (2003). Role of endogenous reverse transcriptase in murine early embryo development. Mol. Reprod. Dev. 66, 225–236. 10.1002/mrd.10349 [DOI] [PubMed] [Google Scholar]
- Rebollo R., Romanish M. T., Mager D. L. (2012). Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu. Rev. Genet. 46, 21–42. 10.1146/annurev-genet-110711-155621 [DOI] [PubMed] [Google Scholar]
- Rodic N., Burns K. H. (2013). Long interspersed element–1 (LINE-1): passenger or driver in human neoplasms? PLoS Genet. 9:e1003402. 10.1371/journal.pgen.1003402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodić N., Sharma R., Sharma R., Zampella J., Dai L., Taylor M. S., et al. (2014). Long Interspersed Element-1 protein expression is a hallmark of many human cancers. Am. J. Pathol. 184, 1280–1286. 10.1016/j.ajpath.2014.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodić N., Steranka J. P., Makohon-Moore A., Moyer A., Shen P., Sharma R., et al. (2015). Retrotransposon insertions in the clonal evolution of pancreatic ductal adenocarcinoma. Nat. Med. 21, 1060–1064. 10.1038/nm.3919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciamanna I., Gualtieri A., Cossetti C., Osimo E. F., Ferracin M., Macchia G., et al. (2013). A tumor-promoting mechanism mediated by retrotransposon- encoded reverse transcriptase is active in human transformed cell lines. Oncotarget 4, 2271–2287. 10.18632/oncotarget.1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciamanna I., Gualtieri A., Piazza P. V., Spadafora C. (2014). Regulatory roles of LINE-1-encoded reverse transcriptase in cancer onset and progression. Oncotarget 5, 8039–8051. 10.18632/oncotarget.2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciamanna I., Landriscina M., Pittoggi C., Quirino M., Marelli C., Beraldi R., et al. (2005). Inhibition of endogenous reverse transcriptase antagonizes human tumor growth. Oncogene 24, 3923–3931. 10.1038/sj.onc.1208562 [DOI] [PubMed] [Google Scholar]
- Sciamanna I., Vitullo P., Curatolo A., Spadafora C. (2011). A reverse transcriptase-dependent mechanism is essential for murine preimplantation development. Genes 2, 360–373. 10.3390/genes2020360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla R., Upton K. R., Muñoz-Lopez M., Gerhardt D. J., Fisher M. E., Nguyen T., et al. (2013). Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell 153, 101–111. 10.1016/j.cell.2013.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinibaldi-Vallebona P., Matteucci C., Spadafora C. (2011). Retrotransposon-encoded reverse transcriptase in the genesis, progression and cellular plasticity of human cancer. Cancers 3, 1141–1157. 10.3390/cancers3011141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin R. K., Martienssen R. (2007). Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8, 272–285. 10.1038/nrg2072 [DOI] [PubMed] [Google Scholar]
- Solyom S., Ewing A. D., Rahrmann E. P., Doucet T., Nelson H. H., Burns M. B., et al. (2012). Extensive somatic L1 retrotransposition in colorectal tumors. Genome Res. 22, 2328–2338. 10.1101/gr.145235.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sottoriva A., Kang H., Ma Z., Graham T. A., Salomon M. P., Zhao J., et al. (2015). A Big Bang model of human colorectal tumor growth. Nat. Genet. 47, 209–216. 10.1038/ng.3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spadafora C. (2008). Sperm-mediated ‘reverse’ gene transfer: a role of reverse transcriptase in the generation of new genetic information. Hum. Reprod. 23, 735–740. 10.1093/humrep/dem425 [DOI] [PubMed] [Google Scholar]
- Spadafora C. (2015). A LINE-1–encoded reverse transcriptase–dependent regulatory mechanism is active in embryogenesis and tumorigenesis. Ann. N.Y. Acad. Sci. 1341, 164–171. 10.1111/nyas.12637 [DOI] [PubMed] [Google Scholar]
- Stella G. M., Senetta R., Cassenti A., Ronco M., Cassoni P. (2012). Cancers of unknown primary origin: current perspectives and future therapeutic strategies. J. Translat. Med. 10:12. 10.1186/1479-5876-10-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh N., Baehner L., Moltzahn F., Melton C., Shenoy A., Chen J., et al. (2010). MicroRNA function is globally suppressed in mouse oocytes and early embryos. Curr. Biol. 20, 271–277. 10.1016/j.cub.2009.12.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X. H., Gudas L. J. (2011). Retinoids, retinoic acid receptors, and cancer. Annu. Rev. Pathol. 6, 345–364. 10.1146/annurev-pathol-011110-130303 [DOI] [PubMed] [Google Scholar]
- Temin H. M. (1971). Guest editorial. The protovirus hypothesis: speculations on the significance of RNA-directed DNA synthesis for normal development and for carcinogenesis. J. Natl. Cancer Inst. 46, III–VI. [PubMed] [Google Scholar]
- Temin H. M., Mizutani S. (1970). RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature 226, 1211–1213. 10.1038/2261211a0 [DOI] [PubMed] [Google Scholar]
- Terasaki N., Goodier J. L., Cheung L. E., Wang Y. J., Kajikawa M., Kazazian H. H., Jr., et al. (2013). In vitro screening for compounds that enhance human L1 mobilization. PLoS ONE 8:e74629. 10.1371/journal.pone.0074629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullu E., Weiner A. M. (1984). Human genes and pseudogenes for the 7SL RNA component of signal recognition particle. EMBO J. 3, 3303–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hurk J. A. J. M., Meij I. C., Seleme M. C., Kano H., Nikopoulos K., Hoefsloot L. H., et al. (2007). L1 retrotransposition can occur early in human embryonic development. Hum. Mol. Genet. 16, 1587–1592. 10.1093/hmg/ddm108 [DOI] [PubMed] [Google Scholar]
- Vitullo P., Sciamanna I., Baiocchi M., Sinibaldi-Vallebona P., Spadafora C. (2012). LINE-1 retrotransposon copies are amplified during murine early embryo development. Mol. Reprod. Dev. 79, 118–127. 10.1002/mrd.22003 [DOI] [PubMed] [Google Scholar]
- Wallace M. R., Andersen L. B., Saulino A. M., Gregory P. E., Glover T. W., Collins F. S. (1991). A de novo Alu insertion results in neurofibromatosis type 1. Nature 353, 864–866. 10.1038/353864a0 [DOI] [PubMed] [Google Scholar]
- Watanabe T., Tomizawa S., Mitsuya K., Totoki Y., Yamamoto Y., et al. (2011). Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science 332, 848–852. 10.1126/science.1203919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Gilbert N., Ooi S. L., Lawler J. F., Ostertag E. M., Kazazian H. H., et al. (2001). Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 21, 1429–1439. 10.1128/MCB.21.4.1429-1439.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A., Goodson M., Goode D. K., Snell P., McEwen G. K., Vavouri T., et al. (2005). Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 3:e7. 10.1371/journal.pbio.0030007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Kazazian H. H., Jr. (2006). L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat. Struc. Mol. Biol. 13, 763–771. 10.1038/nsmb1141 [DOI] [PubMed] [Google Scholar]
- Zhang H., Chen Z., Wang X., Huang Z., He Z., Chen Y. (2013). Long non-coding RNA: a new player in cancer. J. Hemat. Oncol. 6:37. 10.1186/1756-8722-6-37 [DOI] [PMC free article] [PubMed] [Google Scholar]