Abstract
It is increasingly apparent that nature evolved peroxiredoxins not only as H2O2 scavengers but also as highly sensitive H2O2 sensors and signal transducers. Here we ask whether the H2O2 sensing role of Prx can be exploited to develop probes that allow to monitor intracellular H2O2 levels with unprecedented sensitivity. Indeed, simple gel shift assays visualizing the oxidation of endogenous 2-Cys peroxiredoxins have already been used to detect subtle changes in intracellular H2O2 concentration. The challenge however is to create a genetically encoded probe that offers real-time measurements of H2O2 levels in intact cells via the Prx oxidation state. We discuss potential design strategies for Prx-based probes based on either the redox-sensitive fluorophore roGFP or the conformation-sensitive fluorophore cpYFP. Furthermore, we outline the structural and chemical complexities which need to be addressed when using Prx as a sensing moiety for H2O2 probes. We suggest experimental strategies to investigate the influence of these complexities on probe behavior. In doing so, we hope to stimulate the development of Prx-based probes which may spearhead the further study of cellular H2O2 homeostasis and Prx signaling.
Keywords: biosensor, genetically encoded fluorescent probes, hydrogen peroxide, peroxiredoxin
INTRODUCTION
Nature evolved specialized proteins to respond to increased intracellular hydrogen peroxide (H2O2). These proteins typically remove H2O2 by either dismutation or reduction (Winterbourn, 2013). Some of these proteins additionally serve as H2O2 sensors and signal transmitters to inform the cell about changes in intracellular H2O2 levels (Delaunay et al., 2002; Rhee et al., 2012; Sobotta et al., 2015). In principle, it should be possible to use these H2O2-sensing proteins as research tools, namely to exploit them as biosensors for the real-time observation of H2O2 in living cells.
Indeed, the two existing genetically encoded H2O2 probes, HyPer and roGFP2-Orp1, are based on OxyR and Orp1, respectively (Bilan and Belousov, 2015; Schwarzlander et al., 2015). OxyR is a H2O2-sensing transcription factor from bacteria and Orp1 is a member of the glutathione peroxidase family from yeast. Both proteins have a rate constant for the reaction with H2O2 of approximately 105 M−1s−1 (Aslund et al., 1999). However, peroxiredoxins have a significantly higher reactivity towards H2O2 which can be as high as 107 – 108 M−1 s−1 (Rhee and Woo, 2011). This potentially makes them superior H2O2 sensors. Indeed, a side by side comparison between endogenous Prx2 and ectopic roGFP2-Orp1 in mammalian cells showed that Prx2 is many times more sensitive than Orp1 (Sobotta et al., 2013). Nevertheless, little has been done to develop biosensors based on Prx. In the following we will address the question of whether such probes can be developed.
It is interesting to note that endogenous 2-Cys peroxiredoxins have already been utilized as H2O2 reporters, albeit in a biochemical way that imposes many restrictions (Cox et al., 2010; Poynton and Hampton, 2014). Oxidation of 2-Cys peroxiredoxins yields covalent dimers which can be visualized by non-reducing SDS-PAGE and Prx-specific immunoblotting. The Prx dimerization assay is capable of detecting increases in intracellular H2O2 levels associated with physiologically relevant signaling events (Cox et al., 2008; Kumar et al., 2009). Moreover, subcellular information can be obtained by analyzing peroxiredoxins expressed in different organelles (e.g. Prx1 in the cytosol and Prx3 in the mitochondrial). However, the approach is limited in scope (Cox et al., 2008; Kumar et al., 2009), as it doesn’t allow real-time monitoring of the Prx redox state in intact cells. Cell need to be lysed, which inevitably causes unwanted oxidation of peroxiredoxins, unless intact cells are pretreated with sufficient amounts of alkylating agents prior to cell lysis. Finding the best conditions for preserving the endogenous Prx redox state and excluding artifacts is not always a trivial task (Cox et al., 2010; Poynton and Hampton, 2014; Sobotta et al., 2013). Thus, the challenge is to develop genetically encoded probes which do not suffer from these limitations and can be monitored in living cells (Bilan and Belousov, 2015; Schwarzlander et al., 2015).
ENGINEERING PRX-BASED GENETICALLY ENCODED FLUORESCENT PROBES
Genetically encoded Prx-based H2O2 probes will necessarily consist of (at least) two moieties. The first moiety is the Prxs domain and the second a fluorescent reporter protein. In currently used genetically encoded redox probes the fluorescent reporter is either circularly permutated yellow fluorescent protein (cpYFP) or redox-sensitive green fluorescent protein (roGFP) (Fig. 1). The cpYFP reporter responds to a structural perturbation, whereas the roGFP reporter responds to the oxidation of its cysteine pair to a disulfide bridge (Bilan and Belousov, 2015; Schwarzlander et al., 2015). Another potential strategy is the use of a Förster resonance energy transfer (FRET) pair of two fluorescent proteins to sense structural changes imposed by the oxidation of the peroxiredoxin (Barranco-Medina et al., 2009). Thus, based on existing probes at least three different designs can be envisaged.
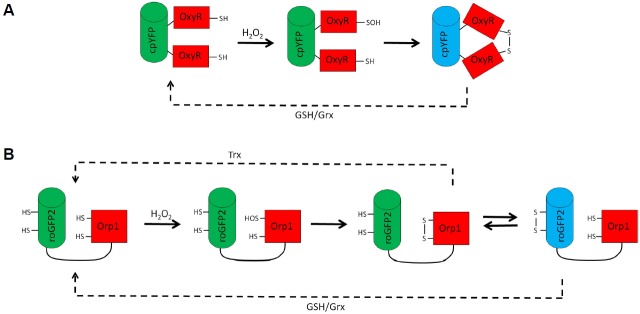
Fig. 1.
The H2O2 sensing mechanism of HyPer (A) and roGFP2-Orp1 (B). The sensing moieties of both probes (OxyR and Orp1 respectively) are colored in red and the fluorescent reporter moiety (i.e. cpYFP and roGFP2) in green or blue. (A) HyPer consists of a cpYFP fused to two halves of the OxyR regulatory domain. Upon the reaction of OxyR with H2O2, an intramolecular disulfide is formed which triggers a conformational change and thus modifies the fluorescence of cpYFP. (B) The roGFP2-Orp1 probe senses H2O2 via Orp1. An intramolecular disulfide is formed in Orp1 which is transferred to roGFP2 by thiol-disulfide exchange. The introduction of the disulfide into roGFP2 changes its fluorescence. Both probes are reversible in cellulo due to the reduction by glutathione/glutaredoxin(GSH/Grx) and/or thioredoxin(Trx) (dashed lines).
The key requirement for a roGFP-based Prx probe is the ability of the peroxiredoxin to mediate oxidation of the roGFP cysteine pair. Recently, it was found that the human 2-Cys peroxiredoxin Prx2 acts as a transmitter of oxidizing equivalents towards the transcription factor STAT3 (Sobotta et al., 2015). Thus, it is conceivable that 2-Cys peroxiredoxins can also transmit oxidation to other proteins, including roGFP (Figs. 2A and 2B). Obviously, various Prx-roGFP or roGFP-Prx fusion proteins would need to be tested and optimized empirically to identify those in which the oxidizing equivalents are transmitted most efficiently to the roGFP moiety.
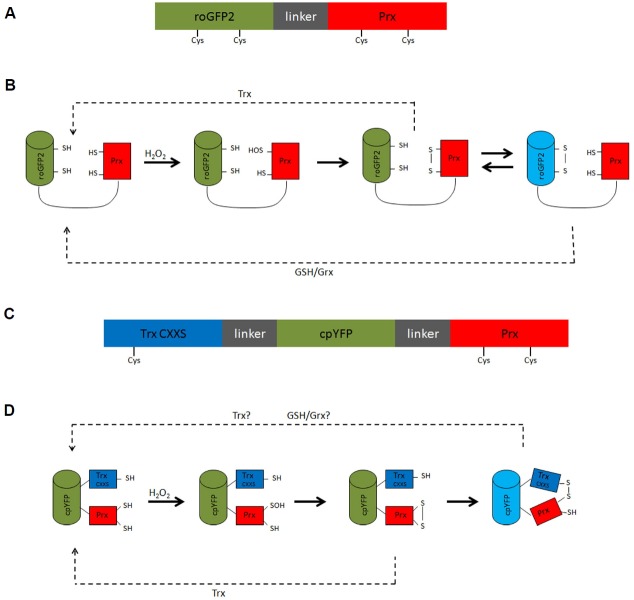
Fig. 2.
Example designs for a roGFP-based (A, B) and a cpYFP-based Prx probe (C, D). Schematic representation of the roGFP-Prx probe (A), and Trx(CXXS)-cpYFP-Prx probe (C) with the reporter moiety in green, flexible linkers in grey and the Prx moiety in red. The Trx moiety lacking its resolving cysteine in the Trx(CXXS)-cpYFP-Prx probe is shown in blue. Cysteine residues involved in the reaction mechanism are denoted. (B, D) Hypothetical reaction scheme of the suggested probes. For simplicity, both schemes are based on a monomeric atypical 2-Cys Prx. Howerer, similar schemes apply to dimeric typical 2-Cys Prxs. Possible reduction pathways are indicated.
When using cpYFP as reporter moiety, Prx oxidation must provoke a conformational change that can be sensed by the cpYFP fluorophore. Peroxiredoxins undergo a local unfolding transition upon reaction with H2O2 (Hall et al., 2011). However, it is not obvious if and how this limited structural change could be transferred to the cpYFP moiety. Alternatively, a cpYFP domain could be positioned to respond to the decamer to dimer transition, which accompanies oxidation of typical 2-Cys peroxiredoxins (Barranco-Medina et al., 2009). Perhaps a cpYFP moiety that is placed between two Prx domains would sense decamer dissociation. Yet another strategy is a fusion protein in which cpYFP is sandwiched between a Prx and a thioredoxin (Trx) domain, the latter lacking its resolving cysteine (Figs. 2C and 2D). The reasoning behind this design is that the mutant Trx domain will covalently trap the oxidized Prx domain thus causing a structural change that is sensed by the cpYFP moiety. This strategy was used previously to generate a methionine sulfoxide sensitive probe in which cpYFP is inserted between a methionine sulfoxide reductase domain and a mutant Trx domain (Tarrago et al., 2015). A major disadvantage of utilizing circular permutated fluorescent proteins (cpFPs) like cpYFP is their intrinsic pH sensitivity and the requirement to use stringent pH controls in each experiment (Bilan and Belousov, 2015). However, a potential advantage is that cpFPs of different color can be used to create spectral probe variants (Ermakova et al., 2014; Zhao et al., 2011).
The use of FRET to investigate 2-Cys peroxiredoxin behavior has already been reported (Muthuramalingam et al., 2009; Seidel et al., 2010). The authors used two (single-step FRET) or three (two-step FRET) Prx-fluorescent protein (FP) pairs to monitor the decamer dissociation upon peroxiredoxin oxidation. Although these Prx-FP fusions showed potential as cellular H2O2 biosensors, no further developments in this direction have been reported.
ANTICIPATING COMPLICATIONS THAT MAY AFFECT PRX-BASED PROBES
The envisaged Prx-based biosensors promise to achieve much higher sensitivity than the current generation of genetically encoded H2O2 sensors (Ezerina et al., 2014). However, peroxiredoxins exhibit rather complex biochemical and structural behavior (Rhee and Woo, 2011; Perkins et al., 2015). This may complicate or compromise their utilization as H2O2 sensors. In the following we discuss these complexities and how they may affect probe behavior. For simplicity, we focus our discussion on roGFP-Prx fusion proteins, but most considerations equally apply to the other design strategies.
The influence of N- or C-terminal fusions
First of all, fusion to a fluorescent protein might already influence Prx behavior. Previously, the presence of an N- or C-terminal tag was found to influence peroxidase activity, chaperone holdase activity, hyperoxidation sensitivity or oligomeric structure of several Prxs (Cao et al., 2007; Konig et al., 2013; Koo et al., 2002; Noichri et al., 2015; Randall et al., 2014; Sayed and Williams, 2004; Seo et al., 2009). Thus, each potential Prx-based probe should be functionally characterized and compared to the unfused Prx.
The influence of substrate specificity
Many Prxs not only react with H2O2, but also show reactivity towards organic peroxides and peroxinitrite (Perkins et al., 2014). For example, human Prx5 and E. coli Tpx are 100- to 200-fold more reactive towards organic peroxides than to H2O2 (Hall et al., 2009; Trujillo et al., 2007). In contrast, other Prx are much more restricted in their substrate specificity, for instance, S. typhimurium AhpC shows a 100-fold higher reactivity with H2O2 as compared to organic peroxides (Parsonage et al., 2008). The implications of substrate preference for probe design will depend on the purpose of the envisaged Prx-based probe. If the purpose of the probe is to report on intracellular H2O2 with the highest possible selectivity, then a correspondingly selective Prx domain should be employed. However, if the probe is used as a reporter for the Prx oxidation state as such, the probe can simply use the particular Prx of interest.
The influence of oligomeric structure
The oligomeric state of different peroxiredoxins can vary from monomers to decamers/docecamers and may change upon oxidation (Barranco-Medina et al., 2009; Hall et al., 2011). In principle, a probe should have a simple design, and thus a monomeric Prx may be preferred as a sensing moiety. However, only a few peroxiredoxins are monomeric. The majority forms dimers and higher oligomeric structures. A typical 2-Cys peroxiredoxin fused to a fluorescent reporter would need to dimerize to be active. This situation raises several questions: Does a probe based on a typical 2-Cys Prx only form probe-probe dimers, or does it also form heterodimers with endogenous Prx molecules? Does the probe form decamers, or is decamerization prevented by the fused reporter protein? If it does form decamers, does it form probe homo-decamers or mixed hetero-decamers with endogenous peroxiredoxins? Furthermore, is decamerization required for the probe to function? Mutations at the A-type interface, either stabilizing or destabilizing decamerization, may be used to investigate this question. For example, Parsonage et al. showed that substitution of Thr77 of Salmonella typhimurium AhpC for isoleucine or aspartic acid disturbs decamer formation (Parsonage et al., 2005). In any case the oligomeric status of Prx-based probes should be investigated.
The influence of hyperoxidation
The inactivation of peroxiredoxins by hyperoxidation has been observed under physiological circumstances (Nelson et al., 2013; Perkins et al., 2014). If a Prx-based probe turns out to be susceptible to hyperoxidation, it may under certain conditions report false results. Fortunately, not all Prx are highly sensitive to hyperoxidation. Bacterial Prxs are in general more robust, whereas some eukaryotic Prxs, including human Prx2, are remarkably sensitive (Nelson et al., 2013; Wood et al., 2003). Thus, bacterial Prxs may be preferable over eukaryotic Prxs when designing a Prx-based probe. Nevertheless, an intrinsically sensitive Prx could be modified to decrease its hyperoxidation sensitivity. Several factors have been described to influence hyperoxidation sensitivity, including two distinct sequence motifs: a GGLG and a C-terminal YF motif (Wood et al., 2003). It has been shown that alteration of these motifs and their surrounding residues modulate hyperoxidation sensitivity (Sayed and Williams, 2004; Wang et al., 2012). One possibility to decrease hyperoxidation sensitivity is the truncation of the C-terminus (Koo et al., 2002; Randall et al., 2014; Sayed and Williams, 2004; Seo et al., 2009). This strategy could be used to increase the robustness of hyperoxidation-sensitive peroxiredoxins, making them more suitable as a sensing moiety for a probe. In any case, the hyperoxidation sensitivity of any Prx-based probe should be determined experimentally since unambiguous predictors of hyperoxidation sensitivity are lacking (Perkins et al., 2014). Immunoblotting with hyperoxidation specific antibodies allows to determine hyperoxidation of Prx-based probes (Fig. 3A).
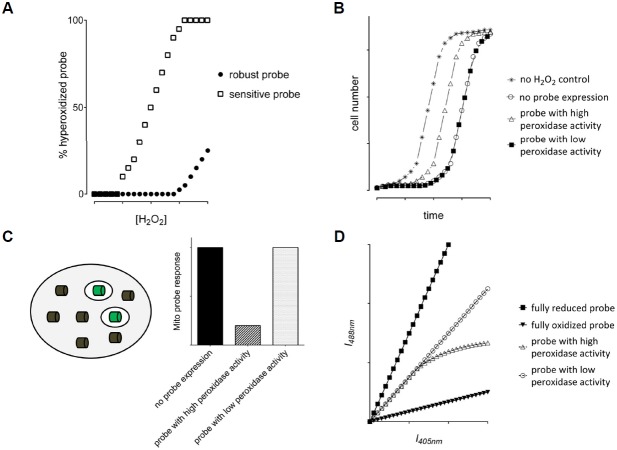
Fig. 3.
Strategies to investigate hyperoxidation sensitivity (A) and physiological impact (B–D) of Prx-based sensors. (A) The fraction of hyper-oxidized probe as a function of the applied H2O2 concentration is determined by hyperoxidation-specific immunoblotting. (B) A growth recovery experiment can be used to determine the influence of the probe expression on cell growth and survival. A theoretical growth curve upon treatment with sublethal concentrations of H2O2 is shown for probes with and without a physiological impact. (C) Investigating the influence of a probe on H2O2 scavenging capacity. A non-fluorescent probe version (grey cylinder) is expressed in the cytosol and its impact on H2O2 homeostasis is measured using a fluorescent probe (green cylinders) expressed in mitochondria. A theoretical result is shown on the right for probes with and without an impact on H2O2 homeostasis. (D) Expected fluorescence intensity scatter plots for a ratiometric probe with low (open circles) and high H2O2 scavenging activity (open triangles).
The influence of post-translational modifications on the Prx domain
Peroxiredoxins are subject to various post-translational modifications which influence their peroxidase activity or other properties. For instance, phosphorylation of Thr90 in human Prx1 and Thr89 in Prx2 attenuates peroxidase activity and promotes chaperone holdase activity whereas Ser32 phosphorylation of Prx1 is reported to increase peroxidase activity (Chae et al., 2012; Chang et al., 2002; Jang et al., 2006; Qu et al., 2007; Zykova et al., 2010). N-terminal acetylation of human Prx2 was found to influence hyperoxidation sensitivity (Seo et al., 2009). Thus, it is conceivable that a Prx-based probe can also undergo post-translational modifications under certain conditions. Such modifications may alter the probe response and lead to the misinterpretation of measurements. For those residues already known to be potentially modified, corresponding probe point mutants (e.g. preventing or mimicking phosphorylation) can be created and studied in parallel. Alternatively or additionally, a probe based on a heterologous Prx may be employed, thus decreasing the probability of its post-translational modification in the host system.
The influence of the cellular reducing systems
A dynamic (i.e. reversible) probe offers the possibility to monitor transient elevations in H2O2. Since the reaction with the peroxide is an irreversible reaction, probe reduction depends on cellular reducing systems (Figs. 1 and 2). Thus, a Prx-based probe is a dual-specificity sensor in the sense that it responds not only to changes in peroxide levels, but also to changes in the activity of the relevant reducing system. The reducing system to which the probe responds will depend on the design of the probe (Fig. 2). If the probe uses a typical 2-Cys Prx moiety, it will likely be reduced by the Trx system, unless the Prx moiety is intentionally altered to avoid reduction by thioredoxin (e.g. by mutation of the resolving cysteine). Designing a Prx-based probe to respond to a particular reducing system may be useful in order to limit or adjust the total H2O2 turnover of the probe. The latter determines how much H2O2 is consumed by the probe inside cells (see next paragraph). In any case, it will be important to characterize in detail how a given Prx-based probe is reduced inside cells. This can be achieved by using selective deletion/silencing of reducing systems inside cells and can additionally be studied in vitro with recombinant proteins.
The effect of introducing additional peroxidase activity to the cell
In principle, the ectopic expression of any H2O2-consuming probe may perturb cellular H2O2 homeostasis (Schwarzlander et al., 2015). The impact of a Prx-based probe on endogenous H2O2 levels will depend on two factors, namely the probe’s expression level and its H2O2 turnover. The latter depends on both the rate of probe oxidation and the rate of probe reduction. Under most conditions, the endogenous peroxidase activity of the cell is highly efficient, not least due to the high reactivity and abundance of endogenous Prxs (Rhee et al., 2012). Thus, it can be expected that ectopic expression of a Prx-based probe at levels not higher than that of endogenous Prxs will not lead to a major impact on endogenous H2O2 homeostasis. Nevertheless, the impact of expressing a H2O2-consuming probe will need to be determined experimentally. A simple and coarse approach would be to follow growth and survival of cells exposed to a range of H2O2 concentrations in the absence or presence of ectopic probe expression. If probe expression does not enhance the overall H2O2 scavenging capacity, the rate and extent of recovery will not be different from cells lacking probe expression (Fig. 3B). However, this experiment does not directly investigate the impact of probe expression on endogenous H2O2 levels and cellular adaptations could obscure the interpretation. More sophisticated experiments to directly investigate the contribution of a Prx-based probe to cellular H2O2 scavenging could be envisaged along the following lines: a non-fluorescent mutant of the probe (i.e. a mutant with identical expression, but lacking a mature fluorophore) is expressed in the cytosol and a fluorescent version of the same or alternative probe is expressed in the mitochondrial matrix. A bolus of H2O2 is given from outside the cell and the amount of H2O2 reaching the mitochondrial matrix is monitored by the mitochondrially targeted probe. This reveals if and how much the cytosolic probe (which is made non-fluorescent in order not to interfere with measurement of the mitochondrial probe) contributes to H2O2 scavenging (Fig. 3C). An alternative approach makes use of the wide distribution of probe expression levels within a cell population upon transient transfection (which reaches from barely detectable expression to very strong overexpression) (Barata and Dick, 2013; Schwarzlander et al., 2015). Any ratiometric probe (like the cpYFP and roGFP based probes) that does not influence its own measurement (i.e. endogenous H2O2 levels) is expected to yield a perfectly straight line on a scatter plot that relates the fluorescent emission intensities at the two excitation wavelengths. Any deviation of this linearity at higher fluorescence intensities (i.e. higher expression levels) would indicate an impact of the probe on H2O2 levels. These assays should also allow screening for probe variants with minimal impact on endogenous H2O2 homeostasis.
The effect of introducing additional Prx chaperone activity to the cell
Chaperon holdase activity is a well-known non-peroxidase function of 2-Cys peroxiredoxins (Rhee and Woo, 2011). The gain of chaperone activity seems to be tightly linked to the loss of peroxidase activity through hyperoxidation or heat stress. It is conceivable that a 2-Cys Prx-based probe introduces additional chaperone activity to the cell, especially if the Prx is sensitive to hyperoxidation. However, the reporter moiety might sterically hinder the chaperone activity. Indeed, the N-terminus of peroxiredoxins is crucial for the binding of client proteins and an N-terminal tag was found to abolish the chaperone activity (Teixeira et al., 2015). The intrinsic chaperone activity of a recombinant Prx-based probe may be determined with commonly used in vitro chaperone assays or with cellular recovery assays to assess the impact of the additional chaperone activity on cell survival (Fig. 3B) (Moon et al., 2013; Teixeira et al., 2015).
In summary, despite the many complexities that need to be addressed, peroxiredoxins promise to become the foundation of the next generation of genetically encoded H2O2 biosensors.
REFERENCES
- Aslund F., Zheng M., Beckwith J., Storz G. Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol-disulfide status. Proc. Natl. Acad. Sci. USA. 1999;96:6161–6165. doi: 10.1073/pnas.96.11.6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barata A.G., Dick T.P. In vivo imaging of H2O2 production in Drosophila. Methods Enzymol. 2013;526:61–82. doi: 10.1016/B978-0-12-405883-5.00004-1. [DOI] [PubMed] [Google Scholar]
- Barranco-Medina S., Lazaro J.J., Dietz K.J. The oligomeric conformation of peroxiredoxins links redox state to function. FEBS Lett. 2009;583:1809–1816. doi: 10.1016/j.febslet.2009.05.029. [DOI] [PubMed] [Google Scholar]
- Bilan D., Belousov V. HyPer family probes: state of the art. Antioxid. Redox Signal. 2015 doi: 10.1089/ars.2015.6586. [Epub ahead of Print] [DOI] [PubMed] [Google Scholar]
- Cao Z., Bhella D., Lindsay J.G. Reconstitution of the mitochondrial PrxIII antioxidant defence pathway: general properties and factors affecting PrxIII activity and oligomeric state. J. Mol. Biol. 2007;372:1022–1033. doi: 10.1016/j.jmb.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Chae H.Z., Oubrahim H., Park J.W., Rhee S.G., Chock P.B. Protein glutathionylation in the regulation of peroxiredoxins: a family of thiol-specific peroxidases that function as antioxidants, molecular chaperones, and signal modulators. Antioxid. Redox Signal. 2012;16:506–523. doi: 10.1089/ars.2011.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T.S., Jeong W., Choi S.Y., Yu S., Kang S.W., Rhee S.G. Regulation of peroxiredoxin I activity by Cdc2-mediated phosphorylation. J. Biol. Chem. 2002;277:25370–25376. doi: 10.1074/jbc.M110432200. [DOI] [PubMed] [Google Scholar]
- Cox A.G., Pullar J.M., Hughes G., Ledgerwood E.C., Hampton M.B. Oxidation of mitochondrial peroxiredoxin 3 during the initiation of receptor-mediated apoptosis. Free Rad. Biol. Med. 2008;44:1001–1009. doi: 10.1016/j.freeradbiomed.2007.11.017. [DOI] [PubMed] [Google Scholar]
- Cox A.G., Winterbourn C.C., Hampton M.B. Measuring the redox state of cellular peroxiredoxins by immunoblotting. Methods Enzymol. 2010;474:51–66. doi: 10.1016/S0076-6879(10)74004-0. [DOI] [PubMed] [Google Scholar]
- Delaunay A., Pflieger D., Barrault M.B., Vinh J., Toledano M.B. A thiol peroxidase is an H2O2 receptor and redoxtransducer in gene activation. Cell. 2002;111:471–481. doi: 10.1016/s0092-8674(02)01048-6. [DOI] [PubMed] [Google Scholar]
- Ermakova Y.G., Bilan D.S., Matlashov M.E., Mishina N.M., Mark-vicheva K.N., Subach O.M., Subach F.V., Bogeski I., Hoth M., Enikolopov G., et al. Red fluorescent genetically encoded indicator for intracellular hydrogen peroxide. Nat. Commun. 2014;5:5222. doi: 10.1038/ncomms6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezerina D., Morgan B., Dick T.P. Imaging dynamic redox processes with genetically encoded probes. J. Mol. Cell. Cardiol. 2014;73:43–49. doi: 10.1016/j.yjmcc.2013.12.023. [DOI] [PubMed] [Google Scholar]
- Hall A., Nelson K., Poole L.B., Karplus P.A. Structure-based insights into the catalytic power and conformational dexterity of peroxiredoxins. Antioxid. Redox Signal. 2011;15:795–815. doi: 10.1089/ars.2010.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A., Sankaran B., Poole L.B., Karplus P.A. Structural changes common to catalysis in the Tpx peroxiredoxin sub-family. J. Mol. Biol. 2009;393:867–881. doi: 10.1016/j.jmb.2009.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H.H., Kim S.Y., Park S.K., Jeon H.S., Lee Y.M., Jung J.H., Lee S.Y., Chae H.B., Jung Y.J., Lee K.O., et al. Phosphorylation and concomitant structural changes in human 2-Cys peroxiredoxin isotype I differentially regulate its peroxidase and molecular chaperone functions. FEBS Lett. 2006;580:351–355. doi: 10.1016/j.febslet.2005.12.030. [DOI] [PubMed] [Google Scholar]
- Konig J., Galliardt H., Jutte P., Schaper S., Dittmann L., Dietz K.J. The conformational bases for the two functionalities of 2-Cysteine peroxiredoxins as peroxidase and chaperone. J. Exp. Bot. 2013;64:3483–3497. doi: 10.1093/jxb/ert184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo K.H., Lee S., Jeong S.Y., Kim E.T., Kim H.J., Kim K., Song K., Chae H.Z. Regulation of thioredoxin peroxidase activity by C-terminal truncation. Arch. Biochem. Biophys. 2002;397:312–318. doi: 10.1006/abbi.2001.2700. [DOI] [PubMed] [Google Scholar]
- Kumar V., Kitaeff N., Hampton M.B., Cannell M.B., Winterbourn C.C. Reversible oxidation of mitochondrial peroxiredoxin 3 in mouse heart subjected to ischemia and reperfusion. FEBS Lett. 2009;583:997–1000. doi: 10.1016/j.febslet.2009.02.018. [DOI] [PubMed] [Google Scholar]
- Moon J.C., Kim G.M., Kim E.K., Lee H.N., Ha B., Lee S.Y., Jang H.H. Reversal of 2-Cys peroxiredoxin oligomerization by sulfiredoxin. Biochem. Biophys. Res. Commun. 2013;432:291–295. doi: 10.1016/j.bbrc.2013.01.114. [DOI] [PubMed] [Google Scholar]
- Muthuramalingam M., Seidel T., Laxa M., Nunes de Miranda S.M., Gartner F., Stroher E., Kandlbinder A., Dietz K.J. Multiple redox and non-redox interactions define 2-Cys peroxiredoxin as a regulatory hub in the chloroplast. Mol. Plant. 2009;2:1273–1288. doi: 10.1093/mp/ssp089. [DOI] [PubMed] [Google Scholar]
- Nelson K.J., Parsonage D., Karplus P.A., Poole L.B. Evaluating peroxiredoxin sensitivity toward inactivation by peroxide substrates. Method Enzymol. 2013;527:21–40. doi: 10.1016/B978-0-12-405882-8.00002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noichri Y., Palais G., Ruby V., D’Autreaux B., Delaunay-Moisan A., Nystrom T., Molin M., Toledano M.B. In vivo parameters influencing 2-Cys Prx oligomerization: The role of enzyme sulfinylation. Redox Biol. 2015;6:326–333. doi: 10.1016/j.redox.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsonage D., Youngblood D.S., Sarma G.N., Wood Z.A., Karplus P.A., Poole L.B. Analysis of the link between enzymatic activity and oligomeric state in AhpC, a bacterial peroxiredoxin. Biochemistry. 2005;44:10583–10592. doi: 10.1021/bi050448i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsonage D., Karplus P.A., Poole L.B. Substrate specificity and redox potential of AhpC, a bacterial peroxiredoxin. Proc. Natl. Acad. Sci. USA. 2008;105:8209–8214. doi: 10.1073/pnas.0708308105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins A., Poole L.B., Karplus P.A. Tuning of peroxiredoxin catalysis for various physiological roles. Biochemistry. 2014;53:7693–7705. doi: 10.1021/bi5013222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins A., Nelson K.J., Parsonage D., Poole L.B., Karplus P.A. Peroxiredoxins: guardians against oxidative stress and modulators of peroxide signaling. Trends Biochem. Sci. 2015;40:435–445. doi: 10.1016/j.tibs.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poynton R.A., Hampton M.B. Peroxiredoxins as biomarkers of oxidative stress. Biochimica Biophys. Acta. 2014;1840:906–912. doi: 10.1016/j.bbagen.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Qu D., Rashidian J., Mount M.P., Aleyasin H., Parsanejad M., Lira A., Haque E., Zhang Y., Callaghan S., Daigle M., et al. Role of Cdk5-mediated phosphorylation of Prx2 in MPTP toxicity and Parkinson’s disease. Neuron. 2007;55:37–52. doi: 10.1016/j.neuron.2007.05.033. [DOI] [PubMed] [Google Scholar]
- Randall L.M., Manta B., Hugo M., Gil M., Batthyany C., Trujillo M., Poole L.B., Denicola A. Nitration transforms a sensitive peroxiredoxin 2 into a more active and robust peroxidase. J. Biol. Chem. 2014;289:15536–15543. doi: 10.1074/jbc.M113.539213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee S.G., Woo H.A. Multiple functions of peroxiredoxins: peroxidases, sensors and regulators of the intracellular messenger H(2)O(2), and protein chaperones. Antioxid. Redox Signal. 2011;15:781–794. doi: 10.1089/ars.2010.3393. [DOI] [PubMed] [Google Scholar]
- Rhee S.G., Woo H.A., Kil I.S., Bae S.H. Peroxiredoxin Functions as a Peroxidase and a Regulator and Sensor of Local Peroxides. J. Biol. Chem. 2012;287:4403–4410. doi: 10.1074/jbc.R111.283432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed A.A., Williams D.L. Biochemical characterization of 2-Cys peroxiredoxins from Schistosoma mansoni. J. Biol. Chem. 2004;279:26159–26166. doi: 10.1074/jbc.M401748200. [DOI] [PubMed] [Google Scholar]
- Schwarzlander M., Dick T.P., Meyer A.J., Morgan B. Dissecting redox biology using fluorescent protein sensors. Antioxid. Redox Signal. 2015 doi: 10.1089/ars.2015.6266. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Seidel T., Seefeldt B., Sauer M., Dietz K.J. In vivo analysis of the 2-Cys peroxiredoxin oligomeric state by two–step FRET. J. Biotechnol. 2010;149:272–279. doi: 10.1016/j.jbiotec.2010.06.016. [DOI] [PubMed] [Google Scholar]
- Seo J.H., Lim J.C., Lee D.Y., Kim K.S., Piszczek G., Nam H.W., Kim Y.S., Ahn T., Yun C.H., Kim K., et al. Novel protective mechanism against irreversible hyperoxidation of peroxiredoxin: Nalpha-terminal acetylation of human peroxiredoxin II. J. Biol. Chem. 2009;284:13455–13465. doi: 10.1074/jbc.M900641200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobotta M.C., Barata A.G., Schmidt U., Mueller S., Millonig G., Dick T.P. Exposing cells to H2O2: a quantitative comparison between continuous low-dose and one-time high-dose treatments. Free Rad. Biol. Med. 2013;60:325–335. doi: 10.1016/j.freeradbiomed.2013.02.017. [DOI] [PubMed] [Google Scholar]
- Sobotta M.C., Liou W., Stocker S., Talwar D., Oehler M., Ruppert T., Scharf A.N., Dick T.P. Peroxiredoxin-2 and STAT3 form a redox relay for H2O2 signaling. Nat. Chem. Biol. 2015;11:64–70. doi: 10.1038/nchembio.1695. [DOI] [PubMed] [Google Scholar]
- Tarrago L., Peterfi Z., Lee B.C., Michel T., Gladyshev V.N. Monitoring methionine sulfoxide with stereospecific mechanism-based fluorescent sensors. Nat. Chem. Biol. 2015;11:332–338. doi: 10.1038/nchembio.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira F., Castro H., Cruz T., Tse E., Koldewey P., Southworth D.R., Tomas A.M., Jakob U. Mitochondrial peroxiredoxin functions as crucial chaperone reservoir in Leishmania infantum. Proc. Natl. Acad. Sci. USA. 2015;112:E616–624. doi: 10.1073/pnas.1419682112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo M., Clippe A., Manta B., Ferrer-Sueta G., Smeets A., Declercq J.P., Knoops B., Radi R. Pre-steady state kinetic characterization of human peroxiredoxin 5: taking advantage of Trp84 fluorescence increase upon oxidation. Arch. Biochem. Biophys. 2007;467:95–106. doi: 10.1016/j.abb.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Wang X., Wang L.K., Wang X., Sun F., Wang C.C. Structural insights into the peroxidase activity and inactivation of human peroxiredoxin 4. Biochem. J. 2012;441:113–118. doi: 10.1042/BJ20110380. [DOI] [PubMed] [Google Scholar]
- Winterbourn C.C. The biological chemistry of hydrogen peroxide. Methods Enzymol. 2013;528:3–25. doi: 10.1016/B978-0-12-405881-1.00001-X. [DOI] [PubMed] [Google Scholar]
- Wood Z.A., Poole L.B., Karplus P.A. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 2003;300:650–653. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Araki S., Wu J., Teramoto T., Chang Y.F., Nakano M., Abdelfattah A.S., Fujiwara M., Ishihara T., Nagai T., et al. An expanded palette of genetically encoded Ca(2)(+) indicators. Science. 2011;333:1888–1891. doi: 10.1126/science.1208592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zykova T.A., Zhu F., Vakorina T.I., Zhang J., Higgins L.A., Urusova D.V., Bode A.M., Dong Z. T-LAK cell-originated protein kinase (TOPK) phosphorylation of Prx1 at Ser-32 prevents UVB-induced apoptosis in RPMI7951 melanoma cells through the regulation of Prx1 peroxidase activity. J. Biol. Chem. 2010;285:29138–29146. doi: 10.1074/jbc.M110.135905. [DOI] [PMC free article] [PubMed] [Google Scholar]