Lin et al. show that Bhlhe40 expression identifies encephalitogenic CD4+ T helper cells and define a pertussis toxin–IL-1–Bhlhe40 pathway active in experimental autoimmune encephalomyelitis, a mouse model of multiple sclerosis.
Abstract
The features that define autoreactive T helper (Th) cell pathogenicity remain obscure. We have previously shown that Th cells require the transcription factor Bhlhe40 to mediate experimental autoimmune encephalomyelitis (EAE), a mouse model of multiple sclerosis. Here, using Bhlhe40 reporter mice and analyzing both polyclonal and TCR transgenic Th cells, we found that Bhlhe40 expression was heterogeneous after EAE induction, with Bhlhe40-expressing cells displaying marked production of IFN-γ, IL-17A, and granulocyte-macrophage colony-stimulating factor. In adoptive transfer EAE models, Bhlhe40-deficient Th1 and Th17 cells were both nonencephalitogenic. Pertussis toxin (PTX), a classical co-adjuvant for actively induced EAE, promoted IL-1β production by myeloid cells in the draining lymph node and served as a strong stimulus for Bhlhe40 expression in Th cells. Furthermore, PTX co-adjuvanticity was Bhlhe40 dependent. IL-1β induced Bhlhe40 expression in polarized Th17 cells, and Bhlhe40-expressing cells exhibited an encephalitogenic transcriptional signature. In vivo, IL-1R signaling was required for full Bhlhe40 expression by Th cells after immunization. Overall, we demonstrate that Bhlhe40 expression identifies encephalitogenic Th cells and defines a PTX–IL-1–Bhlhe40 pathway active in EAE.
Autoreactive CD4+ T helper (Th) cells specific for components of myelin drive experimental autoimmune encephalomyelitis (EAE), a widely used animal model of the human neuroinflammatory disease multiple sclerosis (MS). In the active EAE model in C57BL/6 mice, naive Th cells are primed by subcutaneous immunization with a peptide derived from myelin oligodendrocyte glycoprotein (MOG35-55) emulsified in CFA (Stromnes and Goverman, 2006). Along with MOG/CFA, mice are treated systemically with the co-adjuvant pertussis toxin (PTX), an ADP-ribosylating exotoxin derived from Bordetella pertussis that has been proven necessary for clinical disease in this model (Levine and Sowinski, 1973; Bettelli et al., 2003). Although the target cell types and mechanisms of action of PTX are not fully understood, PTX has been shown to increase blood–brain barrier permeability (Kerfoot et al., 2004; Kügler et al., 2007) and promote the maturation and cytokine production of antigen-presenting cells (Ryan et al., 1998; Bagley et al., 2002). Several studies have shown PTX treatment or B. pertussis infection to induce IL-1β and IL-6 production by myeloid cells (Chen et al., 2007; Zhang et al., 2011; Connelly et al., 2012; Dumas et al., 2014), which, during EAE, could contribute to PTX-mediated effects on regulatory T (T reg) cells (Cassan et al., 2006; Chen et al., 2006) and Th17 cells (Chen et al., 2007; Andreasen et al., 2009).
We and others have previously demonstrated that the transcription factor basic helix–loop–helix family member e40 (Bhlhe40; also known as Dec1, Stra13, Sharp2, and Bhlhb2) is required in a Th cell–intrinsic fashion for susceptibility to EAE (Martínez-Llordella et al., 2013; Lin et al., 2014). Bhlhe40 is a member of the basic helix–loop–helix–Orange subfamily of transcription factors with a recognized role in regulating circadian rhythms, cellular differentiation, and immune cell function (Ow et al., 2014). Bhlhe40-deficient (Bhlhe40−/−) mice resist EAE, and although their Th cells are capable of mounting largely normal antigen-specific responses to immunization, Bhlhe40−/− Th cells show markedly decreased secretion of GM-CSF, an effector cytokine required for EAE (Codarri et al., 2011; El-Behi et al., 2011), and increased secretion of IL-10, a cytokine with immunoregulatory properties (Bettelli et al., 1998; Lin et al., 2014). In vitro, Bhlhe40−/− Th cells differentiate normally in appropriate polarizing conditions into Th1, Th2, and Th17 cells subsets, although in each case Bhlhe40 deficiency results in the abnormal expression of ∼200–300 genes, including Csf2 (encoding GM-CSF) and Il10 (Lin et al., 2014). Bhlhe40 is expressed in all subsets of polarized Th cells in vitro, and is known to be regulated in part through a signal provided by CD28 in combination with TCR signaling (Martínez-Llordella et al., 2013). Nevertheless, the pathways that regulate Bhlhe40 expression in Th cells in vivo during an immune response and the features of Bhlhe40-expressing Th cells during EAE remain unknown.
RESULTS
Bhlhe40GFP Tg mice show Bhlhe40 expression in immune cells
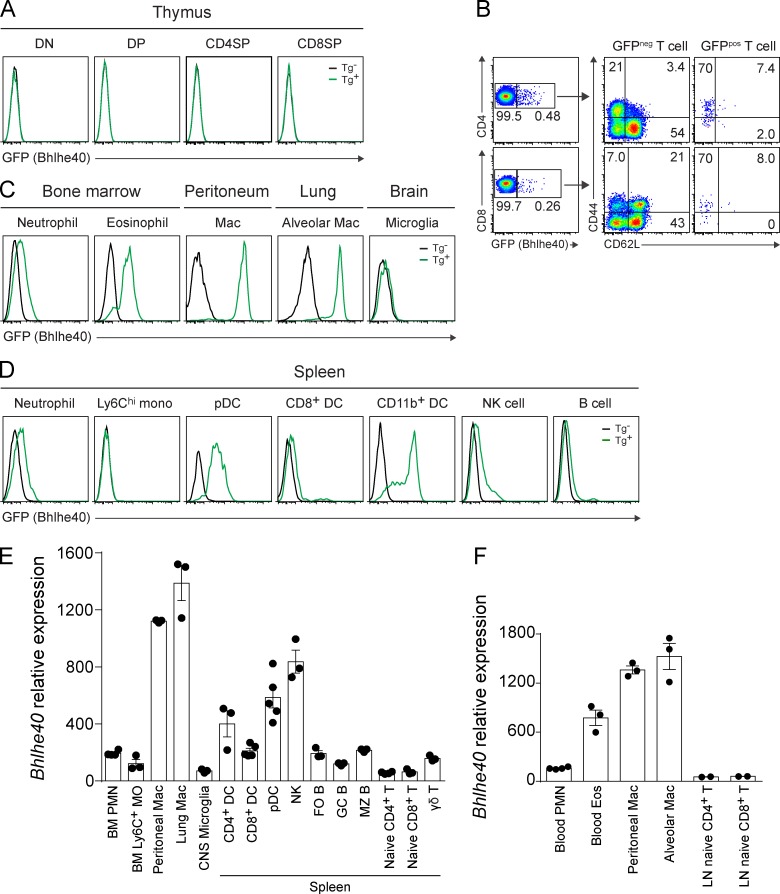
We used bacterial artificial chromosome (BAC) transgenic (Tg) reporter mice generated by the Gene Expression Nervous System Atlas (GENSAT) Project (Schmidt et al., 2013) to identify and study Bhlhe40 expression in Th cells in vivo. Cells from these Bhlhe40GFP mice show Bhlhe40 expression through enhanced GFP in the context of a BAC transgene spanning the 205-kb genomic DNA segment containing Bhlhe40. To determine the steady-state expression of Bhlhe40, we performed flow cytometry on tissues obtained from these reporter mice. Thymocytes of all developmental stages did not express GFP (Fig. 1 A). Splenic CD4+ and CD8+ T cells expressed GFP in >0.5% of cells (Fig. 1 B). These rare GFPpos cells stained as CD44+CD62L−, indicating prior antigen experience. Other immune cells showed a range of GFP expression (Fig. 1, C and D). An overall comparison of our flow cytometry results with the expression of Bhlhe40 in immune cells based on expression microarray datasets from the Immgen Consortium (Heng and Painter, 2008) showed excellent agreement (Fig. 1, E and F). These data indicate that Bhlhe40GFP mice faithfully reveal Bhlhe40 expression.
Figure 1.
Bhlhe40GFP mice show Bhlhe40 expression in immune cells. (A–D) GFP (Bhlhe40) expression in multiple immune cell types in thymus (A), spleen (B and D), and bone marrow, peritoneum, lung, and brain (C) from naive nontransgenic (Tg−) or Bhlhe40GFP (Tg+) reporter mice. DN, double-negative cells; DP, double-positive cells; CD4SP, CD4 single-positive cells; CD8SP, CD8 single-positive cells; mac, macrophages; mono, monocytes; pDC, plasmacytoid DCs. All cell types were analyzed at least two times in at least two independent experiments. (B) GFP (Bhlhe40) expression by splenic CD4+ and CD8+ T cells from naive Bhlhe40GFP mice. CD44 and CD62L expression by GFPneg and GFPpos T cells were analyzed. (E and F) Bhlhe40 expression in immune cells based on microarray datasets from the Immgen Consortium, datasets GSE15907 (E; phase 1) and GSE37448 (F; phase 2). BM, bone marrow; PMN, polymorphonuclear cells; MO, monocytes; DC, dendritic cells; FO B, follicular B cells; GC B, germinal center B cells; MZ B, marginal zone B cells; Eos, eosinophils. Data are mean ± SEM with dots representing individual microarrays performed by Immgen.
Bhlhe40-expressing Th cells are enriched in the CNS and are robust cytokine producers during EAE
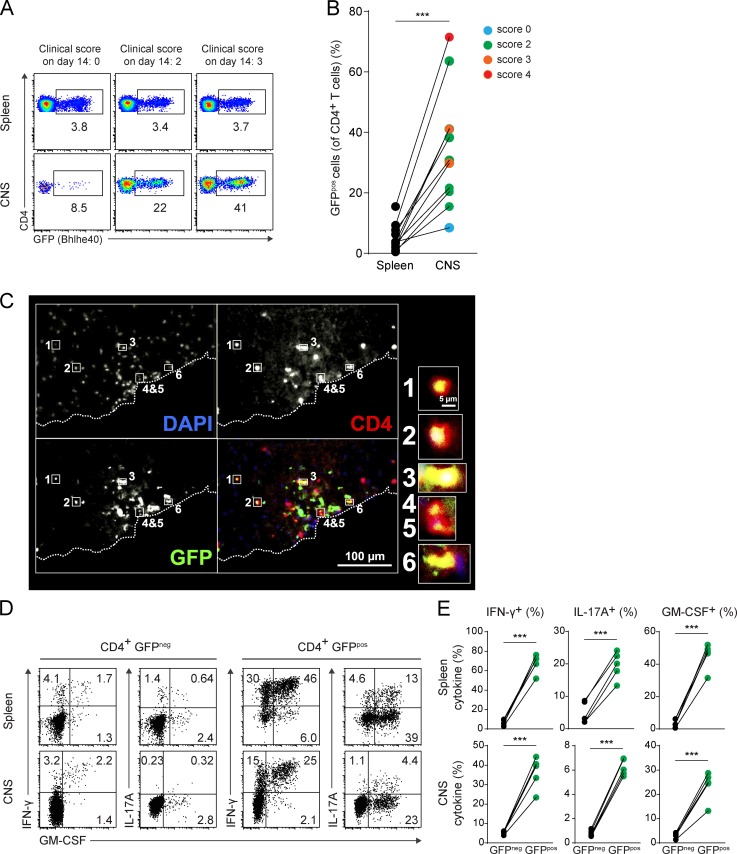
Based on the known cell-intrinsic requirement for Bhlhe40 expression in Th cells for EAE susceptibility (Martínez-Llordella et al., 2013; Lin et al., 2014), we expected that Bhlhe40 would be expressed in effector T cells during EAE. Flow cytometry of Bhlhe40GFP mice at day 13–14 after EAE induction showed that ∼1–10% of splenic and ∼10–70% of CNS-infiltrating CD4+ T cells expressed GFP at this time (Fig. 2, A and B), notably more than in naive mice. In general, the frequency of GFPpos CD4+ T cells in the CNS correlated with disease severity at the day of sacrifice. GFP expression was also notable in CD8+ and γδ T cells during EAE, again at an increased frequency in the CNS relative to the spleen, consistent with these cell types participating in disease pathogenesis (Sutton et al., 2009; Huber et al., 2013; unpublished data). Some GFP expression was also observed in CD11b+ infiltrating myeloid cells during EAE, but not in microglia (unpublished data). Histological examination of spinal cord sections from diseased mice confirmed GFP expression within CD4+ T cells (Fig. 2 C) and CD11b+ myeloid cells (unpublished data) present in inflammatory lesions. Strikingly, intracellular cytokine staining (ICS) showed that Bhlhe40-expressing GFPpos CD4+ T cells produced almost all of the IFN-γ, IL-17A, and GM-CSF in both the spleen and CNS (Fig. 2, D and E), suggesting that Bhlhe40 expression could serve as an identifying feature of encephalitogenic Th cells.
Figure 2.
Bhlhe40-expressing Th cells are enriched in the CNS and are robust cytokine producers during EAE. (A) GFP (Bhlhe40) expression by CD4+ T cells in the spleen and CNS of Bhlhe40GFP mice on day 14 after EAE induction. Clinical scores of individual mice on the day of sacrifice are indicated. One experiment out of four is shown (n = 10 mice total). (B) Quantitation of the percentage of GFPpos cells (of CD4+ T cells) in spleen and CNS from A. Each line represents an individual mouse (n = 10). Data are combined from four independent experiments performed at day 13 or 14 after EAE induction. Clinical scores on the day of sacrifice are indicated by color. (C) Immunofluorescent staining of a spinal cord section at day 15 after EAE induction in a Bhlhe40GFP mouse (clinical score of 2). Dashed line represents the edge of the tissue. Bottom right quadrant is the overlapping image of DAPI (blue), CD4 (red), and GFP (green). High magnification images of selected CD4+ T cells expressing GFP are shown. Data are representative of two independent experiments (n = 2 mice total). (D) ICS plots for IFN-γ, IL-17A, and GM-CSF for GFPneg and GFPpos CD4+ T cells in the spleen and CNS at day 14 after EAE induction in Bhlhe40GFP mice. One experiment out of two is shown (n = 5 mice). (E) Quantitation of the frequency of GFPneg or GFPpos CD4+ T cells secreting IFN-γ, IL-17A, or GM-CSF from D. Each line represents an individual mouse (n = 5). Data are combined from two experiments. Paired two-tailed Student's t test was performed to determine significance. ***, P ≤ 0.001.
PTX serves as an essential co-adjuvant by inducing Bhlhe40 expression in Th cells
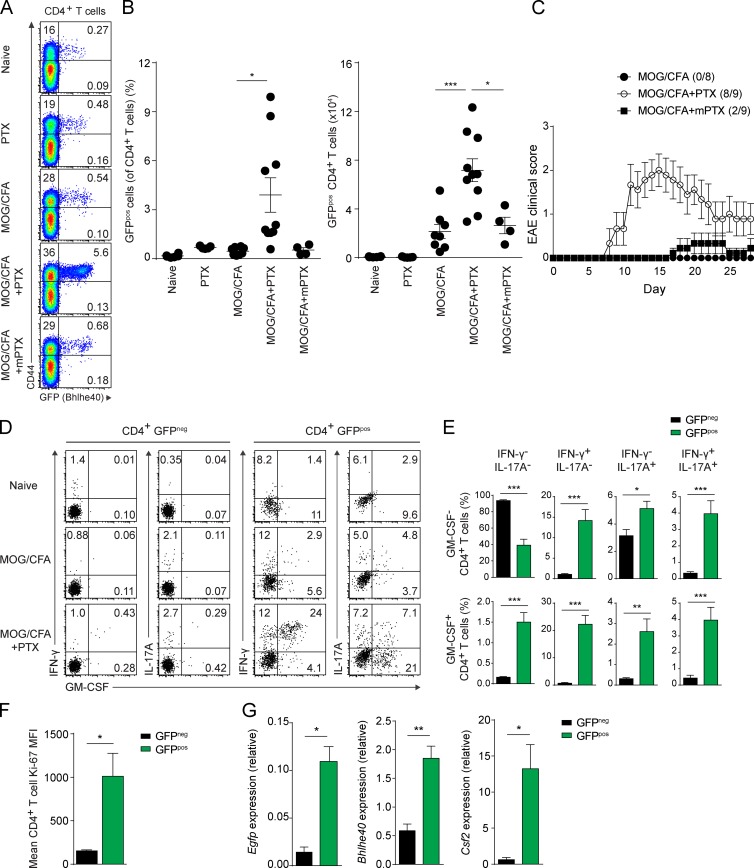
1 wk after immunization of Bhlhe40GFP mice with MOG/CFA, we observed only a very small increase in the abundance of GFP-expressing CD4+ T cells in the DLNs relative to naive mice (Fig. 3, A and B), with almost all GFPpos cells being CD44+. Because the same immunization given with systemic administration of PTX had resulted in robust GFP expression by CD4+ T cells during EAE, we reasoned that the co-adjuvant PTX might serve as a stimulus for Bhlhe40 expression. Indeed, DLNs from mice immunized with MOG/CFA and treated with PTX contained significantly increased numbers of GFPpos CD4+ T cells relative to naive mice, PTX-only–treated mice, or non-PTX–treated MOG/CFA-immunized mice (Fig. 3, A and B), with these GFPpos T cells again bearing CD44. To mediate this effect, the ADP-ribosylating activity of PTX was necessary, as treatment of MOG/CFA-immunized mice with mutant PTX (mPTX) lacking this activity did not result in an increase in the frequency or number of GFPpos CD4+ T cells relative to MOG/CFA-immunized mice given no PTX. In immunized mice followed for the development of clinical EAE, only PTX, but not mPTX, was able to serve as a co-adjuvant for disease induction (Fig. 3 C). ICS showed PTX treatment to be a strong inducer of IFN-γ, IL-17A, and GM-CSF secretion, particularly among GFPpos CD4+ T cells (Fig. 3, D and E), again suggesting that Bhlhe40 expression serves to identify encephalitogenic Th cells. In the setting of MOG/CFA immunization given with PTX treatment, GFPpos CD4+ T cells showed higher expression of Ki-67, indicating that these cells were more proliferative than their GFPneg counterparts (Fig. 3 F). qRT-PCR analysis comparing GFPneg and GFPpos CD4+ T cells from these mice showed that GFPpos cells expressed higher levels of Egfp, Bhlhe40, and Csf2 transcripts (Fig. 3 G).
Figure 3.
PTX serves as an essential co-adjuvant by inducing Bhlhe40 expression in Th cells. (A) GFP (Bhlhe40) and CD44 expression by CD4+ T cells from the DLN of Bhlhe40GFP mice on day 7. Groups of mice were either unimmunized or immunized with MOG/CFA. PTX or mPTX was administered i.p. on days 0 and 2. One experiment out of five is shown (n = 4–10/group). (B) Quantitation of the percentage (left) of GFPpos cells (of CD4+ T cells) and the absolute number (right) of GFPpos CD4+ T cells in the DLNs from (A). Dots represent individual mice. Data are combined from five experiments (n = 4–10/group). (C) Mean EAE scores of WT mice immunized with MOG/CFA. PTX or mPTX, where indicated, were administered i.p. on days 0 and 2. Data are combined from two experiments (n = 8–9). Incidence of clinical disease is indicated. (D) ICS plots for IFN-γ, IL-17A, and GM-CSF for GFPneg and GFPpos CD4+ T cells in the DLN in naive mice or at day 7 after immunization as indicated in Bhlhe40GFP mice. One experiment out of three is shown (n = 9 mice). (E) Quantitation of the frequency of GFPneg and GFPpos CD4+ T cells from MOG/CFA + PTX-immunized mice producing each of the eight combinations of IFN-γ, IL-17A, and GM-CSF. Data are combined from three experiments (n = 9). (F) Mean fluorescence intensity (MFI) of Ki-67 by GFPneg or GFPpos CD4+ T cells from the DLN of Bhlhe40GFP mice on day 7 after immunization with MOG/CFA + PTX. Data are combined from two experiments (n = 7). (G) Quantitative RT-PCR analysis of Egfp, Bhlhe40, and Csf2 expression by sorted GFPneg and GFPpos CD4+ T cells from the DLN of Bhlhe40GFP mice on day 7 after MOG/CFA + PTX immunization (n = 3 mice from a single experiment). Data are mean ± SEM. Unpaired (B) or paired (E–G) two-tailed Student's t tests were performed to determine significance. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
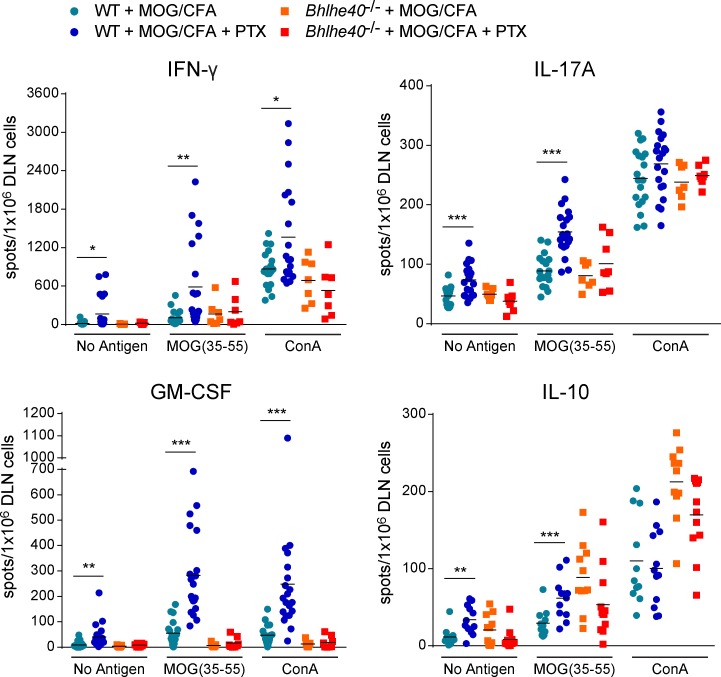
We next tested the hypothesis that PTX co-adjuvanticity was Bhlhe40 dependent. We previously showed by ELISPOT assays that antigen/CFA immunization in the absence of PTX primed generally normal frequencies of antigen-specific IFN-γ and IL-17A responses in the DLNs of Bhlhe40−/− mice (Lin et al., 2014). These mice, however, showed a markedly decreased frequency of antigen-specific GM-CSF–producing T cells, and an increased frequency of IL-10–producing cells relative to WT mice. We repeated these ELISPOT assays in WT and Bhlhe40−/− mice, but included additional groups of mice that were treated with PTX (Fig. 4). In WT mice, PTX treatment significantly increased the frequency of MOG-specific cells producing all four cytokines. Importantly, PTX treatment showed no ability to augment MOG-specific responses in Bhlhe40−/− mice. These data indicate that PTX co-adjuvanticity is Bhlhe40 dependent, and explain, at least in part, why PTX is a required co-adjuvant for clinical EAE induction in C57BL/6 mice.
Figure 4.
PTX co-adjuvanticity is Bhlhe40 dependent. ELISPOT assays for the quantitation of cells secreting IFN-γ, IL-17A, GM-CSF, or IL-10 performed on DLN cells 7 d after MOG/CFA immunization of WT or Bhlhe40−/− mice treated with or without PTX. Data are combined from four experiments (n = 7–13 mice/group). Dots represent individual mice. For clarity, only mean values (lines) are shown. Unpaired two-tailed Student's t test was performed to determine significance. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Bhlhe40 expression identifies the cytokine-producing fraction of autoreactive Th cells
We have shown that GFP expression in CD4+ T cells of Bhlhe40GFP mice identifies cytokine-producing Th cells, whereas GFPneg CD4+ T cells largely lack cytokine production. These results could be explained by either of two nonmutually exclusive possibilities. In one scenario, GFPneg cells may represent CD4+ T cells with TCR specificities unrelated to the ongoing myelin-specific response. Bystander CD4+ T cells have been described in the CNS during EAE (Lees et al., 2010), so it is likely that at least some GFPneg CD4+ T cells in this compartment are bystanders. Alternatively, Bhlhe40 expression may be heterogeneous among autoreactive CD4+ T cells. In this scenario, only those self-reactive T cells expressing Bhlhe40 would serve as encephalitogenic, cytokine-producing effectors.
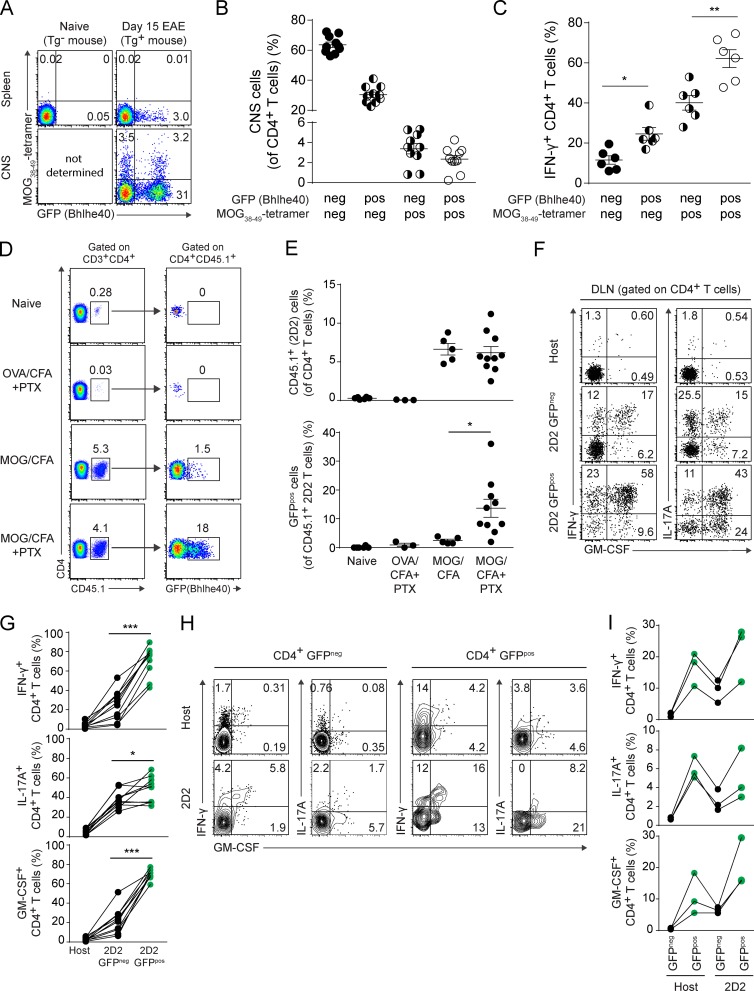
We tested these possibilities by two approaches. First, we identified MOG-specific CD4+ T cells using MOG38-49–I-Ab tetramers at day 15 after EAE induction in Bhlhe40GFP mice. Although we failed to detect tetramerpos cells in the spleen, ∼7% of CNS-infiltrating CD4+ T cells were stained by tetramer, and nearly half of them were GFPpos (Fig. 5, A and B). Among tetramerpos cells, those expressing GFP showed the highest level of IFN-γ production (Fig. 5 C). Interestingly, however, most CNS-infiltrating GFPpos cells did not stain with tetramer. Although this may suggest bystander activation by non-MOG–specific T cells, we feel that this is more consistent with the findings of Sabatino et al. (2011), where most MOG-specific cells after EAE induction had a low affinity for MOG/I-Ab, such that they were negative for tetramer staining. These low-affinity cells were reported to contribute significantly to the pool of IFN-γ–producing T cells in the CNS. Consistent with this, we also found that among tetramerneg cells, a higher frequency of GFPpos cells produced IFN-γ (Fig. 5 C).
Figure 5.
Bhlhe40 expression identifies the cytokine-producing fraction of autoreactive Th cells. (A) Plots of MOG38-49–I-Ab tetramer staining and GFP (Bhlhe40) expression by CD4+ T cells from the spleen and CNS of naive non-Tg and EAE-induced Bhlhe40GFP mice (day 15). One experiment out of two is shown (n = 10 mice). (B) Quantitation of the percentage of CNS CD4+ T cells positive or negative for GFP (Bhlhe40) and/or MOG38-49–I-Ab tetramer staining in the CNS at day 15 after EAE induction. Data are combined from two experiments (n = 10). (C) Quantitation of the frequency of IFN-γ+ cells within the indicated CD4+ T cell subsets (as shown in B). One experiment out of two is shown (n = 6). (D) Plots for (left) CD4 and CD45.1 and (right) CD4 and GFP (Bhlhe40) on DLN cells after the transfer of T cells from 2D2 Bhlhe40GFP CD45.1/CD45.2 mice into WT (CD45.2) recipients. 1 d after cell transfer, mice were left unimmunized or immunized with the indicated peptide (MOG35-55 or OVA323-339) in CFA. Some mice were treated with PTX on days 0 and 2. DLNs were harvested on day 7 after immunization. One experiment out of three is shown (n = 3–10/group). (E) Quantitation of the percentage of (top) CD45.1+ 2D2 cells (of CD4+ T cells) and (bottom) CD45.1+ 2D2 GFPpos cells (of CD45.1+ 2D2 cells) in the DLNs from (D). Dots represent individual mice. Data are combined from three experiments (n = 3–10 mice/group). (F) ICS plots for IFN-γ, IL-17A, and GM-CSF for host CD4+ T cells, GFPneg, or GFPpos 2D2 cells in the DLN from MOG/CFA + PTX-immunized mice from (D). One experiment out of three is shown (n = 10 mice). (G) Quantitation of the frequency of host CD4+ T cells or GFPneg or GFPpos 2D2 cells secreting IFN-γ, IL-17A, or GM-CSF from F. Lines connect data from individual mice (n = 10). Data are combined from three experiments. (H) ICS plots for IFN-γ, IL-17A, and GM-CSF for host GFPneg or GFPpos CD4+ T cells, or GFPneg or GFPpos 2D2 cells in the CNS at day 14 after EAE induction. T cells from 2D2 Bhlhe40GFP CD45.1/CD45.2 mice were transferred into Bhlhe40GFP (CD45.2) recipients 1 d before immunization with MOG/CFA + PTX. One experiment out of two is shown (n = 6 mice). (I) Quantitation of the frequency of host GFPneg or GFPpos CD4+ T cells, GFPneg, or GFPpos 2D2 cells secreting IFN-γ, IL-17A, or GM-CSF from H. Lines connect data from individual mice (n = 3). One experiment out of two is shown. Data are mean ± SEM. Unpaired (E) or paired (C and G) two-tailed Student's t tests were performed to determine significance. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
In our second approach, we crossed 2D2 TCR Tg mice, bearing T cells with a TCR specific for MOG35-55 (Bettelli et al., 2003) to Bhlhe40GFP mice and crossed in one allele of the congenic marker CD45.1 (resulting in hematopoietic cells with co-expression of CD45.1 and CD45.2). Purified CD4+ T cells from these mice (henceforth termed 2D2 Bhlhe40GFP CD45.1/CD45.2) were transferred to WT recipients (bearing only CD45.2), and 1 d later these mice were immunized with MOG/CFA with or without PTX treatment (Fig. 5, D–G). To control for the possible effects of microbial ligands in CFA or the effects of PTX that are independent of cognate antigen stimulation, a separate group of mice that received 2D2 Bhlhe40GFP CD45.1/CD45.2 CD4+ T cells was immunized with an irrelevant peptide antigen (OVA323-339)/CFA and treated with PTX. Immunization with MOG resulted in an expansion of the population of transferred T cells in the DLN 1 wk later, with additional PTX treatment resulting in a markedly increased frequency of GFPpos T cells (Fig. 5, D and E). In MOG/CFA-immunized mice treated with PTX, ICS (Fig. 5, F and G) on DLN cells showed that host-derived polyclonal CD4+ T cells secreted minimal amounts of IFN-γ, IL-17A, and GM-CSF. 2D2 Bhlhe40GFP CD45.1/CD45.2 CD4+ T cells produced all of these cytokines, although cytokine production was significantly increased among 2D2 GFPpos T cells compared with 2D2 GFPneg T cells, particularly for IFN-γ and GM-CSF. Thus, there exists heterogeneity among clonal autoreactive T cells in their expression of Bhlhe40, and expression of this transcription factor identifies those cells with encephalitogenic features. In a variation of the aforementioned experiment, we transferred 2D2 Bhlhe40GFP CD45.1/CD45.2 CD4+ T cells to Bhlhe40GFP reporter mice (CD45.2) to allow for the simultaneous analysis of GFPneg and GFPpos host CD4+ T cells and GFPneg and GFPpos 2D2 CD4+ T cells after EAE induction (Fig. 5, H and I). In these mice, ICS staining showed significantly increased cytokine production by both GFPpos host and GFPpos 2D2 CD4+ T cells, relative to their GFPneg counterparts, supporting the notion that Bhlhe40 expression identified both monoclonal and polyclonal encephalitogenic T cells.
Bhlhe40 is essential for the encephalitogenicity of Th cells in adoptive transfer models of EAE
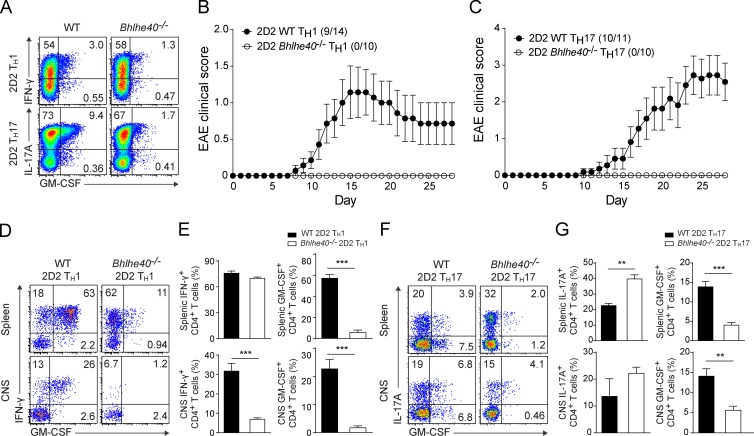
To further test the notion that Bhlhe40-expressing CD4+ T cells are encephalitogenic, we used an adoptive transfer system of EAE that allows in vitro polarization of 2D2 cells without a requirement for immunization to generate MOG-specific cells (Jäger et al., 2009). Because in our immunizations with MOG/CFA + PTX, we found GFPpos CD4+ T cells to produce both IFN-γ and IL-17A, we separately tested both Th1 and Th17 adoptive transfer models, comparing the encephalitogenicity of 2D2 WT (CD45.1) and 2D2 Bhlhe40−/− (CD45.2) cells after transfer to WT recipients (CD45.1/CD45.2). Despite normal Th1 or Th17 polarization before transfer (Fig. 6 A), 2D2 Bhlhe40−/− cells were completely nonencephalitogenic in vivo (Fig. 6, B and C). Nevertheless, 2D2 Bhlhe40−/− cells could be identified in recipient mice at 28–34 d after transfer. Stimulation of these cells recovered from spleens showed that they maintained production of their hallmark cytokines at levels similar to 2D2 WT cells recovered from diseased mice (Fig. 6, D–G). Notably, GM-CSF production by polarized 2D2 Bhlhe40−/− cells was decreased relative to 2D2 WT cells both before transfer and upon recovery, consistent with our previous experiments using polyclonal CD4+ T cells (Lin et al., 2014). These findings confirm a cell-intrinsic requirement for Bhlhe40 in both encephalitogenic Th1 and Th17 cells.
Figure 6.
Bhlhe40 is essential for the encephalitogenicity of Th cells in adoptive transfer models of EAE. (A) ICS plots for the indicated cytokines from polarized WT and Bhlhe40−/− 2D2 Th1 or Th17 cells at day 4 of culture. One experiment out of three is shown (n = 10–14 mice/group). (B and C) Mean EAE scores of WT mice after receiving transfers of WT or Bhlhe40−/− 2D2 Th1 (B) or Th17 (C) cells. Data are combined from four (Th1, n = 10–14) or two (Th17, n = 10–11) experiments. Incidence of clinical disease is indicated. (D–G) ICS plots for the indicated cytokines (D and F) and quantitation of the frequency (E and G) of cytokine-producing WT and Bhlhe40−/− 2D2 Th1 (D and E) or Th17 (F and G) cells in the spleen and CNS on days 28–34 after immunization. (D and E) Data are combined from two independent experiments (n = 7/group). (F and G) One experiment of two is shown (n = 3–4/group). Data are mean ± SEM. Unpaired two-tailed Student's t test was performed to determine significance. **, P ≤ 0.01; ***, P ≤ 0.001.
Bhlhe40 expression negatively correlates with Foxp3 and IL-10 expression during EAE
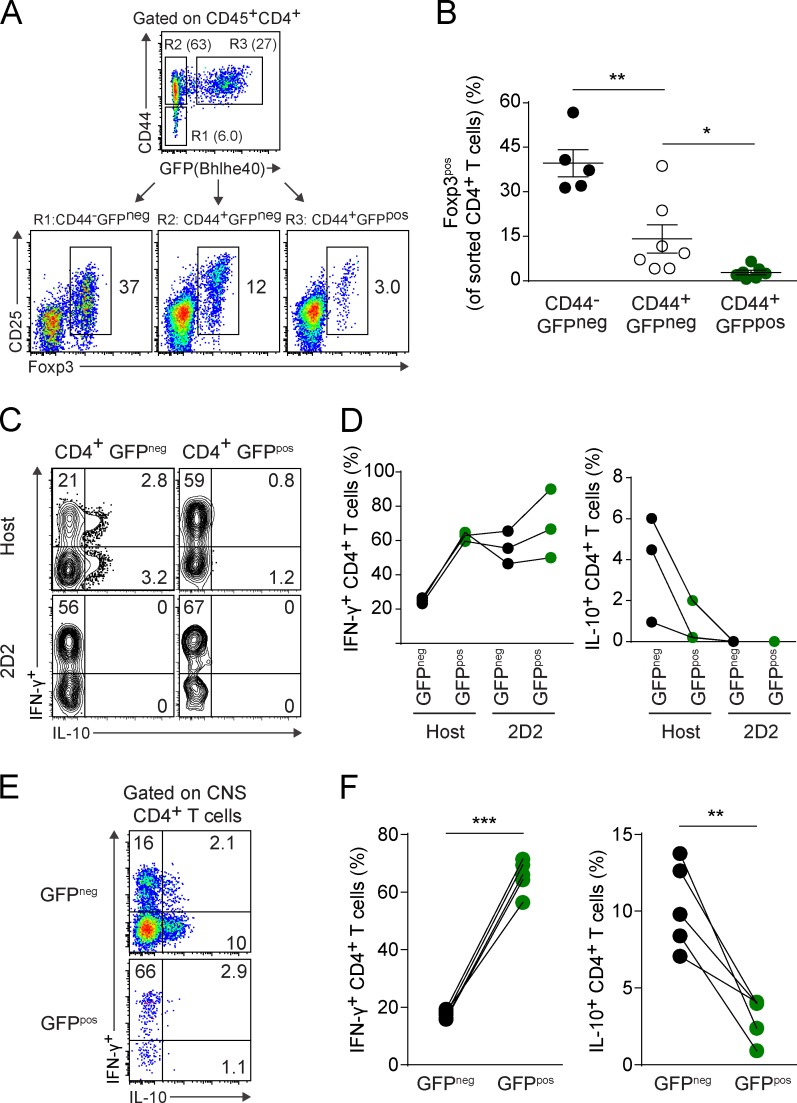
Foxp3pos T reg cells and the suppressive cytokine IL-10 have been recognized to ameliorate disease during EAE (Kohm et al., 2002; Zhang et al., 2004; Lowther and Hafler, 2012). Given our findings that Bhlhe40-expressing CD4+ T cells were robust producers of inflammatory cytokines and that expression of Bhlhe40 was required in adoptive transfer models of EAE, we hypothesized that Bhlhe40-expressing cells in the CNS during EAE would be unlikely to express Foxp3 or IL-10. We performed intracellular staining for Foxp3 on CNS-infiltrating immune cells after EAE induction in Bhlhe40GFP mice, but found that this protocol significantly diminished our ability to discriminate GFPpos CD4+ T cells. To circumvent this problem, we sorted three populations of CNS-infiltrating CD4+ T cells at day 16 after EAE induction in reporter mice based on their expression of CD44 and GFP, and performed subsequent intracellular staining for Foxp3 (Fig. 7 A). Many Foxp3pos CD4+ T cells were present in the CD44−GFPneg (R1) and CD44+GFPneg (R2) gates, whereas within the population of CD44+GFPpos (R3) cells, only a small fraction of T cells were Foxp3pos (Fig. 7, A and B). We are unsure of the identity of these Bhlhe40 and Foxp3 double-positive cells, but they may be akin to populations of T reg cells that express Foxp3 and a second transcription factor, as has been reported for T-betposFoxp3pos T reg cells in EAE (O’Connor et al., 2012; McPherson et al., 2015). We also analyzed IL-10 production by CNS-infiltrating CD4+ T cells in reporter mice that received congenically marked 2D2 Bhlhe40GFP cells to allow us to track both GFPneg and GFPpos host and TCR Tg T cells. Although a high frequency of GFPpos host and GFPpos 2D2 cells produced IFN-γ, IL-10 production came almost entirely from host GFPneg cells (Fig. 7, C and D). These IL-10–producing GFPneg host cells likely represent a mixture of both T reg cells and IFN-γ+IL-10+ type 1 regulatory T cells (Tr1) cells (Zhang et al., 2004; Pot et al., 2009; Apetoh et al., 2010; de Kouchkovsky et al., 2013), whereas GFPpos host and 2D2 cells represent autoreactive effectors. At day 29, during the resolving phase of EAE, GFPneg CD4+ T cells in the CNS continued to express higher levels of IL-10 than GFPpos T cells, with the latter continuing to robustly produce IFN-γ (Fig. 7, E and F).
Figure 7.
Bhlhe40 expression negatively correlates with Foxp3 and IL-10 expression. (A) Strategy for sorting CD44−GFPneg (R1), CD44+GFPneg (R2), and CD44+GFPpos (R3) CD4+ T cells in the CNS at day 16 after EAE induction. Foxp3 and CD25 expression on sorted cells from each population is shown. One experiment out of two is shown (n = 5–7 mice). (B) Quantitation of the frequency of Foxp3pos cells within each of the three populations as shown in A. Data are combined from two experiments (n = 5–7). Data are mean ± SEM. (C) ICS plots for IFN-γ and IL-10 for host GFPneg or GFPpos CD4+ T cells, or GFPneg or GFPpos 2D2 cells in the CNS at day 14 after EAE induction. T cells from 2D2 Bhlhe40GFP CD45.1/CD45.2 mice were transferred into Bhlhe40GFP (CD45.2) recipients 1 d before immunization with MOG/CFA + PTX. One experiment out of two is shown (n = 6 mice). (D) Quantitation of the frequency of host GFPneg or GFPpos CD4+ T cells, GFPneg, or GFPpos 2D2 cells secreting IFN-γ or IL-10 from (C). Lines connect data from individual mice (n = 3). One experiment out of two is shown. (E) ICS plots for IFN-γ and IL-10 for GFPneg and GFPpos CD4+ T cells in the CNS at day 29 after EAE induction in Bhlhe40GFP mice. One experiment out of two is shown (n = 5 mice). (F) Quantitation of the frequency of GFPneg or GFPpos CD4+ T cells secreting IFN-γ (left) or IL-10 (right) from E. Lines connect data from individual mice (n = 5). Data are combined from two experiments. Unpaired (B) or paired (F) two-tailed Student's t tests were performed to determine significance. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Bhlhe40-expressing Th17 cells exhibit a pathogenic molecular signature
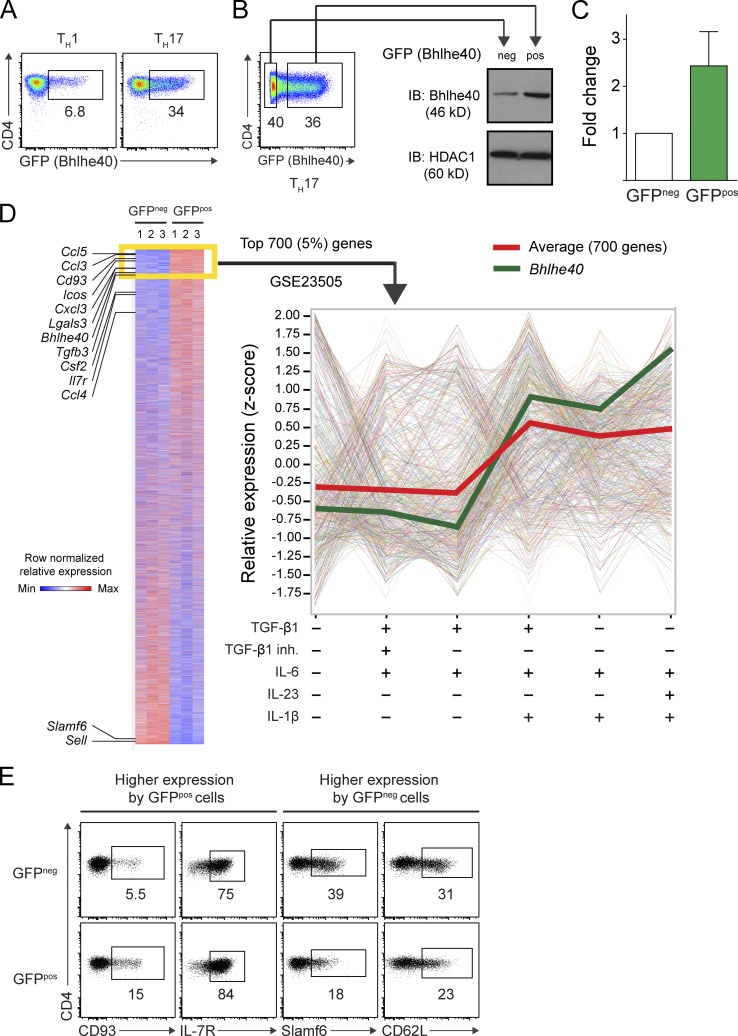
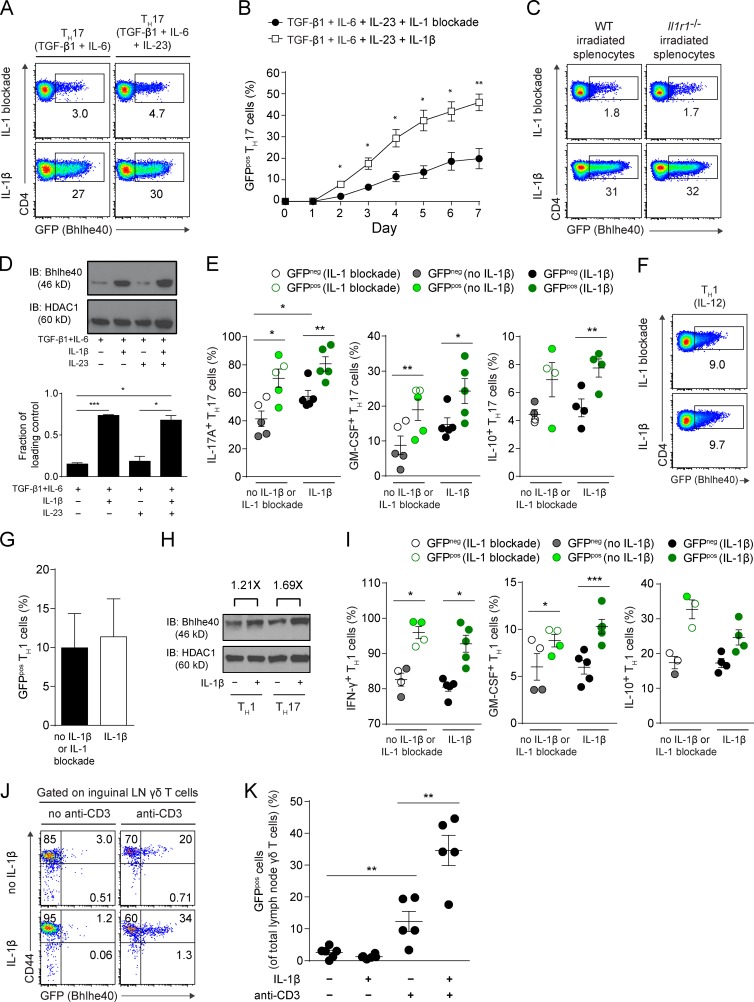
Next, we polarized naive CD4+ T cells from Bhlhe40GFP mice in Th1 (IL-12) or Th17 (TGF-β1, IL-6, IL-23, and IL-1β) conditions in vitro. On day 4, ∼5–10% of Th1 cells and ∼30–40% of Th17 cells expressed GFP (Fig. 8 A). Because of significant GFP expression by Th17 cells and their relevance to the EAE model, we sorted GFPneg and GFPpos Th17 cells for transcriptional analysis. Immunoblotting confirmed that GFPpos cells expressed ∼2.5 times more Bhlhe40 than GFPneg cells (Fig. 8, B and C). Expression microarrays were performed on these cells and differentially expressed transcripts were identified (Fig. 8 D). Strikingly, GFPpos Th17 cells exhibited a pathogenic signature, with higher expression of many genes previously identified as being expressed by encephalitogenic Th17 cells, including Bhlhe40, Csf2, Tgfb3, Il7r, Icos, and several chemokines (Codarri et al., 2011; El-Behi et al., 2011; Walline et al., 2011; Lee et al., 2012; Martínez-Llordella et al., 2013; Lin et al., 2014). We confirmed differential expression for four transcripts that encoded surface proteins by flow cytometry (Fig. 8 E), two with higher expression in GFPpos cells (CD93 and IL-7R) and two with higher expression in GFPneg cells (Slamf6 and CD62L). Lastly, we compared our microarray data to a publically available Gene Expression Omnibus (GEO) dataset (GSE23505) in which microarrays were performed on Th17 cells derived from cultures using a variety of cytokine conditions (Ghoreschi et al., 2010; Fig. 8 D). Using gene set analysis, we tested whether the 700 most differentially expressed genes with higher expression in Bhlhe40-expressing (GFPpos) cells in our experiment (Fig. 8 D, yellow box) showed differential expression in Th17 cells across culture conditions. Interestingly, as a group, these 700 genes showed an overall pattern to suggest that their expression was specifically increased by culture with IL-1β (Fig. 8 D, red line), independent of whether Th17 cells were derived by culture with TGF-β1 and/or IL-23. In these data, Bhlhe40 was also expressed more highly in conditions that included IL-1β (Fig. 8 D, green line). This finding suggested a model in which IL-1β acted through Bhlhe40 to promote encephalitogenicity of Th17 cells and led us to explore whether IL-1β directly regulated the expression of Bhlhe40.
Figure 8.
Bhlhe40-expressing Th17 cells exhibit a pathogenic molecular signature. (A) CD4 and GFP (Bhlhe40) expression by polarized Bhlhe40GFP Th1 or Th17 cells at day 4 of culture. Data are representative of at least five experiments with at least eight mice per condition. (B) Strategy for sorting GFPneg and GFPpos Th17 cells at day 4 of culture. Sorted cells were subjected to immunoblots for Bhlhe40 and HDAC1 (loading control). One experiment out of two is shown (n = 5 mice). (C) Quantitation of the band intensity of Bhlhe40 relative to HDAC1 in sorted GFPneg and GFPpos Th17 cells from (B). Data are expressed as a fold change in intensity comparing sorted cells from the same culture. Data are combined from two experiments (n = 5). Mean ± SEM. (D) Heat map comparing gene expression between sorted GFPneg and GFPpos Th17 cells from three mice. Listed genes have been associated with EAE or represent surface markers amenable to flow cytometry (Cd93, Il7r, Slamf6, and Sell). Shown are the 14,000 probesets with the highest mean expression across all six arrays. The yellow box identifies the 700 genes (5%) that were most highly expressed in GFPpos Th17 cells compared with GFPneg Th17 cells. The relative expression of these 700 genes was analyzed in GEO DataSet GSE23505 (Ghoreschi et al., 2010) in which Th cells had been cultured in different cytokine conditions as indicated (right). The red line indicates the mean expression of these genes. The green line represents the expression of Bhlhe40. (E) CD4 and CD93, IL-7R, Slamf6, or CD62L (encoded by Sell) expression by GFPneg and GFPpos Th17 cells cultured as in A. One experiment out of two is shown (n = 4 mice).
IL-1β increases Bhlhe40 expression in Th17 and γδ T cells
We used our Th17 cell culture system with purified CD4+ T cells from Bhlhe40GFP mice to test whether IL-1β induced the expression of Bhlhe40. Indeed, IL-1β, but not IL-23, induced strong expression of GFP in this system (Fig. 9 A). Expression of GFP was notable by day 2 in these cultures, and increased steadily through day 7 (Fig. 9 B). By performing Th17 cultures with irradiated splenocytes from Il1r1−/− mice, we confirmed that IL-1β acted directly on CD4+ T cells to increase their GFP expression (Fig. 9 C). Immunoblotting for Bhlhe40 also confirmed induction of Bhlhe40 by IL-1β (Fig. 9 D). IL-1β increased both IL-17A and GM-CSF production by Th17 cells, with GFPpos cells displaying the highest production of these cytokines (Fig. 9 E). In contrast with our in vivo results (Fig. 7, C–F), in vitro, GFPpos cells also produced slightly higher levels of IL-10 (Fig. 9 E). IL-10 production derived mainly from IL-17A+ cells (unpublished data). Consistent with the fact that IL-1R is expressed at low levels on Th1 cells (Chung et al., 2009; Guo et al., 2009), IL-1β only minimally increased GFP expression when added to Th1 cultures (Fig. 9, F–H). Nevertheless, a higher frequency of GFPpos Th1 cells expressed IFN-γ, GM-CSF, and IL-10 (Fig. 9 I), with GM-CSF and IL-10 production deriving mainly from IFN-γ+ cells (not depicted). γδ T cells are known to produce GM-CSF in response to IL-1β (Sutton et al., 2009), and we have previously shown that γδ T cells require Bhlhe40 to produce GM-CSF (Lin et al., 2014). Similar to Th17 cells, TCR-activated γδ T cells from Bhlhe40GFP mice also increased their expression of GFP in response to IL-1β (Fig. 9, J and K).
Figure 9.
IL-1β increases Bhlhe40 expression in Th17 and γδ T cells. (A) CD4 and GFP (Bhlhe40) expression by Bhlhe40GFP Th17 cells at day 4 of culture in the indicated conditions. One experiment out of three is shown (n = 4 mice). (B) Kinetics of GFP (Bhlhe40) expression by Bhlhe40GFP Th17 cells cultured in the presence of IL-1 blockade (anti–IL-1β and anti–IL-1R) or IL-1β. Data are combined from two experiments (n = 3–5 mice/condition). (C) CD4 and GFP (Bhlhe40) expression by Bhlhe40GFP Th17 (TGF-β1 + IL-6 + IL-23) cells at day 4 of culture in the presence of IL-1 blockade or IL-1β and with WT or Il1r1−/− irradiated splenocytes. One experiment out of two is shown (n = 3 mice). (D) Immunoblots of Bhlhe40 and HDAC1 in Th17 cells cultured with the indicated cytokines for 4 d. (bottom) Quantitation of the band intensity of Bhlhe40 relative to HDAC1. Data are representative of four experiments (n = 2 mice/condition). (E) Quantitation of the frequency of GFPneg or GFPpos Th17 (TGF-β1 + IL-6 + IL-23) cells secreting IL-17A, GM-CSF, or IL-10 cultured in the absence or presence of IL-1β or IL-1 blockade. Data are combined from four experiments (n = 4–5 mice/condition). (F) CD4 and GFP (Bhlhe40) expression by Bhlhe40GFP Th1 cells at day 4 of culture with IL-1β or IL-1 blockade. One experiment out of three is shown (n = 5 mice). (G) Quantitation of the percentage of GFP (Bhlhe40)-expressing Th1 cells cultured in the presence (IL-1β) or absence of IL-1β (no IL-1β or IL-1 blockade). Data are combined from four experiments (n = 5). (H) Immunoblots of Bhlhe40 and HDAC1 in Th1 cells cultured with or without IL-1β for 4 d. The fold change of band intensity of Bhlhe40 relative to HDAC1 is indicated. Data are representative of two experiments (n = 3 mice). (I) Quantitation of the frequency of GFPneg or GFPpos Th1 cells secreting IFN-γ, GM-CSF, or IL-10 cultured in the absence or presence of IL-1β or IL-1 blockade. Data are combined from four experiments (n = 3–5/condition). (J) CD44 and GFP (Bhlhe40) expression by lymph node γδ T cells from Bhlhe40GFP mice cultured for 3 d in the indicated conditions. One experiment out of three is shown (n = 5 mice). (K) Quantitation of the percentage of GFP (Bhlhe40)-expressing γδ T cells from J. Data are combined from three experiments (n = 5 mice/condition). Data are mean ± SEM. Unpaired (B, D, and K) or paired (G) two-tailed Student's t tests were performed to determine significance. For E and I, p-values represent paired, two-tailed Student's t test when comparing GFPneg to GFPpos cells from the same culture, and represent unpaired two-tailed Student's t test when comparing cultures with no IL-1β or IL-1 blockade vs. IL-1β. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
IL-1R signaling is required in vivo for full Bhlhe40 expression by Th cells
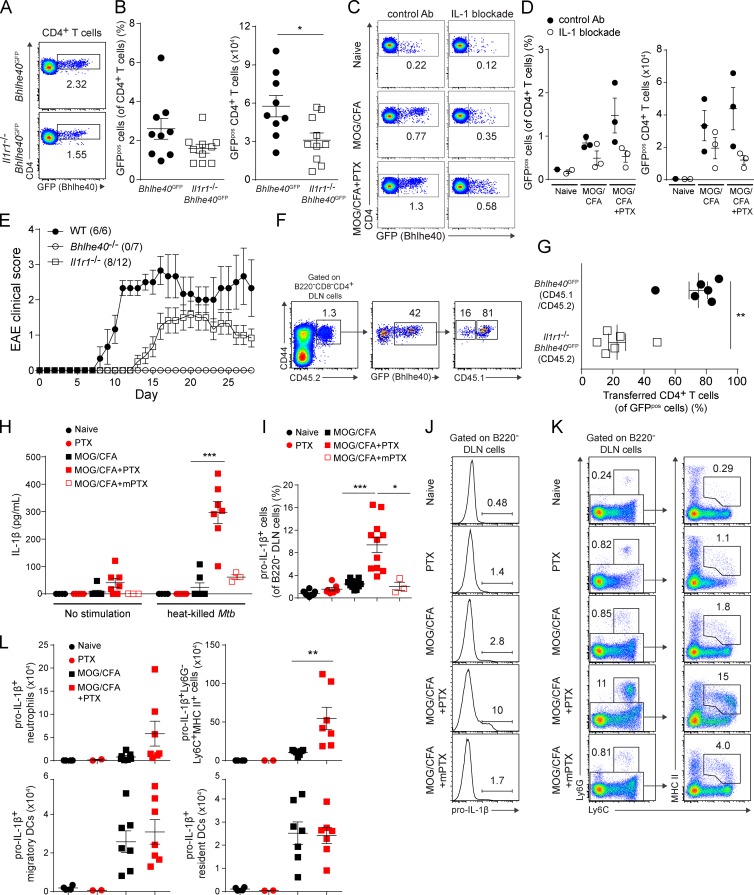
To test whether IL-1 played a role in the induction of Bhlhe40 in CD4+ T cells in vivo, we crossed Bhlhe40GFP reporter mice with Il1r1−/− mice. We immunized these mice with MOG/CFA, along with PTX treatment, and compared their CD4+ T cell responses to control Bhlhe40GFP mice. In the absence of IL-1R signaling, fewer CD4+ T cells in the DLN expressed GFP (Fig. 10, A and B). Similarly, in vivo IL-1 blockade by antibodies in Bhlhe40GFP mice resulted in fewer GFPpos CD4+ T cells after immunization and PTX treatment (Fig. 10, C and D). Consistent with previous studies (Matsuki et al., 2006; Sutton et al., 2006; McCandless et al., 2009; Lukens et al., 2012; Croxford et al., 2015a,b), and the notion that Bhlhe40 expression is partially dependent on IL-1R signaling, we observed a lower incidence of clinical EAE in Il1r1−/− mice compared with WT mice, with a later onset and less severe disease (Fig. 10 E). Bhlhe40−/− mice showed complete EAE resistance, indicating that Bhlhe40 serves a more essential role in the development of EAE than IL-1 does. To define a cell-intrinsic role for IL-1R signaling in the induction of Bhlhe40, we co-transferred CD4+ T cells from Bhlhe40GFP (CD45.1/CD45.2) or Il1r1−/−Bhlhe40GFP (CD45.2) mice that had previously been immunized with MOG/CFA + PTX to WT (CD45.1) recipients. The next day, these mice were immunized with MOG/CFA + PTX, and at day 7 DLNs were examined for GFP expression on the transferred CD4+ T cells, which were found to uniformly express the activation marker CD44 (Fig. 10 F). Within the population of GFPpos cells, IL-1R–sufficient Bhlhe40GFP cells were found at an ∼3:1 ratio over Il1r1−/−Bhlhe40GFP cells (Fig. 10, F and G), confirming a cell-intrinsic role for IL-1R signaling in Bhlhe40 induction.
Figure 10.
IL-1R signaling is required in vivo for full Bhlhe40 expression by Th cells, and systemic PTX induces IL-1β production by lymph node cells. (A) GFP (Bhlhe40) expression by CD4+ T cells from the DLNs of Bhlhe40GFP or Il1r1−/− Bhlhe40GFP mice on day 7 after immunization with MOG/CFA + PTX. One representative experiment out of three is shown (n = 9–10 mice/group). (B) Quantitation of the percentage (left) of GFPpos cells (of CD4+ T cells) and the (right) number of GFPpos CD4+ T cells in the DLNs from A. Data are combined from three experiments (n = 9–10/group). (C) GFP (Bhlhe40) expression by CD4+ T cells from the DLNs of Bhlhe40GFP mice treated with control Ab or IL-1 blockade in vivo on days −1, 1, and 4. Mice were left unimmunized or were immunized with MOG/CFA ± PTX treatment (days 0 and 2). One experiment out of two is shown (n = 1–3 mice/group). (D) Quantitation of the percentage (left) of GFPpos cells (of CD4+ T cells) and the (right) number of GFPpos CD4+ T cells in the DLNs from (C). One experiment out of two is shown (n = 1–3 mice/group). (E) Mean EAE scores of WT, Bhlhe40−/−, and Il1r1−/− mice immunized with MOG/CFA + PTX. Data are combined from two experiments (n = 6–12). Incidence of clinical disease is indicated. (F) CD4+ T cells from the DLN of MOG/CFA + PTX-immunized Bhlhe40GFP (CD45.1/CD45.2) mice and Il1r1−/−Bhlhe40GFP (CD45.2) mice were purified on day 7 and co-transferred to WT (CD45.1) recipients 1 d before immunization with MOG/CFA + PTX. 7 d after this immunization, transferred Bhlhe40GFP or Il1r1−/−Bhlhe40GFP GFPpos cells were identified. One experiment out of two is shown (n = 6). (G) Quantitation of the percentage of transferred Bhlhe40GFP or Il1r1−/−Bhlhe40GFP GFPpos cells (of GFPpos cells) in the DLNs from F. One experiment out of two is shown (n = 6). (H) Groups of WT mice were either unimmunized or immunized with MOG/CFA. PTX or mPTX was administered i.p. on days 0 and 2. DLN cells, collected on day 7, were cultured with or without 50 µg/ml heat-killed Mtb for 2 d and supernatants were analyzed for IL-1β. Data are combined from three experiments (n = 3–7 mice/group). (I) Quantitation of the percentage of pro–IL-1β+ cells (of B220− DLN cells) from WT mice treated as in H. Data are combined from four experiments (n = 3–12/group). (J) ICS for pro–IL-1β expression by B220− DLN cells from representative mice shown in I. (K; left) Ly6G and Ly6C expression on B220− DLN cells from the mice shown in (J). Numbers show the percentage of Ly6G+Ly6C+ neutrophils. (right) MHC class II and Ly6C expression by Ly6G−B220− cells. Numbers show the percentage of monocytes/moDCs. One experiment out of four is shown (n = 3–12/group). (L) Quantitation of the number of pro–IL-1β+ neutrophils, MHC II+Ly6C+ monocytes/moDCs, migratory DCs, and resident DCs in the DLNs from (I-K). Data are combined from two experiments (n = 2–7/group). Data are mean ± SEM. Dots represent individual mice except in E. Unpaired (B, D, H, I, and L) or paired (G) two-tailed Student's t tests were performed to determine significance. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Systemic PTX treatment induces IL-1β production by lymph node cells
Because both PTX treatment and IL-1β regulated Bhlhe40 expression by CD4+ T cells after immunization, we next asked whether PTX could regulate IL-1β production in vivo. DLN cells from MOG/CFA + PTX-immunized mice at day 7 secreted IL-1β in culture (Fig. 10 H). Treatment with heat-killed Mycobacterium tuberculosis (Mtb), a component of CFA, could further augment this secretion. mPTX, which failed to induce Bhlhe40 expression in CD4+ T cells in vivo (Fig. 3, A and B), also was unable to drive IL-1β secretion by lymph node cells when combined with MOG/CFA immunization. Pro–IL-1β was detectable in lymph node cells only in mice that received PTX with immunization (Fig. 10, I and J), and DLNs from these mice contained sizeable populations of Ly6G+ neutrophils and Ly6G−Ly6C+MHC class II+ monocytes/monocyte-derived DCs (moDCs; Fig. 10 K). Neutrophils and Ly6G−Ly6C+MHC class II+ cells, but not migratory or resident DCs, were the major source of PTX-induced IL-1 (Fig. 10 L).
DISCUSSION
We and others have previously demonstrated that the transcription factor Bhlhe40 is required for autoimmune neuroinflammation through its action in Th cells (Martínez-Llordella et al., 2013; Lin et al., 2014). Here, we have used novel Bhlhe40GFP reporter mice to study Bhlhe40-expressing cells during EAE. Bhlhe40-expressing Th cells were abundant in the CNS, where they secreted IFN-γ, IL-17A, and GM-CSF. PTX, when administered systemically with immunization, served as a strong stimulus for Bhlhe40 expression by Th cells. PTX treatment enhanced antigen-specific Th cell cytokine responses, and this effect was abrogated in Bhlhe40−/− mice. IL-1β production by lymph node myeloid cells was markedly augmented by systemic PTX treatment administered with MOG/CFA immunization. IL-1β acted directly on Th17 cells to positively regulate the expression of Bhlhe40, with Bhlhe40-expressing Th17 cells exhibiting an encephalitogenic transcriptional signature.
We find that Bhlhe40 expression is nonuniform in autoreactive CD4+ T cells during EAE. Bhlhe40 is not expressed in naive CD4+ T cells, but is induced upon T cell activation (Sun et al., 2001; Miyazaki et al., 2010). Polyclonal T cells with a spectrum of TCR affinities for MHC class II peptide could tune Bhlhe40 expression levels, although in experiments with monoclonal TCR Tg CD4+ T cells, we also found nonuniform Bhlhe40 expression. Recently, Helmstetter et al. (2015) described intraclonal heterogeneity in the production of IFN-γ by Th1 cells, explained by intrapopulation differences in the expression of the transcription factor T-bet. These authors also studied monoclonal T cells, and concluded that a diverse TCR repertoire was not essential to generate heterogeneity in effector cell responses. Previous work has described a range of TCR signal strength, even in stimulated monoclonal T cells, determined by the stochastic expression of proximal signaling components downstream of the TCR (Feinerman et al., 2008). Therefore, it remains possible that TCR signal strength could contribute to establishing a range of Bhlhe40 expression in activated T cells. Numerous non-TCR signals also likely act in combination to establish a population of T cells with a wide spectrum of transcription factor and effector molecule expression levels. CD28 signaling is known to induce Bhlhe40 expression in CD4+ T cells (Martínez-Llordella et al., 2013), and other positive or negative co-stimulatory signals may also participate. In naive PD-1–deficient mice, for example, CD8+ T central memory phenotype cells express higher levels of Bhlhe40 than the same cells derived from naive WT mice (Charlton et al., 2013). Diverse cytokine receptor signals also likely integrate to shape T cell responses. We found IL-1R signaling to strongly impact on Bhlhe40 expression levels in Th17 cells. IL-1 has been shown to induce Bhlhe40 in primary human gingival epithelial cells through a PI-3K–Akt pathway (Bhawal et al., 2012), and in primary human amnion mesenchymal cells (Li et al., 2011). Recently, IL-1R signals were shown to modulate Th17 cell responses through the repression of SOCS3 and the subsequent strengthening of STAT3 activity (Basu et al., 2015). It is interesting to consider whether these pathways are involved in the induction of Bhlhe40 by IL-1 in Th17 cells.
The development of autoimmune diseases is thought to involve both genetic and environmental factors. In the EAE model, roles for both pathogenic and commensal microbes have been considered to regulate effector and regulatory Th cell responses (Berer and Krishnamoorthy, 2014). Active EAE induction protocols involve immunization with myelin antigens administered with CFA to prime dormant autoreactive Th cells, although in C57BL/6 mice this strategy is insufficient to induce EAE. In these mice, PTX is a required co-adjuvant (Levine and Sowinski, 1973; Bettelli et al., 2003), perhaps functioning as a surrogate for the environmental factors that trigger MS in genetically susceptible hosts. Our work shows that PTX co-adjuvanticity, as read out by an increase in antigen-specific CD4+ T cell cytokine responses, requires Bhlhe40 expression in vivo. Immunization combined with PTX treatment induces Bhlhe40 expression in CD4+ T cells, and this induction of Bhlhe40 is partially IL-1 dependent. PTX was unable to act directly on T cells in vitro to induce Bhlhe40 expression (unpublished data), suggesting that PTX acts indirectly in vivo to promote CD4+ T cell responses.
We have found that immunization combined with PTX treatment induces IL-1β production by neutrophils and monocytes/moDCs. Our data are consistent with two nonmutually exclusive actions for PTX in this process. Evidence exists for the ability of PTX to directly stimulate myeloid cell production of IL-1β. Zhang et al. (2011) showed that macrophages infected with PTX-deficient Bordetella pertussis produced less IL-1β than those infected with WT bacteria. More recently, Dumas et al. (2014) showed that PTX administered by a protocol akin to its use in EAE rapidly triggered IL-1β production by peritoneal macrophages and neutrophils in a pathway dependent on the ADP-ribosylating activity of the toxin and on a pyrin-containing inflammasome. Alternatively, PTX may induce initial GM-CSF production (and perhaps Bhlhe40 expression) by antigen-stimulated Th cells through an undefined mechanism. This GM-CSF may then act directly on lymph node myeloid cells to induce their production of IL-1β, which could feed forward to direct full Bhlhe40 expression and encephalitogenicity of Th cells. Consistent with this proposed positive-feedback loop, a cell-intrinsic role for GM-CSF receptor signaling on CCR2+ monocytes after EAE induction was recently described by Croxford et al. (2015a,b), who concluded that T cell–derived GM-CSF served to regulate IL-1β production by Ly6C+MHC class II+ cells, and that this signaling was required for the development of EAE. Khameneh et al. (2011) have also reported that GM-CSF can synergize with a TLR ligand to enhance IL-1β secretion by bone marrow–derived DCs/macrophages via increased NF-κB activation. In MS, the detection of IL-1 in the cerebrospinal fluid at disease onset has been linked with cortical pathology (Seppi et al., 2014), whereas its detection during remission in relapsing-remitting MS has been shown to correlate with a more severe disease course (Rossi et al., 2014). The role of IL-1 in directly promoting Th cell pathogenicity in MS remains unknown.
Our study demonstrates that among autoreactive Th cells, Bhlhe40 expression identifies those with encephalitogenic features. No data exists on whether human BHLHE40 is expressed in T cells in MS lesions. Some studies have focused on the role of IFN-γ/IL-17 double-producing Th1/17 cells as being the bona fide pathogenic cells in EAE or MS (Kebir et al., 2009; Duhen et al., 2013), which in some cases have also been reported to produce GM-CSF. A requirement for GM-CSF in EAE is well established (Codarri et al., 2011; El-Behi et al., 2011), and newer studies have suggested a role for this cytokine in MS (Hartmann et al., 2014; Noster et al., 2014; Rasouli et al., 2015). IL-1β in conjunction with IL-12 can direct human Th17 cells to differentiate into IFN-γ/IL-17/GM-CSF triple-producing cells (Duhen and Campbell, 2014). We have demonstrated that Bhlhe40 expression in murine Th cells controls GM-CSF production (Lin et al., 2014), identifies encephalitogenic Th17 cells, and is regulated by IL-1. Our study provides impetus for exploring the role of BHLHE40 in autoreactive Th cells in human autoimmune disease.
MATERIALS AND METHODS
Mice
C57BL/6 (Taconic), B6.SJL (CD45.1; Taconic), 2D2 TCR transgenic (The Jackson Laboratory), Il1r1−/− (on a C57BL/6 background; The Jackson Laboratory), and Bhlhe40−/− (Sun et al., 2001; backcrossed 10 generations to the C57BL/6 background) mice were maintained in our SPF facility. The Bhlhe40GFP BAC Tg mouse strain, STOCK Tg(Bhlhe40-EGFP)PX84Gsat/Mmucd, identification number 034730-UCD, was obtained from the Mutant Mouse Regional Resource Center (MMRRC), a National Center for Research Resources-National Institutes of Health (NCRR-NIH) funded strain repository, and was donated to the MMRRC by the National Institute of Neurological Disorders and Stroke (NINDS) funded Gene Expression Nervous System Atlas (GENSAT) BAC transgenic project. Frozen spermatozoa from hemizygotes were obtained and mice were reanimated and backcrossed to the C57BL/6 background (hemizygous mice at backcross 7–11 were used). A strain-specific PCR with primers suggested by the MMRRC was used to type for the presence of the transgene (forward, 5′-GGGCAGCCCTTTCTCAGACTCTAC-3′; reverse, 5′-GGTCGGGGTAGCGGCTGAA-3′). 2D2 TCR transgenic mice were crossed to B6.SJL mice to generate 2D2 CD45.1 mice that were subsequently crossed to Bhlhe40GFP mice or Bhlhe40−/− mice for some experiments. Il1r1−/− mice were crossed to Bhlhe40GFP mice for some experiments. Experiments were performed with mice at 8–20 wk of age. All animal experiments were approved by the Animal Studies Committee of Washington University in St. Louis.
Induction of active EAE and immunizations
For EAE induction, mice were immunized subcutaneously with 100 µg MOG35–55 (CS Bio Co.) emulsified in CFA (made with 5 mg/ml heat-killed Mtb H37Ra [BD] in incomplete Freund’s adjuvant [BD]). PTX (List Biological Laboratories) or mutant PTX (mPTX; mutated at two positions in the S1 subunit [R9K and E129A]; List Biological Laboratories) was injected i.p. (300 ng) on days 0 and 2. Mice were monitored for signs of classical EAE for at least 28 d and graded on a standard 0–5 scale, as previously described (Lees et al., 2010). For analysis of T cell responses in DLNs, mice were immunized in hind footpads with 10 nM MOG35–55 or OVA323-339 peptide emulsified in CFA. Some mice were injected with PTX i.p. (300 ng) on days 0 and 2. To analyze MOG-specific T cell responses, 2 × 106 magnetically purified 2D2 Bhlhe40GFP CD45.1/CD45.2 CD4+ T cells (Dynabeads FlowComp Mouse CD4 kit; Invitrogen) were transferred i.p. to Bhlhe40GFP or WT recipient mice (CD45.2) 1 d before MOG/CFA ± PTX immunization. In some experiments, 1.5 × 106 magnetically purified CD4+ T cells from the DLN of MOG/CFA + PTX-immunized (day 7) Bhlhe40GFP (CD45.1/CD45.2) mice and 1.5 × 106 magnetically purified CD4+ T cells from the DLN of MOG/CFA + PTX-immunized (day 7) Il1r1−/− Bhlhe40GFP (CD45.2) mice were co-transferred into WT (CD45.1) recipients 1 d before immunization with MOG/CFA + PTX. At day 7 after this immunization, DLN cells were collected and analyzed for GFP expression.
In vivo IL-1 blockade
Anti–IL-1β antibody (200 µg/dose; clone B122; Bio X Cell or Leinco Technologies, Inc.) and anti–IL-1R antibody (200 µg/dose; clone JAMA-147; Bio X Cell or Leinco Technologies, Inc.) were combined (IL-1 blockade) in 200 µl of PBS and injected i.p. into mice on days −1, 1, and 4. Controls received 400 µg/dose isotype control Armenian hamster IgG (clone PIP; a gift from R.D. Schreiber, Washington University, St. Louis, MO) on the same days. Mice were left unimmunized or were immunized with MOG/CFA or MOG/CFA + PTX on day 0 and sacrificed on day 7.
Cell preparation
Cells from bone marrow, peritoneum, thymus, inguinal lymph node, and spleen were collected from naive WT and Bhlhe40GFP mice for analysis of GFP expression in immune cells. Lungs were perfused with 10 ml PBS via injection into the right ventricle. Lungs were minced and digested with 4 mg/ml collagenase D (Roche) at 37°C for 40–60 min with stirring, followed by addition of EDTA (5 mM final concentration) with incubation on ice for 5 min. Brains and spinal cords from naive mice, immunized mice, or mice that received adoptive transfers of 2D2 cells were collected after perfusion of the left ventricle with 30 ml PBS, and were digested with 500 µg/ml type I collagenase (Sigma-Aldrich) and 10 µg/ml DNase I (Sigma-Aldrich) in the presence of 0.1 µg/ml TLCK trypsin inhibitor (Sigma-Aldrich) and 10 mM Hepes (pH 7.4) in HBSS at room temperature for 1 h with shaking. Centrifuged cells were resuspended in a 70%/37%/30% Percoll gradient (Sigma-Aldrich) in HBSS and centrifuged for 30 min at 1,200 g. The 30% Percoll layer containing debris was discarded and the cells from the interface of the 30 and 37% layers were collected for subsequent use.
From naive, PTX-treated, and immunized mice, DLNs were digested with 250 µg/ml collagenase B (Roche) and 30 U/ml DNase I (EMD) for 50–60 min at 37°C with stirring in Iscove’s modified Dulbecco’s media (IMDM) containing 10% FCS, l-glutamine, sodium pyruvate, nonessential amino acids, penicillin/streptomycin, and 2-mercaptoethanol (cIMDM). EDTA (5 mM final concentration) was added, and cells were incubated on ice for 5 min. Spleens were mashed and filtered through 70-µm strainers to make single-cell suspensions. From all cell suspensions, erythrocytes were lysed with ACK lysis buffer if necessary, followed by filtration through a 70-µm strainer.
In vitro Th1 and Th17 polarization and adoptive transfer EAE models
Naive CD62L+ CD4+ T cells were magnetically purified from spleens and inguinal lymph nodes using EasySep mouse naive CD4+ T cell isolation kits (StemCell Technologies). Th1 and Th17 cells were polarized by the method of Jäger et al. (2009) with minor modifications. In brief, naive CD4+ T cells were cultured in cIMDM with WT, Bhlhe40−/−, or Il1r1−/− irradiated splenocytes (3,400 rads), 2.5 µg/ml soluble anti-CD3 (clone 145-2C11; Bio X Cell or Leinco Technologies, Inc.), and anti–IL-4 (20 µg/ml; clone 11B11; Bio X Cell or Leinco Technologies, Inc.). Th1 cell cultures also included IL-12 (10 ng/ml; BioLegend). Th17 cells were polarized with the addition of anti–IFN-γ (20 µg/ml; clone XMG1.2 [Bio X Cell] or clone H22 [Leinco Technologies, Inc.]), recombinant human TGF-β1 (3 ng/ml; BioLegend), and recombinant mouse IL-6 (30 ng/ml; BioLegend). In some experiments, recombinant mouse IL-1β (10 ng/ml; BioLegend) or anti–IL-1R (20 µg/ml; clone JAMA-147; Bio X Cell or Leinco Technologies, Inc.) plus anti–IL-1β (20 µg/ml; clone B122; Bio X Cell or Leinco Technologies, Inc.; IL-1 blockade) was added to the culture. Cells were split on day 2 with or without the addition of recombinant mouse IL-2 (10 ng/ml; BioLegend) for Th1 cells or IL-23 (10 ng/ml; BioLegend or R&D Systems) for Th17 cultures. For analysis by flow cytometry or immunoblotting, Th17 cells were harvested on day 4–7. For generation of effector 2D2 T cells, naive WT 2D2 (CD45.1) and Bhlhe40−/− 2D2 (CD45.2) cells were magnetically purified and polarized in Th1 or Th17 conditions. Cells were split on day 2 and cultured with IL-2 (Th1) or IL-23 (Th17) for 4 more days. Polarized cells were harvested, counted, and restimulated with plate bound anti-CD3 (2 µg/ml) and anti-CD28 (2 µg/ml; clone 37.51; Bio X Cell or BioLegend) at a concentration of 2 × 106/ml for 48 h. After restimulation, cells were harvested, counted, and washed. 2 × 106 Th1 or Th17 cells were injected i.p. into WT recipients. Mice were monitored for signs of EAE for at least 28 d.
Lymph node γδ T cell stimulation
Inguinal lymph nodes were collected from naive Bhlhe40GFP mice and cell suspensions were cultured at 5 × 105 cells/well in 200 µl cIMDM in the presence or absence of 2.5 µg/ml soluble anti-CD3 and/or 10 ng/ml IL-1β. At day 3, flow cytometry was performed to analyze GFP expression by γδ T cells.
Flow cytometry
The following anti–mouse antibodies were obtained from BioLegend: FITC (17A2), APC, or APC-Cy7 (145-2C11) anti-CD3e, BV510 or PE-Cy7 anti-CD4 (RM4-5), APC-Cy7 or PerCP-Cy5.5 anti-CD8α (53-6.7), APC or PE-Cy7 anti-CD11b (M1/70), APC-Cy7 anti-CD11c (N418), APC anti-CD25 (PC61), PB anti-CD44 (IM7), APC anti-CD45.2 (104), PE anti-CD62L (MEL-14), APC anti–IL-7Rα (CD127; SB/199), Alexa Fluor 488 anti-B220 (RA3-6B2), FITC, PB, or PerCP-Cy5.5 anti-Ly6C (HK1.4), PE anti-Ly6G (1A8), BV510 or PB anti–I-A/I-E (M5/114.15.2), A647 anti-ICAM2 (3C4), APC anti-NK1.1 (PK136), PerCP-Cy5.5 anti-NKp46 (29A1.4), PerCP-Cy5.5 anti–Siglec H (551), PB anti-TCRβ (H57-597), APC anti-TCR γδ (GL3), PE-Cy7 anti–IFN-γ (XMG1.2), PerCP-Cy5.5 anti–L-17A (TC11-18H10.1), and PE anti–GM-CSF (MP1-22E9). The following anti–mouse antibodies were purchased from BD: V450 anti-CD4 (RM4-5), APC-Cy7, V500, or BV510 anti-CD45 (30-F11), V500 anti-B220 (RA3-6B2), PE anti–Ki-67 (B56), PE anti-NK1.1 (PK136), and PE anti–Siglec F (E50-2440). The following anti–mouse antibodies were obtained from eBioscience: PE-Cy7 anti-CD93 (AA4.1), PB anti-CD49b (DX5), APC anti-Slamf6 (eBio13G3-19D), APC-e780 anti-CD45 (30-F11), PE anti-Foxp3 (FJK-16s), and PerCP-eFluor710 anti-pro–IL-1β (NJTEN3). V450 or PerCP-Cy5.5 anti-CD45.1 (A20), APC-Cy7 anti-CD45.2, and PE-Cy7 anti-Ly6G (1A8) were purchased from Tonbo Biosciences. PO-PRO-1 and 7-AAD obtained from Life Technologies were used for flow cytometry performed on lungs to discriminate live and dead cells.
Fc receptor blocking was performed (clone 93; BioLegend or clone 2.4G2; Bio X Cell) in FACS buffer (0.5% BSA, 2 mM EDTA, and 0.02% sodium azide in PBS) for 5–10 min at 4°C before surface staining (4°C for 20 min). ICS was performed after a 3–4 h stimulation of T cells with PMA (50 ng/ml; Enzo Life Sciences) and ionomycin (1 µM; Enzo Life Sciences) with Brefeldin A (1 µg/ml; Enzo Life Sciences). Cells were fixed in 4% paraformaldehyde (Electron Microscopy Sciences) at room temperature for 20 min, followed by permeabilization with the Cytofix/Cytoperm Fixation and Permeabilization kit (BD). ICS was performed in permeabilization buffer at 4°C for 20 min. APC MOG38–49–I-Ab tetramers were obtained from the National Institutes of Health Tetramer Core Facility. Splenic or CNS cells were incubated with 4.7 µg/ml (1:300) tetramers for 30 min at 37°C. Cells were then used for surface staining. For Ki-67 staining, cells were initially fixed with 2% paraformaldehyde (4°C for 20 min), and then fixed and permeabilized with the eBioscience Foxp3/Transcription Factor Staining Buffer Set. Ki-67 staining was performed for 1 h at 4°C. For Foxp3 staining, sorted CD44−GFPneg, CD44+GFPneg, and CD44+GFPpos CD4+ T cells from the CNS of EAE-induced mice were fixed and permeabilized with the eBioscience Foxp3/Transcription Factor Staining Buffer Set. Foxp3 staining was performed for 30–60 min at room temperature. Flow cytometry was performed on a FACSCanto II (BD) or a FACSAria II (BD). Flow cytometry data were analyzed with FlowJo software (Tree Star).
Fluorescent microscopy
Spinal cords from EAE-induced Bhlhe40GFP mice were dissected, fixed in formalin followed by sucrose, frozen in OCT media, and cryosectioned as 10-µm sections. Tissues were blocked with 0.4% Triton-X 100 in 10% FBS, and then in 10% FBS. Sections were then stained with primary antibody (biotin anti–mouse CD4; clone RM4-5; Bio X Cell) or biotin anti–mouse CD11b (clone M1/70; Bio X Cell; both biotinylated in house) diluted in 10% FBS, washed, and then stained with goat anti–rat IgG (H+L) Alexa Flour 555 (Invitrogen). Sections were mounted with Abcam Fluoroshield Mounting Medium with DAPI and viewed on a Nikon Eclipse E800 epifluorescence scope equipped with a QImaging EXi Blue camera and QCapture software (QImaging).
Enzyme-linked ImmunoSpot assay (ELISPOT)
DLN cells from immunized mice were cultured in Mutiscreen Filter Plates (EMD Millipore) at 0.5–1 × 106 cells/well in cIMDM and stimulated with media, 10 µM MOG35-55 peptide, or 1 µg/ml concanavalin A (ConA; Sigma-Aldrich) at 37°C overnight. IFN-γ and IL-10 ELISPOT pairs were obtained from BD. IL-17A and GM-CSF ELISPOT pairs and streptavidin-alkaline phosphatase were purchased from BioLegend. An Immunospot counter (Cellular Technology Ltd.) was used for spot counting after plate development with NBT/BCIP substrate (Roche).
ELISA
DLN cells from naive, PTX-treated, or immunized mice were cultured at 106 cells/well in 200 µl cIMDM and left unstimulated or stimulated with 50 µg/ml heat-killed Mtb at 37°C for 48 h. Cell-free supernatants were analyzed for mature IL-1β by ELISA (BioLegend). Assays were performed on Nunc Maxisorp plates and developed with streptavidin-HRP (BioLegend) plus TMB substrate (BioLegend). Standard curves were generated with purified IL-1β (BioLegend).
Immgen data analysis and expression microarrays
Data from the Immgen Consortium (GEO accession nos. GSE15907 [phase 1] and GSE37448 [phase 2]) were downloaded and used to determine Bhlhe40 expression in a variety of immune cell types. Arraystar 5 software (DNAstar) was used to normalize data using the robust multiarray analysis method with quantile normalization.
Naive CD4+ T cells from Bhlhe40GFP mice were cultured in Th17 conditions (TGF-β1 + IL-6 + IL-23 + IL-1β) for 4 d and stimulated for 3 h with PMA and ionomycin. GFPneg and GFPpos Th17 cells (CD45.2+CD4+CD8−) were purified (95% purity) by cell sorting before total RNA was isolated (E.Z.N.A. MicroElute Total RNA kit; Omega Bio-Tek). RNA was amplified using the Ovation PicoSL WTA System V2 (Nugen), biotin labeled with the Encore Biotin Module (Nugen), fragmented, and hybridized to Affymetrix Mouse Gene 1.0 ST arrays. Arraystar 5 software was used as described above. Data have been deposited in GEO under accession no. GSE75407. GENE-E software (Broad Institute) was used to determine differential gene expression and generate a heat map for the most expressed 14,000 probesets. The 700 genes (5%) that were most highly expressed in GFPpos Th17 cells compared with GFPneg Th17 cells were identified, and GENE-E software was used to determine the relative expression of these 700 genes in GEO accession no. GSE23505 (arrays performed on Affymetrix Mouse Genome 430 2.0 arrays; Ghoreschi et al., 2010) in which Th cells had been cultured in 6 different cytokine conditions.
Quantitative RT-PCR
GFPneg and GFPpos CD4+ T cells (gated on CD3+B220−CD4+ cells) were sorted from DLNs on day 7 after immunization with MOG/CFA + PTX. RNA was isolated (E.Z.N.A. MicroElute Total RNA kit; Omega Bio-Tek), and then cDNA was synthesized (High Capacity RNA-to-cDNA kit; Invitrogen). Quantitative real-time PCR was performed with Power SYBR Green PCR Master Mix (Applied Biosystems) using a StepOnePlus Real-Time PCR machine (Applied Biosystems). Gene expression was determined relative to Hprt by the ΔCT method. The following primers were used: Hprt, forward 5′-TCAGTCAACGGGGGACATAAA-3′, reverse 5′-GGGGCTGTACTGCTTAACCAG-3′; Egfp, forward 5′-CCTACGGCGTGCAGTGCTTCAGC-3′, reverse 5′-CGGCGAGCTGCACGCTGCGTCCTC-3′; Bhlhe40, forward 5′-ACGGAGACCTGTCAGGGATG-3′, reverse 5′-GGCAGTTTGTAAGTTTCCTTGC-3′; Csf2, forward 5′-GCCATCAAAGAAGCCCTGAA-3′, reverse 5′-GCGGGTCTGCACACATGTTA-3′.
Immunoblotting
Th1 or Th17 cells were counted and lysed at 106/40 µl in Laemmli sample buffer (Bio-Rad Laboratories) containing 2.5% β-mercaptoethanol. Cell lysates were loaded and separated by 12% SDS-PAGE (Bio-Rad Laboratories) and transferred to BioBlot-PVDF membranes (Costar). Blots were incubated with anti-Bhlhe40 (Novus Biologicals; used at 1:1,000) or anti-HDAC1 (Abcam; used at 1:2,000) primary antibodies at 4°C overnight with shaking. Blots were washed at least four times before incubation with anti–rabbit IgG-HRP (clone 5A6-1D10 [light chain specific]; Jackson ImmunoResearch Laboratories) at room temperature for 60 min with shaking. After five washes, Clarity Western ECL substrate (Bio-Rad Laboratories) or SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific) was applied, and blots were placed on Blue basic autoradiography film (GeneMate). Film was developed with a Medical Film Processor (model SRX-101A; Konica Minolta), and scanned films were analyzed with ImageJ software (NIH).
Accession no
The GEO accession no. for the microarray data reported in this paper is GSE75407.
Statistical analysis
Data were analyzed by paired or unpaired two-tailed Student’s t tests (Prism; GraphPad Software, Inc.) as indicated in the figure legends, with P ≤ 0.05 considered significant.
Acknowledgments
We thank Marina Cella and Takeshi Egawa for careful reading of the manuscript.
This work was supported by the NIH (AI113118; B.T. Edelson), a Burroughs Wellcome Fund Career Award for Medical Scientists (B.T. Edelson), an Edward Mallinckrodt, Jr. Foundation Grant (B.T. Edelson), and a Research Grant from the National Multiple Sclerosis Society (J.H. Russell). C.-C. Lin was supported by the McDonnell International Scholars Academy at Washington University. N.N. Jarjour was supported by grant 5T32AI007163 from the NIH. The mouse strain used for this research project, STOCK Tg(Bhlhe40-EGFP)PX84Gsat/Mmucd, identification number 034730-UCD, was obtained from the Mutant Mouse Regional Resource Center (MMRRC), a NCRR-NIH funded strain repository, and was donated to the MMRRC by the NINDS funded GENSAT BAC transgenic project (The GENSAT Project, NINDS Contract # N01NS02331 to the Rockefeller University). We acknowledge the NIH Tetramer Core Facility (contract HHSN272201300006C) for provision of MOG38–49–I-Ab tetramers. Research reported in this publication was supported by the Washington University Institute of Clinical and Translational Sciences grant UL1 TR000448 from the National Center for Advancing Translational Sciences of the NIH. The content is solely the responsibility of the authors and does not necessarily represent the official view of the NIH.
The authors declare no competing financial interests.
Author contributions: C.-C. Lin planned and performed experiments and wrote the manuscript. T.R. Bradstreet, E.A. Schwarzkopf, N.N. Jarjour, C. Chou, A.S. Archambault, and J. Sim performed experiments. B.H. Zinselmeyer analyzed microscopy data. J.A. Carrero and M.N. Artyomov analyzed microarray data. G.F. Wu and J.H. Russell provided advice and edited the manuscript. R. Taneja provided Bhlhe40−/− mice and made helpful suggestions. B.T. Edelson supervised the study and wrote the manuscript.
Footnotes
Abbreviations used:
- BAC
- bacterial artificial chromosome
- EAE
- experimental autoimmune encephalomyelitis
- ICS
- intracellular cytokine staining
- moDC
- monocyte-derived DC
- MOG
- myelin oligodendrocyte glycoprotein
- mPTX
- mutant PTX
- MS
- multiple sclerosis
- Mtb
- Mycobacterium tuberculosis
- PTX
- pertussis toxin
References
- Andreasen C., Powell D.A., and Carbonetti N.H.. 2009. Pertussis toxin stimulates IL-17 production in response to Bordetella pertussis infection in mice. PLoS One. 4:e7079 10.1371/journal.pone.0007079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apetoh L., Quintana F.J., Pot C., Joller N., Xiao S., Kumar D., Burns E.J., Sherr D.H., Weiner H.L., and Kuchroo V.K.. 2010. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat. Immunol. 11:854–861. 10.1038/ni.1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagley K.C., Abdelwahab S.F., Tuskan R.G., Fouts T.R., and Lewis G.K.. 2002. Pertussis toxin and the adenylate cyclase toxin from Bordetella pertussis activate human monocyte-derived dendritic cells and dominantly inhibit cytokine production through a cAMP-dependent pathway. J. Leukoc. Biol. 72:962–969. [PubMed] [Google Scholar]
- Basu R., Whitley S.K., Bhaumik S., Zindl C.L., Schoeb T.R., Benveniste E.N., Pear W.S., Hatton R.D., and Weaver C.T.. 2015. IL-1 signaling modulates activation of STAT transcription factors to antagonize retinoic acid signaling and control the TH17 cell-iTreg cell balance. Nat. Immunol. 16:286–295. 10.1038/ni.3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berer K., and Krishnamoorthy G.. 2014. Microbial view of central nervous system autoimmunity. FEBS Lett. 588:4207–4213. 10.1016/j.febslet.2014.04.007 [DOI] [PubMed] [Google Scholar]
- Bettelli E., Das M.P., Howard E.D., Weiner H.L., Sobel R.A., and Kuchroo V.K.. 1998. IL-10 is critical in the regulation of autoimmune encephalomyelitis as demonstrated by studies of IL-10- and IL-4-deficient and transgenic mice. J. Immunol. 161:3299–3306. [PubMed] [Google Scholar]
- Bettelli E., Pagany M., Weiner H.L., Linington C., Sobel R.A., and Kuchroo V.K.. 2003. Myelin oligodendrocyte glycoprotein-specific T cell receptor transgenic mice develop spontaneous autoimmune optic neuritis. J. Exp. Med. 197:1073–1081. 10.1084/jem.20021603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhawal U.K., Ito Y., Tanimoto K., Sato F., Fujimoto K., Kawamoto T., Sasahira T., Hamada N., Kuniyasu H., Arakawa H., et al. . 2012. IL-1β-mediated up-regulation of DEC1 in human gingiva cells via the Akt pathway. J. Cell. Biochem. 113:3246–3253. 10.1002/jcb.24205 [DOI] [PubMed] [Google Scholar]
- Cassan C., Piaggio E., Zappulla J.P., Mars L.T., Couturier N., Bucciarelli F., Desbois S., Bauer J., Gonzalez-Dunia D., and Liblau R.S.. 2006. Pertussis toxin reduces the number of splenic Foxp3+ regulatory T cells. J. Immunol. 177:1552–1560. 10.4049/jimmunol.177.3.1552 [DOI] [PubMed] [Google Scholar]
- Charlton J.J., Chatzidakis I., Tsoukatou D., Boumpas D.T., Garinis G.A., and Mamalaki C.. 2013. Programmed death-1 shapes memory phenotype CD8 T cell subsets in a cell-intrinsic manner. J. Immunol. 190:6104–6114. 10.4049/jimmunol.1201617 [DOI] [PubMed] [Google Scholar]
- Chen X., Winkler-Pickett R.T., Carbonetti N.H., Ortaldo J.R., Oppenheim J.J., and Howard O.M.. 2006. Pertussis toxin as an adjuvant suppresses the number and function of CD4+CD25+ T regulatory cells. Eur. J. Immunol. 36:671–680. 10.1002/eji.200535353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Howard O.M., and Oppenheim J.J.. 2007. Pertussis toxin by inducing IL-6 promotes the generation of IL-17-producing CD4 cells. J. Immunol. 178:6123–6129. 10.4049/jimmunol.178.10.6123 [DOI] [PubMed] [Google Scholar]
- Chung Y., Chang S.H., Martinez G.J., Yang X.O., Nurieva R., Kang H.S., Ma L., Watowich S.S., Jetten A.M., Tian Q., and Dong C.. 2009. Critical regulation of early Th17 cell differentiation by interleukin-1 signaling. Immunity. 30:576–587. 10.1016/j.immuni.2009.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codarri L., Gyülvészi G., Tosevski V., Hesske L., Fontana A., Magnenat L., Suter T., and Becher B.. 2011. RORγt drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat. Immunol. 12:560–567. 10.1038/ni.2027 [DOI] [PubMed] [Google Scholar]
- Connelly C.E., Sun Y., and Carbonetti N.H.. 2012. Pertussis toxin exacerbates and prolongs airway inflammatory responses during Bordetella pertussis infection. Infect. Immun. 80:4317–4332. 10.1128/IAI.00808-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxford A.L., Lanzinger M., Hartmann F.J., Schreiner B., Mair F., Pelczar P., Clausen B.E., Jung S., Greter M., and Becher B.. 2015a The cytokine GM-CSF drives the inflammatory signature of CCR2+ monocytes and licenses autoimmunity. Immunity. 43:502–514. 10.1016/j.immuni.2015.08.010 [DOI] [PubMed] [Google Scholar]
- Croxford A.L., Spath S., and Becher B.. 2015b GM-CSF in neuroinflammation: Licensing myeloid cells for tissue damage. Trends Immunol. 36:651–662. 10.1016/j.it.2015.08.004 [DOI] [PubMed] [Google Scholar]
- de Kouchkovsky D., Esensten J.H., Rosenthal W.L., Morar M.M., Bluestone J.A., and Jeker L.T.. 2013. microRNA-17-92 regulates IL-10 production by regulatory T cells and control of experimental autoimmune encephalomyelitis. J. Immunol. 191:1594–1605. 10.4049/jimmunol.1203567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhen T., and Campbell D.J.. 2014. IL-1β promotes the differentiation of polyfunctional human CCR6+CXCR3+ Th1/17 cells that are specific for pathogenic and commensal microbes. J. Immunol. 193:120–129. 10.4049/jimmunol.1302734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhen R., Glatigny S., Arbelaez C.A., Blair T.C., Oukka M., and Bettelli E.. 2013. Cutting edge: the pathogenicity of IFN-γ-producing Th17 cells is independent of T-bet. J. Immunol. 190:4478–4482. 10.4049/jimmunol.1203172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas A., Amiable N., de Rivero Vaccari J.P., Chae J.J., Keane R.W., Lacroix S., and Vallières L.. 2014. The inflammasome pyrin contributes to pertussis toxin-induced IL-1β synthesis, neutrophil intravascular crawling and autoimmune encephalomyelitis. PLoS Pathog. 10:e1004150 10.1371/journal.ppat.1004150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Behi M., Ciric B., Dai H., Yan Y., Cullimore M., Safavi F., Zhang G.X., Dittel B.N., and Rostami A.. 2011. The encephalitogenicity of T(H)17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat. Immunol. 12:568–575. 10.1038/ni.2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinerman O., Veiga J., Dorfman J.R., Germain R.N., and Altan-Bonnet G.. 2008. Variability and robustness in T cell activation from regulated heterogeneity in protein levels. Science. 321:1081–1084. 10.1126/science.1158013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoreschi K., Laurence A., Yang X.P., Tato C.M., McGeachy M.J., Konkel J.E., Ramos H.L., Wei L., Davidson T.S., Bouladoux N., et al. . 2010. Generation of pathogenic T(H)17 cells in the absence of TGF-β signalling. Nature. 467:967–971. 10.1038/nature09447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Wei G., Zhu J., Liao W., Leonard W.J., Zhao K., and Paul W.. 2009. IL-1 family members and STAT activators induce cytokine production by Th2, Th17, and Th1 cells. Proc. Natl. Acad. Sci. USA. 106:13463–13468. 10.1073/pnas.0906988106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann F.J., Khademi M., Aram J., Ammann S., Kockum I., Constantinescu C., Gran B., Piehl F., Olsson T., Codarri L., and Becher B.. 2014. Multiple sclerosis-associated IL2RA polymorphism controls GM-CSF production in human TH cells. Nat. Commun. 5:5056 10.1038/ncomms6056 [DOI] [PubMed] [Google Scholar]
- Helmstetter C., Flossdorf M., Peine M., Kupz A., Zhu J., Hegazy A.N., Duque-Correa M.A., Zhang Q., Vainshtein Y., Radbruch A., et al. . 2015. Individual T helper cells have a quantitative cytokine memory. Immunity. 42:108–122. 10.1016/j.immuni.2014.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng T.S., and Painter M.W.. Immunological Genome Project Consortium . 2008. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9:1091–1094. 10.1038/ni1008-1091 [DOI] [PubMed] [Google Scholar]
- Huber M., Heink S., Pagenstecher A., Reinhard K., Ritter J., Visekruna A., Guralnik A., Bollig N., Jeltsch K., Heinemann C., et al. . 2013. IL-17A secretion by CD8+ T cells supports Th17-mediated autoimmune encephalomyelitis. J. Clin. Invest. 123:247–260. 10.1172/JCI63681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger A., Dardalhon V., Sobel R.A., Bettelli E., and Kuchroo V.K.. 2009. Th1, Th17, and Th9 effector cells induce experimental autoimmune encephalomyelitis with different pathological phenotypes. J. Immunol. 183:7169–7177. 10.4049/jimmunol.0901906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebir H., Ifergan I., Alvarez J.I., Bernard M., Poirier J., Arbour N., Duquette P., and Prat A.. 2009. Preferential recruitment of interferon-β-expressing TH17 cells in multiple sclerosis. Ann. Neurol. 66:390–402. 10.1002/ana.21748 [DOI] [PubMed] [Google Scholar]
- Kerfoot S.M., Long E.M., Hickey M.J., Andonegui G., Lapointe B.M., Zanardo R.C., Bonder C., James W.G., Robbins S.M., and Kubes P.. 2004. TLR4 contributes to disease-inducing mechanisms resulting in central nervous system autoimmune disease. J. Immunol. 173:7070–7077. 10.4049/jimmunol.173.11.7070 [DOI] [PubMed] [Google Scholar]
- Khameneh H.J., Isa S.A., Min L., Nih F.W., and Ruedl C.. 2011. GM-CSF signalling boosts dramatically IL-1 production. PLoS One. 6:e23025 10.1371/journal.pone.0023025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohm A.P., Carpentier P.A., Anger H.A., and Miller S.D.. 2002. Cutting edge: CD4+CD25+ regulatory T cells suppress antigen-specific autoreactive immune responses and central nervous system inflammation during active experimental autoimmune encephalomyelitis. J. Immunol. 169:4712–4716. 10.4049/jimmunol.169.9.4712 [DOI] [PubMed] [Google Scholar]
- Kügler S., Böcker K., Heusipp G., Greune L., Kim K.S., and Schmidt M.A.. 2007. Pertussis toxin transiently affects barrier integrity, organelle organization and transmigration of monocytes in a human brain microvascular endothelial cell barrier model. Cell. Microbiol. 9:619–632. 10.1111/j.1462-5822.2006.00813.x [DOI] [PubMed] [Google Scholar]
- Lee Y., Awasthi A., Yosef N., Quintana F.J., Xiao S., Peters A., Wu C., Kleinewietfeld M., Kunder S., Hafler D.A., et al. . 2012. Induction and molecular signature of pathogenic TH17 cells. Nat. Immunol. 13:991–999. 10.1038/ni.2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees J.R., Sim J., and Russell J.H.. 2010. Encephalitogenic T-cells increase numbers of CNS T-cells regardless of antigen specificity by both increasing T-cell entry and preventing egress. J. Neuroimmunol. 220:10–16. 10.1016/j.jneuroim.2009.11.017 [DOI] [PubMed] [Google Scholar]
- Levine S., and Sowinski R.. 1973. Experimental allergic encephalomyelitis in inbred and outbred mice. J. Immunol. 110:139–143. [PubMed] [Google Scholar]
- Li R., Ackerman W.E. IV, Summerfield T.L., Yu L., Gulati P., Zhang J., Huang K., Romero R., and Kniss D.A.. 2011. Inflammatory gene regulatory networks in amnion cells following cytokine stimulation: translational systems approach to modeling human parturition. PLoS One. 6:e20560 10.1371/journal.pone.0020560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.C., Bradstreet T.R., Schwarzkopf E.A., Sim J., Carrero J.A., Chou C., Cook L.E., Egawa T., Taneja R., Murphy T.L., et al. . 2014. Bhlhe40 controls cytokine production by T cells and is essential for pathogenicity in autoimmune neuroinflammation. Nat. Commun. 5:3551 10.1038/ncomms4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowther D.E., and Hafler D.A.. 2012. Regulatory T cells in the central nervous system. Immunol. Rev. 248:156–169. 10.1111/j.1600-065X.2012.01130.x [DOI] [PubMed] [Google Scholar]
- Lukens J.R., Barr M.J., Chaplin D.D., Chi H., and Kanneganti T.D.. 2012. Inflammasome-derived IL-1β regulates the production of GM-CSF by CD4(+) T cells and γδ T cells. J. Immunol. 188:3107–3115. 10.4049/jimmunol.1103308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Llordella M., Esensten J.H., Bailey-Bucktrout S.L., Lipsky R.H., Marini A., Chen J., Mughal M., Mattson M.P., Taub D.D., and Bluestone J.A.. 2013. CD28-inducible transcription factor DEC1 is required for efficient autoreactive CD4+ T cell response. J. Exp. Med. 210:1603–1619. 10.1084/jem.20122387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuki T., Nakae S., Sudo K., Horai R., and Iwakura Y.. 2006. Abnormal T cell activation caused by the imbalance of the IL-1/IL-1R antagonist system is responsible for the development of experimental autoimmune encephalomyelitis. Int. Immunol. 18:399–407. 10.1093/intimm/dxh379 [DOI] [PubMed] [Google Scholar]
- McCandless E.E., Budde M., Lees J.R., Dorsey D., Lyng E., and Klein R.S.. 2009. IL-1R signaling within the central nervous system regulates CXCL12 expression at the blood-brain barrier and disease severity during experimental autoimmune encephalomyelitis. J. Immunol. 183:613–620. 10.4049/jimmunol.0802258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson R.C., Turner D.G., Mair I., O’Connor R.A., and Anderton S.M.. 2015. T-bet expression by Foxp3+ T regulatory cells is not essential for their suppressive function in CNS autoimmune disease or colitis. Front. Immunol. 6:69 10.3389/fimmu.2015.00069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki K., Miyazaki M., Guo Y., Yamasaki N., Kanno M., Honda Z., Oda H., Kawamoto H., and Honda H.. 2010. The role of the basic helix-loop-helix transcription factor Dec1 in the regulatory T cells. J. Immunol. 185:7330–7339. 10.4049/jimmunol.1001381 [DOI] [PubMed] [Google Scholar]
- Noster R., Riedel R., Mashreghi M.F., Radbruch H., Harms L., Haftmann C., Chang H.D., Radbruch A., and Zielinski C.E.. 2014. IL-17 and GM-CSF expression are antagonistically regulated by human T helper cells. Sci. Transl. Med. 6:241ra80 10.1126/scitranslmed.3008706 [DOI] [PubMed] [Google Scholar]
- O’Connor R.A., Floess S., Huehn J., Jones S.A., and Anderton S.M.. 2012. Foxp3+ Treg cells in the inflamed CNS are insensitive to IL-6-driven IL-17 production. Eur. J. Immunol. 42:1174–1179. 10.1002/eji.201142216 [DOI] [PubMed] [Google Scholar]
- Ow J.R., Tan Y.H., Jin Y., Bahirvani A.G., and Taneja R.. 2014. Stra13 and Sharp-1, the non-grouchy regulators of development and disease. Curr. Top. Dev. Biol. 110:317–338. 10.1016/B978-0-12-405943-6.00009-9 [DOI] [PubMed] [Google Scholar]
- Pot C., Jin H., Awasthi A., Liu S.M., Lai C.Y., Madan R., Sharpe A.H., Karp C.L., Miaw S.C., Ho I.C., and Kuchroo V.K.. 2009. Cutting edge: IL-27 induces the transcription factor c-Maf, cytokine IL-21, and the costimulatory receptor ICOS that coordinately act together to promote differentiation of IL-10-producing Tr1 cells. J. Immunol. 183:797–801. 10.4049/jimmunol.0901233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasouli J., Ciric B., Imitola J., Gonnella P., Hwang D., Mahajan K., Mari E.R., Safavi F., Leist T.P., Zhang G.X., and Rostami A.. 2015. Expression of GM-CSF in T Cells Is Increased in Multiple Sclerosis and Suppressed by IFN-β Therapy. J. Immunol. 194:5085–5093. 10.4049/jimmunol.1403243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi S., Studer V., Motta C., Germani G., Macchiarulo G., Buttari F., Mancino R., Castelli M., De Chiara V., Weiss S., et al. . 2014. Cerebrospinal fluid detection of interleukin-1β in phase of remission predicts disease progression in multiple sclerosis. J. Neuroinflammation. 11:32 10.1186/1742-2094-11-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan M., McCarthy L., Rappuoli R., Mahon B.P., and Mills K.H.. 1998. Pertussis toxin potentiates Th1 and Th2 responses to co-injected antigen: adjuvant action is associated with enhanced regulatory cytokine production and expression of the co-stimulatory molecules B7-1, B7-2 and CD28. Int. Immunol. 10:651–662. 10.1093/intimm/10.5.651 [DOI] [PubMed] [Google Scholar]
- Sabatino J.J. Jr., Huang J., Zhu C., and Evavold B.D.. 2011. High prevalence of low affinity peptide-MHC II tetramer-negative effectors during polyclonal CD4+ T cell responses. J. Exp. Med. 208:81–90. 10.1084/jem.20101574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E.F., Kus L., Gong S., and Heintz N.. 2013. BAC transgenic mice and the GENSAT database of engineered mouse strains. Cold Spring Harb. Protoc. 10.1101/pdb.top073692 [DOI] [PubMed] [Google Scholar]
- Seppi D., Puthenparampil M., Federle L., Ruggero S., Toffanin E., Rinaldi F., Perini P., and Gallo P.. 2014. Cerebrospinal fluid IL-1beta correlates with cortical pathology load in multiple sclerosis at clinical onset. J. Neuroimmunol. 270:56–60. 10.1016/j.jneuroim.2014.02.014 [DOI] [PubMed] [Google Scholar]
- Stromnes I.M., and Goverman J.M.. 2006. Active induction of experimental allergic encephalomyelitis. Nat. Protoc. 1:1810–1819. 10.1038/nprot.2006.285 [DOI] [PubMed] [Google Scholar]
- Sun H., Lu B., Li R.Q., Flavell R.A., and Taneja R.. 2001. Defective T cell activation and autoimmune disorder in Stra13-deficient mice. Nat. Immunol. 2:1040–1047. 10.1038/ni721 [DOI] [PubMed] [Google Scholar]
- Sutton C., Brereton C., Keogh B., Mills K.H., and Lavelle E.C.. 2006. A crucial role for interleukin (IL)-1 in the induction of IL-17-producing T cells that mediate autoimmune encephalomyelitis. J. Exp. Med. 203:1685–1691. 10.1084/jem.20060285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton C.E., Lalor S.J., Sweeney C.M., Brereton C.F., Lavelle E.C., and Mills K.H.. 2009. Interleukin-1 and IL-23 induce innate IL-17 production from gammadelta T cells, amplifying Th17 responses and autoimmunity. Immunity. 31:331–341. 10.1016/j.immuni.2009.08.001 [DOI] [PubMed] [Google Scholar]
- Walline C.C., Kanakasabai S., and Bright J.J.. 2011. IL-7Rα confers susceptibility to experimental autoimmune encephalomyelitis. Genes Immun. 12:1–14. 10.1038/gene.2010.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Koldzic D.N., Izikson L., Reddy J., Nazareno R.F., Sakaguchi S., Kuchroo V.K., and Weiner H.L.. 2004. IL-10 is involved in the suppression of experimental autoimmune encephalomyelitis by CD25+CD4+ regulatory T cells. Int. Immunol. 16:249–256. 10.1093/intimm/dxh029 [DOI] [PubMed] [Google Scholar]
- Zhang X., Hester S.E., Kennett M.J., Karanikas A.T., Bendor L., Place D.E., and Harvill E.T.. 2011. Interleukin-1 receptor signaling is required to overcome the effects of pertussis toxin and for efficient infection- or vaccination-induced immunity against Bordetella pertussis. Infect. Immun. 79:527–541. 10.1128/IAI.00590-10 [DOI] [PMC free article] [PubMed] [Google Scholar]