Abstract
Peroxiredoxins are cysteine-dependent peroxide reductases that group into 6 different, structurally discernable classes. In 2011, our research team reported the application of a bioinformatic approach called active site profiling to extract active site-proximal sequence segments from the 29 distinct, structurally-characterized peroxiredoxins available at the time. These extracted sequences were then used to create unique profiles for the six groups which were subsequently used to search GenBank(nr), allowing identification of ∼3500 peroxiredoxin sequences and their respective subgroups. Summarized in this minireview are the features and phylogenetic distributions of each of these peroxiredoxin subgroups; an example is also provided illustrating the use of the web accessible, searchable database known as PREX to identify subfamily-specific peroxiredoxin sequences for the organism Vitis vinifera (grape).
Keywords: active site profiling, bioinformatics, disulfide reductase, peroxide reductase, thiol peroxidase
INTRODUCTION
Although they were previously either unknown or underappreciated, peroxiredoxins (Prxs) have risen through the ranks in the past decade and a half to become recognized as arguably the most important peroxide scavengers, alongside glutathione peroxidases, in many biological systems and conditions (Adimora et al., 2010; Parsonage et al., 2005; Winterbourn, 2008). Their highly conserved active site contains an essential Cys, surrounded by absolutely conserved Pro and Arg residues, as well as a highly conserved Thr (replaced only by Ser in a low percent of Prxs). These residues are exquisitely arranged to activate the bound peroxide and catalyze O-O bond scission, while promoting attack of the Cys thiolate on the terminal hydroxyl of the substrate (Ferrer-Sueta et al., 2011; Hall et al., 2010; 2011). Thus this Pxxx(T/S)xxC, with the conserved Arg contributed by a different part of the sequence, is an essential feature of the active sites of Prxs.
With our rapidly expanding knowledge of genomic sequences from an ever-increasing number of organisms, we are poised to learn much more about the evolution and determinants of structure and function in families and superfamilies of proteins like Prxs, but the proper annotation of new sequences being added to the databases daily is lagging far behind and is largely dependent on automated processes for functionally annotating new sequences as they become available. Unfortunately, such processes can be inherently error prone, and “overannotation” wherein annotations are transferred from one sequence or group of sequences to another without sufficient supporting evidence is a huge problem (Leuthaeuser et al., 2015; Schnoes et al., 2009). As elaborated in in 2011 by Nelson et al., Prxs have not been plagued as much by over-annotation or misannotation, as only a small percentage of Prx sequences have been assigned to either the incorrect Prx subfamily or annotated with another incorrect, specific function. Rather, the most common problems for the Prx subfamily arise from a lack of annotation, with many sequences identified as either hypothetical or unknown proteins or identified in very vague terms such as peroxidase or antioxidant proteins. While bioinformatics approaches have been able to identify and tease apart distinct subgroups of Prxs based on sequences and/or structural features of these proteins (Copley et al., 2004; Hall et al., 2011; Knoops et al., 2007; Nelson et al., 2011), this level of Prx annotation is frequently absent in the sequence databases and many names such as thioredoxin peroxidase, atypical 2-Cys Prx, and 1-Cys Prx can refer to multiple proteins belonging to different Prx subfamilies. Thus, our group embarked on a project to establish a bioinformatics approach that would provide reliable identification and classification of Prxs centering on sequence information most critical to their function, and in 2011 we reported the implementation of this approach and the information gained by its use (Nelson et al., 2011). In addition, we prepared a database of the results from these analyses and provided web-accessible search capabilities to aid in extracting the information gathered in this manner for use by Prx researchers worldwide (Soito et al., 2011). The approaches taken and a sampling of results that can be obtained using these latter tools, as well as the characteristics associated with the different classes of Prxs thus identified, are summarized in this minireview.
DASP, A BIOINFORMATICS TOOL FOR EXTRACTING FUNCTIONALLY-RELEVANT SEQUENCE INFORMATION
Pure sequence-based analyses using full sequence, particularly when conducted iteratively, provide considerable information about conserved domains, residues and, in favorable cases, functions, but this latter extrapolation is often a “leap” in the absence of structural and/or functional characterization of a very similar orthologous protein. One approach to begin honing in on the most important regions of sequence, known as active site profiling (using the “Deacon Active Site Profiler” tool, or DASP), has been designed to enable extraction of functionally “rich” information from databases beginning with structural information for a group (e.g. family or superfamily) of functionally-characterized proteins and some knowledge of their key residues around a function site (typically the active site) (Cammer et al., 2003). DASP requires the manual selection of “key residues” that define a functional site within a structurally characterized protein (typically the active site); for example, key residues used for peroxiredoxins included the three important residues in the PxxxT/SxxC motif and a conserved active-site Trp /Phe residue (Trp81 in Salmonella typhimurium AhpC). DASP then extracts the sequence fragments within 10 Å of the key residues (Fig. 1). These sequence motifs are concatenated to form an Active Site Signature which can be aligned with those of the other members of the family or class to form a subfamily-specific active site profile (ASP). A position specific scoring matrix created from this alignment can then be used to search sequence databases to identify new members (Fig. 1). Output from a DASP search includes a prioritized list of the best matches from a sequence database like GenBank(nr), clarifying relationships between protein sequences while taking advantage of the ever-deeper pool of sequence information available.
Fig. 1.
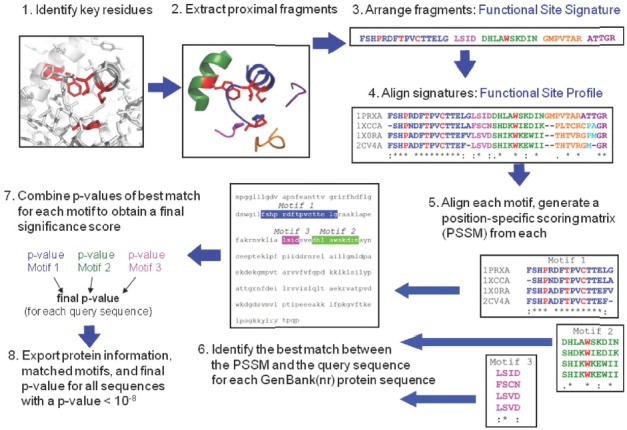
Identification of peroxiredoxin sequences using the Deacon Active Site Profiling (DASP) tool. (1) The active site of human Prx6 (PDB identifier 1prx) is shown with the four key residues highlighted in red. (2) Structural segments located within 10 Å of the center of geometry of the key catalytic residues are identified (each segment shown in a different color) and extracted from the global structure. (3) The sequence fragments are then combined to form a functional site signature (residue colors correspond to the color of structure segments in 2; key residues are highlighted in red). (4) Functional-site signatures for structurally characterized members of the Prx6 subfamily are aligned using ClustalW (Larkin et al., 2007; Thompson et al., 1994) to create a functional site profile. (5) Motifs are identified within any fragments that contain at least three residues and position specific scoring matrices (PSSM) (Bailey and Gribskov, 1998b) are created for each motif. (6) For each sequence in a user-selected sequence database, the PSSM for each motif is used to find and score the segment within a query sequence which best matches the motif. (7) Each time a motif is matched to a position in the protein sequence, a p-value is calculated that represents the probability of finding a match as good as the observed match within a random sequence. The p-values for all motifs in a single sequence are then combined using QFAST to obtain the final statistical significance score (final p-value) (Bailey and Gribskov, 1998a). (8) The protein information (including accession numbers, annotations, and species), final p-value, and sequence fragments matched to each queried motif are exported for all sequences with a final p-value more significant than a user-selected p-value. This figure was adapted from Soito et al. (2011).
BIOINFORMATIC ANALYSES OF PEROXIREDOXINS
As alluded to above, in 2011, our group reported the application of this structure-centered sequence analysis approach to Prxs, a ubiquitous family or superfamily of proteins representing discrete subgroups or isoforms (based on a set of structural and functional features described in more detail below) (Hall et al., 2011; Nelson et al., 2011). We extracted the active site signatures for all Prx proteins in the PDB database and then, based on structural and biochemical analysis assigned them to one of six subgroups of Prxs designated Prx1/AhpC (abbreviated as Prx1), Prx5, Prx6, Tpx, BCP/PrxQ (abbreviated PrxQ), and AhpE (named for canonical representatives of each) based on previously published structural characterizations (Hall et al., 2011). Subfamily-specific active site profiles were then used to identify 3,516 Prx sequences from the January 2008 Gen-Bank database that could be unambiguously assigned to one of the six subgroups. This allowed for subsequent detailed analysis of several characteristics within and between each Prx subfamily, including (a) species distribution, (b) resolving cysteine location and prevalence, and (c) residue conservation at each position within the Prx active site [see Fig. 3 in (Nelson et al., 2011)].
Fig. 3.

Sequence alignments of V. vinifera Prxs of interest generated by PREX (A) or by multiple sequence alignments of all six true Prxs (B). (A) shows two of the automatically-generated sequence alignments, accessed by clicking on an individual sequence signature in the output from a PREX search such as that depicted in Fig. 2B. Note that the PREX query sequence (the signature that was selected) is shown aligned with several other members of the same subgroup as well as one representative from each of the other 5 subgroups. (B) depicts a section around the conserved active site sequence of the multiple sequence alignment of all six bona fide Prxs from V. vinifera, listed by their uniprot designation and aligned using Clustal W (Larkin et al., 2007; Thompson et al., 1994).
Recognizing the huge value in providing such subgroup specific information to the community of thiol peroxidase researchers, as well as those not as familiar with the Prxs, we set up a web-accessible, searchable database (http://csb.wfu.edu/PREX) that allows the user to quickly and easily find the subfamily assignment for any Prx protein identified utilizing the methods presented in the associated 2011 paper (Nelson et al., 2011; Soito et al., 2011). We also provide distinguishing features and typical characteristics for each of the six Prx subfamilies. Since publication, the database has been further updated to take advantage of newly deposited Prx structures in forming the active site profiles (this was particularly important for the PrxQ subfamily) and to account for changes in the November 2010 and October 2011 versions of GenBank(nr) database. The current version of PREX provides 8108 validated Prx sequences, each of which have been assigned to a single Prx subfamily.
PREX allows researchers worldwide to search for Prxs within any genus or species and provides a clearer understanding of the number and types of Prxs present in a particular organism, particularly where nomenclature is confusing. For example, plants have a variety of Prxs in groups Prx1, Prx5, Prx6 and PrxQ, and often multiple examples of each of these (in part due to the multiple organelles including chloroplasts with specialized photosynthetic functions). Historically, a variety of names have been coined to describe some of these classes (e.g. PrxQ, PrxIIA-E, PrxD, etc.), but the connections between these and the specific structural and functional subfamily is not obvious. As an example, we searched for “peroxiredoxin” proteins in “Vitis vinifera” (grape) using text-based searching in both Uniprot (http://uniprot.org) and PREX (http://csb.wfu.edu/PREX) databases (although BLAST analyses using protein sequences as the query are available at both websites as well). The Uniprot output demonstrates the confusing nature of the annotations for the returned sequences, whereas the PREX output provides clarity in this regard (Table 1 and Fig. 2). Moreover, the output from PREX includes the actual signature motifs extracted from the database, which helps clarify which of the sequences may be redundant. For a given sequence signature, PREX generates an automated sequence alignment of the query sequence along with a few other representatives of that subgroup, plus a single canonical member of each of the other (non-query) subgroups (Fig. 3). Looking deeper into the Uniprot output, we find that 3 out of the 9 sequences returned are not Prxs at all, even though they are annotated as such (Table 1). Two of these bear no identifiable Prx-like regions at all (D7TQA7 and A5BWD1), and the third (G1JT82) is similar within regions flanking the expected PxxxTxxC motif, but apparently lacks this essential active site region altogether (perhaps due to either the expression of a splice variant or to one or more mistakes in the sequence analysis). Thus, the annotation provided in the PREX output is more reliable and gives clarity with respect to the subfamily to which each predicted protein belongs. A more detailed and comprehensive analysis of the accuracy and specificity of this method can be found in (Hall et al., 2011; Nelson et al., 2011).
Table 1.
Peroxiredoxins identified from Vitis vinifera using text-based searches of Uniprot and PREX databases.
| Uniprot Entry | Protein name (Uniprot) | Gene name | Length | Subfamily (PREX) | DASP Signature from PREXa |
|---|---|---|---|---|---|
| D7TBK8 | Peroxiredoxin | VITISV_023716 | 162 | Prx5(a) | FGVPGAFTPTCSVKHVPlvsvnVMKAWAK-TYPDlgtrsrrfEAGGE |
| D7TCA6 | Peroxiredoxin Q | VIT_11s0016g00560 | 214 | PrxQ | YFYPADETPGCTKQACgisgSHKAFAKKYyvldkkg |
| D7T674 | 1-Cys peroxiredoxin | VIT_05s0020g00600 | 219 | Prx6 | FSHPGDFTPVCTTELlscdQSHKEWIKDIEastgr |
| D7T6T0 | Type II peroxiredoxin F | VIT_05s0020g02850 | 201 | Prx5(b) | gtdlvFGLPGAYTGVCSAQHVPcvavnTLNAWAEKLEAlg prshrw |
| D7TQA7 | Type II peroxiredoxin 2 | VIT_08s0040g03130 | 254 | Not a Prx | - |
| G1JT83 | 2-Cys peroxiredoxin | VITISV_025619 | 274 | Prx1 | FFYPLDFTFVCPTEvsidSH-LAWVQTDRsgglgQGVALRGsmk |
| A5BWD1 | Type II peroxiredoxin 1 | VITISV_040398 | 256 | Not a Prx | - |
| G1JT87 | Type II peroxiredoxin E | VITISV_042154 | 212 | Prx5(c) | FAVPGAFTPTCSQKHLPcisvnVMKAWKADL-KIlgvrsrryEEGGA |
| G1JT82 | 1-Cys peroxiredoxin 03 | 183 | Missing PxxxTxxC | - |
Lower and upper case letters are used to distinguish different motifs within the PREX sequence signatures. The essential active site motif of Prxs is underlined in each active site signature.
Fig. 2.
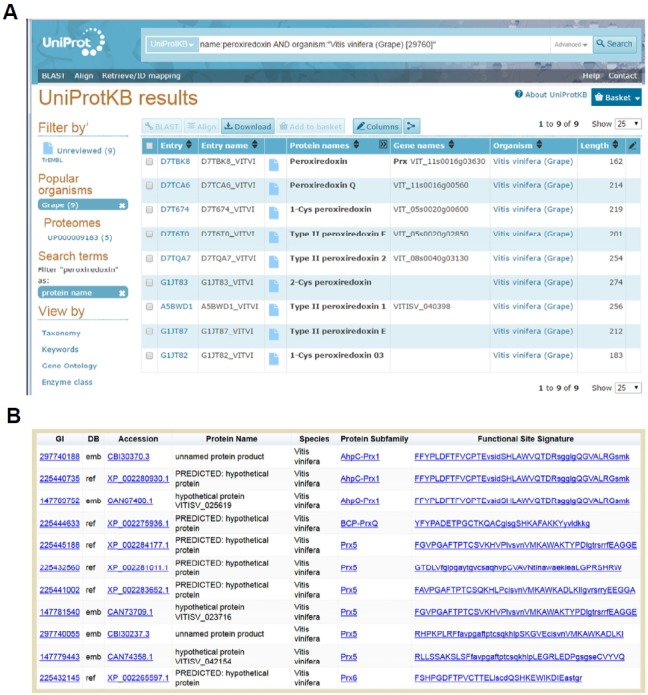
Output from two web tools used to search for peroxiredoxins present in Vitis vinifera (grape). (A) shows the output from a search for “peroxiredoxin” and the organism name at http://uniprot.org. (B) shows the output from a parallel search conducted using the PREX database at http://csb.wfu.edu/PREX.
SPECIES DISTRIBUTION OF PRX CLASSES
With accurate and abundant sequence information for each Prx subfamily, we can learn a great deal about how different Prx subfamilies are distributed across species. As illustrated in Fig. 4, the subclasses are not uniformly distributed; two of the groups, Tpx and AhpE, are found nearly exclusively in eubacteria (with the single exceptions postulated to be caused by lateral gene transfer) (Nelson et al., 2011). Notably, all six classes show up in eubacteria, whereas four of the subfamilies, PrxQ, Prx1, Prx5 and Prx6, are found in fungi and plants. There are no Prx5 orthologues in archaea, and no PrxQ representatives are found in animals, although they are well represented in plants. PrxQ, which is somewhat heterogeneous compared to other groups, has been described as the most representative of the ancestral Prx (Hall et al., 2011). In this regard, it is striking that complex metazoans have dispensed with this class of Prx which, at least in plants, presumably became more specialized to function within the unique redox environment of the chloroplast (Dietz, 2011).
Fig. 4.
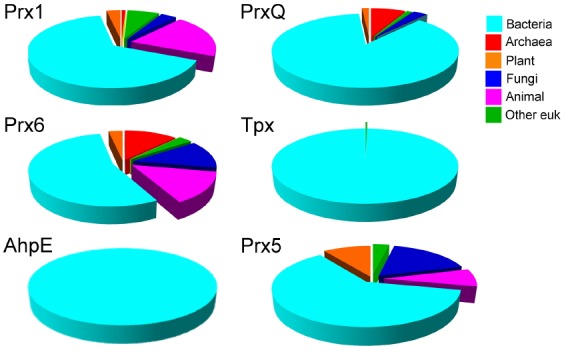
Phylogenetic distribution of individual Prx groups across all species. The organism name for each Prx sequence was first extracted from the DASP output file and the complete lineage of each organism was obtained from the NCBI Taxonomy databases. This information was used to calculate the fraction of sequences within each subfamily that belong to the indicated biological subdivision. Each species was only counted once in each subfamily even if multiple protein sequences were identified. To prevent results being biased by oversampling of sequences from multiple bacterial strains, multiple strains of the same species were only counted once for each subfamily. The prevalence of cyan in the figure reflects the huge number of bacterial species compared to archaeal and eukaryotic species.
FEATURES OF DISTINCT PRX CLASSES
What is it that is particularly distinct about these different groups of Prxs? When Prxs were first identified as a widespread group of antioxidant proteins in the early 1990’s, they were recognized to possess a single, absolutely conserved Cys residue (Chae et al., 1994a). It was also realized that a second catalytically important Cys residue near the C-terminus was present in some but not all of the Prxs, leading to the designation of 1-Cys and 2-Cys Prxs. As the mechanistic details of these and additional Prxs were explored over the next decade, it became clear that the single conserved Cys bears the sulfur that attacks the peroxide substrate (and is therefore called the peroxidatic Cys, or CP), generating a Cys sulfenic acid (R-SOH) in the process (Chae et al., 1994b; Ellis and Poole, 1997). The second Cys residue then “resolves” the nascent sulfenic acid, forming a disulfide bond, and is referred to as the resolving Cys (CR); in 1-Cys Prx proteins, this role is fulfilled by a small molecule thiol or a cysteine residue arising from another protein. The first Prx proteins characterized were Prx1 group members, which have since been shown to almost exclusively generate an intersubunit disulfide bond with the CR being located in the C-terminus of a partner subunit (a “typical 2-Cys” Prx). As more Prxs were identified and characterized, it became clear that Cys residues located in other parts of the Prx protein could also serve as resolving cysteines; these proteins were grouped together under the designation of “atypical 2-Cys Prxs”. To date, we recognize five positions within the Prx fold in which resolving Cys residues can reside (Fig. 5); those in the extreme N- or C-termini make intersubunit disulfide bonds, whereas those present in α2, α3 or α5 helices form intrasubunit disulfide bonds with the peroxidatic Cys. In early examples of Prxs, it appeared that the absence or location of the resolving Cys might be a defining feature of different subclasses of Prxs. However, with the abundance of subfamily-specific sequences, we now know that this view is too simplistic. In fact, there are examples of Prxs lacking a resolving Cys in all six subfamilies, although such proteins are most prevalent in the Prx6 and Prx5 subfamilies (Nelson et al., 2011). In addition, only about two thirds of the members in the PrxQ group appear to possess resolving Cys residues (Perkins et al., 2015). For the AhpE group, there are too few representatives to draw many conclusions yet, however members of this subfamily include both 1-Cys proteins and those with a CR in helix α5. Members of the bacterial Tpx group, like those in the Prx1 group, are more homogeneous in this respect, with > 95% having their resolving Cys in helix α3. Presence of a Cys at this position is not, however, diagnostic of a Tpx group member; ∼6% of PrxQ group members also have a resolving Cys in this position while another ∼60% have their resolving Cys located in the same helix (α2) as the peroxidatic Cys (Perkins et al., 2015). This diversity of locations implies that the resolving Cys has arisen multiple times in evolution, even within a given subfamily.
Fig. 5.
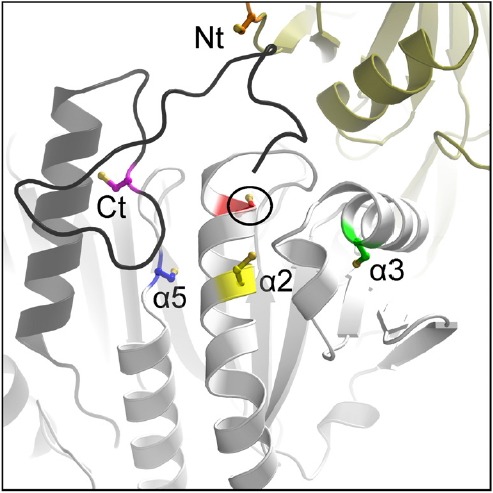
Variable locations of the resolving Cys (CR). Shown are the various positions of the peroxiredoxin CR (colored sidechains) in relation to the active site peroxidatic Cys (CP, circled and in red). Intramolecular CP-CR disulfides are formed for the α2 (yellow), α3 (green), and α5 (blue) types, and intermolecular disulfides are formed for the N-terminal (Nt, orange CR in the gold chain) and C-terminal (Ct, magenta CR in the black chain) types. (CR residues are mapped onto a composite structure based on S. typhimurium AhpC, Protein Databank Identifier 4MA9). Reproduced with permission from (Perkins et al., 2015).
Structural distinctions between subfamily members are also notable, some of which can be identified from sequences alone. For example, the α2 helix of Prx 5 proteins has an amino acid insertion that creates a characteristic “bulge” (or pi helix) (Perkins et al., 2015). Additionally, Prx1 and Prx6 group members have an extended C-terminus relative to the other groups. Studies of Prx1 group member interactions with other proteins have uncovered a significant role for the extended C-terminal tail in these proteins for promoting interactions with important redox partners. For eukaryotic Prx1 proteins which can be oxidatively damaged by high peroxide levels, repair of the hyperoxidized Cys at the active site is mediated by the protein sulfiredoxin (Srx) which interacts with the Prx “client” through an “embrace” during which the C-terminal tail of one monomer wraps around the Srx protein catalyzing repair of the other subunit of the dimer (Jönsson et al., 2008). In recent studies detailing interactions of a bacterial Prx known as AhpC with its electron-donating partner, AhpF, the extended C-terminus of AhpC from E. coli was also found to play an important role in mediating the protein-protein interactions needed for electron transfer between these proteins (Dip et al., 2014).
In contrast, a major structural feature distinguishing certain Prx subgroups from others, their oligomeric interface(s), is not particularly evident from their sequences alone. While some PrxQ members are monomeric, nearly all Prxs form homodimers (Perkins et al., 2015). Prx1, Prx6 and AhpE dimers are formed through interactions at the edges of their central β-sheets (called the B interface), whereas Prx5, PrxQ and Tpx dimers form through interactions at an alternate or “backside” interface (called the A interface) (Fig. 6). Using the A interface, Prx1 and Prx6 dimers can come together to form (α2)5 decamers (or occasionally dodecamers comprised of six dimers). Interestingly, in some representative Prx1 proteins, interactions at the A interface are redox sensitive, with oxidation to form disulfide-linked dimers promoting dissociation (Barranco-Medina et al., 2008; Wood et al., 2002). The biological significance of this redox-linked change in oligomeric state is as yet unclear.
Fig. 6.
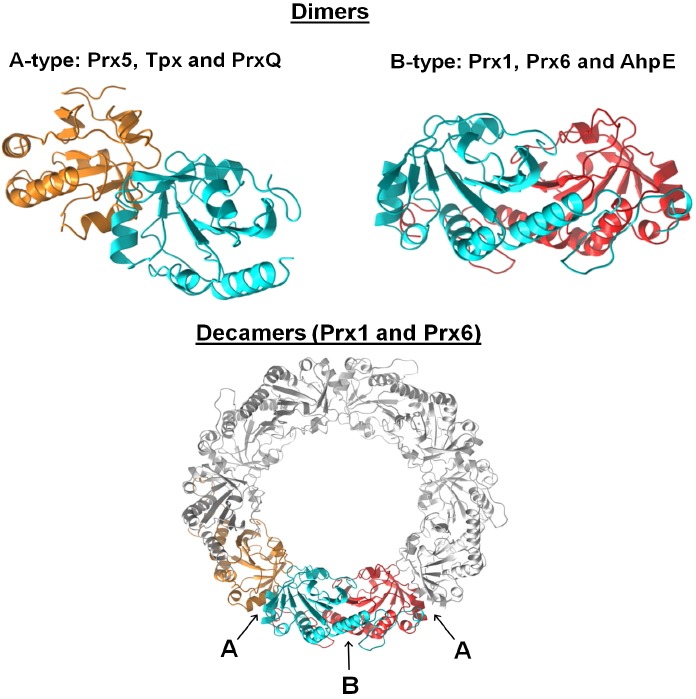
Dimeric interfaces and quaternary structures of Prxs. Homodimeric complexes are formed using either an A-type interface, where the monomers interact near helix α3, or B-type dimers where the interaction is through the β-strands, generating an extended 10–14 strand β-sheet. Further interactions at the A-interfaces of some Prx1 and Prx6 members generate (α2)5 decamers [or in rare cases (α2)6 dodecamers]. The blue subunit is displayed at approximately the same orientation in each of the structures to illustrate these interaction interfaces that together build the decamer. For a number of Prx1 members, the structural change upon disulfide bond formation destabilizes the A-type dimer interface, and the decamer dissociates to B-type dimers. The structures depicted are: Aeropyrum pernix PrxQ (A-type dimer, Protein Data Bank Identifier 4GQF), and wild type S. typhimurium AhpC (B-type dimer and decamer, Protein Data Bank Identifier 4MA9). Adapted from (Perkins et al., 2015).
Kinetic distinctions between classes are less clear as much remains to be discovered about how a wide range of Prxs function in terms of their substrate specificities and steady state and rapid reaction kinetic profiles. One obvious distinction in some cases is the reductant used to reduce the disulfide bond in oxidized Prxs and recycle the catalytic cysteine. Thioredoxins often function as the primary Prx reductant, although glutaredoxins are increasingly recognized as serving in this role for an array of Prxs. In addition, some Prxs have evolved significant specificity for their electron donors. Two excellent examples of this are from the Prx1 group. The first example includes the AhpC proteins from E. coli and Salmonella typhimurium which are reduced specifically by the specialized flavoprotein AhpF, mentioned above. Notably, the structural gene encoding AhpF is nearly always found within several hundred base pairs downstream of that encoding AhpC, and these AhpC-AhpF pairs are found in both Gram negative and Gram positive bacteria. Another case exhibiting specialization toward reductant is tryparedoxin peroxidase (TryP), another Prx1 group member, which is recycled in vivo by the thioredoxin-like reductant known as tryparedoxin. Tryparedoxins contain a CXXC motif and are found exclusively in kinetoplastids where they serve a similar role to thioredoxin and reduce tryparedoxin peroxidase, ribonucleotide reductase, and other regulatory proteins (Flohé, 2010). These organisms have unique redox systems in that they do not rely on glutathione as their prominent low molecular weight thiol, but rather a conjugated form of glutathione with two of the tri-peptides connected by a spermidine linker (Poole, 2015). This thiol, known as trypanothione, receives its electrons from NADPH via another specialized flavoprotein, trypanothione reductase, and is the source of electrons for recycling tryparedoxin, and thus TryP.
In conclusion, the ability to readily determine the subfamily to which a given Prx belongs is important in providing a more complete understanding of that protein’s biochemical and structural features. It is therefore important to continue to develop improved bioinformatic tools and annotations in our sequence databases to support continued work on thiol peroxidases and other important enzyme families.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (R01 GM050389). The authors are grateful for the assistance and guidance of Jacquelyn Fetrow and Stacy Knutson, as well as the extensive effort by Laura Soito to set up the PREX database and implement updates from the GenBank(nr), and by Arden Perkins to generate Figs. 5 and 6.
REFERENCES
- Adimora N.J., Jones D.P., Kemp M.L. A model of redox kinetics implicates the thiol proteome in cellular hydrogen peroxide responses. Antioxid. Redox Signal. 2010;13:731–743. doi: 10.1089/ars.2009.2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L., Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998a;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- Bailey T.L., Gribskov M. Methods and statistics for combining motif match scores. J. Comput. Biol. 1998b;5:211–221. doi: 10.1089/cmb.1998.5.211. [DOI] [PubMed] [Google Scholar]
- Barranco-Medina S., Kakorin S., Lazaro J.J., Dietz K.J. Thermodynamics of the dimer-decamer transition of reduced human and plant 2-cys peroxiredoxin. Biochemistry. 2008;47:7196–7204. doi: 10.1021/bi8002956. [DOI] [PubMed] [Google Scholar]
- Cammer S.A., Hoffman B.T., Speir J.A., Canady M.A., Nelson M.R., Knutson S., Gallina M., Baxter S.M., Fetrow J.S. Structure-based active site profiles for genome analysis and functional family subclassification. J. Mol. Biol. 2003;334:387–401. doi: 10.1016/j.jmb.2003.09.062. [DOI] [PubMed] [Google Scholar]
- Chae H.Z., Robison K., Poole L.B., Church G., Storz G., Rhee S.G. Cloning and sequencing of thiol-specific antioxidant from mammalian brain: alkyl hydroperoxide reductase and thiol-specific antioxidant define a large family of antioxidant enzymes. Proc. Natl. Acad. Sci. USA. 1994a;91:7017–7021. doi: 10.1073/pnas.91.15.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae H.Z., Uhm T.B., Rhee S.G. Dimerization of thiol-specific antioxidant and the essential role of cysteine 47. Proc. Natl. Acad. Sci. USA. 1994b;91:7022–7026. doi: 10.1073/pnas.91.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copley S.D., Novak W.R., Babbitt P.C. Divergence of function in the thioredoxin fold suprafamily: evidence for evolution of peroxiredoxins from a thioredoxin-like ancestor. Biochemistry. 2004;43:13981–13995. doi: 10.1021/bi048947r. [DOI] [PubMed] [Google Scholar]
- Dietz K.J. Peroxiredoxins in plants and cyanobacteria. Antioxid. Redox Signal. 2011;15:1129–1159. doi: 10.1089/ars.2010.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dip P.V., Kamariah N., Nartey W., Beushausen C., Kostyuchenko V.A., Ng T.S., Lok S.M., Saw W.G., Eisenhaber F., Eisenhaber B., et al. Key roles of the Escherichia coli AhpC C-terminus in assembly and catalysis of alkylhydroperoxide reductase, an enzyme essential for the alleviation of oxidative stress. Biochim. Biophys. Acta. 2014;1837:1932–1943. doi: 10.1016/j.bbabio.2014.08.007. [DOI] [PubMed] [Google Scholar]
- Ellis H.R., Poole L.B. Roles for the two cysteine residues of AhpC in catalysis of peroxide reduction by alkyl hydroperoxide reductase from Salmonella typhimurium. Biochemistry. 1997;36:13349–13356. doi: 10.1021/bi9713658. [DOI] [PubMed] [Google Scholar]
- Ferrer-Sueta G., Manta B., Botti H., Radi R., Trujillo M., Denicola A. Factors affecting protein thiol reactivity and specificity in peroxide reduction. Chem. Res. Toxicol. 2011;24:434–450. doi: 10.1021/tx100413v. [DOI] [PubMed] [Google Scholar]
- Flohé L. Changing paradigms in thiology from antioxidant defense toward redox regulation. Methods Enzymol. 2010;473:1–39. doi: 10.1016/S0076-6879(10)73001-9. [DOI] [PubMed] [Google Scholar]
- Hall A., Parsonage D., Poole L.B., Karplus P.A. Structural evidence that peroxiredoxin catalytic power is based on transition-state stabilization. J. Mol. Biol. 2010;402:194–209. doi: 10.1016/j.jmb.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A., Nelson K., Poole L.B., Karplus P.A. Structure-based insights into the catalytic power and conformational dexterity of peroxiredoxins. Antioxid. Redox Signal. 2011;15:795–815. doi: 10.1089/ars.2010.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jönsson T.J., Johnson L.C., Lowther W.T. Structure of the sulphiredoxin-peroxiredoxin complex reveals an essential repair embrace. Nature. 2008;451:98–101. doi: 10.1038/nature06415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops B., Loumaye E., Van der Eecken V. Evolution of the peroxiredoxins: taxonomy, homology and characterization. In: Flohé L., Harris J.R., editors. In peroxiredoxin systems. New York: Springer; 2007. pp. 27–40. [DOI] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Leuthaeuser J.B., Knutson S.T., Kumar K., Babbitt P.C., Fetrow J.S. Comparison of topological clustering within protein networks using edge metrics that evaluate full sequence, full structure, and active site microenvironment similarity. Protein Sci. 2015;24:1423–1439. doi: 10.1002/pro.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K.J., Knutson S.T., Soito L., Klomsiri C., Poole L.B., Fetrow J.S. Analysis of the peroxiredoxin family: Using active-site structure and sequence information for global classification and residue analysis. Proteins. 2011;79:947–964. doi: 10.1002/prot.22936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsonage D., Youngblood D.S., Sarma G.N., Wood Z.A., Karplus P.A., Poole L.B. Analysis of the link between enzymatic activity and oligomeric state in AhpC, a bacterial peroxiredoxin. Biochemistry. 2005;44:10583–10592. doi: 10.1021/bi050448i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins A., Nelson K.J., Parsonage D., Poole L.B., Karplus P.A. Peroxiredoxins: guardians against oxidative stress and modulators of peroxide signaling. Trends Biochem. Sci. 2015;40:435–445. doi: 10.1016/j.tibs.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole L.B. The basics of thiols and cysteines in redox biology and chemistry. Free Radic. Biol. Med. 2015;80:148–157. doi: 10.1016/j.freeradbiomed.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnoes A.M., Brown S.D., Dodevski I., Babbitt P.C. Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comput. Biol. 2009;5:e1000605. doi: 10.1371/journal.pcbi.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soito L., Williamson C., Knutson S.T., Fetrow J.S., Poole L.B., Nelson K.J. PREX: PeroxiRedoxin classification indEX, a database of subfamily assignments across the diverse peroxiredoxin family. Nucleic Acids Res. 2011;39:D332–337. doi: 10.1093/nar/gkq1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterbourn C.C. Reconciling the chemistry and biology of reactive oxygen species. Nat. Chem. Biol. 2008;4:278–286. doi: 10.1038/nchembio.85. [DOI] [PubMed] [Google Scholar]
- Wood Z.A., Poole L.B., Hantgan R.R., Karplus P.A. Dimers to doughnuts: redox-sensitive oligomerization of 2-cysteine peroxiredoxins. Biochemistry. 2002;41:5493–5504. doi: 10.1021/bi012173m. [DOI] [PubMed] [Google Scholar]


