Abstract
Steroid hormones regulate physiological processes in species ranging from plants to humans. A wide range of steroid hormones exist, and their contributions to processes, such as growth, reproduction, development, and aging, is almost always complex. Understanding the biosynthetic pathways that generate steroid hormones and the signaling pathways that mediate their effects is thus of fundamental importance. In this work, we review recent advances in (i) the biological role of steroid hormones in the roundworm Caenorhabditis elegans and (ii) the development of novel methods to facilitate the detection and identification of these molecules. Our current understanding of steroid signaling in this simple organism serves to illustrate the challenges we face moving forward. First, it seems clear that we have not yet identified all of the enzymes responsible for steroid biosynthesis and/or degradation. Second, perturbation of steroid signaling affects a wide range of phenotypes, and subtly different steroid molecules can have distinct effects. Finally, steroid hormone levels are critically important, and minute variations in quantity can profoundly impact a phenotype. Thus, it is imperative that we develop innovative analytical tools and combine them with cutting-edge approaches including comprehensive and highly selective liquid chromatography coupled to mass spectrometry based on new methods such as supercritical fluid chromatography coupled to mass spectrometry (SFC-MS) if we are to obtain a better understanding of the biological functions of steroid signaling.
Keywords: aging, steroid signaling pathway, Caenorhabditis elegans, analytical chemistry, steroid hormones
Introduction
Since its first detailed description by Sydney Brenner in 1974, the nematode Caenorhabditis elegans has become a key model organism for biological research (1). Many early studies on fundamental processes, such as apoptosis, development, and aging, were performed using C. elegans. The many practical advantages offered by C. elegans include fixed and known cell lineage, short development time, and translucent body [reviewed in detail elsewhere (2)]. A normal C. elegans life cycle consists of hatching from eggs, four larval stages, L1–L4, and a fertile adult stage, at which point the individual worm is composed of exactly 959 somatic cells and a germline. The hermaphroditic nature of the worm makes it easy to maintain large populations of isogenic animals. The relative simplicity of C. elegans and its straightforward life cycle has therefore driven scientists to use this animal model to study various biological processes and uncover important signaling and metabolic pathways.
Caenorhabditis elegans possesses a functional steroid signaling pathway that in many ways resembles that found in mammals (3). Although the pathway’s role in development is clear, its impact on longevity is still not well understood. Information gathered to date suggests that the regulation of steroid signaling is complex, and we propose that completely deciphering its function and regulation will require a combination of genetic manipulation of the enzymatic pathways coupled with the development of sophisticated analytical chemistry techniques capable of capturing the molecules. Indeed, many of the small steroid precursor molecules and the genes encoding the key steroidogenic enzymes remain to be formally identified.
Here, we review current knowledge of the components, regulation, and actions of this signaling pathway, as well as the analytical chemical methods that have been employed to date. Finally, we will introduce some promising new tools for the study of steroid signaling, including the use of steroidomics as an untargeted approach to steroid analysis.
Biological Actions of C. elegans Steroids
Functions during Development
The role of steroid signaling during C. elegans development is clearly established. During the larval stages, the worm can sense the environment and make developmental decisions accordingly. When conditions are challenging (food scarcity, high temperature, or crowded environment), young larvae enter into a stress-resistant, non-reproductive Dauer stage (Dauer, German for enduring). Dauer larvae can live for months and retain the capacity to resume development into reproductive adulthood when conditions improve. The steroid signaling pathway is instrumental in this decision-making process. Under favorable conditions, the insulin and TGF-β signaling pathways are activated, resulting in production of the two steroid hormone Δ4- and Δ7-dafachronic acid [often referred to as dafachronic acid (DA)] by the cytochrome P450 DAF-9 (4). DA then binds to the nuclear hormone receptor (NHR) DAF-12 (5) that is no longer repressed by the corepressor DIN-1S/SHARP under these conditions (6). Activation of DAF-12-dependent transcription prevents entry into the Dauer stage and promotes development to reproductive adulthood. When conditions are unfavorable, however, DA is not produced and DAF-12 remains bound to DIN-1S/SHARP, thereby repressing transcription and leading to larval arrest at the Dauer stage. As a result, animals carrying strong daf-9 mutant alleles are Dauer constitutive while daf-12 null mutants are Dauer defective. Interestingly, daf-12 mutants carrying mutations in their ligand-binding domain (LBD) are Dauer constitutive (class 6), in agreement with an instructive role for DAF-12 in the Dauer decision (7). It is important to note that several pathways appear to independently affect the decision to enter into Dauer. For example, daf-2(e1370) mutants, which harbor a defective insulin receptor, are Dauer constitutive. However, overexpression of daf-9 in this strain only partially suppresses their tendency to enter the Dauer stage, and addition of DA induces these animals to arrest at the L3 stage rather than to continue development into gravid adults (5).
Steroid signaling also governs developmental timing. In response to environmental conditions, production of DA and activation of DAF-12-dependent transcription increase the expression of the let-7 microRNA family (8). These microRNAs are components of the heterochronic pathway, which controls the timing of developmental events in C. elegans (9, 10). Mutants defective in daf-12 or carrying mutations in the microRNAs mir-48, mir-241, and mir-84 exhibit disorganized developmental progression, with specific cell divisions being reiterated or skipped (8). The steroid signaling pathway is therefore capable of responding to the environment to control developmental timing through a number of mechanisms. Clearly, this pathway is a central regulator of the C. elegans developmental machinery. Since different levels of the steroid hormones may trigger qualitatively distinct signals, analytical methods capable of measuring DA levels with great precision will be necessary to unravel the intricate relationship between steroid production and physiological outcome.
Functions in Adult Lifespan
Steroid signaling affects the lifespan of adult C. elegans through various mechanisms reviewed here below.
Temperature-Dependent Modulation of Lifespan
Early experiments showed that C. elegans strains incapable of producing DA [i.e., strong daf-9 mutants (also called class 2 alleles)] were long-lived at low (15°C) but not at warmer (20 or 25°C) temperatures (11–13). This phenotype is not observed in animals carrying weaker daf-9 alleles, such as rh50 (class 1 allele), which generally shorten lifespan at 25°C but have no effect at 15 or 20°C. Lifespan extension observed in strong daf-9 mutants at 15°C is completely dependent on expression of functional DAF-12 (13). Moreover, complementation of daf-9 mutants with the DAF-12 ligand restores the wild-type lifespan at 15°C, confirming that the long-lived phenotype required DA signaling via DAF-12 (12). Interestingly, either addition of DA to mutants solely during development or expression of daf-9 in the hypodermis or in neuron-like cells (called XXX cells) during development is sufficient for the wild-type lifespan at 15°C to be restored (12). These results suggest that steroid-dependent regulation of lifespan begins during development.
The extended lifespan of daf-9 null mutants at 15°C is associated with increased stress resistance. Animals carrying either of the e1406 or dh6 daf-9 null alleles are resistant to both thermal (35°C) and oxidative (hydrogen peroxide) stress (12), and this resistance is also reversed by addition of DA or introduction of a daf-12 mutation. Interestingly, animals with low insulin signaling (i.e., mutation of the insulin receptor gene daf-2) are also stress resistant; however, in this case, addition of DA is without effect, indicating that the daf-9 and daf-2 mutations confer resistance through distinct mechanisms (12). This is consistent with the observation that overexpression of daf-9 does not affect the long lifespan of daf-2 mutants (14). However, daf-9-dependent stress resistance is partly dependent on DAF-16, the FOXO transcription factor downstream of the insulin signaling pathway. These results show that although longevity and development are both affected by steroid signaling, they involve different regulatory mechanisms.
Several studies have shed light on the involvement of the steroid signaling pathway in temperature-sensitive lifespan regulation. Lee and Kenyon showed that thermosensory defects (ablation of the AFD thermosensory neurons or mutation of ttx-1 and ttx-3, the transcription factors involved in thermosensation) decrease the C. elegans lifespan at high temperature (25°C) (15). However, this is not observed in hypomorphic daf-9(rh50) mutants or daf-12 null mutants (15). This finding is in accordance with the observation that steroid signaling is required for lifespan regulation by thermosensory neurons at high temperature. Consistent with this, overexpression of daf-9 in ttx-1 mutants restores the wild-type lifespan (15). Finally, recent work has described a role for daf-41, the C. elegans homolog of p23 cochaperone/prostaglandin E synthase-3, in temperature-dependent modulation of lifespan. The lifespan of these mutants is virtually temperature insensitive since, compared with wild-type animals, their lifespan is longer at 25°C, shorter at 15°C, and no different at 20°C. Interestingly, a daf-12 null mutation reverses the short lifespan of daf-41 mutants at 15°C but has no effect at 20 or 25°C (16). The role of steroid signaling in temperature-dependent regulation of lifespan is obviously complex, and much work remains to fully elucidate its involvement.
Germline-Mediated Longevity
Steroid signaling controls longevity in germline-less C. elegans. In 1999, Hsin and Kenyon showed that laser ablation of the germline precursor cells at the first larval stage extends the lifespan by ~60%, and this extension depends on the somatic gonad, the FOXO transcription factor DAF-16, and the NHR DAF-12 (17). In addition to DAF-12, several other components of the DA biosynthetic pathway are required for lifespan extension though ablation of the germline, including the Rieske-like protein, DAF-36 (18), the cytochrome P450, DAF-9 (12), and the 3-hydroxysteroid dehydrogenase, DHS-16 (18). Although the cytochrome P450 oxidoreductase EMB-8 and the alternative 3-hydroxysteroid dehydrogenase HSD-1 have been identified as enzymes in the steroid signaling pathway, the requirement for germline-dependent lifespan extension of EMB-8 has not yet been tested (19), and HSD-1 is not required (20). These differences of how enzymes involved in the steroid biosynthetic pathway impact the lifespan of germline-less animals may point to the involvement of other steroid hormones, such as the precursors or derivatives of DA. Analytical techniques capable of identifying and quantifying very low levels of steroid hormones will be indispensable for such studies.
Interestingly, animals that lack the entire gonad (germline and somatic gonad) are not long-lived unless they are provided with exogenous DA and possess functional DAF-12 and DAF-16 proteins (21). One possible interpretation for these data is that DA is produced in response to germline ablation, possibly in the somatic gonad itself or in tissues controlled by the somatic gonad. However, direct measurements of DA after germline ablation have produced conflicting results depending on the analytical method used. Compared with wild-type animals, DA levels were either increased (22) or unchanged (23) in glp-1(e2141ts) mutants, which are unable to produce a germline. Because glp-1 mutants are morphologically different from wild-type animals but similar to gonad-less animals, a better approach to resolve this question might be to directly compare DA levels after ablation of the somatic gonad versus the germline. The development of analytical techniques capable of detecting steroid hormones in a tissue-specific manner would be a great advance in the field. In any event, if the lack of DA production in response to germline ablation is confirmed, this might suggest that DAF-12 responsiveness to its ligand is heightened under these conditions (23). How that might occur remains to be determined.
Of note, steroid signaling regulates germline-mediated longevity through molecular strategies similar to those employed during development. For example, daf-12-dependent production of the microRNAs mir-81 and mir-241 is required for both the heterochronic pathway during development and lifespan extension in response to germline ablation (8, 22). Recently, it was shown that DAF-12 activation in response to germline ablation induces expression of mir-81 and mir-241, which activate DAF-16 by inhibiting the expression of AKT-1 (22). However, the transcriptional activities of the steroid and insulin signaling pathways appeared to be largely independent (24).
Lifespan Extension through Dietary Restriction
A recent study used two independent models to show that DA is produced in response to dietary restriction; these were adult wild-type worms subjected to complete bacterial deprivation, and eat-2 mutants, which have reduced food intake due to a pharyngeal pumping defect (25). Interestingly, two isoforms of DA (Δ4 and Δ7) were augmented by dietary restriction, an observation made possible by the use of carefully refined analytical techniques (26). Although daf-9 mRNA levels were elevated under these conditions, how DA synthesis is activated by dietary restriction is not clear. Several potential DA biosynthetic pathways exist (27), suggesting that the precise route to DA production can only be resolved if all cholesterol-derived metabolites are measured under the conditions in which DA is produced. Elevated daf-9 mRNA levels are also observed in daf-16 and daf-7 mutants subjected to dietary restriction, suggesting that, in contrast to its regulation during development, steroid signaling in adult C. elegans is not controlled by the insulin and TGF-β signaling pathways. Interestingly, activation of the steroid signaling pathway is required for dietary restriction-induced lifespan extension, since weak daf-9 mutants subjected to dietary restriction do not live longer. Surprisingly, DAF-12 does not appear to be involved in this response; instead, the closely related NHR-8 has been implicated (25). NHR-8 is a master regulator of cholesterol homeostasis in C. elegans, but its role in longevity determination is not well understood (28).
In wild-type C. elegans, nutrient limitation leads to a large reduction in the size of the germline, but this is not observed in steroid signaling pathway mutants. When germline size reduction is re-established in steroid mutants by mutations in the Notch or TGF-β signaling pathways, which control germline stem cell proliferation by different mechanisms, lifespan extension is also restored (25). Taken together, these results suggest that steroid signaling is capable of affecting the germline, depending on the nutritional status of the animal, and that germline dynamics play a role in dietary restriction-mediated lifespan extension. It is interesting to note that although steroid signaling regulates reproduction in response to nutritional cues in both adult and developing C. elegans, the mechanisms involved are profoundly different.
Male-Induced Loss of Fitness and Shortening of the Lifespan
Caenorhabditis elegans exists predominantly as self-fertilizing hermaphrodites, and males are relatively rare. Under conditions of stress, however, the frequency of males increases markedly. The Murphy and Brunet labs independently reported that mating of hermaphroditic worms with males induces a drastic loss of fitness in the hermaphrodites, characterized by body shrinkage and precocious death (29, 30). Interestingly, Shi and Murphy showed that shrinking and early death are the direct result of mating. DAF-9 and DAF-12 are required for body shrinking and, to a lesser extent, for the rapid decay leading to death, suggesting that shrinking and short lifespan can be uncoupled in this context. They also found that daf-9 levels in the spermatheca were significantly decreased in response to mating.
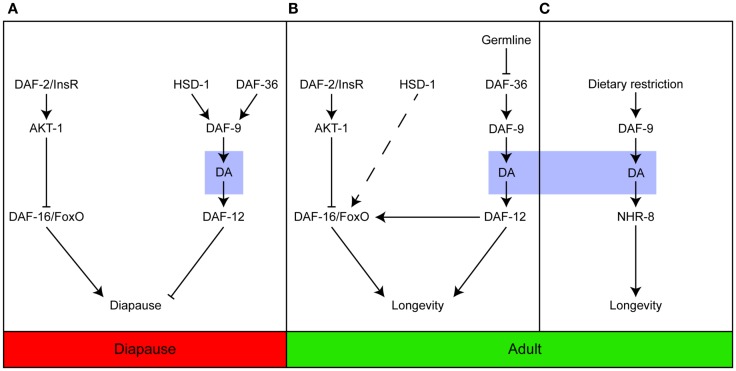
Collectively, the studies described here indicate that the steroid signaling pathway plays an important role in controlling the response of developing and adult C. elegans to their environments. The pathway regulates the timing of key events that allow the animals to develop and grow and age at a pace in keeping with the challenges imposed by the prevailing conditions, most notably temperature, nutritional availability, and reproductive capacity (Figure 1).
Figure 1.
Dafachronic acid plays important roles in developing and adult worms. Steroid signaling is important for entry into the Dauer diapause (A); longevity in response to germline ablation (B); and the response to dietary restriction (C). In contrast to (A,B), which are dependent on the activation of DAF-12, dietary restriction induces longevity via NHR-8. Adapted from Dumas et al. (31).
Current Knowledge of Dafachronic Acid Biosynthesis
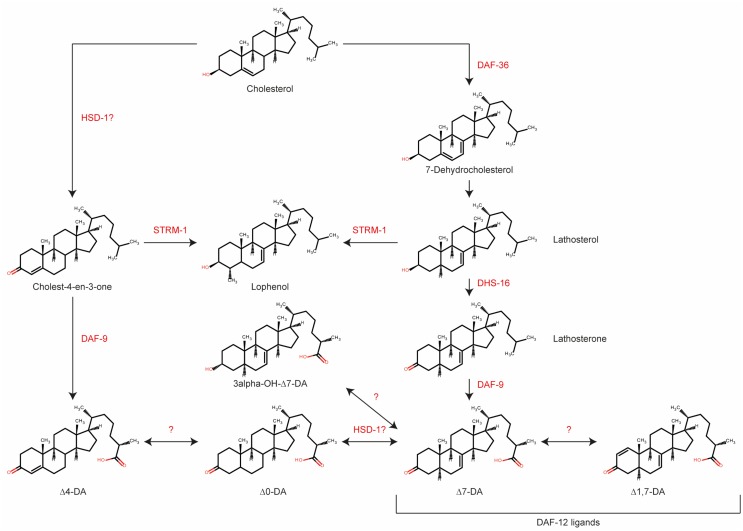
Although many enzymes taking part in DA production have been identified, we do not yet know the precise reaction sequence starting with cholesterol. It is important to note that DA actually refers to two molecules, Δ4- and Δ7-DA, which could potentially have different biosynthetic pathways. Motola et al. first identified Δ4- and Δ7-DA as products of the DAF-9-catalyzed reaction and showed that both hormones are direct ligands of the NHR DAF-12 (5). Additional DA biosynthetic enzymes were subsequently identified, as mentioned above, including the Rieske-like oxygenase DAF-36 (18), a C. elegans ortholog of 3β-hydroxysteroid dehydrogenase/Δ5-Δ4 isomerase, HSD-1 (19), a 3-hydroxysteroid dehydrogenase, DHS-16, and a NADPH–cytochrome P450 oxidoreductase, EMB-8 (27).
Although the enzyme STRM-1 does not take part in DA biosynthesis directly, it may play important roles in regulating their levels (32). This enzyme adds a methyl group to position 4 in the A ring of DA precursors. As a result, the methylated molecules can no longer serve as substrates of DAF-9, and DAs therefore cannot be produced. Hannich et al. proposed that STRM-1 could regulate Dauer formation by controlling the sterol pool for DA synthesis (32). Notably, although the derived 4-methyl sterols were no longer available for DA biosynthesis, they were sufficient for the nematode molting process (32).
The most recent and comprehensive analysis of enzymes and intermediates in DA biosynthesis was reported in 2014 by Mahanti et al., who used a metabolomics approach (33). This study described several novel forms of DA, including 3-hydroxy derivatives of Δ7-DA, and Δ1,7-DA and Δ0-DA, which have two or no double bonds, respectively. The new molecules were identified after extensive activity-guided fractionation, based on an in vitro luciferase assay in HEK293T cells and an in vivo Dauer rescue assay, followed by comparison of the metabolomes of daf-22 and daf-9 daf-12 animals using nuclear magnetic resonance (NMR) spectroscopy. In this study, daf-22 mutants were employed as ligand-rich positive controls since the DAF-22 homolog, SCPx, is involved in steroid side chain breakdown, and daf-9 daf-12 animals were the ligand-deficient negative controls (32). Interestingly, Δ4-DA could not be detected in any samples, suggesting that this isoform may be present at very low concentrations compared with Δ7-DA. In vitro and in vivo testing of synthetic molecules showed that Δ7- and Δ1,7-DA were very potent DAF-12 ligands, and both were able to rescue daf-9 phenotypes and restore longevity in glp-1;daf-9 double mutants. Lastly, the authors examined the effects of mutation of putative biosynthetic enzymes. The results suggested that HSD-1 is involved in the synthesis of Δ7-DA, in contrast to an earlier finding (18). strm-1 mutants contained higher level of DAs than did wild-type animals, which is also in line with previous results (33).
Recent results suggest that DA signaling might be more complex than previously anticipated (19, 27, 33). Based on current knowledge, we propose the putative biosynthetic pathway shown in Figure 2. Major gaps in our knowledge still exist; for example, the enzyme converting 7-dehydrocholesterol to lathosterol is not known, although it should theoretically have Δ5 reductase activity. Interestingly, HSD-1 has two paralogs, HSD-2 and -3, but their involvement in DA synthesis has not been rigorously examined to date. Better integration of analytical chemistry and metabolomics tools will facilitate the identification of new enzymes, intermediates, and products in the DA biosynthetic pathway.
Figure 2.
Biosynthetic pathway for various dafachronic acids based on current knowledge. Several additional steroids and enzymes may exist. Adapted from Mahanti et al. (33).
Every description of a new gene in the DA biosynthetic pathway suggests the existence of new intermediate products. Table 1 summarizes the DAs and steroid molecules related to DA biosynthesis that, based on current knowledge, could be present in C. elegans. The molecules can be roughly divided into three classes: DAs, DA precursors, and other sterols. From a chemical viewpoint, the molecules within a class are very similar, differing only in the position of a double bond or the functional group at position 3, and they have only very small differences in the octanol–water partition coefficient (logP). Furthermore, some molecules in each class have the same exact mass (or both molecules, in the case of “other sterols”). However, even small differences may have significant impacts on the biological effects and potencies of the molecules. Sharma et al. synthesized a number of DA variants with different double bond positions (none, Δ4, Δ5, Δ7) and different stereochemistries at C25 (R or S), and tested their effects in two assays: transactivation of the DAF-12 LBD in HEK293 cells and rescue of the daf-9 Dauer phenotype (34). They demonstrated that Δ7-DA is the most potent activator and that the double bond position is more important than the stereochemistry at C25 (34). Molecular simulations showed that interaction of the DAF-12 LBD with the 25R isomer is energetically more favorable (35). New molecules are expected to exhibit properties based on the similarity of their chemical structures. Therefore, novel and highly selective and/or efficient separation methods that will facilitate a comprehensive analysis of the C. elegans steroidome content are needed.
Table 1.
Molecular properties of dafachronic acids and known precursor substances.
| Name | Exact mass | Formula | LogP | ChEBI ID | Class |
|---|---|---|---|---|---|
| (25S)-Δ1,7-DA | 412.29774 | C27H40O3 | 6.25 | CHEBI:83137 | DA |
| (25S)-Δ7-DA | 414.31339 | C27H42O3 | 6.26 | CHEBI:78699 | |
| 3α-OH-(25S)-Δ7-DA | 416.32904 | C27H44O3 | 6.05 | ||
| (25S)-Δ4-DA | 414.31339 | C27H42O3 | 6.62 | CHEBI:71560 | |
| (25S)-Δ0-DA | 416.32904 | C27H44O3 | 6.66 | CHEBI:78701 | |
| 3β-OH-(25S)-Δ7-DA | 416.32904 | C27H44O3 | 6.05 | ||
| Cholesterol | 386.35486 | C27H46O | 7.11 | CHEBI:16113 | DA precursor |
| 7-Dehydrocholesterol | 384.33921 | C27H44O | 6.71 | CHEBI:17759 | |
| Lathosterol | 386.354866 | C27H46O | 7.11 | CHEBI:17168 | |
| Lathosterone | 384.33921 | C27H44O | 7.32 | CHEBI:71550 | |
| 4-Cholesten-3-one | 384.33921 | C27H44O | 7.68 | CHEBI:16175 | |
| 4α-Methyl-5α-cholest-7-en-3β-ol (Lophenol) | 400.37051 | C28H48O | 7.48 | CHEBI:18378 | Others |
| 4α-Methyl-5α-cholest-8(14)-en-3β-ol | 400.37051 | C28H48O | 7.43 | CHEBI:71581 |
Exact mass is important for mass spectrometric detection.
LogP = water/octanol partition coefficient.
New Approaches for the Analysis of Known Steroids and Identification of Unknown Steroids in C. elegans
Analytical Approaches for the Analysis of C. elegans Steroids
To date, only targeted methods, based mostly on gas chromatography (GC) or liquid chromatography (LC) coupled to mass spectrometry (GC-MS and LC-MS, respectively), have been employed for analysis of C. elegans steroids. Direct analysis of substrate and product levels is crucial for assessing enzymatic activities. The analytical techniques for steroids range from simple fractionation using solid phase extraction materials or 2D-thin layer chromatography to highly specific technologies, such as LC-MS/MS or 2D-NMR. Furthermore, chemical synthesis is essential for confirming the identity of newly isolated compounds. GC is the method of choice for molecules that are volatile or can be made volatile by appropriate derivatization, whereas LC is preferable for more polar steroids such as bile acids. Chromatography plays a key role in the analysis of C. elegans steroids because several molecules have the same exact mass (Table 1), making separation critical. Because many compounds have similar logP values, the separation methods must also be highly selective.
Methods for the Quantification of C. elegans Steroids
Determining the quantity of different steroids in vivo is of paramount importance for understanding regulatory circuits, but the identity of the steroids must be known in order to quantify them. Both GC-MS/MS and LC-MS/MS have been used to analyze steroids in C. elegans to substantiate results obtained with genetic tools. Here, we discuss some of these methods.
The method used by Broué et al. for steroid analysis is based on extensive fractionation using solid phase extraction and HPLC, followed by derivatization with heptafluorobutyric acid anhydride and GC-MS/MS analysis. A large quantity of worms (5 × 103 to 3 × 105) are required for this procedure but it has good sensitivity (lower fg) (36). However, the authors did not measure either DAs or their precursors.
Motola et al. first identified the DAs using a single quadrupole LC-MS system for chemical analysis. Separation was achieved by a C18 column with a methanol (MeOH) to MeOH/acetonitrile (ACN)/water gradient and selected ion monitoring (SIM) analysis of different masses corresponding to 4-cholesten-3-one, 1,4 cholesten-3-one, and 3-keto-4-cholestenoic acid in positive mode and 3-keto-4 cholestenoic acid in negative mode (5).
Wollam et al. used a combination of LC-MS/MS and GC-MS/MS to analyze their C. elegans samples. For LC-MS/MS, a triple quadrupole instrument was employed to follow levels of lathosterone, lathosterol, 7-dehydrocholesterol, and 4-cholesten-3-one. Separation was achieved by a C18 column with a 90–100% MeOH gradient. DA levels were also analyzed using GC-MS/MS (27). Overall, GC-MS appears to be the preferred method for analysis of C. elegans steroids since it was also employed by Shen et al. (22) and Mahanti et al. (33). For this technique, samples must be derivatized to increase their volatility. Shen et al. used multiple reaction monitoring on a GC-MS/MS system for selective detection. Multiple reaction monitoring is based on the selection of a mass of interest, followed by fragmentation of the molecule in a collision cell, and detection of specific fragments to be obtained and quantified. This is usually performed with a triple quadrupole or quadrupole time of flight (Q-ToF) instrument.
Using SIM, Mahanti et al. were able to follow production of Δ7-DA, Δ1,7-DA, Δ0-DA, Δ4-DA, 3α- and 3β-OH-Δ7DA, and 4-cholesten-3-one (33). Quantification of the different DAs was performed using response factors determined from synthetic standards, and the MS response was linear between 10 pg and 5 ng. Interestingly, these authors were unable to detect Δ4-DA in their samples, although 4-cholesten 3-one was detected in amounts similar to the Δ7-DA precursor lathosterone (33).
Liquid chromatography coupled to mass spectrometry is an interesting alternative to GC-MS because it does not require prior derivatization. In particular, bile acids such as DAs are amenable to electrospray ionization (ESI) due to the carboxylic acid functionality. ESI is used to ionize molecules in the LC effluent. Li et al. described an even more sensitive method for quantification of certain DAs, in which samples were derivatized with 2-picolylamine to further decrease the detection limit (37). The authors evaluated the method with respect to recovery after extraction with chloroform/MeOH and various quantification methods for their precision, accuracy, and limit of detection (LOD). The best on-column LOD (1 pg) was obtained with high-resolution SIM. As proof-of-principle, Li et al. applied their method to detect DAs at different developmental stages of wild-type, daf-9, daf-12, and daf-36 animals. DAs were undetectable in the double mutant compared with wild-type animals, and levels in the daf-36 mutants were intermediate between the two. DA levels were also measured in two different daf-2 alleles, daf-7, and daf-11 mutants (37). Although the method is very sensitive, its major drawback is that it cannot distinguish between the Δ4- and Δ7-DA isomers.
Sardella et al. used a click quinidine-based chiral stationary phase to separate C25 epimers of DAs (38). Retention was based on anion exchange, whereas separation of stereoisomers was based on stereoselective H-bonding. Detection was carried out at 220 nm. Although the method was qualitative and only tested with chemical standards, the eluent they used is MS compatible and the method could potentially be used for quantification in biological samples. However, this remains to be determined (38).
We recently published a method for the separation of Δ4- and Δ7-DAs using a novel C18 sub-2 μm core–shell particle column in combination with an acetonitrile (ACN) gradient, which allowed independent quantification of the two isomers. We tested a number of gradients for separation of the two isomers and found that a focused gradient with a 65% isocratic step was the best solution. The method was evaluated for extraction efficiency and LOD (26). In contrast to Li et al., we used 100% MeOH for extraction but obtained similar extraction recoveries. Although our LOD was slightly higher than that of Li et al. at ~5 pg per isomer, the major advantage of our method is separation of the isomers. Moreover, we successfully applied this method to quantify changes in both DA isomers in animals subjected to bacterial deprivation (25, 26).
GC-MS and LC-MS each have advantages and disadvantages. Although GC-MS enables high-resolution separation with chromatographic peak widths of several seconds, its drawbacks include derivatization is necessary to increase volatility, separations are carried out at temperatures exceeding 200°C, and columns show only restricted selectivities. In addition, to achieve good separation long runtimes >45 min are needed. GC-MS has been used to analyze steroids for decades and is a well-established method readily available to many labs. By contrast, LC-MS offers a wide variety of column materials with various selectivities for the separation of different, structurally closely related, isomers. No derivatization is needed, but it can be used to increase the sensitivity for weakly ionizing molecules. However, despite increasingly widespread use, LC-MS is not accessible to all labs and dedicated methods must be developed for demanding separations, as is the case for Δ4 and Δ7-DA. Compared to GC-MS, LC-MS run times are generally shorter with around 30–40 min.
New Analytical Approaches for the Discovery and Analysis of C. elegans Steroids
Numerous methods have been described for targeted analysis of known C. elegans steroids. However, there clearly exists a need not only for improved, faster, and more sensitive methods but also for methods to identify new and potentially bioactive intermediates or products. Non-targeted metabolomics – the systematic analysis of all small molecules in a system or organism – therefore holds great promise for the discovery of novel steroid metabolites and conjugates. Mahanti et al. have already provided remarkable proof-of-concept for this approach using NMR (33). The major drawback of NMR is its low sensitivity compared to MS, and the large sample quantities required can be a serious limitation for large-scale analysis. Non-targeted MS-based metabolomics approaches have been used in C. elegans research for a variety of purposes; for example, metabolic profiling of daf-2 mutants (39) and the development of a comprehensive method for analysis of lipid content (40). Direct infusion ion cyclotron resonance Fourier transform mass spectrometry (DI-ICR-FT/MS), which has very low sample consumption and ultra-high resolution and sensitivity, is a novel tool for analysis of small molecules and holds great promise for non-targeted metabolomics in C. elegans (41). In conjunction with sophisticated data analysis tools, this technology enables identification of novel metabolites in small samples (42, 43). Despite its sensitivity, DI-ICR-FT/MS does have some limitations since it cannot discriminate between isomers with the same molecular formula and exact mass because it does not employ chromatographic separation.
Steroidomics, the non-targeted analysis of steroids, is a sub-category of metabolomics or lipidomics. Similar to the other “omics,” its goal is to comprehensively detect and quantify all steroids in a cell or even an organism. LC-MS using reversed-phase separation is currently the most employed technique (42, 43), but other methods will eventually have to be developed to accommodate the growing complexity and number of isomeric structures to be identified and quantified. Similar to non-targeted metabolomics and lipidomics, steroidomics employs generic separation methods to allow detection of the maximal number of metabolites. However, generic methods could fail to separate isomers exhibiting subtle structural differences. A reversed-phase method based on a water–ACN gradient with formic acid additive is routinely used in our lab for metabolic profiling, and although it can detect DAs, it cannot separate the Δ4 and Δ7 isomers (our unpublished work). Thus, novel approaches, such as improved stationary phases, are required to facilitate separation of isomers during profiling.
A promising tool for future steroidomics applications is supercritical fluid chromatography (SFC), which is currently experiencing a revival. Although this technique has been known for decades, it has been largely overshadowed by HPLC and GC. However, SFC combines the best features of both methodologies: the different column selectivities of HPLC and the efficiency of GC. SFC uses supercritical CO2 as the eluent, which has low viscosity and high diffusivity. Furthermore, supercritical CO2 is miscible with a wide range of solvents, including MeOH and iProH, and a number of additives (e.g., ammonium hydroxide and ammonium acetate) can be used to fine-tune the separation, peak shapes, and sensitivity. In 2013, Taguchi et al. reported a supercritical fluid chromatography coupled to mass spectrometry (SFC-MS)-based method for the analysis of conjugated and unconjugated bile acids using a sub-2 μm amide column (44). They detected a total of 25 bile acids in rat plasma that was directly injected without extraction or further sample preparation (44). Similarly, SFC coupled to Q-ToF–MS was previously used for non-targeted analysis of rat and dog bile (45). Here, this technique was established as a viable alternative for metabolic profiling. Compared to GC- and LC-MS, SFC-MS yields short runtimes below 15 min. Therefore, SFC-MS holds great promise for use in C. elegans research as a superior method for resolving structurally closely related steroids.
These non-targeted analysis techniques allow detecting a wider range of molecules, including some that may not be detectable by targeted or focused analysis. These methods can also lead to the identification of yet unknown molecules (not described in any database), although this is a tedious and often time-consuming task. It requires expert analysis of tandem MS or 1D- or 2D-NMR-spectra, purification or synthesis of a reference standard, and testing of biological activity. One example is the discovery of novel dafachronic acid species by Mahanti et al. (33). Total time until final verification and biological testing took over 9 months (Franck Schroeder, personal communication). In addition, these techniques usually require specialized equipment and data analysis strategies. This is especially true for SFC-MS, not as widespread as GC- or LC-MS, where special pumps, auto sampler, and backpressure regulators are employed. Additionally, the separation mechanism can be different from LC or GC leading to counterintuitive separation results, which has be analyzed and evaluated carefully by a specialist.
Genetic evidence strongly suggests that many steroidogenic enzymes have yet to be identified and characterized. Furthermore, genetic studies have also raised the possibility that steroid synthesis pathways may operate in a tissue-specific manner. C. elegans XXX cells have been hypothesized to be a potential site for DA production, but DAF-9, the enzyme responsible for DA production, is also found in the somatic gonad, spermatheca, and hypoderma. Tissue-specific analysis of steroids is therefore required to address this. One potential method is imaging MS; for example, using matrix-assisted laser desorption ionization (MALDI) in combination with high resolving mass spectrometers, such ICR-FT/MS (46). Although imaging of steroid compounds has already been described using MALDI-ICR-FT/MS (47), current commercially available instruments are unable to produce a detailed analysis of the worm’s morphology with satisfactory resolution. Several proof-of-principle papers have been described (46–49), but they had limited resolution with no mean to finely analyze metabolites levels within the animals. In this regard, two obvious methodological advances needed are new sample preparation methods to enable visualization of metabolites within the worm and its tissues, and novel detection instruments aimed at smaller spatial resolutions of <5 μm (50, 51).
New analytical techniques hold great promise to identify new steroid signaling molecules, better understand biosynthesis, and enable precise quantification of more molecules. However, most recent tools, such as SFC, are available to only a few laboratories specialized in this field. In order to truly democratize such technique, an effort will be necessary in the future to make it available to more laboratories.
Conclusion
Although steroid hormones and their biological roles have been studied for over a century, we do not yet have a complete picture of their synthesis or function. With its central role during development and in adulthood, the steroid signaling pathway is of great interest for understanding complex biological traits such as aging. C. elegans is an excellent model organism for the development of new techniques that should significantly improve our ability to study steroid signaling and its biological roles. Recent publications show that DA, the worm-specific bile acid, can exist as various isoforms, suggesting that it has more complex functions than anticipated. Furthermore, there are undoubtedly enzymes, substrates, and products still to be identified. Genetics and analytical chemistry are extremely powerful tools individually, but in combination they enable an approach to identify and characterize the functions of steroid molecules that is currently beyond reach. Steroid signaling often relies on subtle perturbations in the levels of specific molecules, and current detection methods do not have the sensitivity to record such variations. Novel approaches, such as steroidomics and SFC-based analyses, will therefore take us nearer to our goal of obtaining a complete picture of the diverse biological roles of steroids.
Author Contributions
HA, PF, and MW wrote the manuscript. MW was more involved in the analytical chemistry part while HA and PF were in charge of the biological part.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to all authors whose work was not cited due to space limitation and thank all members of the Aguilaniu lab and Anne O’Rourke for helpful comments on the manuscript. This work was funded by the ARC (PJA 20141201697) and an ERC consolidator grant (H2020/2014-2019-N°647003) to HA.
References
- 1.Brenner S. The genetics of Caenorhabditis elegans. Genetics (1974) 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hulme SE, Whitesides GM. Chemistry and the worm: Caenorhabditis elegans as a platform for integrating chemical and biological research. Angew Chem Int Ed Engl (2011) 50:4774–807. 10.1002/anie.201005461 [DOI] [PubMed] [Google Scholar]
- 3.Antebi A. Nuclear receptor signal transduction in C. elegans. WormBook (2015):1–49. 10.1895/wormbook.1.64.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antebi A. Steroid regulation of C. elegans diapause, developmental timing, and longevity. Curr Top Dev Biol (2013) 105:181–212. 10.1016/B978-0-12-396968-2.00007-5 [DOI] [PubMed] [Google Scholar]
- 5.Motola DL, Cummins CL, Rottiers V, Sharma KK, Li T, Li Y, et al. Identification of ligands for DAF-12 that govern Dauer formation and reproduction in C. elegans. Cell (2006) 124:1209–23. 10.1016/j.cell.2006.01.037 [DOI] [PubMed] [Google Scholar]
- 6.Ludewig AH, Kober-Eisermann C, Weitzel C, Bethke A, Neubert K, Gerisch B, et al. A novel nuclear receptor/coregulator complex controls C. elegans lipid metabolism, larval development, and aging. Genes Dev (2004) 18:2120–33. 10.1101/gad.312604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antebi A, Yeh WH, Tait D, Hedgecock EM, Riddle DL. Daf-12 encodes a nuclear receptor that regulates the Dauer diapause and developmental age in C. elegans. Genes Dev (2000) 14:1512–27. 10.1101/gad.14.12.1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bethke A, Fielenbach N, Wang Z, Mangelsdorf DJ, Antebi A. Nuclear hormone receptor regulation of microRNAs controls developmental progression. Science (2009) 324:95–8. 10.1126/science.1164899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science (1984) 226:409–16. 10.1126/science.6494891 [DOI] [PubMed] [Google Scholar]
- 10.Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell (1997) 88:637–46. 10.1016/S0092-8674(00)81906-6 [DOI] [PubMed] [Google Scholar]
- 11.Gerisch B, Weitzel C, Kober-Eisermann C, Rottiers V, Antebi A. A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev Cell (2001) 1:841–51. 10.1016/S1534-5807(01)00085-5 [DOI] [PubMed] [Google Scholar]
- 12.Gerisch B, Rottiers V, Li D, Motola DL, Cummins CL, Lehrach H, et al. A bile acid-like steroid modulates Caenorhabditis elegans lifespan through nuclear receptor signaling. Proc Natl Acad Sci U S A (2007) 104:5014–9. 10.1073/pnas.0700847104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia K, Albert PS, Riddle DL. DAF-9, a cytochrome P450 regulating C. elegans larval development and adult longevity. Development (2002) 129:221–31. [DOI] [PubMed] [Google Scholar]
- 14.Mak HY, Ruvkun G. Intercellular signaling of reproductive development by the C. elegans DAF-9 cytochrome P450. Development (2004) 131:1777–86. 10.1242/dev.01069 [DOI] [PubMed] [Google Scholar]
- 15.Lee S-J, Kenyon C. Regulation of the longevity response to temperature by thermosensory neurons in Caenorhabditis elegans. Curr Biol (2009) 19:715–22. 10.1016/j.cub.2009.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horikawa M, Sural S, Hsu A-L, Antebi A. Co-chaperone p23 regulates C. elegans lifespan in response to temperature. PLoS Genet (2015) 11:e1005023. 10.1371/journal.pgen.1005023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsin H, Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature (1999) 399:362–6. 10.1038/20694 [DOI] [PubMed] [Google Scholar]
- 18.Rottiers V, Motola DL, Gerisch B, Cummins CL, Nishiwaki K, Mangelsdorf DJ, et al. Hormonal control of C. elegans Dauer formation and life span by a Rieske-like oxygenase. Dev Cell (2006) 10:473–82. 10.1016/j.devcel.2006.02.008 [DOI] [PubMed] [Google Scholar]
- 19.Patel DS, Fang LL, Svy DK, Ruvkun G, Li W. Genetic identification of HSD-1, a conserved steroidogenic enzyme that directs larval development in Caenorhabditis elegans. Development (2008) 135:2239–49. 10.1242/dev.016972 [DOI] [PubMed] [Google Scholar]
- 20.Dumas KJ, Guo C, Wang X, Burkhart KB, Adams EJ, Alam H, et al. Functional divergence of dafachronic acid pathways in the control of C. elegans development and lifespan. Dev Biol (2010) 340:605–12. 10.1016/j.ydbio.2010.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamawaki TM, Berman JR, Suchanek-Kavipurapu M, McCormick M, Gaglia MM, Lee S-J, et al. The somatic reproductive tissues of C. elegans promote longevity through steroid hormone signaling. PLoS Biol (2010) 8:e1000468. 10.1371/journal.pbio.1000468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen Y, Wollam J, Magner D, Karalay O, Antebi A. A steroid receptor-microRNA switch regulates life span in response to signals from the gonad. Science (2012) 338:1472–6. 10.1126/science.1228967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li T-M, Liu W, Lu S, Zhang Y-P, Jia L-M, Chen J, et al. No significant increase in the δ4- and δ7-dafachronic acid concentration in the long-lived glp-1 mutant, nor in the mutants defective in Dauer formation. G3 (Bethesda) (2015) 5:1473–9. 10.1534/g3.115.018812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCormick M, Chen K, Ramaswamy P, Kenyon C. New genes that extend Caenorhabditis elegans’ lifespan in response to reproductive signals. Aging Cell (2012) 11:192–202. 10.1111/j.1474-9726.2011.00768.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thondamal M, Witting M, Schmitt-Kopplin P, Aguilaniu H. Steroid hormone signalling links reproduction to lifespan in dietary-restricted Caenorhabditis elegans. Nat Commun (2014) 5:4879. 10.1038/ncomms5879 [DOI] [PubMed] [Google Scholar]
- 26.Witting M, Rudloff H-C, Thondamal M, Aguilaniu H, Schmitt-Kopplin P. Fast separation and quantification of steroid hormones δ4- and δ7-dafachronic acid in Caenorhabditis elegans. J Chromatogr B Analyt Technol Biomed Life Sci (2014) 978-979C:118–21. 10.1016/j.jchromb.2014.12.005 [DOI] [PubMed] [Google Scholar]
- 27.Wollam J, Magner DB, Magomedova L, Rass E, Shen Y, Rottiers V, et al. A novel 3-hydroxysteroid dehydrogenase that regulates reproductive development and longevity. PLoS Biol (2012) 10:e1001305. 10.1371/journal.pbio.1001305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Magner DB, Wollam J, Shen Y, Hoppe C, Li D, Latza C, et al. The NHR-8 nuclear receptor regulates cholesterol and bile acid homeostasis in C. elegans. Cell Metab (2013) 18:212–24. 10.1016/j.cmet.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi C, Murphy CT. Mating induces shrinking and death in Caenorhabditis mothers. Science (2014) 343:536–40. 10.1126/science.1242958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maures TJ, Booth LN, Benayoun BA, Izrayelit Y, Schroeder FC, Brunet A. Males shorten the life span of C. elegans hermaphrodites via secreted compounds. Science (2014) 343:541–4. 10.1126/science.1244160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dumas KJ, Guo C, Shih H-J, Hu PJ. Influence of steroid hormone signaling on life span control by Caenorhabditis elegans insulin-like signaling. G3 (Bethesda) (2013) 3:841–50. 10.1534/g3.112.005116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hannich JT, Entchev EV, Mende F, Boytchev H, Martin R, Zagoriy V, et al. Methylation of the sterol nucleus by STRM-1 regulates Dauer larva formation in Caenorhabditis elegans. Dev Cell (2009) 16:833–43. 10.1016/j.devcel.2009.04.012 [DOI] [PubMed] [Google Scholar]
- 33.Mahanti P, Bose N, Bethke A, Judkins JC, Wollam J, Dumas KJ, et al. Comparative metabolomics reveals endogenous ligands of DAF-12, a nuclear hormone receptor, regulating C. elegans development and lifespan. Cell Metab (2014) 19:73–83. 10.1016/j.cmet.2013.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma KK, Wang Z, Motola DL, Cummins CL, Mangelsdorf DJ, Auchus RJ. Synthesis and activity of dafachronic acid ligands for the C. elegans DAF-12 nuclear hormone receptor. Mol Endocrinol (2009) 23:640–8. 10.1210/me.2008-0415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alvarez LD, Mañez PA, Estrin DA, Burton G. The Caenorhabditis elegans DAF-12 nuclear receptor: structure, dynamics, and interaction with ligands. Proteins (2012) 80:1798–809. 10.1002/prot.24076 [DOI] [PubMed] [Google Scholar]
- 36.Broué F, Liere P, Kenyon C, Baulieu E-E. A steroid hormone that extends the lifespan of Caenorhabditis elegans. Aging Cell (2007) 6:87–94. 10.1111/j.1474-9726.2006.00268.x [DOI] [PubMed] [Google Scholar]
- 37.Li T-M, Chen J, Li X, Ding X-J, Wu Y, Zhao L-F, et al. Absolute quantification of a steroid hormone that regulates development in Caenorhabditis elegans. Anal Chem (2013) 85:9281–7. 10.1021/ac402025c [DOI] [PubMed] [Google Scholar]
- 38.Sardella R, Carotti A, Gioiello A, Lisanti A, Ianni F, Lindner W, et al. Chromatographic separation of free dafachronic acid epimers with a novel triazole click quinidine-based chiral stationary phase. J Chromatogr A (2014) 1339:96–102. 10.1016/j.chroma.2014.02.080 [DOI] [PubMed] [Google Scholar]
- 39.Castro C, Krumsiek J, Lehrbach NJ, Murfitt SA, Miska EA, Griffin JL. A study of Caenorhabditis elegans DAF-2 mutants by metabolomics and differential correlation networks. Mol Biosyst (2013) 9:1632–42. 10.1039/c3mb25539e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Witting M, Maier TV, Garvis S, Schmitt-Kopplin P. Optimizing a ultrahigh pressure liquid chromatography-time of flight-mass spectrometry approach using a novel sub-2μm core-shell particle for in depth lipidomic profiling of Caenorhabditis elegans. J Chromatogr A (2014) 1359:91–9. 10.1016/j.chroma.2014.07.021 [DOI] [PubMed] [Google Scholar]
- 41.Witting M, Lucio M, Tziotis D, Wägele B, Suhre K, Voulhoux R, et al. DI-ICR-FT-MS-based high-throughput deep metabotyping: a case study of the Caenorhabditis elegans-Pseudomonas aeruginosa infection model. Anal Bioanal Chem (2015) 407:1059–73. 10.1007/s00216-014-8331-5 [DOI] [PubMed] [Google Scholar]
- 42.Walker A, Lucio M, Pfitzner B, Scheerer MF, Neschen S, de Angelis MH, et al. Importance of sulfur-containing metabolites in discriminating fecal extracts between normal and type-2 diabetic mice. J Proteome Res (2014) 13:4220–31. 10.1021/pr500046b [DOI] [PubMed] [Google Scholar]
- 43.Tziotis D, Hertkorn N, Schmitt-Kopplin P. Kendrick-analogous network visualisation of ion cyclotron resonance Fourier transform mass spectra: improved options for the assignment of elemental compositions and the classification of organic molecular complexity. Eur J Mass Spectrom (Chichester, Eng) (2011) 17:415–21. 10.1255/ejms.1135 [DOI] [PubMed] [Google Scholar]
- 44.Taguchi K, Fukusaki E, Bamba T. Simultaneous and rapid analysis of bile acids including conjugates by supercritical fluid chromatography coupled to tandem mass spectrometry. J Chromatogr A (2013) 1299:103–9. 10.1016/j.chroma.2013.05.043 [DOI] [PubMed] [Google Scholar]
- 45.Jones MD, Rainville PD, Isaac G, Wilson ID, Smith NW, Plumb RS. Ultra high resolution SFC-MS as a high throughput platform for metabolic phenotyping: application to metabolic profiling of rat and dog bile. J Chromatogr B Analyt Technol Biomed Life Sci (2014) 966:200–7. 10.1016/j.jchromb.2014.04.017 [DOI] [PubMed] [Google Scholar]
- 46.Sun N, Ly A, Meding S, Witting M, Hauck SM, Ueffing M, et al. High-resolution metabolite imaging of light and dark treated retina using MALDI-FTICR mass spectrometry. Proteomics (2014) 14:913–23. 10.1002/pmic.201300407 [DOI] [PubMed] [Google Scholar]
- 47.Cobice DF, Mackay CL, Goodwin RJA, McBride A, Langridge-Smith PR, Webster SP, et al. Mass spectrometry imaging for dissecting steroid intracrinology within target tissues. Anal Chem (2013) 85:11576–84. 10.1021/ac402777k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Menger RF, Clendinen CS, Searcy LA, Edison AS, Yost RA. MALDI mass spectrometric imaging of the nematode Caenorhabditis elegans. Curr Metabolomics (2015) 3:130–7. 10.2174/2213235X03666150525223412 [DOI] [Google Scholar]
- 49.Patti GJ, Tautenhahn R, Johannsen D, Kalisiak E, Ravussin E, Brüning JC, et al. Meta-analysis of global metabolomic data identifies metabolites associated with life-span extension. Metabolomics (2014) 10:737–43. 10.1007/s11306-013-0608-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kimura Y, Sugiura Y, Hayasaka T, Setou M. Visualizing the spatial distribution of biomolecules in C. elegans by imaging mass spectrometry (2015). Available from: http://www.wormbook.org/wbg/articles/volume-18-number-1/visualizing-the-spatial-distribution-of-biomolecules-in-c-elegans-by-imaging-mass-spectrometry/ [Google Scholar]
- 51.Guenther S, Römpp A, Kummer W, Spengler B. AP-MALDI imaging of neuropeptides in mouse pituitary gland with 5 μm spatial resolution and high mass accuracy. Int J Mass Spectrom (2011) 305:228–37. 10.1016/j.ijms.2010.11.011 [DOI] [Google Scholar]