Abstract
Drought stress is a severe environmental factor that greatly restricts plant distribution and crop production. Recently, we have found that overexpressing AtWRKY57 enhanced drought tolerance in Arabidopsis thaliana. In this study, we further reported that the Arabidopsis WRKY57 transcription factor was able to confer drought tolerance to transgenic rice (Oryza sativa) plants. The enhanced drought tolerance of transgenic rice was resulted from the lower water loss rates, cell death, malondialdehyde contents and relative electrolyte leakage while a higher proline content and reactive oxygen species-scavenging enzyme activities was observed during stress conditions. Moreover, further investigation revealed that the expression levels of several stress-responsive genes were up-regulated in drought-tolerant transgenic rice plants, compared with those in wild-type plants. In addition to the drought tolerance, the AtWRKY57 over-expressing plants also had enhanced salt and PEG stress tolerances. Taken together, our study indicates that over-expressing AtWRKY57 in rice improved not only drought tolerance but also salt and PEG tolerance, demonstrating its potential role in crop improvement.
Keywords: AtWRKY57, drought tolerance, Oryza sativa, Stress, ROS
Introduction
Drought is a critical abiotic stress that severely restricts crop production (Zhu, 2002). With the process of evolution, plants have gained a variety of strategies with the purpose of avoiding drought stress by reducing water loss or increasing water uptake. Nevertheless, other strategies need to protect plant cells from damage when water is exhausted and tissue dehydration unavoidable (Verslues et al., 2006). Additionally, the molecular, cellular, and whole-plant levels strategies should be coordinated to adapt to drought stress (Yu et al., 2008).
Under drought- or salt-stress conditions, plants accumulate reactive oxygen species (ROS) (Verslues et al., 2006). In living cells, ROS such as superoxide, hydrogen peroxide (H2O2), and hydroxyl radicals are generated as harmful substances via aerobic metabolism. Through partially reduced or activated derivatives of oxygen, ROS can destroy DNA, proteins and carbohydrates, resulting in cell death (Mittler et al., 2004). A master level of ROS gives rise to the oxidation of biomolecules, such as lipids, nucleic acids and proteins, which caused cellular damage. When CO2 fixation is restricted under environmental stress conditions, the photosynthetic electron transport system generates ROS (Asada, 1999). To defend oxidative stress, organisms have evolved an effective system to protect themselves. For example, numerous stress-related genes were induced by ROS in response to oxidative stress in these defensive systems (Demple and Amabile-Cuevas, 1991; Gasch et al., 2000; Desikan et al., 2002). As higher plants have the ability to coordinately regulate multiple antioxidant genes, they are much tolerant to oxidative stresses.
Normally, the maintenance of routine homeostasis is achieved through the ROS-scavenging system in plant cells, which is mainly mediated by enzymatic defenses, including superoxide dismutase (SOD), catalase (CAT), and peroxidases (POX) (Mittler et al., 2004). Generally, SODs, which catalyze the dismutation of superoxide into oxygen and H2O2, provide the first line of defense against ROS in various subcellular compartments, such as chloroplast, mitochondria and cytosol (Raychaudhuri and Deng, 2000). The physiological role of CAT is to break down H2O2 in the cell (Scandalios, 2002). Therefore, increased CAT activity would result in H2O2 degradation. PODs are a group of enzymes that catalyze the oxidation of many substrates (e.g., phenolic compounds) at the expense of H2O2 (Asada, 1987). The increased activity of these enzymes would decrease ROS levels. Recent reports have demonstrated that transgenic rice plants with enhanced ROS-scavenging abilities had improved drought tolerance (Ouyang et al., 2010; Zhang et al., 2011). For example, OsSIK1 functions in stress signaling through scavenging and detoxification of ROS. In OsSIK1-overexpressing plants, the high levels of POD and CAT enzymes resulted to low levels of H2O2 (Ouyang et al., 2010). Similarly, the improved drought tolerance of HRF1-overexpressing transgenic rice plants was partially resulted from the increased ROS-scavenging activities (Zhang et al., 2011).
Under environment stress conditions, the stress-related proteins not only function in protecting cells from damage but also regulate the expression of downstream genes for signal sensing, perception and transduction (Kreps et al., 2002; Seki et al., 2002). These proteins can be classified into two groups. The first group protein plays a crucial role to avoid cellular injury, such as detoxification enzymes, Late Embryogenesis Abundant (LEA) proteins, and the key enzymes for osmolyte biosynthesis (Kreps et al., 2002; Seki et al., 2002). The second group includes numerous transcription factors involved in further regulation of transcriptional control and signal transduction. The CBF/DREB factor, Basic Leucine Zipper families, CUC transcription factor, NAM, plant nuclear factor Y (NF-Y) B subunits, zinc finger and ATAF, belong to this group (Umezawa et al., 2006; Nelson et al., 2007; Takasaki et al., 2010). Studies on these transcription factors will contribute to uncover the respect for commercially improving drought tolerance in crops through genetic engineering.
The WRKY family consists of 74 and 102 members in Arabidopsis thaliana and Oryza sativa, respectively (Eulgem et al., 2000; Wu et al., 2005); and majority of them play critical roles in biotic and abiotic stress responses (Eulgem and Somssich, 2007; Miller et al., 2008). Recently, increasing evidences confirmed that numerous of WRKY genes are involved in drought stress. For example, ABO3/WRKY63 plays a key role in plant responses to ABA and drought stress (Ren et al., 2010). Overexpression of a stress-induced OsWRKY45 significantly confer drought tolerance in Arabidopsis and rice (Qiu and Yu, 2008; Tao et al., 2011). Especially, our previous study demonstrated that overexpression of AtWRKY57 improved drought tolerance by directly targeting the promoter sequences of NCED3 to increase the content of ABA in Arabidopsis (Jiang et al., 2012). These evidences give us a hypothesis that the improvement of plant drought tolerance might be realized through gene manipulation approaches. To test this hypothesis, we further over-expressed AtWRKY57 in rice and demonstrated that the stress tolerance of the transgenic rice under drought conditions was significantly improved. 3,3′-Diaminobenzidine (DAB) and nitro blue tetrazolium (NBT) staining analyses showed that the ROS levels in transgenic lines were lower than in control plants after drought-stress treatment. Consistent with the low ROS levels, the antioxidative enzyme activities were also enhanced in the transgenic lines. Moreover, high expression levels of stress-responsive genes also supported the drought tolerance in transgenic lines. Overall, our results indicated that the over-expression of AtWRKY57 in rice conferred the adaptation of rice to drought tolerance by reducing ROS damage and up-regulating the expression of stress-responsive genes.
Materials and Methods
Construction and Transformation of AtWRKY57 in Rice
The full-length cDNA sequence of AtWRKY57 was obtained from Arabidopsis using the same method as described in our previous study (Jiang et al., 2012). The full coding sequence of AtWRKY57 was cloned into pUN1301 in the sense orientation behind the Ubiquitin promoter. Then the T-DNA was transformed into ZH11 (Oryza sativa L. ssp. japonica cv. Zhonghua11) via the Agrobacterium tumefaciens-mediated method (Hiei et al., 1994). After transformation, the calli were selected from half-strength (MS) medium containing 100 μg/ml hygromycin. Seedlings with hygromycin-resistant were transplanted to soil in a growth chamber.
Plant Growth Conditions
The sterilized Oryza seeds sowed on medium and kept in a growth chamber at 22°C under long-day conditions [16 h light/8 h dark cycles]. One week generation, seedlings were then transplanted in soil and half-strength MS medium supplemented with 1.5% (W/V) sucrose for drought stress, NaCl and PEG treatments. The soils are commonly used loam, mixed 50% humus soil, 30% coconut tree branny, 20% red clay.
Drought-Tolerance Assays
Drought-tolerance assays were performed using 4-week-old plants. The transgenic rice and control seedlings were transplanted in the same pot and treated with drought stress by withholding water for 20 days. Three independent pots repeated at the same time and a representative result displayed. Three independent experimental replications were conducted.
To evaluate the water loss rates, flag leaves were detached from the plants and weighed at designated time intervals at room temperature. The proportion of fresh weight lost was calculated based on the initial plant weight. At least three biological replicates for each sample were used for the calculation.
Trypan Blue, DAB and NBT Staining
For DAB staining, leaf sections of approximately 5 cm in length were cut and soaked in a 1% solution of DAB in 50 mM Tris-HCl buffer (pH 6.5). After 30 min vacuum infiltrating, the immersed leaves were incubated in the dark for 20 h at room temperature. And then the leaves were bleached by bath in boiling ethanol until the brown spots appeared clearly. The area of brown spots are represented the DAB reaction degree to H2O2.
Leaf sections of approximately 5 cm in length were excised to detect superoxide accumulation by a 0.1% solution of NBT in 10 mM potassium phosphate buffer (pH 7.8) as described previously (Fitzgerald et al., 2004). After 15 min vacuum infiltrating, the immersed leaves were incubated overnight at room temperature. After incubation, the leaves were fixed and cleared in alcoholic lacto-phenol (2:1:1, 95% ethanol:lactic acid:phenol) at 65°C for 30 min, rinsed with 50% ethanol, and then rinsed with water. When NBT interacts with superoxide, a blue precipitate forms is visible in leaves.
Proline (Pro) Content, Malondialdehyde (MDA) Content, and Electrolyte Leakage Measurements
The proline concentration was determined as described (Bates, 1973). Approximately 0.5 g of transgenic and control leaf segments were homogenized in 10 ml 3% aqueous sulfosalicylic acid and centrifuged at 3,000 × g for 20 min. 2 ml of supernatant was reacted with 2 ml acid ninhydrin and 2 ml glacial acetic acid in a test tube at 100°C for 1 h, cooled on ice, and the absorbance at 520 was measured. L-Pro was used as a standard to calculate the proline concentration.
The MDA content was determined as described (Heath and Packer, 1968) with slight modifications. Approximately 1 g of transgenic and control leaf segments were homogenized in 10 ml of 10% trichloroacetic (v/v) and centrifuged at 5,000 × g for 10 min. 2 ml of supernatant was reacted with 2 ml thiobarbituric acid in a test tube at 100°C for 15 min, quickly cooled on ice, and the absorbance at 532 was measured. The MDA content was confirmed using the extinction coefficient of 155 nM-1 cm-1, and expressed as nmol g-1 FW.
The relative ion leakage was checked following the method of Clarke et al. (2004). For the above assays, each data point is the average of three replicates. At least three experiments were performed, and the results are consistent. The result from one set of experiments is presented here.
Oxidative Enzyme Activity Measurements
The leaves of 4-week-old rice seedlings were dehydrated for 2 h, and then homogenized in a solution of 50 mM sodium phosphate buffer (pH 7.8) containing 1% polyvinylpyrrolidone and 10 mM β-mercaptoethanol in an ice-cold mortar. After centrifugation (13,000 × g, 15 min) at 4°C, the supernatant was used to identify SOD, POD and CAT activity levels. U min-1 mg-1 protein was represented the enzyme activity of SOD, POD, and CAT.
The ability to inhibit the photochemical reduction of NBT chloride was used for the determination of the total SOD activity as described by Beauchamp and Fridovich (1971). The reduction of NBT by 50% of the quantity of enzyme required was defined as one unit of SOD activity.
The activity of POD was determined as described by Maehly and Chance (1954). Three milliliter of reaction mixture contained 30 μl enzyme extract, 5.4 mM guaiacol, 50 mM sodium acetate buffer (pH 5.6), and 15 mM H2O2. The oxidation of guaiacol to tetraguaiacol was contributed to the increase in absorbance monitored at 470 nm. A 0.01 absorbance increase per min at 470 nm was defined as one unit of POD activity.
The activity of CAT was measured following the method of Cakmak and Marschner (1992) by determining the rate of H2O2 disappearance at 240 nm. Three milliliter of reaction mixture contained 30 μl enzyme extract, 10 mM H2O2 and 50 mM phosphate buffer (pH 7.0). A 0.01 absorbance decrease per min at 240 nm was defined as one unit of CAT activity.
For each enzyme’s activity, the data points are the average of three replicates. Three experiments were performed, and the results are consistent. The result from one set of experiments is presented here.
Salt and Osmotic Tolerance Assays
For salt tolerance assays, 2-week-old seedlings grown on half-strength MS agar medium were transferred into half-strength MS liquid medium for 2 weeks growth and then transferred into half-strength MS liquid medium supplemented with 175 mM NaCl and incubated at 22°C under long-day conditions for 2 days. After 2 days of NaCl treatment, the seedlings were transferred into half-strength MS liquid medium for 7 days recovery.
For PEG tolerance assays, 2-week-old seedlings grown on half-strength MS agar medium were transferred into half-strength MS liquid medium for 2 weeks of growth and then transferred into half-strength MS liquid medium supplemented with 25% PEG6000 (m/v) and incubated at 22°C under long-day conditions for 4 days. After 4 days of PEG treatment, the seedlings were transferred into half-strength MS liquid medium for 7 days recovery.
The transgenic rice and control seedlings were transplanted in the same pot for NaCl and PEG treatments. Three independent pots repeated at the same time and a representative result displayed in the manuscript. Three independent experimental replications were conducted.
Real-Time RT-PCR Analysis
For the real-time RT-PCR analysis, the same method was used as described in our previous studies (Jiang et al., 2012, 2014). We conducted three independent experiments (three biological replications and three technological replications in every independent experiment) and one representative result was displayed. All of the primer sequences used in real-time RT-PCR analysis were listed in Supplementary Table 1.
Statistical Analysis
Statistically significant differences (∗P < 0.05) based on the Student’s test computed by the SigmaPlot10.0. Data are the means ± SE of three independent experiments (3 biological replications and three technological replications in every independent experiment).
Results
Constitutive Expression of AtWRKY57 in Transgenic Rice Lines
In our previous study, we confirmed that overexpression of AtWRKY57 significantly enhanced drought tolerance in Arabidopsis (Jiang et al., 2012). To explore whether AtWRKY57 plays an important role in improving the agronomic traits through gene manipulation approaches, we introduced this gene to rice. More than 20 transgenic lines were generated and five lines were randomly selected to check AtWRKY57 expression by northern blotting (Supplementary Figure 1). And then two lines, Line 3 and Line 5, were chosen for further analysis (Supplementary Figure 1). There were no significant differences in morphology between the control and transgenic plants (Figure 1A).
FIGURE 1.
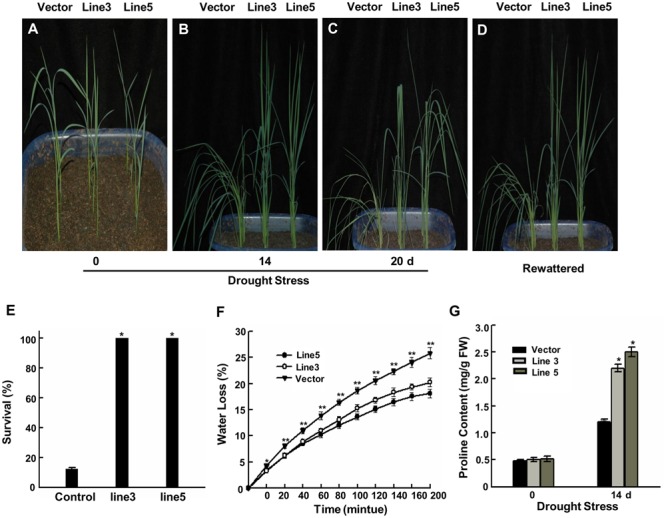
Improved drought tolerance in AtWRKY57 transgenic rice. (A) Before drought treatment. (B) Drought for 14 days. (C) Drought for 20 days. (D) Recovery for 7 days after 20 days drought treatment. Drought stress was imposed on 4-week-old T3 transgenic seedlings in greenhouse. Drought experiments were repeated three times and at least 40 plants for each individual lines were used in each repeated experiment and one representative picture was shown. (E) Survival rate after 20 days drought stress. Values are mean ± SE (n = 40 plants, ∗P < 0.05). (F) Rate of water loss by detached leaves from control and transgenic plants. Values are the mean ± SE (n = 6 plants, ∗∗P < 0.01). (G) Proline content in the leaves of 4-week-old transgenic and control plants with or without drought treatments. Values are the mean ± SE of three independent experiments (∗P < 0.05). FW, Fresh weight.
Improved Drought Tolerance in AtWRKY57 Transgenic Plants
The transgenic lines and control seeds were germinated simultaneously on half-strength MS agar medium containing 2% sucrose with or without hygromycin at 100 μg/ml and then planted in soil after 1 week. Four-week-old plants were treated with natural drought stress (not supplied with water). The control plants showed wilting symptoms 6 days before the transgenic lines. After 14 days treatment, the transgenic plants did not display any drought-stress symptoms, while the wild-type plants exhibited severe drought symptoms (Figure 1B). Up to 20 days of treatment, the control showed obvious drought-stress symptoms (Figure 1C). When plants were re-watered, only 12.3% of control plants were survived and most of them never recovered; however, all of the transgenic rice plants survived (Figures 1D,E). These results suggested that these transgenic rice plants acquired significantly improved drought tolerance. Soil moisture contents and their dynamics showed in Supplementary Figure 2.
Transpiration water loss is an important factor related to drought tolerance. Flag leaves were detached and the changes of fresh weight were determined over a 200-min period to assess the water loss rate of transgenic and control plants. A slower water loss rate was displayed in the transgenic lines’ leaves than the control’ (Figure 1F). The reduced water loss rate is favorable for an increased drought tolerance in the transgenic lines. In response to drought stress, stomata often close to limit water loss by transpiration. Given that water loss rate were lower in two transgenic lines than in control plants, we further investigated whether stomata density and/or stomata aperture affects this progress. White nail polish blotting was used to count the stomata density and measure stomata aperture. The ratio of stomatal width to length indicated the degree of stomatal closure. The results showed that the stomata density didn’t displayed significant difference between control and two transgenic plants leaf adaxial surface (Supplementary Figures 3A,B). However, two transgenic lines’ stomata showed more quick closure than control’ under dehydration treatments (Supplementary Figure 3C). These results suggest that more quick closure of stomata in two transgenic lines result in the lower water loss rate, which may be critical for transgenic plants to adapt to drought stress.
The accumulation of proline in plant is associated with adaptation to environmental stress through metabolic adjustments (Ábrahám et al., 2003). We also checked the proline contents of transgenic and control plants under normal growth and drought-stress conditions to characterize the physiological basis for the improved stress tolerance. No differences in the proline contents were observed in the leaves of transgenic and control plants under normal conditions (Figure 1G). However, under drought conditions, transgenic plants began to accumulate proline after 14 days and further accumulated an up to fourfold higher proline content compared with the levels prior to drought stress, whereas control plants showed a low increase in proline. This result demonstrates that the proline accumulation corresponded to the increased drought tolerance of transgenic plants.
Decreasing the ROS Damage in AtWRKY57 Transgenic Plants
Leaves of control plants began to produce brownish lesions after 14 days of drought stress (Figure 2A). In contrast, none of the transgenic plants exhibited lesion formation grown under the same conditions. Lesion formation was accompanied by significant trypan blue staining that indicates cell death in the control leaves (Figure 2B).
FIGURE 2.
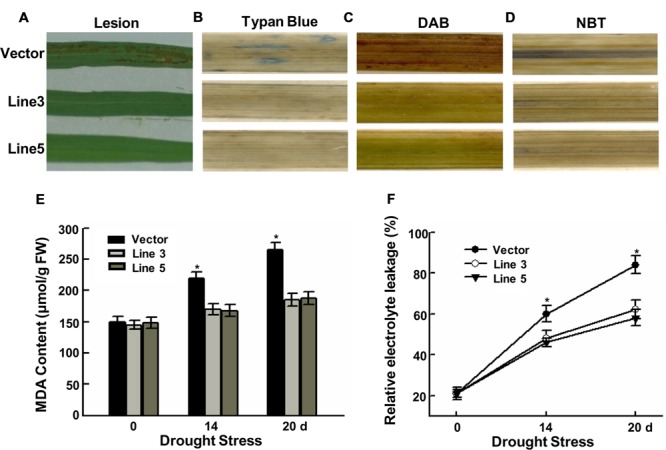
Decreased the ROS damage in transgenic rice. Phenotype of brownish lesions (A), Typan blue staining (B), DAB staining (C) and NBT staining (D) after 14 days drought stress. (E) MDA content in the leaves of control and transgenic plants after 0, 14, and 20 days drought stress. Values are the mean ± SE of three independent experiments (∗P < 0.05). FW, Fresh weight. (F) Relative electrolyte leakage in the leaves of control and transgenic plants after 0, 14, and 20 days drought stress. Values are the mean ± SE of three independent experiments (∗P < 0.05).
Stress usually causes damage via oxidative damage in plants including the generation of ROS, represented as H2O2 and superoxide (Zhu, 2001; Mittler, 2002; Xiong and Zhu, 2002). As activation of AtWRKY57 enhanced the drought tolerance of transgenic rice plants, we further determined whether AtWRKY57 is involved in drought tolerance via ROS detoxification. Transgenic rice and control seedlings were subjected to DAB staining and NBT staining to detect H2O2 and superoxides in their leaves. After 14 days of drought stress, the transgenic plants had very few brown H2O2 and superoxide spots within the leaf segments, whereas more than half of the leaf area of the control plants became brown (Figures 2C,D). The leave segments of control plants displayed more brown areas than compared transgenic plants. These results confirmed that over-expressing AtWRKY57 in rice could efficiently remove the H2O2 and superoxide produced during drought stress.
Malondialdehyde, acting as a biomarker for lipid peroxidation, is an effect of oxidative damage deriving from decomposition product of polyunsaturated fatty acid hydroperoxides. The MDA contents in transgenic lines and controls were similar under normal growth conditions, but there was a significant difference after drought stress. Then, the MDA contents of two AtWRKY57 over-expressing lines were significantly lower than those of control plants (Figure 2E).
Electrolyte leakage, an indicator of membrane damage, was also measured following drought stress. The results showed that the leaves of two AtWRKY57 over-expressing lines exhibited significantly lower electrolyte leakage levels, compared to those of control leaves (Figure 2F). After 14 days of drought stress, more than 60% of the ions leaked from cells in control plants, whereas the ion leakage of AtWRKY57 over-expressing lines was less than 50%.
Overall, these results indicated that the over-expressing AtWRKY57 gene in rice increased the tolerance to drought stress by decreasing ROS damage.
The Enhanced ROS-Scavenging Ability and High-Level Expression of Oxidative Enzyme Genes in AtWRKY57 Transgenic Plants
A decreased cell viability and even cell death was resulted from the over-accumulation of ROS; therefore, scavenging ROS avoids or alleviates the harmful effects on plant under stress conditions. In the ROS-scavenging mechanisms of plants, POD, SOD, and CAT are key enzymes (Mittler, 2002; Xiong and Zhu, 2002; Apel and Hirt, 2004), and are involved in the H2O2 elimination. Following drought stress, the enzyme activities of seedlings were subjected to measurement. Under normal growth conditions, POD, SOD, and CAT activity levels were not different; however, after 14 days of drought stress, the activities of the antioxidative enzymes were all significantly enhanced in the AtWRKY57-overexpressing plants compared with those in the control plants (Figures 3A–C). These results suggested that over-expression of AtWRKY57 gene may enhance the ROS-scavenging ability, which decreases ROS damage.
FIGURE 3.
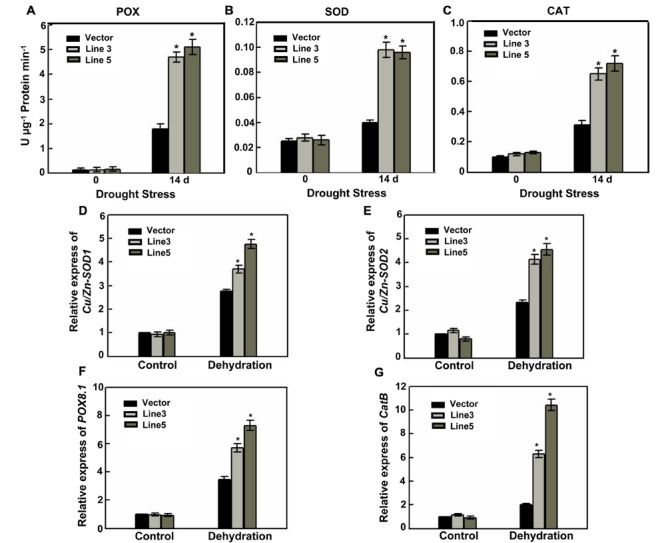
Enhanced the ROS-scavenging ability and the expression of oxidative enzymes genes in transgenic rice. (A–C) POX, SOD, and CAT activities in the leaves of 4-week-old transgenic and control plants before and after drought stress. Values are the mean ± SE of three independent experiments (∗P < 0.05). FW, Fresh weight. Relative expression of oxidative enzymes genes Cu/Zn-SOD1 (D), Cu/Zn-SOD2 (E), POX8.1 (F), and CatB (G) in the leaves of 4-week-old transgenic and control plants before and after drought stress. Values are the mean ± SE of three independent experiments (∗P < 0.05).
To test whether drought stress modifies transcript levels, the expression levels of several antioxidant genes were measured. Consistent with the increase of antioxidative enzymes activities, control and two transgenic plants up-regulated the transcript levels of OsCAT B, OsCu/Zn-SOD1, OsCu/Zn-SOD2 and OsPOD in response to drought stress, with a greater increase in the transgenic plants (Figures 3D–G). This was enhancing the capacity to decompose H2O2 and superoxide in the leaves.
High-Level Expression of Stress-Response Genes in AtWRKY57 Transgenic Plants
To better understand the mechanisms of drought tolerance conferred by over-expressing AtWRKY57, the expressions of several stress-related genes were investigated. As shown in Figure 4A, the expression levels of a pyrroline-5-carboxylate synthesis gene (OsP5CS; Igarashi et al., 1997) were strongly induced in transgenic lines under drought stress compared with those in control plants. This higher expression level of OsP5CS was consistent with the higher proline content in two AtWRKY57 overexpressing lines (Figure 1G). However, the expression level of an ABA synthesis gene (OsNCED5) was not significantly different in transgenic lines and control plants after drought stress (Figure 4B). Dehydration-responsive element-binding (DREB) transcription factors play key important roles in plant-stress responses. DREB proteins encoding by OsDREB1A and OsDREB2A were strongly up-regulated in drought stress, with a greater increase in the transgenic plants compared to the control plants (Figures 4C,D). We also checked another two well-characterized drought resistance-related genes (OsRab21 and OsRab16D), and found that they were significantly affected by water stress. Their expression levels were obviously higher in transgenic plants than in control after drought treatment (Figures 4E,F).
FIGURE 4.
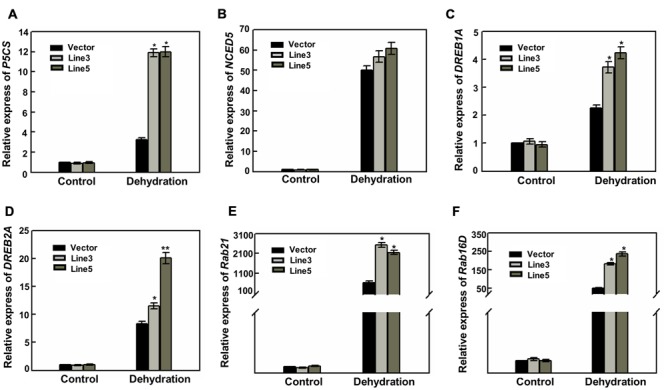
High level expression of stress-responsive genes in transgenic rice. (A–F) Relative expression levels of P5CS, NCED5, DREB1A, DREB2A, Rab21 and Rab16D in the leaves of 4-week-old transgenic and control plants before and after drought stress. Values are the mean ± SE (∗P < 0.05) of three independent experiments.
These results indicated that over-expressing AtWRKY57 gene in rice may enhance the expression of some stress-response genes and finally increase the tolerance to drought stress.
Improved NaCl and PEG Tolerance in AtWRKY57 Transgenic Plants
Our results revealed that the constitutive expression of AtWRKY57 enhanced the drought tolerance in rice (Figure 1). Given the function of WRKY-type regulators in abiotic stress, we further explored the functions of AtWRKY57 in NaCl and PEG stress conditions. We tested the survival rates of transgenic and control plants on MS medium additionally added with 175 mM NaCl. The control plants displayed more severe phenotype, including leaf curves and dehydration, than the transgenic lines after 2 days of NaCl treatment (Figures 5A,B). When plants were recovered in fresh MS medium, none of the control plants survived but most of the transgenic lines reversed (Figure 5C; Supplementary Figure 4). We also tested the survival rate of transgenic lines and control plants on MS medium supplemented with 25% PEG6000. The control plants showed more severe phenotype than transgenic lines after 4 days PEG treatments (Figures 5D,E). When plants were recovered in fresh MS medium, none of the control plants survived, but all of the transgenic lines lived (Figures 5F,G; Supplementary Figure 5).
FIGURE 5.

Improved NaCl and PEG tolerance in transgenic rice. (A) Before NaCl treatment. (B) NaCl treatment for 2 days. (C) Recovery for 7 days after 2 days NaCl treatment. (D) Before PEG treatment. (E) PEG treatment for 4 days. (F,G) Recovery for 7 days after 4 days PEG treatment. NaCl and PEG stress was imposed on 4-week-old T3 transgenic seedlings under water culture conditions in greenhouse. Experiments were repeated three times and at least 30 plants for each individual lines were used in each repeated experiment and one representative picture was shown.
These results showed that consecutively expressing AtWRKY57 enhanced not only drought tolerance but also the NaCl and PEG stress tolerance in rice.
Discussion
Combination of abiotic and biotic stresses used to limit the production of crop. Drought severely restricts crop production as the most important abiotic stress (Boyer, 1982; Rockstrom and Falkenmark, 2000). In a previous study, we confirmed that over-expressing AtWRKY57 significantly conferred drought tolerance in Arabidopsis (Jiang et al., 2012). These results suggested that AtWRKY57 may improve crops’ drought adaptability using gene manipulation. In this study, we evaluated the role of AtWRKY57 in transgenic rice after drought stress.
The drought-tolerance phenotype of AtWRKY57 transgenic rice plants were the result of a collection of physiological indexes observed in the over-expressing plants. AtWRKY57 overexpressing plants displayed higher survival rates most likely because the water loss was reduced in these plants compared to control plants under drought conditions (Figure 1F). P5CS, catalyzing proline biosynthesis, is critical for the increasing of osmotolerance. Drought, salt, and abscisic acid induce the expression of OsP5CS and the conferred osmotolerance is reuslting from an up-regulated expression of OsP5CS which increases proline content in transgenic plants (Xiang et al., 2007). The significantly higher transcript levels of OsP5CS were consistent with the high proline content in transgenic plants after drought stress (Figures 1G and 4A). These results confirmed that the transgenic plants’ adaptation to drought stress was associated with mechanisms of dehydration avoidance through proline metabolic adjustments. Programmed cell death (PCD) and lesion formation in some lesion mimic mutants, such as lsd1, was mainly caused from the elevated levels of extracellular superoxide (Jabs et al., 1996). We ovserved high levels of superoxide, H2O2 and cell death in control plants than in transgenic lines. We believe that the lesion formation in control plants results from their reduced capability to detoxify ROS compared with transgenic plants (Figure 2A). These results are similar to those of Yang et al. (2004), who reported that salicylic acid-deficient transgenic rice contains elevated levels of superoxide and H2O2 and exhibits spontaneous lesion formation in an age- and light-dependent manner.
Drought or salt-stress conditions promoted the accumulation of ROS in plants. MDA is often considered as a reflection of cellular membrane degradation or dysfunction and is also an important intermediary agent in ROS scavenging. Thus, high level of MDA causes PCD and induces toxicity to plant cells (Apel and Hirt, 2004; Hou et al., 2009). High ability of ROS-scavenging enzymes decreased over-accumulated ROS levels which induces PCD in plants (Mittler, 2002; Apel and Hirt, 2004; Chaves and Oliveira, 2004; Farooq et al., 2009; Hou et al., 2009). In AtWRKY57 transgenic plants, lower levels of PCD, DAB and NBT staining, MDA content and relative electrolyte leakage were detected (Figures 2B–F), but increased SOD, POD, and CAT activity levels (Figures 3A–C) and elevated oxidative enzyme genes’ transcript levels (Figures 3D–G) were detected after drought stress, demonstrating that they were better protected from oxidative damages through the enhanced capability to scavenge ROS.
The transcript levels for several stress-tolerant genes were more elevated in AtWRKY57 transgenic rice than in control plants under drought-stress conditions (Figure 4). It is interesting that the relative transcript levels of OsNCED5 and the ABA content were not significantly changed (Figure 4B; Supplementary Figure 6) in AtWRKY57 transgenic plants under drought-stress conditions, which may demonstrate that there were different regulatory mechanisms in transgenic Arabidopsis and transgenic rice. Our previous study revealed that the activated expression of AtWRKY57 conferred Arabidopsis transgenic plants drought tolerant by elevating the ABA contents through directly binding the promoter sequence of AtNCED3 (Jiang et al., 2012). In this study, we found that the enhanced capability to scavenge ROS was important for AtWRKY57 overexpressing transgenic rice plants to tolerate drought stress (Figures 2 and 3). Interestingly, there are increasing studies demonstrated that the same gene may have different regulatory functions and/or mechanisms when overexpressed in different plants species, such as in rice, cotton and Arabidopsis. For example, OsWRKY45 overexpressing transgenic rice showed sensitivity to drought stress (Tao et al., 2011); however, heterologous overexpression of OsWRKY45 in Arabidopsis conferred plants drought tolerant mainly resulting from the reduction of transpiration rate (Qiu and Yu, 2008). Overexpression of OsSNAC1 enhanced drought tolerance of transgenic rice plants by targeting genes that control ROS homeostasis and stomatal closure (Yu et al., 2013), whereas overexpressing OsSNAC1 rendered transgenic cotton plants more drought tolerance by reducing transpiration rate and enhancing root development (Liu et al., 2014). Heterologous expression of the AtDREB1A gene in peanut conferred transgenic plants drought and NaCl tolerance by upregulating proline synthesis to better osmotic adjustments (Sarkar et al., 2014), while the AtDREB1A transgenic Arabidopsis enhanced drought by activating some stress-related genes expression (Liu et al., 1998). Thus, it’s possible that there may be diversified regulatory functions and/or mechanisms for one protein to regulate different physiological processes in different species under stress conditions.
WRKY transcription factors belong to a large family that functions under a variety of abiotic stresses. Our results provided evidences that overexpressing AtWRKY57 also increased the tolerance to salt and PEG stresses (Figures 1 and 5), demonstrating that this is a potential candidate gene for crop improvement. Recently, several studies confirmed that overexpression of some stress-related genes may enhance drought tolerance in rice (Dubouzet et al., 2003; Park et al., 2005; Chen et al., 2008; Hou et al., 2009; Huang et al., 2009; Cui et al., 2011; Gao et al., 2011; Yang et al., 2012; Zou et al., 2012). However, a persistent problem is that the constitutive over-expression of stress-related genes often result in abnormal development and thus reduces crop productivity (Kasuga et al., 1999; Hsieh et al., 2002; Dubouzet et al., 2003; Nakashima et al., 2007; Priyanka et al., 2010). The improvement in drought tolerance should be perfectible without limitation in plant growth and production (Cattivelli et al., 2008). Yu’s study confirmed that the heterologous expression of AtEDT1/HDG11 in rice significantly improved its drought tolerance and also simultaneously increased the grain yield under both normal and drought-stress conditions (Yu et al., 2013). In our study, AtWRKY57 transgenic plants underwent normal development compared with controls (Figure 1A), but we have not statistically analyzed the effects on productivity. Further studies should focus on the grain yield of transgenic plants under drought-stress conditions.
Author Contributions
YJ designed and performed experiments, interpreted data, and wrote the article. DY designed experiments, and edited the article. YQ and YH interpreted data and edited the article. Both authors read and approved the final article.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Natural Science Foundation of China (31300252), the program for Innovative Research Team of Yunnan Province (2014HC017) and the West Light Foundation of Chinese Academy of Sciences.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00145
References
- Ábrahám E., Rigó G., Székely G., Nagy R., Koncz C., Szabados L. (2003). Light dependent induction of proline biosynthesis by abscisic acid and salt stress in inhibited by brassinosteroid in Arabidopsis. Plant Mol. Biol. 51 363–372. 10.1023/A:1022043000516 [DOI] [PubMed] [Google Scholar]
- Apel K., Hirt H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55 373–399. 10.1146/annurev.arplant.55.031903.141701 [DOI] [PubMed] [Google Scholar]
- Asada K. (1987). The water-water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 601–639. 10.1146/annurev.arplant.50.1.601 [DOI] [PubMed] [Google Scholar]
- Asada K. (1999). The water-water cycle in chloroplasts: scavenging of active oxygen and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 601–639. 10.1146/annurev.arplant.50.1.601 [DOI] [PubMed] [Google Scholar]
- Bates L. S. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39 205–207. 10.1016/j.dental.2010.07.006 [DOI] [Google Scholar]
- Beauchamp C., Fridovich I. (1971). Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 44 276–287. 10.1016/0003-2697(71)90370-8 [DOI] [PubMed] [Google Scholar]
- Boyer J. S. (1982). Plant productivity and environment. Science 218 443–448. 10.1126/science.218.4571.443 [DOI] [PubMed] [Google Scholar]
- Cakmak I., Marschner H. (1992). Magnesium deficiency and high light intensity enhance activities of superoxide dismutase, ascorbate peroxidase, and glutathione reductase in bean leaves. Plant Physiol. 98 1222–1227. 10.1104/pp.98.4.1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattivelli L., Rizza F., Badeck F. W., Mazzucotelli E., Mastrangelo A. M., Francia E., et al. (2008). Drought tolerance improvement in crop plants: an integrated view from breeding to genomics. Field Crops Res. 105 1–14. 10.3389/fpls.2015.00563 [DOI] [Google Scholar]
- Chaves M. M., Oliveira M. M. (2004). Mechanisms underlying plant resilience to water deficits: prospects for water-saving agriculture. J. Exp. Bot. 55 2365–2384. 10.1093/jxb/erh269 [DOI] [PubMed] [Google Scholar]
- Chen J. Q., Meng X. P., Zhang Y., Xia M., Wang X. P. (2008). Over-expression of OsDREB genes lead to enhanced drought tolerance in Rice. Biotechnol. Lett. 30 2191–2198. 10.1007/s10529-008-9811-5 [DOI] [PubMed] [Google Scholar]
- Clarke S. M., Mur L. A. J., Wood J. E., Scott I. M. (2004). Salicylic acid dependent signaling promotes basal thermotolerance but is not essential for acquired thermotolerance in Arabidopsis thaliana. Plant J. 33 432–447. 10.1111/j.1365-313X.2004.02054.x [DOI] [PubMed] [Google Scholar]
- Cui M., Zhang W., Zhang Q., Xu Z., Zhu Z., Duan F., et al. (2011). Induced over-expression of the transcription factor OsDREB2A improves drought tolerance in rice. Plant Physiol. Biochem. 49 1384–1391. 10.1016/j.plaphy.2011.09.012 [DOI] [PubMed] [Google Scholar]
- Demple B., Amabile-Cuevas C. F. (1991). Redox redux: the control of oxidative stress responses. Cell 67 837–839. 10.1016/0092-8674(91)90355-3 [DOI] [PubMed] [Google Scholar]
- Desikan R., Griffiths R., Hancock J. T., Neill S. (2002). A new role for an old enzyme: nitrate reductase-mediated nitric oxide generation is required for abscisic acid-induced stomatal closure in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 99 16314–16318. 10.1073/pnas.252461999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouzet J. G., Sakuma Y., Ito Y., Kasuga M., Dubouzet E. G., Miura S., et al. (2003). OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 33 751–763. 10.1046/j.1365-313X.2003.01661.x [DOI] [PubMed] [Google Scholar]
- Eulgem T., Rushton P. J., Robatzek S., Somssich I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5 199–206. 10.1016/S1360-1385(00)01600-9 [DOI] [PubMed] [Google Scholar]
- Eulgem T., Somssich I. E. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10 366–371. 10.1016/j.pbi.2007.04.020 [DOI] [PubMed] [Google Scholar]
- Farooq M., Wahid A., Lee D. J., Ito O., Siddique K. H. M. (2009). Advances in drought resistance of rice. Crit. Rev. Plant Sci. 28 199–217. 10.1080/07352680902952173 [DOI] [Google Scholar]
- Fitzgerald H. A., Chern M.-S., Navarre R., Ronald P. C. (2004). Overexpression of (At)NPR1 in rice leads to a BTH- and environment-induced lesion-mimic/cell death phenotype. Mol. Plant Microbe Interact. 17 140–151. 10.1094/MPMI.2004.17.2.140 [DOI] [PubMed] [Google Scholar]
- Gao T., Wu Y., Zhang Y., Liu L., Ning Y., Wang D., et al. (2011). OsSDIR1 overexpression greatly improves drought tolerance in transgenic rice. Plant Mol. Biol. 76 145–156. 10.1007/s11103-011-9775-z [DOI] [PubMed] [Google Scholar]
- Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., Storz G., et al. (2000). Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11 4241–4257. 10.1091/mbc.11.12.4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath R. L., Packer L. (1968). Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 125 189–198. 10.1016/0003-9861(68)90654-1 [DOI] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komarl T., Kumashiro T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of boundaries of the T-DNA. Plant J. 6 271–282. 10.1046/j.1365-313X.1994.6020271.x [DOI] [PubMed] [Google Scholar]
- Hou X., Xie K., Yao J., Qi Z., Xiong L. (2009). A homolog of human skiinteracting protein in rice positively regulates cell viability and stress tolerance. Proc. Natl. Acad. Sci. U.S.A. 106 6410–6415. 10.1073/pnas.0901940106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T. H., Lee J. T., Charng Y. Y., Chan M. T. (2002). Tomato plants ectopically expressing Arabidopsis CBF1 show enhanced resistance to water deficit stress. Plant Physiol. 130 618–626. 10.1104/pp.006783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. Y., Chao D. Y., Gao J. P., Zhu M. Z., Shi M., Lin H. X. (2009). A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev. 23 1805–1817. 10.1101/gad.1812409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi Y., Yoshida Y., Sanada Y., Yamaguchi-Shinozaki K., Wada K., Shinozaki K. (1997). Characterization of the gene for D1-pyrroline-5-carboxylate synthetase and correlation between the expression of the gene and salt tolerance in Oryza sativa L. Plant Mol. Biol. 33 857–865. 10.1023/A:1005702408601 [DOI] [PubMed] [Google Scholar]
- Jabs T., Dietrich R. A., Dangl J. L. (1996). Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science 273 1853–1856. 10.1126/science.273.5283.1853 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Liang G., Yang S., Yu D. (2014). Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid- and auxin-mediated signals in jasmonic acid-induced leaf senescence. Plant Cell 26 230–245. 10.1105/tpc.113.117838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y. J., Liang G., Yu D. Q. (2012). Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Mol. Plant 5 1375–1388. 10.1093/mp/sss080 [DOI] [PubMed] [Google Scholar]
- Kasuga M., Liu Q., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17 287–291. 10.1038/7036 [DOI] [PubMed] [Google Scholar]
- Kreps J. A., Wu Y., Chang H. S., Zhu T., Wang X., Harper J. F. (2002). Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 130 2129–2141. 10.1104/pp.008532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Jin S., Liu X., Zhu L., Nie Y., Zhang X. (2014). Overexpression of rice NAC gene SNAC1 improves drought and salt tolerance by enhancing root development and reducing transpiration rate in transgenic cotton. PLoS ONE 28:e86895 10.1371/journal.pone.0086895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., et al. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10 1391–1406. 10.2307/3870648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehly A., Chance B. (1954). The assay of catalases and peroxidases. Methods Biochem. Anal. 1 357–424. 10.1002/9780470110171.ch14 [DOI] [PubMed] [Google Scholar]
- Miller G., Shulaev V., Mittler R. (2008). Reactive oxygen signaling and abiotic stress. Physiol. Plant. 133 481–489. 10.1111/j.1399-3054.2008.01090.x [DOI] [PubMed] [Google Scholar]
- Mittler R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7 405–410. 10.1016/S1360-1385(02)02312-9 [DOI] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Gollery M., Breusegem F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9 490–498. 10.1016/j.tplants.2004.08.009 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Tran L. S., Nguyen D., Fujita M., Maruyama K., Todaka D., et al. (2007). Functional analysis of a NAC-type transcription factor OsNAC 6 involved in abiotic and biotic stress responsive gene expression in rice. Plant J. 51 617–630. 10.1111/j.1365-313X.2007.03168.x [DOI] [PubMed] [Google Scholar]
- Nelson D. E., Repetti P. P., Adams T. R., Creelman R. A., Wu J., Warner D. C., et al. (2007). Plant nuclear factor Y(NF–Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc. Natl. Acad. Sci. U.S.A. 104 16450–16455. 10.1073/pnas.0707193104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S. Q., Liu Y. F., Liu P., Lei G., He S. J., Ma B., et al. (2010). Receptor-like kinase OsSIK1 improves drought and salt stress tolerance in rice (Oryza sativa) plants. Plant J. 62 316–329. 10.1111/j.1365-313X.2010.04146.x [DOI] [PubMed] [Google Scholar]
- Park S., Li J. S., Pittman J. K., Berkowitz G. A., Yang H. B., Undurraga S., et al. (2005). Up-regulation of a H+-pyrophosphatase (H+-PPase) as a strategy to engineer drought-resistant crop plants. Proc. Natl. Acad. Sci. U.S.A. 102 18830–18835. 10.1073/pnas.0509512102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyanka B., Sekhar K., Reddy V. D., Rao K. V. (2010). Expression of pigeonpea hybrid-proline-rich protein encoding gene (CcHyPRP) in yeast and Arabidopsis affords multiple abiotic stress tolerance. Plant Biotechnol. J. 8 76–87. 10.1111/j.1467-7652.2009.00467.x [DOI] [PubMed] [Google Scholar]
- Qiu Y. P., Yu D. Q. (2008). Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis. Environ. Exp. Bot. 65 35–47. 10.1016/j.envexpbot.2008.07.002 [DOI] [Google Scholar]
- Raychaudhuri S. S., Deng X. W. (2000). The role of superoxide dismutase in combating oxidative stress in higher plants. Bot. Rev. 66 89–98. 10.1007/BF02857783 [DOI] [Google Scholar]
- Ren X. Z., Chen Z. Z., Liu Y., Zhang H. R., Zhang M., Liu Q., et al. (2010). ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 63 417–429. 10.1111/j.1365-313X.2010.04248.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockstrom J., Falkenmark M. (2000). Semiarid crop production from a hydrological perspective: gap between potential and actual yields. Crit. Rev. Plant Sci. 19 319–346. 10.1080/07352680091139259 [DOI] [Google Scholar]
- Sarkar T., Thankappan R., Kumar A., Mishra G., Dobaria J. R. (2014). Heterologous expression of the atdreb1a gene in transgenic peanut-conferred tolerance to drought and salinity stresses. PLoS ONE 9:e110507 10.1371/journal.pone.0110507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scandalios J. G. (2002). The rise of ROS. Trends Biochem. Sci. 27 483–486. 10.1016/S0968-0004(02)02170-9 [DOI] [PubMed] [Google Scholar]
- Seki M., Narusaka M., Ishida J., Nanjo T., Oono Y., Kamiya A., et al. (2002). Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 31 279–292. 10.1046/j.1365-313X.2002.01359.x [DOI] [PubMed] [Google Scholar]
- Takasaki H., Maruyama K., Kidokoro S., Ito Y., Fujita Y., Shinozaki K., et al. (2010). The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol. Genet. Genomics 284 173–183. 10.1007/s00438-010-0557-0 [DOI] [PubMed] [Google Scholar]
- Tao Z., Kou Y. J., Liu H. B., Li X. H., Xiao J. H., Wang S. P. (2011). OsWRKY45 alleles play different roles in abscisic acid signalling and salt stress tolerance but similar roles in drought and cold tolerance in rice. J. Exp. Bot. 62 4863–4874. 10.1093/jxb/err144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T., Fujita M., Fujita Y., Yamaguchi-Shinozaki K., Shinozaki K. (2006). Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Curr. Opin. Biotechnol. 17 113–122. 10.1016/j.copbio.2006.02.002 [DOI] [PubMed] [Google Scholar]
- Verslues P. E., Agarwal M., Katiyar-Agarwal S., Zhu J. H., Zhu J. K. (2006). Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 45 523–539. 10.1111/j.1365-313X.2005.02593.x [DOI] [PubMed] [Google Scholar]
- Wu K. L., Guo Z. J., Wang H. H., Li J. (2005). The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 12 9–26. 10.1093/dnares/12.1.9 [DOI] [PubMed] [Google Scholar]
- Xiang Y., Huang Y., Xiong L. (2007). Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol. 144 1416–1428. 10.1104/pp.107.101295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L., Zhu J. K. (2002). Molecular and genetic aspects of plant responses to osmotic stress. Plant Cell Environ. 25 131–139. 10.1046/j.1365-3040.2002.00782.x [DOI] [PubMed] [Google Scholar]
- Yang A., Dai X. Y., Zhang W. H. (2012). A R2R3-type MYB gene. OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 63 2541–2556. 10.1093/jxb/err431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. N., Qi M., Mei C. S. (2004). Endogenous salicylic acid protects rice plants from oxidative damage caused by aging as well as biotic and abiotic stress. Plant J. 40 909–919. 10.1111/j.1365-313X.2004.02267.x [DOI] [PubMed] [Google Scholar]
- Yu H., Chen X., Hong Y. Y., Wang Y., Xu P., Ke S. D., et al. (2008). Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell 20 1134–1151. 10.1105/tpc.108.058263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L. H., Chen X., Wang Z., Wang S. M., Wang Y. P., Zhu Q. S., et al. (2013). Arabidopsis enhanced drought tolerance1/HOMEODOMAIN GLABROUS11 confers drought tolerance in transgenic rice without yield penalty. Plant Physiol. 162 1378–1391. 10.1104/pp.113.217596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Xiao S. S., Li W., Feng W., Li J., Wu Z. D., et al. (2011). Overexpression of a Harpin-encoding gene hrf1 in rice enhances drought tolerance. J. Exp. Bot. 62 4229–4238. 10.1093/jxb/err131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. K. (2001). Plant salt tolerance. Trends Plant Sci. 6 66–72. 10.1016/S1360-1385(00)01838-0 [DOI] [PubMed] [Google Scholar]
- Zhu J. K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53 247–273. 10.1146/annurev.arplant.53.091401.143329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Liu C. F., Liu A. L., Zou D., Chen X. B. (2012). Overexpression of OsHsp17.0 and OsHsp23.7 enhances drought and salt tolerance in rice. J. Plant Physiol. 169 628–635. 10.1016/j.jplph.2011.12.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


