Abstract
A challenge in the redox field is the elucidation of the molecular mechanisms, by which H2O2 mediates signal transduction in cells. This is relevant since redox pathways are disturbed in some pathologies. The transcription factor OxyR is the H2O2 sensor in bacteria, whereas Cys-based peroxidases are involved in the perception of this oxidant in eukaryotic cells. Three possible mechanisms may be involved in H2O2 signaling that are not mutually exclusive. In the simplest pathway, H2O2 signals through direct oxidation of the signaling protein, such as a phosphatase or a transcription factor. Although signaling proteins are frequently observed in the oxidized state in biological systems, in most cases their direct oxidation by H2O2 is too slow (101 M−1s−1 range) to outcompete Cys-based peroxidases and glutathione. In some particular cellular compartments (such as vicinity of NADPH oxidases), it is possible that a signaling protein faces extremely high H2O2 concentrations, making the direct oxidation feasible. Alternatively, high H2O2 levels can hyperoxidize peroxiredoxins leading to local building up of H2O2 that then could oxidize a signaling protein (floodgate hypothesis). In a second model, H2O2 oxidizes Cys-based peroxidases that then through thiol-disulfide reshuffling would transmit the oxidized equivalents to the signaling protein. The third model of signaling is centered on the reducing substrate of Cys-based peroxidases that in most cases is thioredoxin. Is this model, peroxiredoxins would signal by modulating the thioredoxin redox status. More kinetic data is required to allow the identification of the complex network of thiol switches.
Keywords: H2O2, Peroxiredoxin, signal transduction, thiol, thiol-disulfide exchange, thioredoxin
INTRODUCTION
H2O2, an endogenous oxidant present in all aerobic cells, was traditionally viewed as a toxic substance (Winterbourn, 2008). However, research during the last two decades built a compelling case supporting the role of H2O2 as a diffusible signaling messenger that controls many cellular processes by modulating the redox state of cysteine residues in proteins (Ying et al., 2007). Accordingly, disruption of signaling pathways mediated by H2O2 and other oxidants would be a mechanism underlying diseases (Jones, 2006).
H2O2 occurs in aerobic cells typically at steady state concentration in the 10 nM range; and NADPH oxidases and/or complex III of the mitochondrial respiratory chain are frequently considered as the major sources, whereas catalase and multiple Cys-based peroxidases exert a fine control of its levels (reviewed by Marinho et al., 2014; Sies, 2014). H2O2 are neutral molecules that can cross biological membranes, although its diffusivity can be modulated by the lipid composition of biological membranes (Branco et al., 2004; Pedroso et al., 2009) and aquaporins provide facilitated transport channels across bilayers (Miller et al., 2010).
A challenge in the field is the elucidation of the molecular mechanisms by which H2O2 is sensed and how this signal is transduced in cells. Furthermore, several chemical and biological aspects appear to be contradictory (Winterbourn, 2008). Elucidation of these aspects is very relevant since redox regulated proteins are implicated in diverse pathologies. For instance, redox medicine is emerging as an innovative field supported by molecular evidence showing deregulated redox signaling pathways dependent on H2O2 to be at the basis of several pathologies, including obesity (Kaszubska et al., 2002) and cardiovascular diseases (Schröder et al., 2012). Furthermore, peroxiredoxins (Prxs) and its partner proteins, thioredoxins (Trxs), have been associated to cancer (Irwin et al., 2013), neurodegenerative (Krapfenbauer et al., 2003), cardiovascular (Ahsan et al., 2009), and metabolic diseases (Abbasi et al., 2014). In principle, understanding the mechanisms of H2O2 sensing and signal transduction may provide the basis for therapeutic interventions in redox-related diseases (Harald et al., 2015; Jones, 2006).
MODELS OF H2O2 SENSING AND SIGNALING
Prxs are highly abundant Cys-based peroxidases with crucial roles in maintaining genome stability, protecting cells against cancer and promoting longevity (Nyström et al., 2012; Rhee and Woo, 2011). Prxs are subdivided into subfamilies that present very divergent amino acid sequences but uniformly possess at least one conserved cysteine residue at the active site, the so-called peroxidatic cysteine, CP (Nelson et al., 2011). Reaction of CP with H2O2 is very fast, with second-order rate constants ranging from 106 to 108 M−1s−1 (Ogusucu et al., 2007; Parsonage et al., 2005; Peskin et al., 2007; Toledo et al., 2011; Trujillo et al., 2007). This represents an extraordinary catalytic power, since free Cys reacts with hydroperoxides with a rate constant equivalent to 101 M−1s−1 (Winterbourn and Metodiewa, 1999).
Remarkably for a protein involved in signal transduction, Prx rapidly reacts with hydroperoxides, but not with other electrophiles such as iodoacetamides or chloroamines (Peskin et al., 2007). Specificity is a desirable feature for a sensor protein. High reactivity and specificity are related to the fact that Prxs stabilize the transition state of a SN2 type of nucleophilic substitution reaction through the activation of not only CP, but also H2O2 (Hall et al., 2011). Besides reactivity and specificity, Prxs are abundant proteins so they present several features appropriate for a sensor protein.
Aspects related to how the oxidation signal is transmitted to other proteins are less understood. It is well known that most Prxs transmit oxidizing equivalents to Trxs, especially in the case of typical 2-Cys Prx, which are Prxs with intermolecular disulfide bonds (in the context of this proposal, we refer to typical 2-Cys Prx as the proteins that belong to the AhpC/Prx1 subgroup, according to Nelson et al., 2011). For some typical 2-Cys Prx, dual function (moonlight property) was reported as thiol peroxidase and chaperone (holdase) (Jang et al., 2004; Rhee and Woo, 2011).
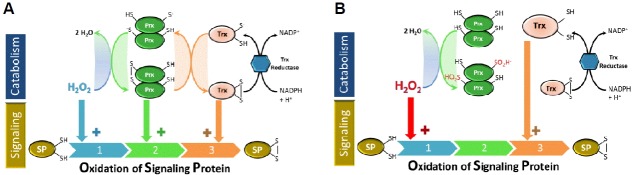
In principle, Prxs may mediate H2O2 signaling through three mechanisms that are not mutually exclusive (Fig. 1A) : (1, blue) H2O2 reacts directly with the target signaling protein (SP), and Prxs control this process by regulating H2O2 tone in cells; (2, green) H2O2 reacts with a Prx which is oxidized and then relays this oxidation to the target signaling protein (SP, for example a transcription factor or a phosphatase); and (3, orange) H2O2 reacts with a Prx which is oxidized, then this Prx is reduced back by its partner thioredoxin (Trx), and it is the oxidized Trx that relays the oxidation to the target SP. Some variations occur when Prx enzymes are hyperoxidized, as will be further described below. For instance, mechanism (1) also encompasses the so-called floodgate hypothesis (Rhee and Woo, 2011; Wood et al., 2003). In this case, when H2O2 accumulates at high levels, CP could be hyperoxidized to sulfinic or sulfonic states (Fig. 1B), allowing accumulation of H2O2, which could then attack other targets. Noteworthy, in this case levels of reduced Trx also increase. Adding more complexity to these mechanisms, it was found recently that glutaredoxin 1 (Grx 1) can also sustain the peroxidase activity of Prxs by reducing the glutathionylated form of Prx formed in the presence of H2O2 and glutathione (GSH) (Peskin et al. 2015). Such pathway may, in addition, prevent the hyperoxidation of Prxs.
Fig. 1.
Mechanisms of H2O2 sensing. In (A) the three mechanisms investigated in these proposal are shown: (1) direct oxidation of the signaling protein (SP) by H2O2; oxidation of SP is mediated by (2) a peroxiredoxin (Prx) or by (3) a thioredoxin (Trx). In (B) the overoxidation of a Prx is shown, which leads to H2O2 accumulation.
Concerning the mechanisms described in Fig. 1, there are evidences supporting them, and most probably all three mechanism operate for different signal transduction proteins and probably depend on several conditions such as second-order rate constants, pH, as well as concentration of oxidants, reductants, SP and Prx, among other factors. In principle, these three mechanistic models are not mutually exclusive, but obviously they should agree with chemical and biological data.
H2O2 SIGNALING THOURGH DIRECT OXIDATION OF SIGNALING PROTEINS (SP)
Reactivities of H2O2 towards SP, like the phosphatases Cdc25B, SHP-2 and protein tyrosine phosphatase 1B (PTB1B), are five to seven orders of magnitude lower than the reactivity towards Prxs or glutathione peroxidases (GPxs) (Winterbourn and Hampton, 2008). In addition, Prxs are much more abundant than SP like phosphatases or transcription factors. The consequence is that SP cannot outcompete with Prxs or GPxs for H2O2.
Noteworthy, other thiol proteins than Prxs and Gpxs can also react faster with H2O2 than ordinary Cys residues present in most SP. Two cases deserve special attention here: glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and OxyR. OxyR is a transcription factor with highly reactive Cys residues capable to react with H2O2 with a second order rate constant of 105 M−1 s−1 (Aslund et al., 1999; Lee et al., 2004). Upon oxidation of reactive Cys, OxyR conformation changes and activates transcription of target genes. Due to these properties, OxyR is considered the H2O2 sensor in bacteria (reviewed by Marinho et al., 2014). GAPDH can be reversible inhibited by H2O2 at more moderate rates (500 M−1 s−1) (Little and O’Brien, 1969; Winterbourn and Hampton, 2008; Peralta et al., 2015), still more reactive than free Cys; and GAPDH is highly abundant (∼240 μM) (Sneider, 2013). Furthermore, most cells contain high amounts of GSH that can also outcompete for H2O2 (Winterbourne and Hampton, 2008).
Therefore, there are multiple competitive routes that can detour H2O2 from the direct reaction toward ordinary thiols present in most SP. However, SP like PTP1B and PTEN, which display low reactivity towards H2O2 (Denu and Tanner, 1998), are found oxidized in biological tissues (Kwon et al., 2004; Lee et al., 1998; Mahadev et al., 2001; Meng et al., 2002). Therefore, attempts were made to reconcile both biological data (showing that Cys-based proteins are oxidized in cellular systems) and chemical information (low reactivity and low abundance of these Cys-based proteins). The so- called “floodgate hypothesis” is one of the most popular models (Rhee and Woo, 2011; Wood et al., 2003) that could be considered a variation to pathway 1, and it is schematically represented in (Fig. 1B, blue pathway). First observation that gave support to this hypothesis, was that typical 2-Cys Prx can be divided into sensitive and robust enzymes and that this property is related with a “gain of function” (Wood et al., 2003). Sensitive Prx are enzymes that are more easily hyperoxidized to sulfinic acids (CP-SO2H−), inactivating these peroxidases and allowing the accumulation of H2O2 in cells. At higher levels, H2O2 would be able to attack other targets that in basal conditions would be outcompeted by sensitive Prx. The build-up of H2O2 is possible because sulfinic acid is not reducible by standard reductants such as Trx and Grx.
The identification of an enzyme capable to specifically reduce typical 2-Cys Prx sulfinic acid at the CP also supported the signaling role of their oxidative inactivation (Biteau et al., 2003). Besides hyperoxidation of human Prx2, phosphorylation of Prx1 also leads to its inactivation (Rawat et al., 2013; Woo et al., 2010), proving an alternative pathway for H2O2 accumulation inside cells. Further experimental evidences for the pathway described in Fig. 1B are presented by Rhee et al. (2012). A problem with this hypothesis is that even in the absence of highly reactive peroxidases, cells still have high amounts of GSH (mM levels) and GAPDH that on chemical grounds can outcompete PTP-1B and PTEN for H2O2 (Winterbourne and Hampton, 2008).
Another attempt to reconcile the low reactivity of H2O2 with SP and redox signaling is based on an extended kinetic analysis that focused not only on the routes of H2O2 elimination but also on the transmission of the oxidation signal. The key notion is whether the fraction of H2O2 that escapes the action of Prxs, glutathione peroxidases and also GSH is sufficient to alter the oxidation state of low-reactive SP, thus transmitting the oxidation signal to these target proteins (Marinho et al., 2014). The H2O2 concentration needed to oxidized 50% of these targets within 5 minutes and in competition with all cellular H2O2-removing systems was estimated in the range 14 to 120 μM for Cdc25B and PTP1B, respectively (Marinho et al. 2014). These H2O2 concentrations are much higher than the sub-micromolar concentrations usually assumed to occur intracellularly, but they may be reached in the vicinity of SP during signaling dependent on the local activation of NADPH oxidases (Chen, 2008; Mishina, 2011; Paulsen, 2012). This analysis was extended by fitting PTP1B and SHP-2 oxidation profiles obtained experimentally to a mathematical model that included not only the oxidation component of redox signaling, but also the reductive part, in which target SPs are reduced back, switching-off the oxidation signal, thus taken into account all possible cellular biochemical reactions. The results obtained suggest that while for PTP1B the redox signaling is compatible with a direct reaction between H2O2 and PTP1B, for SHP-2 an alternative mechanism is more likely (Brito and Antunes, 2014). Thus, pathway 1 (Fig. 1, blue pathway) may operate in vivo under some conditions, namely under relative high local H2O2 concentrations, even if SP display low reactivity towards H2O2.
H2O2 SIGNALING THROUGH MESSENGER PROTEIN OXIDIZED BY PRX
Most of the knowledge related to pathway 2 (Fig. 1A, green pathway) was initially gained through studies on the activation mechanism of Yap1 transcription factors from Saccharomyces cerevisiae (Boronat et al., 2014). Like OxyR, the transcriptional activity of Yap1 is redox regulated. However, apparently Cys residues of Yap1 are not highly reactive and therefore Yap1 does not react directly with H2O2, but instead undergo oxidative activation through thiol-disulfide reshuffling mediated by a Cys-based peroxidase: glutathione peroxidase 3 (Gpx3), also called “Oxidant Receptor Protein” (Orp1) (Delaunay et al., 2002). In this signaling pathway, H2O2 oxidizes Gpx3/Orp1 into a sulfenic acid (Cys-SOH) that then condenses with a Cys residue of Yap1, giving rise to a transient, mixed disulfide between the two proteins. Through thiol – disulfide exchange reactions, an intramolecular disulfide is formed in Yap1. This oxidized form of Yap1 has an altered structure that cannot leave the nucleus. The nuclear accumulation of Yap1 facilitates its ability to induce the transcription of target genes, such as Prx and catalase genes (Delaunay et al., 2002). Ybp1 is a third protein involved in this pathway that physically interacts with Yap1 and thereby stimulates nuclear accumulation of this transcription factor in response to H2O2 (Veal et al., 2003). Interestingly, in strains where Ybp1 gene is mutated, Tsa1 (by far, the most abundant Cys-based peroxidase in yeast) and not Gpx3/Orp1 is the protein that senses H2O2 and activates Yap1 through thiol-disulfide exchange reactions (Tachibana et al., 2009). These studies represented a breakthrough in the field, since they provide a pathway to redox regulate a SP presenting low-reactive thiols through a protein oxidation relay by a Cys-based peroxidase.
Therefore, in contrast to bacteria, Cys-based peroxidases and not transcription factors are the major sensors of H2O2 in yeast. Indeed, a yeast strain deleted for the five Prx genes and three Gpx genes is still viable, but lost its capacity to transcriptionally respond to H2O2 stimuli (Fomenko et al., 2011), suggesting that green pathway (Fig. 1A) is preponderant in the adaptive response of Saccharomyces cerevisiae to H2O2.
According to this mechanism, H2O2 signaling is mediated through sequential transfers of oxidizing equivalents. Upon oxidation, all Prx enzymes are oxidized to sulfenic acids (Cys-SOH) that in most cases can be converted to intra- or inter-molecular disulfides. Oxidized Prxs (Cys-SOH or Cys-SS-Cys) probably selectively transfers oxidizing equivalents to downstream regulatory proteins through specific protein-protein interactions and thiol-disulfide exchange reactions (Gutscher et al., 2009; Netto et al., 2015).
So far, the number of cases in which a high-reactive peroxidase was found to selectively transfer oxidizing equivalents to a SP is relatively low, but the descriptions of new cases are increasing. Remarkably, Prx1 from mammals was shown to physically interact in a redox dependent manner with SP such as: (1) PTEN, a protein with phosphatase activity (Cao et al., 2009); (2) apoptosis signaling kinase1 (ASK1) (Jarvis et al., 2012); and (3) MAPK phosphatases (Turner-Ivey et al., 2013).
Recently, the activation of a transcription factor (STAT3) by mammalian peroxiredoxin2 (Prx2) was also reported and present several features similar to the Gpx3/Orp1 –Yap1 pathway (Sobotta et al., 2015). In all cases, the transfer of oxidizing equivalents involves physical interaction and thiol-disulfide exchange between the two partners.
TRX-PRX MODEL
A lot of attention has been centered on H2O2 in the signaling pathways that Prx take place. In contrast, the roles play by Prx enzymes in regulating the redox state of its other substrate (Trx) has been somehow overlooked. This is surprising, since it is well established for a long time that several signal transduction pathways are activated by the oxidized but not by the reduced form of Trx (reviewed by Berndt et al., 2007). For instance, only reduced Trx1 and Trx2 bind Ask-1, thereby inhibiting its kinase activity. The oxidation of Trx1 leads to the physical dissociation of the Trx1-Ask1 complex and, consequently, to the activation of Ask-1 (Saitoh et al., 1998). Another example of redox regulation is the activation of NF-κB by Trx1. The binding of NF-κB subunit p50 to its target DNA sequence requires the reduction of a single cysteinyl residue by Trx1 (Hayashi et al., 1993; Matthews et al., 1992). Indeed, Trx enzymes recognize the oxidized form of their target proteins with higher selectivity, than their corresponding reduced forms (Palde and Carroll, 2015). It should be referred that Trx itself reacts slowly with H2O2 (Chae et al., 1994; Winterbourne and Hampton, 2008). Therefore, typical 2-Cys Prxs that catalyze the oxidation of Trx by hydroperoxides (Netto et al., 1996; Tairum et al., 2012) can impact redox regulation by modulating the redox status of Trx.
Therefore, another model can be envisaged, in which the redox pathway between H2O2 and the SP would be mediated not only by Prx, but also by Trx (Fig. 1A, pathway 3, red). According to this Prx-Trx signaling pathway, the oxidizing equivalent from H2O2 would be transferred to Prx and then to Trx and finally to the SP. Alternatively, Prx could oxidize Trx, and thereby avoid the interaction of this thiol-disulfide oxidoreductase with a SP.
More examples of redox-regulation by Prx-Trx pathway came from studies using fission yeast. In these organisms, two transcription factors have been found to be redox regulated by H2O2: Pap1 (Brown et al., 2013; Calvo et al., 2013) and Msn2 (Boisnard et al., 2009). Relevant for the adaptive response of fission yeast to H2O2, the formation of 2-Cys Prx disulfide triggers a signaling cascade that activates the otherwise unresponsive Pap1 protein, but only when Trx enzymes become transiently depleted (Brown et al., 2013; Calvo et al., 2013).
As described for the model presented in the section 2.2 (green pathway, Fig. 1), a feature underlying these pathways is the specificity provided by protein-protein interactions and thiol-disulfide exchange reactions. Proteins and their corresponding partners require to share complementary physico-chemical properties (i.e. hydrophobicity, polarity) in order to interact. As a matter of fact, yeast Trx reductase can only reduce yeast but not bacterial or mammalian Trx, due to complementary properties (Oliveira et al., 2010). Noteworthy, a protuberance (containing residues Glu50, Arg146 and Cys170 of yeast Tsa1) present only in the oxidized form of 2-Cys Prx is involved in its recognition by yeast Trx1 (Tairum et al., 2012).
Therefore, physical proximity between proteins is a requirement to allow the transfer of oxidizing equivalents through pathways 2 and 3. The construction of artificial quimeras between Cys-based proteins and redox sensitive GFP, Gutscher et al. (2009) elegantly demonstrated the “proximity-based Protein Thiol Oxidation by H2O2-scavenging peroxidases” concept that also form the basis of several genetically encoded fluorescent probes.
This Prx-Trx model also applies in conditions where cells are exposed to high levels of H2O2, provoking Prx hyperoxidation (Fig. 1B, pathway 3; red). Because TrxR (thioredoxin reductase) levels are limiting, several proteins (including disulfide form of 2-Cys Prx; transcription factors and methionine sulfoxide reductase) compete for reduced Trx1. For instance, the hyperoxidation of Tpx1 (2-Cys Prx from Schizosaccharomyces pombe) allowed accumulation of reduced Trx (since this oxidoreductase is not consumed by Prx in the disulfide form) that could be then used to repair other damaged proteins (Day et al., 2012). In these conditions, the activity of enzymes such as methionine sulfoxide reductase and ribonucleotide reductase would not be limited by shortage of reduced Trx. Remarkably, when Tpx1 is hyperoxidized, Pap1 (a transcription factor) is no longer activated possibly because there is enough Trx to keep this transcription factor in the reduced state and consequently out of the nucleus (reviewed by Veal et al., 2014).
Other observations that supported the notion that Prx participates in redox signaling by regulating the Trx status were gained by studies with Saccharomyces cerevisiae. For instance, mutations that destroy cytosolic TrxR activity rescue the growth of tsa1Δ rad51Δ double mutant and also suppress the genomic instability phenotype of tsa1Δ mutants (Ragu et al., 2014). Furthermore, a mutant of S. cerevisiae for cytosolic TrxR suppressed the growth defect of tsa1Δ mutants in zinc-deficient conditions (MacDiarmid et al., 2013). These studies are consistent with the idea that deletion of Prx genes decreases the rates of Trx oxidation, whereas deletion of TrxR genes decreases rates of Trx reduction and thereby compensates the absence of Cys-based peroxidase by keeping the Trx redox state similar to the levels observed in the wild type strain. These studies suggest that the main role of Prx enzymes at least in some cases may be regulating Trx redox levels, rather than reducing hydroperoxides (Veal et al., 2014).
Probably the redox status of Trx is probably not affected by 2-Cys Prx peroxidase activity in bacteria, because in these microorganism there is a reducing system (AhpF) that is specifically dedicated to reduce this Cys-based peroxidase (Veal et al., 2014). This phenomenon is probably related to the fact that most 2-Cys Prx in bacteria are robust, meaning that they resist to hyperoxidation, even when exposed to high H2O2 levels. In contrast, in eukaryotes, a selection pressure towards the evolution of 2-Cys Prxs that are sensitive to inactivation by H2O2 arose and according to this model could be involved to maintain Trx in the reduced form, available to mediate repair damaged proteins under acute stress conditions (Veal et al., 2014).
Not many examples of the Prx-Trx model are available in mammals. But noteworthy, HsTrx1 (Thioredoxin 1 from humans) contains five Cys residues: Cys32 and Cys35 are the two residues that belong to the conserved CGPC motif, whereas Cys62, Cys69 and Cys73 are three non-conserved residues. HsPrx1 (2-Cys Prx from Homo sapiens) specifically oxidizes Cys32 and Cys35 into an intramolecular disulfide. In contrast, Cys62 and Cys69 is an additional dithiol-disulfide redox center. When a disulfide is formed between Cys62 and Cys69, the ability of HsTrx1 to bind a target is impaired (Watson et al., 2003). Indeed, Cys62-Cys69 non-active site disulfide is not a substrate for HsTrxR1, but to the GSH/HsGrx1 system, whereas the Cys62-Cys69 dithiol can be oxidized by HsPrx1 (Du et al., 2013). These multiple oxidative states of HsTrx1 involving its five Cys residues play a role in the fine tune of the redox regulation of SPs such as Ask1 (kinases), PTEN (phosphatases) and Ref-1 (transcription factor) in redox pathways that 2-Cys Prx enzymes also take place (reviewed by Lee et al., 2013; Meyer et al., 2009).
CONCLUSION
A long way has come since H2O2 was seen as a toxicant, and today thiol switches are established cellular partners in signaling networks. Although the identification of the thiol redox proteome is advancing quickly, there is a massive lack of quantification, namely kinetic parameters. These parameters entail dynamic information that helps to identify in the complex network of thiol switches, those that operate during physiology and those whose disruption is responsible for a pathological state. The reactivity with H2O2 is known for only a few thiol SP (Ferrer-Sueta et al., 2011; Tanner et al., 2011); rate constants for the thiol-disulfide exchange reactions in which a peroxidase transfers oxidizing equivalents to a SP are virtually unknown; the reduction of the oxidized form of thiol SPs, switching-off the oxidation signal (Dagnell et al., 2013), is known only for a couple of interactions (Parsons and Gates, 2013). Determining true rate constants for these processes entails experiments with purified systems that are complex to preform and cannot cope with the rapid identification of new thiol switches provided by large-scale thiol proteomic experiments. As an alternative, the determination of apparent kinetic parameters under the typical redox experiments with intact cells or tissues (Brito and Antunes, 2014) may provide a more practical approach so that quantitative knowledge can follow suit the rapidly expanding redox proteome landscape, establishing the basis for redox kinectomics.
Acknowledgments
Supported by Fundação de Amparo a Pesquisa no Estado de São Paulo (FAPESP), Brazil, by grant 13/07937-8. Supported by Fundação para a Ciência e a Tecnologia (FCT), Portugal, through grant UID/MULTI/00612/2013.
REFERENCES
- Abbasi A., Corpeleijn E., Gansevoort R.T., Gans R.O.B., Struck J., Schulte J., Hillege H.L., van der Harst P., Stolk R.P., Navis G., et al. Circulating peroxiredoxin 4 and type 2 diabetes risk: the Prevention of Renal and Vascular Endstage Disease (PREVEND) study. Diabetologia. 2014;57:1842–1849. doi: 10.1007/s00125-014-3278-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahsan M.K., Lekli I., Ray D., Yodoi J., Das D.K. Redox regulation of cell survival by the thioredoxin superfamily: an implication of redox gene therapy in the heart Antioxid. Redox Signal. 2009;11:2741–2758. doi: 10.1089/ars.2009.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslund F., Zheng M., Beckwith J., Storz G. Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol-disulfide status. Proc. Natl. Acad. Sci. USA. 1999;96:6161–6165. doi: 10.1073/pnas.96.11.6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt C., Lillig C.H., Holmgren A. Thiol-based mechanisms of the thioredoxin and glutaredoxin systems: implications for diseases in the cardiovascular system. Am. J. Physiol. Heart Circ. Physiol. 2007;292:H1227–H1236. doi: 10.1152/ajpheart.01162.2006. [DOI] [PubMed] [Google Scholar]
- Biteau B., Labarre J., Toledano M.B. ATP-dependent reduction of cysteine–sulphinic acid by S. cerevisiae sulphiredoxin. Nature. 2003;425:980–984. doi: 10.1038/nature02075. [DOI] [PubMed] [Google Scholar]
- Boisnard S., Lagniel G., Garmendia-Torres C., Molin M., Boy-Marcotte E., Jacquet M., Toledano M.B., Labarre J., Chedin S. H2O2 activates the nuclear localization of Msn2 and Maf1 through thioredoxins in Saccharomyces cerevisiae. Eukaryot. Cell. 2009;8:1429–1438. doi: 10.1128/EC.00106-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boronat S., Domènech A., Paulo E., Calvo I.A., García-Santamarina S., García P., Encinar Del Dedo J., Barcons A., Serrano E., Carmona M., et al. Thiol-based H2O2 signalling in microbial systems. Redox Biol. 2014;2:395–399. doi: 10.1016/j.redox.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco M.R., Marinho H.S., Cyrne L., Antunes F. Decrease of H2O2 plasma membrane permeability during adaptation to H2O2 in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:6501–6506. doi: 10.1074/jbc.M311818200. [DOI] [PubMed] [Google Scholar]
- Brito P.M., Antunes F. Estimation of kinetic parameters related to biochemical interactions between hydrogen peroxide and signal transduction proteins. Front Chem. 2014;2:82. doi: 10.3389/fchem.2014.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.D., Day A.M., Taylor S.R., Tomalin L.E., Morgan B.A., Veal E.A. A peroxiredoxin promotes H2O2 signaling and oxidative stress resistance by oxidizing a thioredoxin family protein. Cell Rep. 2013;5:1425–1435. doi: 10.1016/j.celrep.2013.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo I.A., Boronat S., Domènech A., García-Santamarina S., Ayté J., Hidalgo E. Dissection of a redox relay: H2O2-dependent activation of the transcription factor Pap1 through the peroxidatic Tpx1-thioredoxin cycle. Cell Rep. 2013;5:1413–1424. doi: 10.1016/j.celrep.2013.11.027. [DOI] [PubMed] [Google Scholar]
- Cao J., Schulte J., Knight A., Leslie N.R., Zagozdzon A., Bronson R., Manevich Y., Beeson C., Neumann C.A. Prdx1 inhibits tumorigenesis via regulating PTEN/AKT activity. EMBO J. 2009;28:1505–1517. doi: 10.1038/emboj.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae H.Z., Chung S.J., Rhee S.G. Thioredoxin-dependent peroxide reductase from yeast. J. Biol. Chem. 1994;269:27670–27678. [PubMed] [Google Scholar]
- Chen K., Kirber M.T., Xiao H., Yang Y., Keaney J.F. Regulation of ROS signal transduction by NADPH oxidase 4 localization. J. Cell Biol. 2008;181:1129–1139. doi: 10.1083/jcb.200709049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagnell M., Frijhoff J., Pader I., Augsten M., Boivin B., Xu J., Mandal P.K., Tonks N.K., Hellberg C., Conrad M., et al. Selective activation of oxidized PTP1B by the thioredoxin system modulates PDGF-receptor tyrosine kinase signaling. Proc. Natl. Acad. Sci. USA. 2013;110:13398–13403. doi: 10.1073/pnas.1302891110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day A.M., Brown J.D., Taylor S.R., Rand J.D., Morgan B.A., Veal E.A. Inactivation of a peroxiredoxin by hydrogen peroxide is critical for thioredoxin-mediated repair of oxidized proteins and cell survival. Mol. Cell. 2012;45:398–408. doi: 10.1016/j.molcel.2011.11.027. [DOI] [PubMed] [Google Scholar]
- Delaunay A., Pflieger D., Barrault M. B., Vinh J., Toledano M. B. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell. 2002;111:471–481. doi: 10.1016/s0092-8674(02)01048-6. [DOI] [PubMed] [Google Scholar]
- Denu J.M., Tanner K.G. Specific and reversible inactivation of protein tyrosine phosphatases by hydrogen peroxide: evidence for a sulfenic acid intermediate and implications for redox regulation. Biochemistry. 1998;37:5633–5642. doi: 10.1021/bi973035t. [DOI] [PubMed] [Google Scholar]
- Du Y., Zhang H., Zhang X., Lu J., Holmgren A. Thioredoxin 1 is inactivated due to oxidation induced by peroxiredoxin under oxidative stress and reactivated by the glutaredoxin system. J. Biol. Chem. 2013;288:32241–32247. doi: 10.1074/jbc.M113.495150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Sueta G., Manta B., Botti H., Radi R., Trujillo M., Denicola A. Factors affecting protein thiol reactivity and specificity in peroxide reduction. Chem. Res. Toxicol. 2011;24:434–450. doi: 10.1021/tx100413v. [DOI] [PubMed] [Google Scholar]
- Fomenko D.E., Koc A., Agisheva N., Jacobsen M., Kaya A., Malinouski M., Rutherford J.C., Siu K.L., Jin D.Y., Winge D.R., et al. Thiol peroxidases mediate specific genome-wide regulation of gene expression in response to hydrogen peroxide. Proc. Natl. Acad. Sci. USA. 2011;108:2729–2734. doi: 10.1073/pnas.1010721108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutscher M., Sobotta M.C., Wabnitz G.H., Ballikaya S., Meyer A.J., Samstag Y., Dick T.P. Proximity-based protein thiol oxidation by H2O2-scavenging peroxidases. J. Biol. Chem. 2009;284:31532–31540. doi: 10.1074/jbc.M109.059246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A., Nelson K., Poole L.B., Karplus P.A. Structure-based insights into the catalytic power and conformational dexterity of peroxiredoxins. Antioxid. Redox Signal. 2011;15:795–815. doi: 10.1089/ars.2010.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Ueno Y., Okamoto T. Oxidoreductive regulation of nuclear factor kappa B. Involvement of a cellular reducing catalyst thioredoxin. J. Biol. Chem. 1993;268:11380–11388. [PubMed] [Google Scholar]
- Harald H.H.W., Schmidt R.S., Vollbracht C., Paulsen G., Riley D., Daiber A., Cuadrado A. Antioxidants in translational medicine. Antioxid. Redox Signal. 2015;10:1130–1143. doi: 10.1089/ars.2015.6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin M.E., Rivera-Del Valle N., Chandra J. Redox control of leukemia: from molecular mechanisms to therapeutic opportunities Antioxid. Redox Signal. 2013;18:1349–1383. doi: 10.1089/ars.2011.4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H.H., Lee K.O., Chi Y.H., Jung B.G., Park S.K., Park J.H., Lee J.R., Lee S.S., Moon J.C., Yun J. W., et al. Two enzymes in one: two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell. 2004;117:625–635. doi: 10.1016/j.cell.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Jarvis R.M., Hughes S.M., Ledgerwood E.C. Peroxiredoxin 1 functions as a signal peroxidase to receive, transduce, and transmit peroxide signals in mammalian cells. Free Radic. Biol. Med. 2012;53:1522–1530. doi: 10.1016/j.freeradbiomed.2012.08.001. [DOI] [PubMed] [Google Scholar]
- Jones D.P. Redefining oxidative stress. Antioxid. Redox Signal. 2006;8:1865–1879. doi: 10.1089/ars.2006.8.1865. [DOI] [PubMed] [Google Scholar]
- Kaszubska W., Falls H.D., Schaefer V.G., Haasch D., Frost L., Hessler P., Kroeger P.E., White D.W., Jirousek M.R., Trevillyan J.M. Protein tyrosine phosphatase 1B negatively regulates leptin signaling in a hypothalamic cell line. Mol. Cell. Endocrinol. 2002;195:109–118. doi: 10.1016/s0303-7207(02)00178-8. [DOI] [PubMed] [Google Scholar]
- Krapfenbauer K., Engidawork E., Cairns N., Fountoulakis M., Lubec G. Aberrant expression of peroxiredoxin subtypes in neurodegenerative disorders. Brain Res. 2003;967:152–160. doi: 10.1016/s0006-8993(02)04243-9. [DOI] [PubMed] [Google Scholar]
- Kwon J., Lee S.R., Yang K.S., Ahn Y., Kim Y.J., Stadtman E.R., Rhee S.G. Reversible oxidation and inactivation of the tumor suppressor PTEN in cells stimulated with peptide growth factors. Proc. Natl. Acad. Sci. USA. 2004;101:16419–16424. doi: 10.1073/pnas.0407396101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.R., Kwon K.S., Kim S.R., Rhee S.G. Reversible inactivation of protein-tyrosine phosphatase 1B in A431 cells stimulated with epidermal growth factor. J. Biol. Chem. 1998;273:15366–15372. doi: 10.1074/jbc.273.25.15366. [DOI] [PubMed] [Google Scholar]
- Lee C., Lee S.M., Mukhopadhyay P., Kim S.J., Lee S.C., Ahn W.S., Yu M.H., Storz G., Ryu S.E. Redox regulation of OxyR requires specific disulfide bond formation involving a rapid kinetic reaction path. Nat. Struct. Mol. Biol. 2004;11:1179–1185. doi: 10.1038/nsmb856. [DOI] [PubMed] [Google Scholar]
- Lee S., Kim S.M., Lee R.T. Thioredoxin and thioredoxin target proteins: from molecular mechanisms to functional significance. Antioxid. Redox Signal. 2013;18:1165–1207. doi: 10.1089/ars.2011.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little C., O’Brien P.J. Mechanism of peroxide-inactivation of the sulphydryl enzyme glyceraldehyde-3-phophate dehydrogenase. Eur. J. Biochem. 1969;10:533–538. doi: 10.1111/j.1432-1033.1969.tb00721.x. [DOI] [PubMed] [Google Scholar]
- MacDiarmid C.W., Taggart J., Kerdsomboon K., Kubisiak M., Panascharoen S., Schelble K., Eide D.J. Peroxiredoxin chaperone activity is critical for protein homeostasis in zinc-deficient yeast. J. Biol. Chem. 2013;288:31313–31327. doi: 10.1074/jbc.M113.512384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadev K., Zilbering A., Zhu L., Goldstein B. J. Insulin-stimulated hydrogen peroxide reversibly inhibits protein-tyrosine phosphatase 1b in vivo and enhances the early insulin action cascade. J. Biol. Chem. 2001;276:21938–21942. doi: 10.1074/jbc.C100109200. [DOI] [PubMed] [Google Scholar]
- Marinho H.S., Real C., Cyrne L., Soares H., Antunes F. Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol. 2014;2:535–562. doi: 10.1016/j.redox.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews J.R., Wakasugi N., Virelizier J.L., Yodoi J., Hay R.T. Thioredoxin regulates the DNA binding activity of NF-κB by reduction of a disulphide bond involving cysteine 62. Nucleic Acids Res. 1992;20:3821–3830. doi: 10.1093/nar/20.15.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng T., Fukada T., Tonks N.K. Reversible oxidation and inactivation of protein tyrosine phosphatases in vivo. Mol. Cell. 2002;9:387–399. doi: 10.1016/s1097-2765(02)00445-8. [DOI] [PubMed] [Google Scholar]
- Meyer Y., Buchanan B.B., Vignols F., Reichheld J.P. Thioredoxins and glutaredoxins: unifying elements in redox biology. Annu. Rev. Genet. 2009;43:335–367. doi: 10.1146/annurev-genet-102108-134201. [DOI] [PubMed] [Google Scholar]
- Miller E.W., Dickinson B.C., Chang C.J. Aquaporin-3 mediates hydrogen peroxide uptake to regulate downstream intracellular signaling. Proc. Natl. Acad. Sci. USA. 2010;107:15681–15686. doi: 10.1073/pnas.1005776107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina N.M., Tyurin-Kuzmin P.A., Markvicheva K.N., Vorotnikov A.V., Tkachuk V.A., Laketa V., et al. Does cellular hydrogen peroxide diffuse or act locally? Antioxid. Redox Signal. 2011;14:1–7. doi: 10.1089/ars.2010.3539. [DOI] [PubMed] [Google Scholar]
- Nelson K.J., Knutson S.T., Soito L., Klomsiri C., Poole L.B., Fetrow J.S. Analysis of the peroxiredoxin family: using active-site structure and sequence information for global classification and residue analysis. Proteins. 2011;79:947–964. doi: 10.1002/prot.22936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netto L.E.S., Chae H.Z., Kang S.W., Rhee S.G., Stadtman E.R. Removal of hydrogen peroxide by thiol-specific antioxidant enzyme (TSA) is involved with its antioxidant properties. TSA possesses thiol peroxidase activity. J. Biol. Chem. 1996;271:15315–15321. doi: 10.1074/jbc.271.26.15315. [DOI] [PubMed] [Google Scholar]
- Netto L.E.S., Oliveira M.A., Tairum-Jr C., da Silva Neto J.F. Conferring specificity in redox pathways by enzymatic thiol/disulfide exchange reactions. Free Radic. Res. 2015;16:1–99. doi: 10.3109/10715762.2015.1120864. [DOI] [PubMed] [Google Scholar]
- Nyström T., Yang J., Molin M. Peroxiredoxins, gerontogenes linking aging to genome instability and cancer. Genes Dev. 2012;26:2001–2008. doi: 10.1101/gad.200006.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogusucu R., Rettori D., Munhoz D.C., Netto L.E.S., Augusto O. Reactions of yeast thioredoxin peroxidases I and II with hydrogen peroxide and peroxynitrite: rate constants by competitive kinetics. Free Radic. Biol. Med. 2007;42:326–334. doi: 10.1016/j.freeradbiomed.2006.10.042. [DOI] [PubMed] [Google Scholar]
- Oliveira M.A., Discola K.F., Alves S. V., Medrano F. J., Guimarães B.G., Netto L.E.S. Insights into the specificity of thioredoxin reductase-thioredoxin interactions. A structural and functional investigation of the yeast thioredoxin system. Biochemistry. 2010;49:3317–3326. doi: 10.1021/bi901962p. [DOI] [PubMed] [Google Scholar]
- Palde P.B., Carroll K.S. A universal entropy-driven mechanism for thioredoxin-target recognition. Proc. Natl. Acad. Sci. USA. 2015;112:7960–7965. doi: 10.1073/pnas.1504376112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsonage D., Youngblood D.S., Sarma G.N., Wood Z.A., Karplus P.A., Poole L.B. Analysis of the link between enzymatic activity and oligomeric state in AhpC, a bacterial peroxiredoxin. Biochemistry. 2005;44:10583–10592. doi: 10.1021/bi050448i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons Z.D., Gates K.S. Thiol-dependent recovery of catalytic activity from oxidized protein tyrosine phosphatases. Biochemistry. 2013;52:6412–6423. doi: 10.1021/bi400451m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen C.E., Truong T.H., Garcia F.J., Homann A., Gupta V., Leonard S.E., Carrol K.S. Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. Nat. Chem. Biol. 2012;8:57–64. doi: 10.1038/nchembio.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedroso N., Matias A.C., Cyrne L., Antunes F., Borges C., Malhó R., de Almeida R.F.M., Herrero E., Marinho H.S. Modulation of plasma membrane lipid profile and microdomains by H2O2 in Saccharomyces cerevisiae. Free Radic. Biol. Med. 2009;46:289–298. doi: 10.1016/j.freeradbiomed.2008.10.039. [DOI] [PubMed] [Google Scholar]
- Peralta D., Bronowska A.K., Morgan B., Dóka É., Van Laer K., Nagy P., Gräter F., Dick T.P. A proton relay enhances H2O2 sensitivity of GAPDH to facilitate metabolic adaptation. Nat. Chem. Biol. 2015;11:156–163. doi: 10.1038/nchembio.1720. [DOI] [PubMed] [Google Scholar]
- Peskin A.V., Low F.M., Paton L.N., Maghzal G.J., Hampton M.B., Winterbourn C.C. The high reactivity of peroxiredoxin 2 with H2O2 is not reflected in its reaction with other oxidants and thiol reagents. J. Biol. Chem. 2007;282:11885–11892. doi: 10.1074/jbc.M700339200. [DOI] [PubMed] [Google Scholar]
- Peskin A. V., Pace P.E., Behring J.B., Paton L. N., Soethoudt M., Bachschmid M.M., Winterbourn C.C. Glutathionylation of the active site cysteines of peroxiredoxin 2 and recycling by glutaredoxin. J. Biol. Chem. 2016 doi: 10.1074/jbc.M115.692798. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat S.J., Creasy C.L., Peterson J.R., Chernoff J. The tumor suppressor Mst1 promotes changes in the cellular redox state by phosphorylation and inactivation of peroxiredoxin-1 protein. J. Biol. Chem. 2013;288:8762–8771. doi: 10.1074/jbc.M112.414524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragu S., Dardalhon M., Sharma S., Iraqui I, Buhagiar-Labarchède G., Grondin V., Kienda G., Vernis L, Chanet R., Kolodner R.D., et al. Loss of the thioredoxin reductase Trr1 suppresses the genomic instability of peroxiredoxin tsa1 mutants. PLoS One. 2014;9:e108123. doi: 10.1371/journal.pone.0108123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee S. G., Woo H. A. Multiple functions of peroxiredoxins: peroxidases, sensors and regulators of the intracellular messenger H2O2, and protein chaperones. Antioxid. Redox Signal. 2011;15:781–794. doi: 10.1089/ars.2010.3393. [DOI] [PubMed] [Google Scholar]
- Rhee S.G., Woo H.A., Kil I.S., Bae S.H. Peroxiredoxin functions as a peroxidase and a regulator and sensor of local peroxides. J Biol. Chem. 2012;287:4403–4410. doi: 10.1074/jbc.R111.283432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh M., Nishitoh H., Fujii M., Takeda K., Tobiume K., Sawada Y., Kawabata M., Miyazono K., Ichijo H. Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) EMBO J. 1998;17:2596–2606. doi: 10.1093/emboj/17.9.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder K., Zhang M., Benkhoff S., Mieth A., Pliquett R., Kosowski J., Kruse C., Luedike P., Michaelis U.R., Weissmann N., et al. Nox4 is a protective reactive oxygen species generating vascular NADPH oxidase. Circ. Res. 2012;110:1217–1225. doi: 10.1161/CIRCRESAHA.112.267054. [DOI] [PubMed] [Google Scholar]
- Sies H. Role of metabolic H2O2 generation: redox signaling and oxidative stress. J. Biol. Chem. 2014;289:8735–8741. doi: 10.1074/jbc.R113.544635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidler N.W. GAPDH: Biological Properties and Diversity. Vol. 985. Springer; 2013. [Google Scholar]
- Sobotta M.C., Liou W., Stöcker S., Talwar D., Oehler M., Ruppert T., Scharf A.N., Dick T.P. Peroxiredoxin-2 and STAT3 form a redox relay for H2O2 signaling. Nat. Chem. Biol. 2015;11:64–70. doi: 10.1038/nchembio.1695. [DOI] [PubMed] [Google Scholar]
- Tachibana T., Okazaki S., Murayama A., Naganuma A., Nomoto A., Kuge S. A major peroxiredoxin-induced activation of Yap1 transcription factor is mediated by reduction-sensitive disulfide bonds and reveals a low level of transcriptional activation. J. Biol. Chem. 2009;284:4464–4472. doi: 10.1074/jbc.M807583200. [DOI] [PubMed] [Google Scholar]
- Tairum C.A., Jr., de Oliveira M.A., Horta B.B., Zara F.J., Netto L.E.S. Disulfide biochemistry in 2-cys peroxiredoxin: requirement of Glu50 and Arg146 for the reduction of yeast Tsa1 by thioredoxin. J. Mol. Biol. 2012;424:28–41. doi: 10.1016/j.jmb.2012.09.008. [DOI] [PubMed] [Google Scholar]
- Tanner J.J., Parsons Z.D., Cummings A.H., Zhou H., Gates K.S. Redox regulation of protein tyrosine phosphatases: structural and chemical aspects. Antioxid. Redox Signal. 2011;15:77–97. doi: 10.1089/ars.2010.3611. [DOI] [PubMed] [Google Scholar]
- Toledo J.C., Audi R., Ogusucu R., Monteiro G., Netto L.E.S., Augusto O. Horseradish peroxidase compound I as a tool to investigate reactive protein-cysteine residues: from quantification to kinetics. Free Radic. Biol. Med. 2011;50:1032–1038. doi: 10.1016/j.freeradbiomed.2011.02.020. [DOI] [PubMed] [Google Scholar]
- Trujillo M., Clippe A., Manta B., Ferrer-Sueta G., Smeets A., Declercq J.P., Knoops B., Radi R. Pre-steady state kinetic characterization of human peroxiredoxin 5: taking advantage of Trp84 fluorescence increase upon oxidation. Arch. Biochem. Biophys. 2007;467:95–106. doi: 10.1016/j.abb.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Turner-Ivey B., Manevich Y., Schulte J., Kistner-Griffin E., Jezierska-Drutel A., Liu Y., Neumann C.A. Role for Prdx1 as a specific sensor in redox-regulated senescence in breast cancer. Oncogene. 2013;32:5302–5314. doi: 10.1038/onc.2012.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veal E.A., Ross S.J., Malakasi P., Peacock E., Morgan B.A. Ybp1 is required for the hydrogen peroxide-induced oxidation of the Yap1 transcription factor. J. Biol. Chem. 2003;278:30896–30904. doi: 10.1074/jbc.M303542200. [DOI] [PubMed] [Google Scholar]
- Watson W.H., Pohl J., Montfort W.R., Stuchlik O., Reed M.S., Powis G., Jones D.P. Redox potential of human thioredoxin 1 and identification of a second dithiol/disulfide motif. J. Biol. Chem. 2003;278:33408–33415. doi: 10.1074/jbc.M211107200. [DOI] [PubMed] [Google Scholar]
- Winterbourn C.C. Reconciling the chemistry and biology of reactive oxygen species. Nat. Chem. Biol. 2008;4:278–286. doi: 10.1038/nchembio.85. [DOI] [PubMed] [Google Scholar]
- Winterbourn C.C., Hampton M.B. Thiol chemistry and specificity in redox signaling. Free Radic. Biol. Med. 2008;45:549–561. doi: 10.1016/j.freeradbiomed.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Winterbourn C.C., Metodiewa D. Reactivity of biologically important thiol compounds with superoxide and hydrogen peroxide. Free Radic. Biol. Med. 1999;27:322–328. doi: 10.1016/s0891-5849(99)00051-9. [DOI] [PubMed] [Google Scholar]
- Woo H.A., Yim S.H., Shin D.H., Kang D., Yu D.Y., Rhee S.G. Inactivation of peroxiredoxin I by phosphorylation allows localized H2O2 accumulation for cell signaling. Cell. 2010;140:517–528. doi: 10.1016/j.cell.2010.01.009. [DOI] [PubMed] [Google Scholar]
- Wood Z.A., Poole L.B., Karplus P.A. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 2003;300:650–653. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- Ying J., Clavreul N., Sethuraman M., Adachi T., Cohen R.A. Thiol oxidation in signaling and response to stress: detection and quantification of physiological and pathophysiological thiol modifications. Free Radic. Biol. Med. 2007;43:1099–1108. doi: 10.1016/j.freeradbiomed.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]