The miR-23∼27∼24 clusters control differentiation of effector T cells. In particular, miR-24 targets IL-4 and miR-27 targets GATA3, thus collaborating in the control of Th2 immunity.
Abstract
Coordinated repression of gene expression by evolutionarily conserved microRNA (miRNA) clusters and paralogs ensures that miRNAs efficiently exert their biological impact. Combining both loss- and gain-of-function genetic approaches, we show that the miR-23∼27∼24 clusters regulate multiple aspects of T cell biology, particularly helper T (Th) 2 immunity. Low expression of this miRNA family confers proper effector T cell function at both physiological and pathological settings. Further studies in T cells with exaggerated regulation by individual members of the miR-23∼27∼24 clusters revealed that miR-24 and miR-27 collaboratively limit Th2 responses through targeting IL-4 and GATA3 in both direct and indirect manners. Intriguingly, although overexpression of the entire miR-23 cluster also negatively impacts other Th lineages, enforced expression of miR-24, in contrast to miR-23 and miR-27, actually promotes the differentiation of Th1, Th17, and induced regulatory T cells, implying that under certain conditions, miRNA families can fine tune the biological effects of their regulation by having individual members antagonize rather than cooperate with each other. Together, our results identify a miRNA family with important immunological roles and suggest that tight regulation of miR-23∼27∼24 clusters in T cells is required to maintain optimal effector function and to prevent aberrant immune responses.
Upon microbial insult, T cells differentiate into effector T helper (Th) cells to generate protective immune responses. Recently, a class of short regulatory noncoding RNAs, the so-called microRNAs (miRNAs), known for their role in tissue development, cellular differentiation, and function, have been demonstrated to be pivotal in regulating immune responses (O’Connell et al., 2010; Lee et al., 2014). Selective expression of a defined set of miRNAs in each T cell lineage suggested miRNAs play distinct roles in controlling different aspects of T cell immunity (Kuchen et al., 2010). However, an emerging view purports that miRNAs, rather than enacting drastic gene changes, primarily reinforce preexisting transcriptional programs or buffer against stochastic fluctuations in gene expression (Ebert and Sharp, 2012). Indeed, despite complex biological phenotypes observed in mice with total T cell– or regulatory T (T reg) cell–specific inactivation of the entire miRNA pathway (Cobb et al., 2006; Chong et al., 2008; Liston et al., 2008; Zhou et al., 2008), the analysis of individual miRNA contribution to specific T cell responses has been largely restricted to a select few whose deficiency resulted in pronounced perturbation of immune cell function (Kroesen et al., 2015).
Many known miRNAs exist in clusters and paralogs with high degrees of evolutionary conservation, suggesting a means for increasing miRNA's impact on gene regulation and resultant biology. The miR-17∼92 cluster, for example, controls immune responses through several cluster members that either target the same gene or different components of common biological pathways (Ventura et al., 2008; Baumjohann et al., 2013; Kang et al., 2013; Simpson et al., 2014). Like the miR-17∼92 family, the miR-23∼27∼24 family contains multiple members and two paralogs: miR-23a∼27a∼24-2 (miR-23a cluster) on chromosome (chr) 8 (chr 19 in human) and miR-23b∼27b∼24-1 (miR-23b cluster) on chr 13 (chr 9 in human). Mature sequences of miR-23a and miR-27a differ by just one nucleotide in comparison to their corresponding paralogs miR-23b and miR-27b, whereas miR-24-1 and miR-24-2 share the same mature sequences. However, despite their distinct expression patterns in T cells, studies of the miR-23 clusters have primarily focused on their role in tumorigenesis (Mertens-Talcott et al., 2007; Chintharlapalli et al., 2009; Guttilla and White, 2009; Zhang et al., 2011; Majid et al., 2012; Wang et al., 2013). Even when in silico target analysis of the individual miRNAs within the miR-23 clusters has suggested an important role for this miRNA family in controlling T cell responses (Chhabra et al., 2010), direct experimental evidence in this direction remains limited (Guerau-de-Arellano et al., 2011; Chandran et al., 2014; Lin et al., 2014).
In this study, by using both gain- and loss-of-function genetic approaches, we investigated the roles of the miR-23 clusters, as well as each miRNA member within this miRNA family in T cell biology. Enforced expression of this miRNA family in T cells resulted in dysregulated T cell activation and autoimmune inflammation, whereas its ablation in T cells led to reduced proliferation and activation even in response to immune challenges. Moreover, in addition to having a general impact on T cell activation, the miR-23 clusters play a central role in T cell differentiation. In particular, different members of the miR-23 family cooperate to potently control Th2 immunity by targeting IL-4 and GATA3 in both direct and indirect manners. Mice harboring T cells devoid of miR-23 clusters developed exacerbated airway eosinophilic inflammation and mucus hypersecretion upon allergen sensitization and challenge. Interestingly, in modulating the differentiation of other Th lineages, we also found that different members of this miRNA family could antagonize rather than cooperate with each other. Together, our data identify a new immune regulatory miRNA family that plays a diverse role in controlling T cell immunity.
RESULTS
Enforced expression of the miR-23 cluster in T cells leads to spontaneous lymphohyperactivation phenotypes
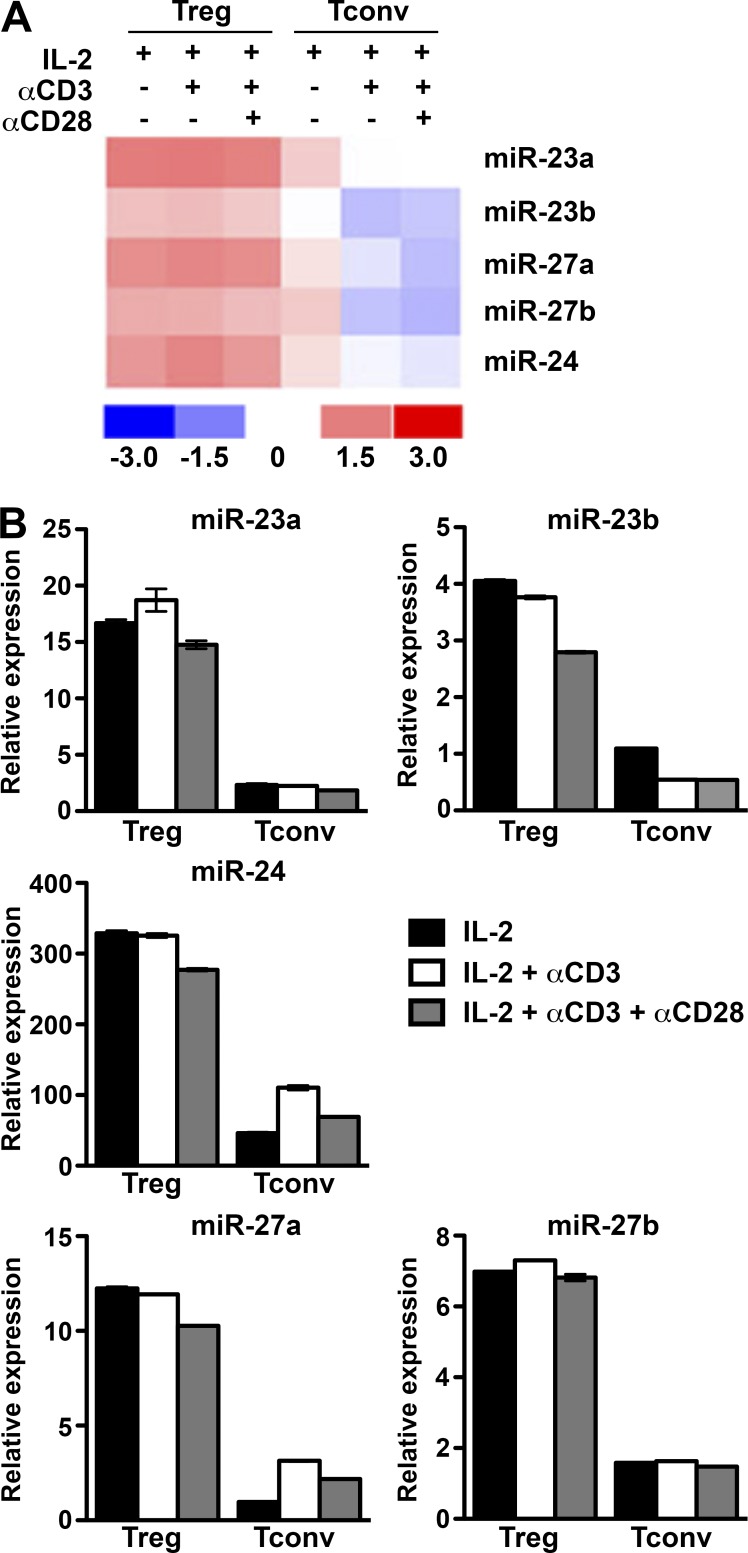
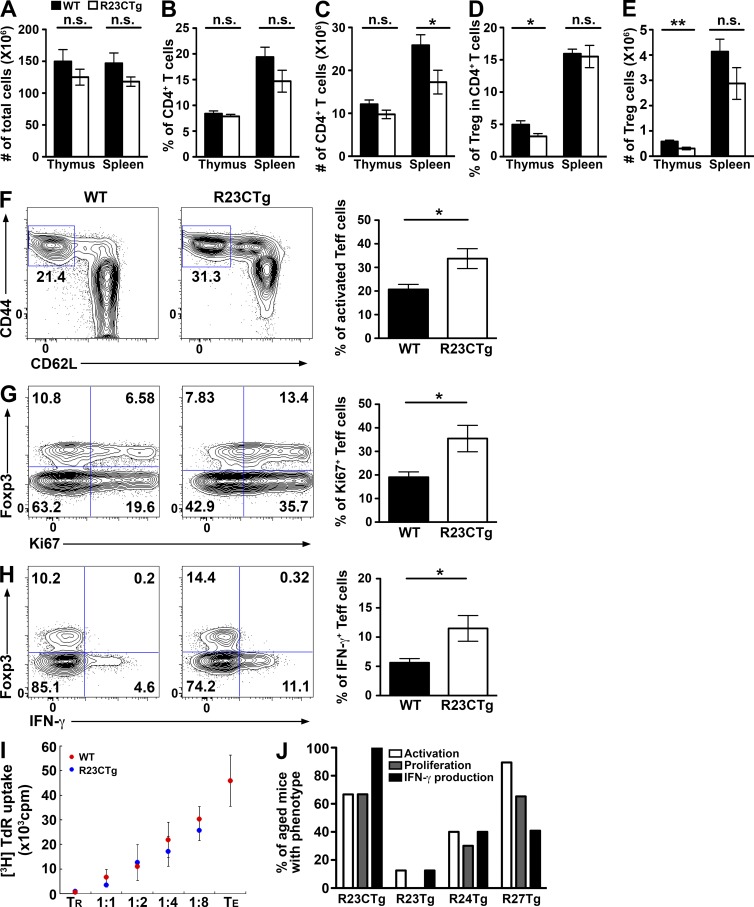
To determine the role of the miR-23 clusters in T cell immunity, we first confirmed the expression levels of both miR-23 clusters in T cells by comparative miRNA expression profiling (Table S1). Consistent with previous studies (Cobb et al., 2006; Jeker et al., 2012), higher levels of miR-23 family members were detected in T reg cells compared with conventional T (T conv) cells, regardless of activation status (Fig. 1). A recent work has shown that down-regulation of miR-23a promotes effector CD8 T cell differentiation and cytotoxicity (Lin et al., 2014). Moreover, it was previously demonstrated that global miRNA down-regulation in T cells resulted in the de-repression of genes critical for T cell differentiation and effector functions (Bronevetsky et al., 2013). Therefore, it is possible that the miR-23 clusters are expressed at necessarily diminished levels to confer normal effector function to T conv cells. To determine the impact of exaggerated miR-23 cluster regulation on T cell immunity, we generated mice that selectively overexpress the whole miR-23a cluster (R23CTg) or individual members (R23Tg, R24Tg, or R27Tg) in T cells (Fig. S1). As shown in Fig. 2 (A–E), significantly reduced thymic T reg cell numbers were detected in R23CTg mice at 6–8 wk of age, whereas no significant alteration in peripheral T reg cell frequencies nor in total thymic or splenic cellularities could be observed. These results suggested that although the miR-23 clusters are highly expressed in T reg cells, enforced expression of this miRNA family before T reg cell differentiation might impair their development in the thymus.
Figure 1.
Distinct expression patterns of miR-23 clusters in T reg versus T conv cells. miRNA array (A) and qPCR analysis (B) of the expression of individual miRNA members in miR-23 clusters in T reg cells versus T conv cells under different culture conditions. Data are representative of two independent experiments. n = 6–15.
Figure 2.
Enforced expression of the miR-23 family in T cells resulted in dysregulated T cell activation and cytokine production in mice. (A–E) Cellularity of the thymus and spleen and the proportions and absolute numbers of thymic and splenic total CD4+ T cells and Foxp3+CD4+ T reg cells in R23CTg mice and control littermates are shown. FACS analysis of CD44hiCD62Llo subset (F), Ki67+ (G), and IFN-γ–secreting cells (H) in Foxp3−CD4+ effector T cells in spleen from 6–8-wk-old R23CTg mice and control littermates were shown. Data are representative of four independent experiments. n = 6–10. *, P < 0.05; **, P < 0.01. (I) T reg cells (TR) isolated from R23CTg or WT control littermates were subjected to in vitro suppression analysis at indicated ratios of responder T cells (TE). Data are representative of three independent experiments. n = 6. (J) Percentages of mice (∼5 mo of age) with overexpression of the whole cluster or individual members harboring effector T cells displaying increased activation, proliferative activity, and production of IFN-γ. n = 4–17.
Interestingly, R23CTg mice exhibited unrestrained activation of effector T cells with greater Ki67 staining, increased CD44hiCD62Llo cell subset, and elevated IFN-γ production (Fig. 2, F–H), despite harboring normal numbers of peripheral T reg cells with comparable suppressor function (Fig. 2 I). These results support our initial hypothesis that expression of miR-23 clusters at a particular reduced level is required to maintain proper effector T cell activation and function. Finally, as mice aged, the T cell activation phenotypes in R23CTg mice could be mostly recapitulated when miR-27 alone and, to a lesser degree, when miR-24 alone were overexpressed, whereas effector T cell responses in R23Tg mice were largely unaltered (Fig. 2 J).
miR-23 family controls the differentiation of multiple Th cell lineages
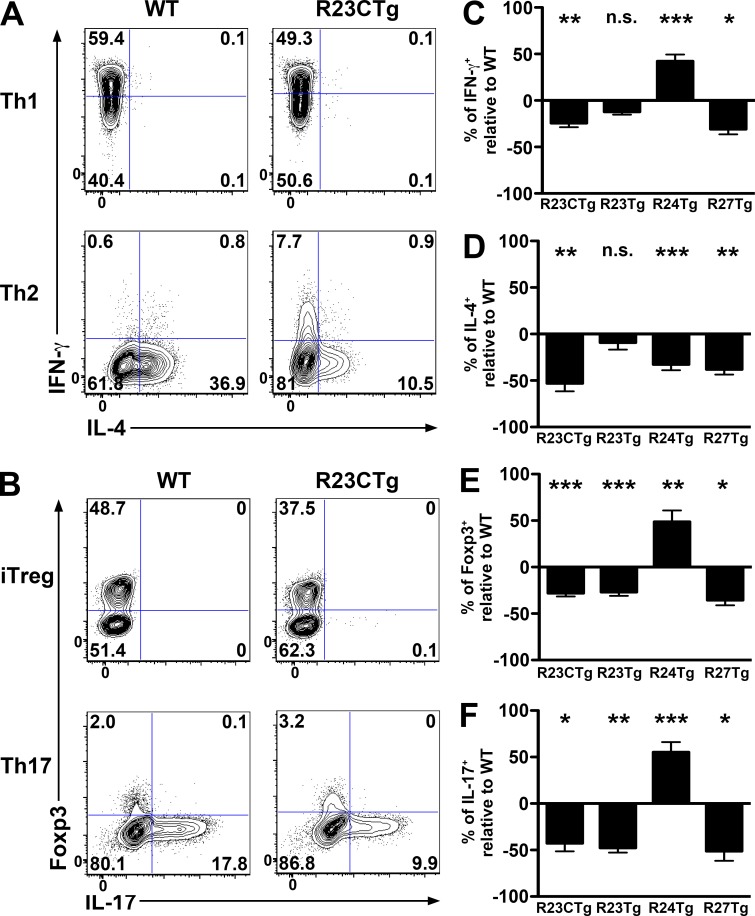
Having observed miR-23 cluster-dependent impairment in T cell immunity under steady state, we examined the effects of overexpression of the miR-23 clusters on effector T cell differentiation. Interestingly, despite T cell hyperactivation in R23CTg mice, overexpression of the miR-23 cluster negatively impacted the differentiation of every Th lineage tested (Fig. 3, A and B). Cluster overexpression also compromised the generation of TGFβ-induced T reg (iT reg) cells, just as it did thymic T reg development (Fig. 2, D and E). Further studies demonstrated that miR-27 controls the generation of all Th lineages in the same manner as the whole miR-23 cluster (Fig. 3, C–F). However, overexpression of miR-23 only led to impaired iTreg and Th17 cell induction without any significant effect on Th1/Th2 polarization. Finally, overexpression of miR-24 reduced Th2 but promoted Th1, Th17, and iT reg differentiation (Fig. 3, C–F). Collectively, our studies suggested that although individual miR-23 family members can cooperatively regulate the same biological processes, they can also antagonize each other under certain conditions to fine tune the biological effects resulting from the regulation of the entire miRNA family.
Figure 3.
miR-23 family controls the differentiation of multiple T cell lineages. Naive T cells isolated from R23CTg mice and WT littermates were cultured in vitro under Th1/Th2-polarizing (A) and Th17/iTreg differentiation (B) conditions, respectively. IFN-γ, IL-4, IL-17, and Foxp3 staining were assessed by FACS analysis. Percentages of cytokine-specific intracellular staining of the T cells isolated from mice with overexpression of the whole miR-23 cluster, as well as individual members under (C) Th1-, (D) Th2-, (E) iTreg-, and (F) Th17-polarizing conditions. Data are representative of three independent experiments. n = 3–6. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
miR-24 and miR-27 regulate Th2 differentiation and function through targeting IL-4 and GATA3
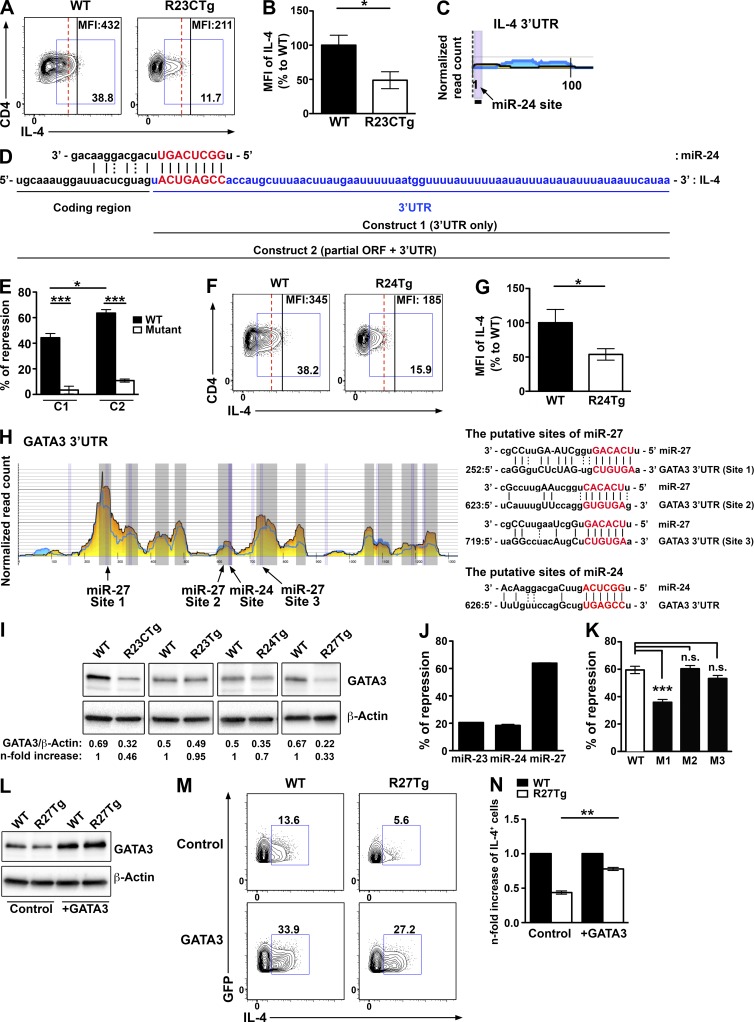
Given that only Th2 cells were not controlled contrastingly by individual members of the miR-23 family, we decided to further investigate the molecular mechanisms underlying miR-23 cluster-dependent regulation of Th2 immunity. The fact that overexpression of the miR-23 cluster in T cells reduced not only the frequency of IL-4–secreting cells but also the amount of IL-4 being produced on a per cell basis suggested that IL-4 itself might be a direct target of this miRNA family (Fig. 4, A and B). Analysis of previous results from high-throughput sequencing of RNAs isolated by cross-linking immunoprecipitation (HITS-CLIP) showed a miR-24–binding site on the relatively short 3′ UTR of the IL-4 transcript (Loeb et al., 2012; Fig. 4 C). More interestingly, this putative binding site is located just 1 nt after the stop codon, strongly suggesting the 3′ end of miR-24 would interact with sequences in the coding region of IL-4 mRNA (Fig. 4 D). Although most of the miRNA studies to date have been focused on the regulatory effect of miRNAs in the 3′ UTR of the target genes, it was well documented that miRNAs could also inhibit their target protein translation potentially through interfering with ribosome movement (Hausser et al., 2013; Brümmer and Hausser, 2014). Our luciferase reporter results confirmed that IL-4 is indeed a direct target of miR-24. Co-transfection of miR-24 with the IL-4 3′ UTR alone or extending partly into the IL-4 coding region resulted in appreciable repression of reporter activity, whereas mutation of the miR-24–binding site abolished this repression (Fig. 4 E). Moreover, the consistent increase in repression by miR-24 detected when the construct included part of the IL-4 coding region (C2) compared with the one without (C1) further suggested that interaction between miR-24 and the IL-4 coding region positively contributed to miR-24–mediated gene regulation (Fig. 4 E). Consistent with these findings, R24Tg T cells produced diminished amounts of IL-4 similar to what was observed in R23CTg T cells (Fig. 4, F and G). Yet negative regulation of IL-4 by miR-24 could not explain the reduced IL-4 production and impaired Th2 responses observed in T cells that overexpressed miR-27 alone. Further analysis of HITS-CLIP results revealed that GATA3, a central transcription factor required for Th2 differentiation and cytokine production, is a potential target of miR-27. The 3′ UTR of GATA3 contains three putative binding sites for miR-27, as well as one for miR-24 (Fig. 4 H). Consistent with this finding, compared with WT, GATA3 protein levels were significantly reduced in CD4+ T cells from R23CTg and R27Tg mice, and to a lesser extent from R24Tg mice, but not from R23Tg mice (Fig. 4 I). Interestingly, luciferase reporter assay demonstrated that GATA3 is directly repressed by miR-27 but not miR-24, and that only the first out of three putative miR-27 binding sites in the GATA3 3′ UTR is functional (Fig. 4, J and K). It is likely the reduced GATA3 expression detected in R24Tg T cells is the consequence of reduced autocrine IL-4 signaling (Scheinman and Avni, 2009). These findings also show that not all binding sites revealed by HITS-CLIP are biologically relevant. Finally, we were able to significantly restore the ability of miR-27–overexpressing T cells to produce IL-4 under Th2-polarizing condition by overexpressing GATA3 (Fig. 4, L–N). Together, these results demonstrated that the miR-23 clusters could suppress Th2 responses by directly repressing IL-4 production, as well as by inhibiting the GATA3-dependent Th2 differentiation program in miR-24– and miR-27–dependent manners.
Figure 4.
miR-24 and miR-27 target IL-4 and GATA3, respectively. FACS analysis (A) and ratios of mean fluorescence intensity (MFI; B) of IL-4 from R23CTg CD4+ T cells compared with WT CD4+ T cells under Th2-polarizing condition. HITS-CLIP analysis (C) and sequence alignment (D) of the putative site of miR-24 in IL4. (E) Ratios of repressed luciferase activity of cells with IL-4 3′ UTR only (C1) or 3′ UTR plus partial coding region containing miR-24–interacting sequences (C2) with or without mutations in the seed sequences in the presence of miR-24 compared with cells transfected with empty vector were shown. FACS analysis (F) and ratios (G) of MFI of IL-4 from R24Tg CD4+ T cells compared with WT CD4+ T cells under Th2-polarizing condition. (H) HITS-CLIP analysis and sequence alignment of putative sites of miR-27 and miR-24 in the 3′ UTR of GATA3. (I) Immunoblot analysis of GATA3 expression in T cells cultured for 4 d under Th2-polarizing condition. Densitometric GATA3 expression values were normalized to β-actin expression values and n-fold increase on the basis of each corresponding WT. Ratios of repressed luciferase activity of cells in the presence of WT GATA3 3′ UTR transfected with indicated miRNA (J) or WT or mutated GATA3 3′ UTR with miR-27 compared with cells transfected with empty vector (K). Immunoblot analysis of GATA3 expression (L) and FACS analysis of IL-4 production (M) in GFP+ R27Tg CD4+ T cells transduced with GATA3-expressing or control vector with a GFP reporter under Th2-polarizing condition. (N) n-fold increase (on the basis of corresponding WT controls) of IL-4+ cells in GFP+ R27Tg CD4+ T cells. All data are representative of three independent experiments. n = 3–6. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
miR-24 and miR-27 collaboratively modulate Th2 gene network, but predominantly repress their targets at the protein level
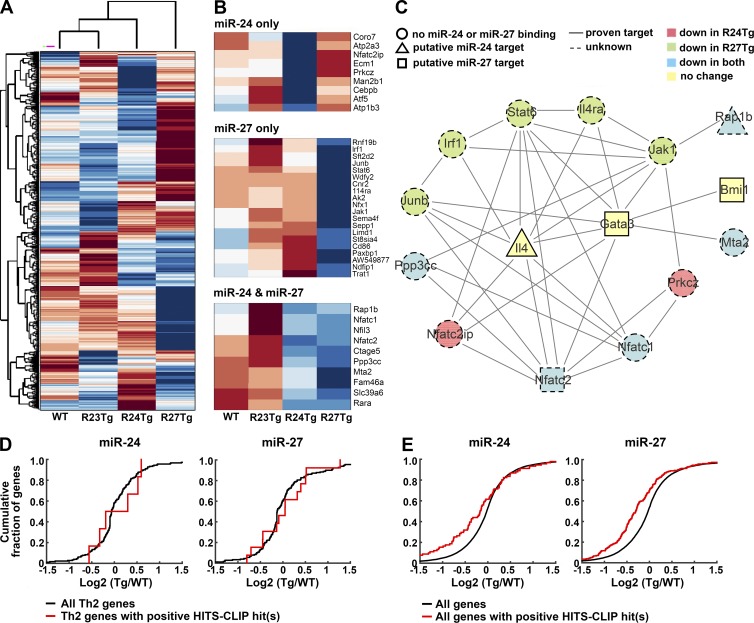
To gain further molecular insights into miR-23 cluster-dependent regulation of Th2 immunity, we performed transcriptome analysis of T cells with overexpression of individual miR-23 family members. CD25−CD44loCD62Lhi naive CD4+ T cells were FACS sorted for RNA sequencing to minimize the secondary effects from miR-24– and miR-27–dependent IL-4 and GATA3 repression in differentiated Th2 cells. Consistent with our ex vivo observations in the aforementioned different miR-23 family Tg mouse lines (Fig. 2 J), whereas the global gene expression profile of R23Tg T cells was largely comparable to that of WT, it was most distinct in R27Tg T cells, with R24Tg T cells exhibiting an intermediate pattern (Fig. 5 A). Additional screening of genes associated with Th2 immune responses (Stubbington et al., 2015; and Gene Ontology Consortium annotation 0045064) further revealed that a substantial proportion (∼30%; 39/132) of Th2-related genes was already down-regulated in nonpolarized T cells overexpressing miR-24 or miR-27 (Fig. 5 B). Next, we performed String analysis to identify potential physical and/or functional interactions between genes that were modulated by miR-24 and/or miR-27 (Fig. 5 C). To this end, many Th2 genes that were down-regulated in R24Tg T cells and/or R27Tg T cells were shown to be highly connected to IL-4 and GATA3. Interestingly, GATA3 and IL-4, as well as Bmi1, a known miR-27 target that could stabilize GATA3 (Guerau-de-Arellano et al., 2011), were in fact not down-regulated in the corresponding Tg T cells. Among genes that were down-regulated, only two, Rap1b and Nfatc2, were predicted to be miR-24 and miR-27 targets, respectively, by HITS-CLIP analysis (Fig. 5 C). Whereas the functional connection between Rap1b and Th2 cells has not been established, apart from its elevated expression in Th2 cells (Stubbington et al., 2015), the role of Nfatc2 in promoting IL-4 production and Th2 differentiation has long been recognized (Diehl et al., 2002; Rengarajan et al., 2002). Therefore, it is plausible that miR-27–mediated Nfatc2 repression could also play an important role in restricting Th2 differentiation and effector function. Nevertheless, the fact that we could only identify one additional miR-23 cluster target that could contribute to the observed Th2 biology by our RNA-seq study and the HITS-CLIP analysis suggested that genes associated with Th2 immune responses are typically not regulated by miR-24 or miR-27 at the mRNA level. In agreement with this hypothesis, cumulative distribution frequency (CDF) plot analysis revealed that there was no detectable difference in the expression of Th2-related genes regardless the presence or the absence of HITS-CLIP–identified miR-24– or miR-27–binding sites in the respective Tg T cell populations compared with control WT T cells (Fig. 5 D). Surprisingly, when all genes were subjected to CDF plot analysis, the ones with HITS-CLIP–identified sites were down-regulated when the corresponding miRNA was overexpressed (Fig. 5 E). These results demonstrated that although miR-23 family might, in general, control their targets at the mRNA level, they predominantly repress genes associated with Th2 responses at the protein level.
Figure 5.
Overexpression of miR-24 or miR-27 negatively impacts Th2 gene network. (A) Clustering of RNA-seq results from R23Tg, R24Tg, R27Tg, and WT naive CD4+ T cells based on total gene expression. (B) Th2-associated genes that were down-regulated in R24Tg T cells only, R27Tg T cells only, or both were shown. (C) String analysis of potential physical and/or functional interactions between Th2 genes that were proven miR-24 or miR-27 targets or down-regulated by miR-24 and/or miR-27 as shown from the RNA-seq results. Putative targets were defined by having perfect seed complementarity between positions 2 and 7 of the corresponding miRNA with positive Argonaute-binding peaks in the HITS-CLIP dataset. Cumulative distribution frequency plots depicting the effect of overexpression of miR-24 or miR-27 on mRNA expression of Th2-associated (D) or all genes (E). Levels of mRNAs of Th2-associated or all genes bearing predicted binding sites of miR-24 or miR-27 (red line) as defined above were compared with mRNAs of total Th2-associated or all genes (black line).
Enhanced Th2 differentiation, albeit reduced activation in T cells devoid of miR-23 clusters, led to Th2-associated immunopathology during airway allergic reaction
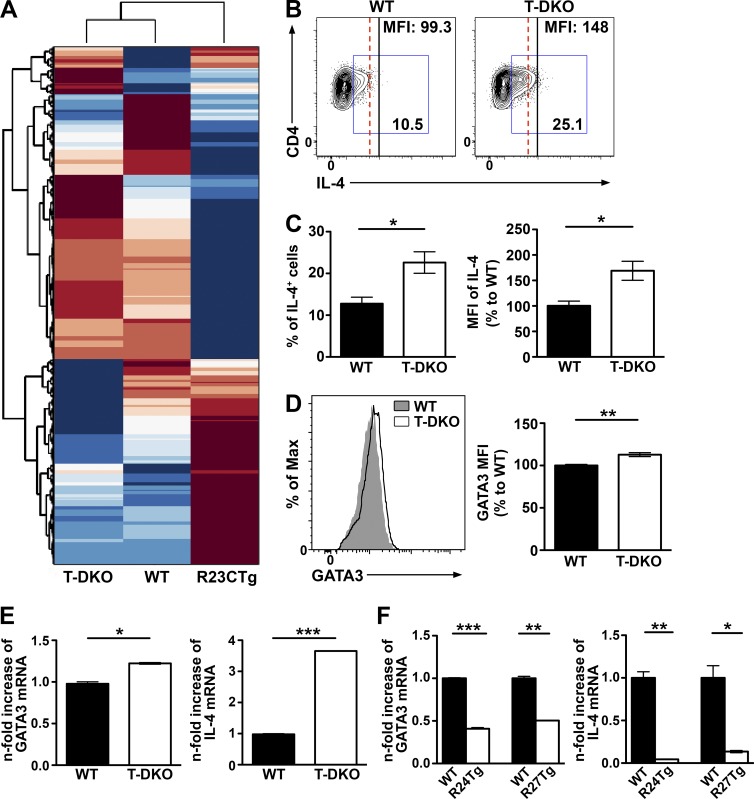
Thus far, we have shown that exaggerated regulation by the miR-23 clusters in T cells led to impaired Th2 responses. However, it is unclear as to whether elimination of miR-23 cluster-mediated gene regulation would result in enhanced Th2 immunity. To directly address this question and to further study the biological role of the miR-23 clusters, we generated mice harboring either a conditional allele of the miR-23a (miR-23aCfl) or the miR-23b cluster (miR-23bCfl; Fig. S2). Despite a lack of aberrant phenotype in mice with T cell–specific ablation of both miR-23 clusters (T-DKO) at steady state (Fig. 6), unbiased transcriptome analysis revealed ∼94% of genes displaying contrasting expression patterns between T cells isolated from T-DKO versus R23CTg mice (Fig. 7 A). Moreover, when T-DKO T cells were subject to Th2 polarization, significantly more IL-4–producing cells, with increased expression of IL-4 and GATA3 on a per cell basis, were detected compared with WT (Fig. 7, B–D), in agreement with our gain-of-function studies. Finally, although IL-4 and GATA3 were not down-regulated in any of our naive Tg T cells by RNA-seq analysis, we could clearly detect increased mRNA levels of IL-4 and GATA3 in T-DKO T cells and decreased mRNA levels of IL-4 and GATA3 in both R24Tg- and R27Tg-differentiated T cells upon Th2 polarization (Fig. 7, E and F). Nevertheless, these observed changes in the mRNA levels of IL-4 and GATA3 might be influenced by not only themselves and each other during Th2 differentiation, but also potentially many other genes targeted by miR-24 or miR-27 in T cells.
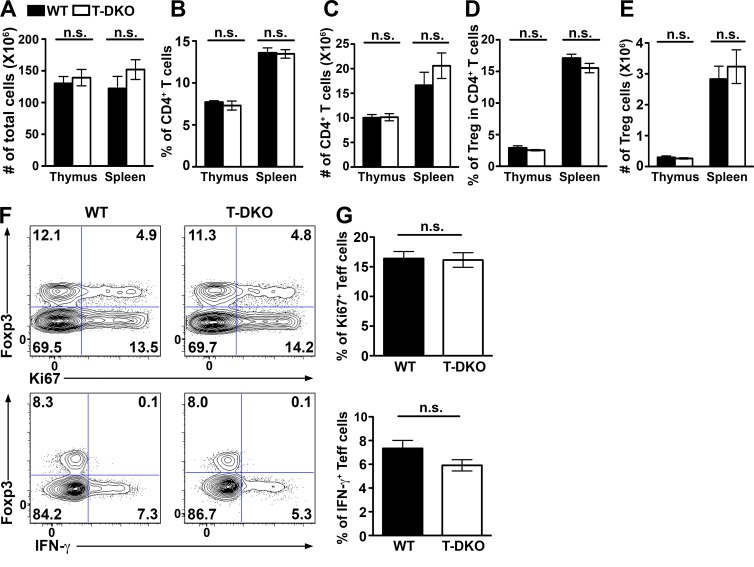
Figure 6.
No detectable immune phenotypes in mice with T cell–specific deletion of both miR-23 clusters at steady state. (A–E) Cellularity of the thymus and spleen and the proportions and absolute numbers of thymic and splenic total CD4+ T cells and Foxp3+CD4+ T reg cells in T-DKO and control littermates are shown. FACS analysis of Ki67+ (F) and IFN-γ-secreting (G) cells in splenic CD4+ T cells isolated from indicated mice are shown. Data are representative of four independent experiments. n = 10.
Figure 7.
Deletion of both miR-23 clusters in T cells resulted in increased Th2 differentiation and IL-4 expression. (A) Clustering of RNA-seq results from T-DKO, R23CTg, and WT naive CD4+ T cells based on total gene expression. FACS analysis (B), frequencies (C), and MFI of IL-4 in CD4+ T cells isolated from T-DKO mice or WT littermates cultured under Th2-polarizing conditions. (D) FACS analysis and MFI of GATA3 from T-DKO CD4+ T cells compared with WT CD4+ T cells. qPCR analysis of IL-4 and GATA3 expression in T cells isolated from T-DKO mice (E) and R24Tg or R27Tg mice (F) upon Th2 polarization. All data are representative of two independent experiments. n = 3–6. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
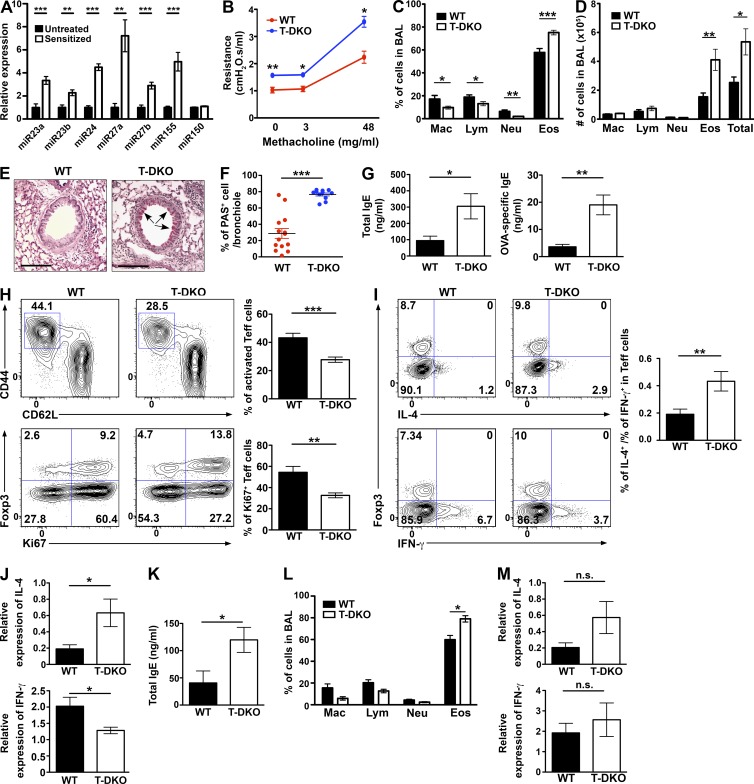
Next, we sought to test whether mice harboring miR-23 cluster-deficient T cells would exhibit dysregulated Th2 immunity in vivo when challenged to mount a Th2 response. To this end, we first used a model of OVA-induced airway allergic asthma in which the role of Th2 cells in driving lung inflammation, airway hyperresponsiveness, and obstruction has been well established (Wakashin et al., 2008). As shown in Fig. 8 A, we could detect increased expression of every member of the miR-23 family in T cells upon OVA challenges. These results suggested that miR-23 family–mediated gene regulation in T cells might serves as a mechanism of negative-feedback regulation of Th2 responses during allergic reaction. Indeed, 1 d after the last OVA challenge, compared with WT littermates, T-DKO mice exhibited significantly higher pulmonary resistance (Fig. 8 B). Selective increases in the percentage and the absolute number of eosinophils in bronchoalveolar lavage (BAL) fluid were also easily detected (Fig. 8, C and D). Histopathological analyses showed much greater goblet cell hyperplasia in the bronchioles and more severe eosinophilia in the perivascular and peribronchial regions of lungs from T-DKO mice than from WT controls (Fig. 8, E and F; and not depicted). Exacerbated lung pathology was accompanied by further elevated production of serum total and OVA-specific IgE, a major feature of allergic asthma, in sensitized T-DKO mice (Fig. 8 G). It should be noted that the observed phenotypes in T-DKO mice were not caused by unrestrained T cell activation. In fact, lung-infiltrating T cells exhibited diminished proliferation and reduced activation in OVA-challenged T-DKO mice compared with those in control animals (Fig. 8 H), complementing our previous observations of enhanced proliferation and activation of T cells with miR-23 cluster overexpression. Though fewer cells were activated, the lung-infiltrating T cells still produced increased IL-4 and reduced IFN-γ (Fig. 8 I), which is consistent with our in vitro polarization studies. Similarly, higher IL-4 and lower IFN-γ expression levels were also detected in lung tissues harvested from OVA-challenged T-DKO mice (Fig. 8 J). Finally, similar to our findings in mice with OVA challenges, we could also detect elevated total IgE levels in the serum and increased eosinophils in BAL fluid when T-DKO mice were challenged with a real allergen, house dust mite (HDM; Fig. 8, K and L). There was also more (albeit not statistically significant) IL-4 in lung tissues of T-DKO mice (Fig. 8 M). However, unlike the OVA model, we did not detect any decrease in IFN-γ production in T-DKO mice, likely because of the complex nature of whole HDM extracts compared with a single purified protein used in the OVA study. Altogether, these results confirmed that the miR-23 clusters in T cells play a key role in restraining Th2 responses and associated immune pathology particularly in the setting of allergic airway inflammation.
Figure 8.
Exacerbated eosinophilic airway inflammation in mice with T cell–specific ablation of both miR-23 clusters. (A) qPCR analysis of different miRNA expression in lung-infiltrating T cells harvested from OVA-challenged mice. miR-155, but not miR-150, has been shown to be up-regulated during OVA challenges and were used as positive and negative controls, respectively (Malmhäll et al., 2014). (B) Pulmonary function test was determined 24 h after last challenge in response to increasing doses of methacholine (0, 3, and 48 mg/ml). Airway resistance of T-DKO mice and WT control littermates. (C) Frequencies and (D) absolute cell numbers of different cell populations in BAL. Mac, macrophages; Lym, lymphocytes; Neu, neutrophils; Eos, eosinophils. (E) PAS staining of lung sections from sensitized control and T-DKO mice are shown (bar, 200 µm), and (F) percentages of PAS+ mucus-secreting cells in per bronchiole were quantified. (G) Serum levels of total IgE and OVA-specific IgE levels were determined by ELISA. (H) FACS analysis and frequencies of CD44hiCD62Llo-activated T cells and Ki67+ T cells from lung in OVA-challenged T-DKO mice and WT control littermates are shown. (I) Frequencies and the ratios of IL-4 and IFN-γ–secreting cells in lung-infiltrating CD4+ T cells isolated from indicated mice were shown. (J) qPCR analysis of IL-4 and IFN-γ mRNA expression in lung tissues harvested from OVA-challenged T-DKO mice or control littermates. Data are represented of three independent experiments. n = 10–14. 24 d after HDM intranasal challenges, serum levels of total IgE levels were determined by ELISA (K) and frequencies cell numbers of different cell populations in BAL were shown (L). (M) qPCR analysis of IL-4 and IFN-γ mRNA expression in lung tissues harvested from HDM-challenged T-DKO mice or control littermates. Data are representative of two independent experiments. n = 8–10. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DISCUSSION
There is little doubt that miRNAs function as important gene regulators in controlling almost every aspect of biology. However, because many miRNAs can be deleted without resulting in any apparent phenotype, there is also a growing consensus that most miRNAs do not play an indispensable role in dictating the outcomes of biological responses (Miska et al., 2007). Nonetheless, there are exceptions to the current idea of miRNAs as mere fine-tuners of biology, particularly among those miRNAs that target genes involved in positive-feedback regulatory circuits. Self-reinforcing loops can amplify even the subtlest changes in gene expression introduced by miRNAs, leading to greater physiological consequences. Moreover, miRNAs can increase their biological impact through binding multiple sites on a gene target or repressing a set of genes that are in a shared pathway or protein complex (Ebert and Sharp, 2012). The aforementioned scenarios can be further exploited by the existence of evolutionarily conserved miRNA clusters where members of a given miRNA cluster often target the same gene or different components of a common biological process (Liang et al., 2014). Additionally, duplication of such miRNA clusters in paralogs ensures rigid control of their targets. In this study, we have provided direct genetic evidence to demonstrate how miR-23 clusters and paralogs can exert their regulatory effects on important biology, particularly in controlling T cell immunity. Specifically, we have shown that miR-24 and miR-27 collaboratively control Th2 immunity through targeting IL-4 and GATA3, respectively.
Interestingly, whereas HITS-CLIP analysis revealed putative binding of both miRNAs to the 3′ UTR of GATA3, only miR-27 but not miR-24 seems to be functionally active in directly repressing GATA3. To this end, as IL-4 itself is a direct target of miR-24, exaggerated miR-24 regulation likely contributes to impaired IL-4 production and a further disruption of GATA3-dependent Th2 cell differentiation program considering the role of autocrine IL-4 signaling in potentiating GATA3 expression (Scheinman and Avni, 2009). However, it should be noted that the miR-24–binding site in mice is not conserved in humans, raising questions about the biological relevance of miR-24–mediated IL-4 repression beyond mouse studies. Nevertheless, miR-24 has been previously shown to be able to control multiple genes with no canonical target seed sequences (Lal et al., 2009). Similar observations in seedless genes targeted by miR-146a and miR-155 were also previously reported (Lu et al., 2010; Loeb et al., 2012). It was shown that the complimentary sequences outside the seed region could contribute to the noncanonical miRNA-mediated repression, albeit at relatively moderate levels compared with the canonical targeting (Loeb et al., 2012). The fact that the 3′ end of miR-24 could interact with more conserved sequences in the IL-4 coding region, as this putative binding site is located just 1 nt after the stop codon of mouse IL-4 gene, suggested that it might be possible that miR-24 could also control human IL-4 expression, even in the absence of canonical seed sequences. Our findings of increased repression in the luciferase reporter assay with the construct containing the extended sequences from the IL-4 coding region where miR-24 could interact further supported this scenario. Alternatively, miR-24 could negatively impact human IL-4 expression in an indirect manner similar to what was reported in miR-27-mediated GATA3 regulation in humans through targeting Bmi1, a molecule that stabilizes GATA3 by blocking its proteasome-dependent degradation (Guerau-de-Arellano et al., 2011). Indeed, our RNA-seq study revealed that many Th2-associated genes were already down-regulated in naive T cells upon overexpression of miR-24 or miR-27. Although it is uncertain as to how many of them were directly controlled by miR-24 or miR-27 because these two miRNAs predominantly repress their respective targets related to Th2 responses at the protein rather than the mRNA level, nevertheless, our results demonstrated that the miR-23 clusters, particularly miR-24 and miR-27, are able to efficiently restrain Th2 differentiation and effector function through directly and indirectly targeting multiple components within the Th2-associated gene network.
Such coordinated repression by miR-23 clusters is not likely to be limited to Th2 regulation. Given that almost every facet of T cell immunity is controlled by this miRNA family, one would imagine that a similar regulatory mechanism could be used by the miR-23 clusters to confer proper immune function to other T cell lineages. For example, we have shown that both miR-23 and miR-27 limit the differentiation of Th17 and iTreg cell populations. As TGFβ signaling plays an integral part in the induction of both T cell lineages, it is probably not a coincidence that several key molecules downstream of TGFβ signaling have been shown to be repressed by these two miRNAs (Rogler et al., 2009). Alternatively, we have found that contrary to miR-23 and miR-27, miR-24 promoted iTreg and Th17 responses. This surprising finding was interesting, but not completely unexpected, as HITS-CLIP analysis has identified miR-24 as the only miR-23 family member that could target Smad7, a negative regulator of TGFβ signaling, suggesting that miR-24 could exert an effect on iTreg and Th17 cells opposite to the other cluster members through augmenting TGFβ signaling (Loeb et al., 2012). Nevertheless, the fact that different members of the same miRNA cluster can antagonize each other in controlling a specific biological process seems to be counterintuitive and raises a question as to how such an unproductive feature in miRNA-mediated gene regulation could be retained evolutionarily. One possibility is that in some situations, one member of the miRNA cluster could exert an opposite function to fine tune the regulatory effects of the entire miRNA family. Another probable clue for this puzzle was provided by a previous study in which Bmp2, a member of TGFβ superfamily, was shown to promote adipocytic differentiation from mesenchymal stem cells by specifically up-regulating miR-24 but not miR-23 or miR-27 (Sun et al., 2009). It is intriguing to speculate that, whereas in most cases individual members of miR-23 family would function cooperatively to maximize their impact, under certain circumstances, a given miR-23 family member could be differentially expressed to control a specific type of T cell response. Finally, beyond regulating T cell immunity, it has been previously documented that miR-23a cluster promotes the differentiation of myeloid cells from hematopoietic progenitors while inhibiting B cell development (Kong et al., 2010). In dendritic cells, up-regulation of either miR-23b or miR-27a was shown to confer tolerogenic activities, allowing them to induce more T reg cells (Min et al., 2012; Zheng et al., 2012). Future studies will continue to unravel the role of this miRNA family not only in T cell immunity but also in regulating other aspects of the immune system.
MATERIALS AND METHODS
Mice
Foxp3GFP (Fontenot et al., 2005) and Foxp3GFPKO mice (Gavin et al., 2007) were described previously. The targeting constructs for miR-23bFBfl, miR-23aCfl, and miR-23bCfl were generated using recombineering. For detailed information see the National Cancer Institute recombineering website (http://redrecombineering.ncifcrf.gov/). Mice on a B6 genetic background with the germ-line transmission were bred to Flp deleter mice to remove the neomycin resistance cassette. T cell–specific deletion of both miR23 clusters were achieved by breeding miR23 a/bCfl/fl mice to CD4-cre mice, respectively. The targeting constructs for R23CTg, R23Tg, R24Tg, and R27Tg were generated similar to what was described previously (Xiao et al., 2007). Mice with T cell–specific overexpression of the entire miR-23 family and individual members were obtained by breeding the aforementioned mice to CD4-cre mice. All mice were maintained and handled in accordance with the Institutional Animal Care and Use Guidelines of UCSD and National Institutes of Health Guidelines for the Care and Use of Laboratory Animals and the ARRIVE guidelines.
miRNA expression profiling and quantitative PCR analysis
Foxp3GFP+ T reg cells and naive CD62Lhi Foxp3GFP− T conv cells from Foxp3GFP mice were sorted on FACSAria (Becton Dickinson), followed by 1 d culture with 100 U/ml IL-2 with or without plate-bound 1 µg/ml αCD3 and/or 1 µg/ml αCD28. Isolated RNA from harvested cells (duplicates of each sample) was subject to miRNA expression profiling analysis using miRCURY LNA microRNA Arrays (Exiqon). For confirming the expression levels of miR-23 clusters by qPCR, TaqMan (Thermo Fisher Scientific) stem-loop real-time RT-PCR was performed as demonstrated previously (Lu et al., 2010).
For detecting cytokine levels in sensitized lungs, total RNA of lung was extracted by using miRNeasy kit (QIAGEN), cDNAs were generated by iScript cDNA synthesis kit (Bio-Rad Laboratories), and real-time PCR was performed using SYBR Green PCR kits (Applied Biosystems). Primers are as follows: IL-4, 5′-TTGAACGAGGTCACAGGAGA-3′ (F) and 5′-AAATATGCGAAGCACCTTGG-3′ (R); IFN-γ, 5′-GCGTCATTGAATCACACCTG-3′ (F) and 5′-GAGCTCATTGAATGCTTGGC-3′ (R); Gapdh, 5′-CGTCCCGTAGACAAAATGGT-3′ (F) and 5′-TCAATGAAGGGGTCGTTGAT-3′ (R).
Ex vivo phenotyping and flow cytometry
Single-cell suspensions of thymus and spleen were prepared by slide mechanical grind. To isolate lymphocytes in lung and lamina propria, tissues were cut and washed in plain RPMI-1640, and epithelial cells were removed (5 mM EDTA and 1 mM DTT, lamina propria only), followed by enzymatic digestion (0.16 U/ml Liberase TL; Roche) and centrifugation with 47% Percoll gradient to enrich lymphocytes. Cell were stained in FACS buffer (5% FBS in PBS) containing Fixable Viability Dye eFluor 780 or 450 (eBioscience) with the following antibodies for surface staining: CD4, CD8, CD44, and CD62L (eBioscience). To detect cytokine production, cells were stimulated in a 96-well plate with 50 ng/ml PMA, 0.5 µg/ml ionomycin, and 1 µg/ml Brefeldin A (all from Sigma-Aldrich) solution for 4 hat 37°C before staining. Intracellular staining of Ki67, Foxp3, IL-4, IL-17A, and IFN-γ was performed after fixation and permeabilization according to the manufacturer’s instructions. Data were analyzed with FlowJo software (Tree Star).
In vitro Th differentiation
CD62L+CD25− naive T cells in spleen were sorted by FACSAria flow cytometers (BD) from 6–8-wk-old mice, and were stimulated for 4 d with 1 µg/ml anti-CD3 and mitomycin C (Sigma-Aldrich)–treated APC, and 50 U/ml recombinant human IL-2 under Th1-, Th2-, iTreg-, or Th17-polarizing conditions. Th cell polarization mediums were supplemented as follows: for Th1 differentiation, 2 U/ml IL-12 and 10 µg/ml αIL-4; for Th2 differentiation, 20 ng/ml IL-4 (in T-DKO study, 10 ng/ml was used), 10 µg/ml αIL-12, and 10 µg/ml αIFN-γ; for iTreg differentiation, 1 ng/ml human TGF-β; and for TH17 differentiation, 2 ng/ml hTGF-β and 20 ng/ml IL-6. All cytokines were obtained from PeproTech and all blocking Abs were obtained from Bio-XCell. Surface and intracellular cytokines were stained and analyzed as previously described.
In vitro suppression assay
FACS purified 4 × 104 naive CD4+CD25−CD62Lhi T cells and CD4+CD25hi T reg cells isolated from R23CTg mice or WT control littermates were mixed at the indicated ratios and stimulated with 1 µg/ml αCD3 antibody in the presence of irradiated (2,000 rads) splenocytes. T cell proliferation was assessed by with 3H-TdR incorporation (cpm) in triplicate cultures during the last 8 h of culture.
Luciferase reporter assay
The 3′ UTR region of GATA3 or IL-4, as well as the 3′ UTR plus partial sequences in the coding region of IL-4, were cloned into psiCheck2 (Promega). miR-23a, miR-24, and miR-27a sequences were respectively cloned into pMDH-PGK-EGFP, similar to what was described previously (Lu et al., 2009). To generate GATA3 or IL-4 3′ UTR mutants, site-directed mutagenesis was performed (Agilent). 1 d before transfection, HEK293T cells were plated at 6.5 × 104 cells per well on a 24-well plate. PsiCheck2 bearing WT 3′ UTR or corresponding mutant 3′ UTR were transfected to HEK293T cells with control vector or miRNA expressing plasmid using Fugene 6 (Promega). Luciferase activities were assessed at 24 h after transfection using Dual-Luciferase Reporter assay system (Promega) according to the manufacturer’s protocol.
Immunoblotting
Naive CD4 T cells (1 × 106 cells/ml) were cultured in the 24-well plate coated with 2 µg/ml of αCD3 and αCD28 under the Th2-polarizing condition. Cells were harvested 4 d later and subjected to lysis with RIPA buffer supplemented with 1 mM PMSF for 20 min. Cell lysates were separated by SDS-PAGE and transferred to PVDF membrane (Bio-Rad Laboratories). Antibodies against GATA3 (BioLegend) and β-Actin (Sigma-Aldrich) were used to visualize the corresponding proteins. The proteins were quantified with ImageJ (National Institutes of Health).
Retrovirus transduction
pMIG-mGATA3, a gift from Z. Werb (University of California, San Francisco, La Jolla, CA; Addgene plasmid 21629; Kouros-Mehr et al., 2008), was used as a template to subclone into pMIR-RI. To generate retrovirus, pMIR-R1 or pMIR-R1-GATA3 (without 3′ UTR) was transfected with pCL-Eco into HEK293T cell using Fugene 6 (Promega). Retroviral supernatants were collected at 48 h after transfection. Naive CD4 T cells (1 × 106 cells/ml) were stimulated with 2 µg/ml of αCD3 and 2 µg/ml of αCD28 for 24 h, followed by spin infection for 90 min at 2,000 rpm in the presence of 8 µg/ml of polybrene (Millipore). Cells were cultured in the Th2-polarizing condition and harvested at 4 d after retrovirus transduction.
Gene expression profiling analysis
CD25−CD44loCD62Lhi naive CD4+ T cells were FACS sorted from 6 wk WT, R23Tg, R24Tg, R27Tg, R23CTg, and T-DKO mice, and poly-A RNA-sequencing was performed using three biological replicates for each cell population. Approximately 30 million uniquely alignable reads per sample reads per sample were generated. Reads were aligned to the mouse genome (mm9, NCBI37) using STAR (Dobin et al., 2013). RNA-seq experiments were normalized and gene expression values were generating for RefSeq annotated transcripts using HOMER (Heinz et al., 2010). Gene expression clustering was performed using Cluster 3.0 and visualized using Java TreeView. For the cumulative distribution function (CDF) plots, target sites were restricted to perfect seed complementarity between positions 2 and 7 of the corresponding miRNA with positive Argonaute binding peaks in the HITS-CLIP dataset (Loeb et al., 2012). Empirical cumulative distributions were computed using Matlab (R2014b) to display the log2(miRNA Tg/WT) against the cumulative frequency of Th2-associated (Stubbington et al., 2015; Gene Ontology Consortium annotation 0045064) or all genes. String analysis was performed to identify potential physical and/or functional interactions between Th2 genes that were proven miR-24 or miR-27 targets or down-regulated by miR-24 and miR-27 as shown in the RNA-seq results. The interaction map was reconstructed manually with additional information obtained from the HITS-CLP data analysis. RNA-seq data are available from NCBI under accession no. GSE75909.
In vivo allergic airway inflammation models
Mice were sensitized by two intraperitoneal injections of 50 µg of OVA (Worthington) that had been emulsified with 0.8 mg of aluminum hydroxide (Thermo Fisher Scientific) in 200 µl PBS on days 0 and 12. Mice were then challenged on days 24, 26, and 28 by intranasal injection of 20 µg (or 10 µg as low-dose group) of OVA. 1 d after the last OVA challenge, mice were anesthetized with 100 mg/kg ketamine and 10 mg/kg xylazine via intraperitoneal injection. Airway reactivity to methacholine was measured in intubated and ventilated mice (flexiVent ventilator; Scireq). Mice were exposed to nebulized PBS and, subsequently, to increasing concentrations (3 and 48 mg/ml) of nebulized methacholine (Sigma-Aldrich) in PBS. The dynamic airway resistance was determined by using Scireq software. After pulmonary function test, the lungs of the mice were lavaged with 0.7 ml of normal saline. The BAL cells were centrifuged onto slides at 1,000 rpm for 3 min in a Shandon Cytospin (Thermo Fisher Scientific). The cells were then stained with Wright-Giemsa stain (Protocol) for 3 min. 200 cells per slide were counted under a microscope, and those with different morphology were noted. In some experiments, HDM extract (Dermatophagoides pteronyssinus; Greer Laboratories) was used instead of OVA. In brief, mice were exposed intranasally to 100 µg in 50 µl PBS on days 0, 7, 14, and 21, and were sacrificed on day 24 for further assessment similar to the work described in the OVA study.
Histology
To assess immunopathology, different tissues were removed and immediately fixed in 10% formalin solution for hematoxylin and eosin staining of sections embedded in paraffin. Inflammation was examined and scored blindly by a UCSD pathologist. For additional assessment of lung pathology during airway allergic reaction, periodic acid-Schiff (PAS) stain was performed to detect goblet cell hyperplasia. To quantitate the level of goblet cell hyperplasia in the airway, the percentage of PAS+ epithelial cells in individual bronchioles was counted, and at least 10 bronchioles were determined.
ELISA
The concentrations of total and OVA-specific IgE in serum were evaluated with ELISA kits (BioLegend) according to the manufacturer’s instructions.
Statistical analyses
Unpaired Student’s t test was performed using Prism software (GraphPad). *, P < 0.05; **, P < 0.01; and ***, P < 0.001 in all data.
Online supplemental material
Fig. S1 shows conditional overexpression of miR-23a cluster in T cells. Fig. S2 shows generation of mice with conditional alleles of miR-23a cluster or miR-23b cluster. Table S1 shows miRNA expression profiling analysis in T reg versus effector T cells under different culture conditions. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20150990/DC1.
Supplementary Material
Acknowledgments
We thank R. Levenzon and B. Nizamova for superb technical assistance, and all members of our laboratory for discussions. We thank Dr. N. Varki for help with pathological evaluation of samples.
This work was supported by National Institutes of Health grants AI089935, AI103646, and AI108651 (to L.-F. Lu), by a grant from Ministry of Science and Technology, Taiwan (ROC: NSC 102-2811-B-182-014; to M.-L. Kuo), and by a grant from Ministry of Education, Taiwan (ROC: EMRPD1C0131; to C.-J. Wu). T. Yasuda is supported by a JSPS Postdoctoral Fellowship for Research Abroad and by the Astellas Foundation for Research on Metabolic Disorders. H.-M. Lee is an Irvington Fellow of the Cancer Research Institute. A.Y. Rudensky is an investigator with the Howard Hughes Medical Institute. L.-F. Lu is a Kimmel Scholar and a Hellman Fellow.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- BAL
- bronchoalveolar lavage
- HDM
- house dust mite
- iT reg
- induced T reg cell
- MFI
- mean fluorescence intensity
- miRNA
- microRNA
- T conv cell
- conventional T cell
References
- Baumjohann D., Kageyama R., Clingan J.M., Morar M.M., Patel S., de Kouchkovsky D., Bannard O., Bluestone J.A., Matloubian M., Ansel K.M., and Jeker L.T.. 2013. The microRNA cluster miR-17∼92 promotes TFH cell differentiation and represses subset-inappropriate gene expression. Nat. Immunol. 14:840–848. 10.1038/ni.2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronevetsky Y., Villarino A.V., Eisley C.J., Barbeau R., Barczak A.J., Heinz G.A., Kremmer E., Heissmeyer V., McManus M.T., Erle D.J., et al. . 2013. T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire. J. Exp. Med. 210:417–432. 10.1084/jem.20111717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brümmer A., and Hausser J.. 2014. MicroRNA binding sites in the coding region of mRNAs: extending the repertoire of post-transcriptional gene regulation. BioEssays. 36:617–626. 10.1002/bies.201300104 [DOI] [PubMed] [Google Scholar]
- Chandran P.A., Keller A., Weinmann L., Seida A.A., Braun M., Andreev K., Fischer B., Horn E., Schwinn S., Junker M., et al. . 2014. The TGF-β-inducible miR-23a cluster attenuates IFN-γ levels and antigen-specific cytotoxicity in human CD8+ T cells. J. Leukoc. Biol. 96:633–645. 10.1189/jlb.3A0114-025R [DOI] [PubMed] [Google Scholar]
- Chhabra R., Dubey R., and Saini N.. 2010. Cooperative and individualistic functions of the microRNAs in the miR-23a~27a~24-2 cluster and its implication in human diseases. Mol. Cancer. 9:232 10.1186/1476-4598-9-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintharlapalli S., Papineni S., Abdelrahim M., Abudayyeh A., Jutooru I., Chadalapaka G., Wu F., Mertens-Talcott S., Vanderlaag K., Cho S.D., et al. . 2009. Oncogenic microRNA-27a is a target for anticancer agent methyl 2-cyano-3,11-dioxo-18beta-olean-1,12-dien-30-oate in colon cancer cells. Int. J. Cancer. 125:1965–1974. 10.1002/ijc.24530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong M.M., Rasmussen J.P., Rudensky A.Y., and Littman D.R.. 2008. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J. Exp. Med. 205:2005–2017. 10.1084/jem.20081219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb B.S., Hertweck A., Smith J., O’Connor E., Graf D., Cook T., Smale S.T., Sakaguchi S., Livesey F.J., Fisher A.G., and Merkenschlager M.. 2006. A role for Dicer in immune regulation. J. Exp. Med. 203:2519–2527. 10.1084/jem.20061692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl S., Chow C.W., Weiss L., Palmetshofer A., Twardzik T., Rounds L., Serfling E., Davis R.J., Anguita J., and Rincón M.. 2002. Induction of NFATc2 expression by interleukin 6 promotes T helper type 2 differentiation. J. Exp. Med. 196:39–49. 10.1084/jem.20020026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert M.S., and Sharp P.A.. 2012. Roles for microRNAs in conferring robustness to biological processes. Cell. 149:515–524. 10.1016/j.cell.2012.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot J.D., Rasmussen J.P., Williams L.M., Dooley J.L., Farr A.G., and Rudensky A.Y.. 2005. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 22:329–341. 10.1016/j.immuni.2005.01.016 [DOI] [PubMed] [Google Scholar]
- Gavin M.A., Rasmussen J.P., Fontenot J.D., Vasta V., Manganiello V.C., Beavo J.A., and Rudensky A.Y.. 2007. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 445:771–775. 10.1038/nature05543 [DOI] [PubMed] [Google Scholar]
- Guerau-de-Arellano M., Smith K.M., Godlewski J., Liu Y., Winger R., Lawler S.E., Whitacre C.C., Racke M.K., and Lovett-Racke A.E.. 2011. Micro-RNA dysregulation in multiple sclerosis favours pro-inflammatory T-cell-mediated autoimmunity. Brain. 134:3578–3589. 10.1093/brain/awr262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttilla I.K., and White B.A.. 2009. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J. Biol. Chem. 284:23204–23216. 10.1074/jbc.M109.031427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausser J., Syed A.P., Bilen B., and Zavolan M.. 2013. Analysis of CDS-located miRNA target sites suggests that they can effectively inhibit translation. Genome Res. 23:604–615. 10.1101/gr.139758.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., and Glass C.K.. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38:576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeker L.T., Zhou X., Gershberg K., de Kouchkovsky D., Morar M.M., Stadthagen G., Lund A.H., and Bluestone J.A.. 2012. MicroRNA 10a marks regulatory T cells. PLoS One. 7:e36684 10.1371/journal.pone.0036684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S.G., Liu W.H., Lu P., Jin H.Y., Lim H.W., Shepherd J., Fremgen D., Verdin E., Oldstone M.B., Qi H., et al. . 2013. MicroRNAs of the miR-17∼92 family are critical regulators of T(FH) differentiation. Nat. Immunol. 14:849–857. 10.1038/ni.2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong K.Y., Owens K.S., Rogers J.H., Mullenix J., Velu C.S., Grimes H.L., and Dahl R.. 2010. MIR-23A microRNA cluster inhibits B-cell development. Exp. Hematol. 38:629–640. 10.1016/j.exphem.2010.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouros-Mehr H., Bechis S.K., Slorach E.M., Littlepage L.E., Egeblad M., Ewald A.J., Pai S.Y., Ho I.C., and Werb Z.. 2008. GATA-3 links tumor differentiation and dissemination in a luminal breast cancer model. Cancer Cell. 13:141–152. 10.1016/j.ccr.2008.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroesen B.J., Teteloshvili N., Smigielska-Czepiel K., Brouwer E., Boots A.M., van den Berg A., and Kluiver J.. 2015. Immuno-miRs: critical regulators of T-cell development, function and ageing. Immunology. 144:1–10. 10.1111/imm.12367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchen S., Resch W., Yamane A., Kuo N., Li Z., Chakraborty T., Wei L., Laurence A., Yasuda T., Peng S., et al. . 2010. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity. 32:828–839. 10.1016/j.immuni.2010.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A., Pan Y., Navarro F., Dykxhoorn D.M., Moreau L., Meire E., Bentwich Z., Lieberman J., and Chowdhury D.. 2009. miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat. Struct. Mol. Biol. 16:492–498. 10.1038/nsmb.1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.M., Nguyen D.T., and Lu L.F.. 2014. Progress and challenge of microRNA research in immunity. Front. Genet. 5:178 10.3389/fgene.2014.00178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang T., Yu J., Liu C., and Guo L.. 2014. An exploration of evolution, maturation, expression and function relationships in mir-23 approximately 27 approximately 24 cluster. PLoS One. 9:e106223 10.1371/journal.pone.0106223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Chen L., Chen G., Hu C., Jiang S., Sevilla J., Wan Y., Sampson J.H., Zhu B., and Li Q.J.. 2014. Targeting miR-23a in CD8+ cytotoxic T lymphocytes prevents tumor-dependent immunosuppression. J. Clin. Invest. 124:5352–5367. 10.1172/JCI76561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston A., Lu L.F., O’Carroll D., Tarakhovsky A., and Rudensky A.Y.. 2008. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J. Exp. Med. 205:1993–2004. 10.1084/jem.20081062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb G.B., Khan A.A., Canner D., Hiatt J.B., Shendure J., Darnell R.B., Leslie C.S., and Rudensky A.Y.. 2012. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol. Cell. 48:760–770. 10.1016/j.molcel.2012.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L.F., Thai T.H., Calado D.P., Chaudhry A., Kubo M., Tanaka K., Loeb G.B., Lee H., Yoshimura A., Rajewsky K., and Rudensky A.Y.. 2009. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 30:80–91. 10.1016/j.immuni.2008.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L.F., Boldin M.P., Chaudhry A., Lin L.L., Taganov K.D., Hanada T., Yoshimura A., Baltimore D., and Rudensky A.Y.. 2010. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 142:914–929. 10.1016/j.cell.2010.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majid S., Dar A.A., Saini S., Arora S., Shahryari V., Zaman M.S., Chang I., Yamamura S., Tanaka Y., Deng G., and Dahiya R.. 2012. miR-23b represses proto-oncogene Src kinase and functions as methylation-silenced tumor suppressor with diagnostic and prognostic significance in prostate cancer. Cancer Res. 72:6435–6446. 10.1158/0008-5472.CAN-12-2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmhäll C., Alawieh S., Lu Y., Sjöstrand M., Bossios A., Eldh M., and Rådinger M.. 2014. MicroRNA-155 is essential for TH2-mediated allergen-induced eosinophilic inflammation in the lung. J. Allergy Clin. Immunol. 133:1429–38; 1438.e1–7 10.1016/j.jaci.2013.11.008 [DOI] [PubMed] [Google Scholar]
- Mertens-Talcott S.U., Chintharlapalli S., Li X., and Safe S.. 2007. The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res. 67:11001–11011. 10.1158/0008-5472.CAN-07-2416 [DOI] [PubMed] [Google Scholar]
- Min S., Li L., Zhang M., Zhang Y., Liang X., Xie Y., He Q., Li Y., Sun J., Liu Q., et al. . 2012. TGF-β-associated miR-27a inhibits dendritic cell-mediated differentiation of Th1 and Th17 cells by TAB3, p38 MAPK, MAP2K4 and MAP2K7. Genes Immun. 13:621–631. 10.1038/gene.2012.45 [DOI] [PubMed] [Google Scholar]
- Miska E.A., Alvarez-Saavedra E., Abbott A.L., Lau N.C., Hellman A.B., McGonagle S.M., Bartel D.P., Ambros V.R., and Horvitz H.R.. 2007. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 3:e215 10.1371/journal.pgen.0030215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell R.M., Rao D.S., Chaudhuri A.A., and Baltimore D.. 2010. Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 10:111–122. 10.1038/nri2708 [DOI] [PubMed] [Google Scholar]
- Rengarajan J., Mowen K.A., McBride K.D., Smith E.D., Singh H., and Glimcher L.H.. 2002. Interferon regulatory factor 4 (IRF4) interacts with NFATc2 to modulate interleukin 4 gene expression. J. Exp. Med. 195:1003–1012. 10.1084/jem.20011128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogler C.E., Levoci L., Ader T., Massimi A., Tchaikovskaya T., Norel R., and Rogler L.E.. 2009. MicroRNA-23b cluster microRNAs regulate transforming growth factor-beta/bone morphogenetic protein signaling and liver stem cell differentiation by targeting Smads. Hepatology. 50:575–584. 10.1002/hep.22982 [DOI] [PubMed] [Google Scholar]
- Scheinman E.J., and Avni O.. 2009. Transcriptional regulation of GATA3 in T helper cells by the integrated activities of transcription factors downstream of the interleukin-4 receptor and T cell receptor. J. Biol. Chem. 284:3037–3048. 10.1074/jbc.M807302200 [DOI] [PubMed] [Google Scholar]
- Simpson L.J., Patel S., Bhakta N.R., Choy D.F., Brightbill H.D., Ren X., Wang Y., Pua H.H., Baumjohann D., Montoya M.M., et al. . 2014. A microRNA upregulated in asthma airway T cells promotes TH2 cytokine production. Nat. Immunol. 15:1162–1170. 10.1038/ni.3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbington M.J., Mahata B., Svensson V., Deonarine A., Nissen J.K., Betz A.G., and Teichmann S.A.. 2015. An atlas of mouse CD4+ T cell transcriptomes. Biol. Direct. 10:14 10.1186/s13062-015-0045-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Wang J., Pan Q., Yu Y., Zhang Y., Wan Y., Wang J., Li X., and Hong A.. 2009. Characterization of function and regulation of miR-24-1 and miR-31. Biochem. Biophys. Res. Commun. 380:660–665. 10.1016/j.bbrc.2009.01.161 [DOI] [PubMed] [Google Scholar]
- Ventura A., Young A.G., Winslow M.M., Lintault L., Meissner A., Erkeland S.J., Newman J., Bronson R.T., Crowley D., Stone J.R., et al. . 2008. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 132:875–886. 10.1016/j.cell.2008.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakashin H., Hirose K., Maezawa Y., Kagami S., Suto A., Watanabe N., Saito Y., Hatano M., Tokuhisa T., Iwakura Y., et al. . 2008. IL-23 and Th17 cells enhance Th2-cell-mediated eosinophilic airway inflammation in mice. Am. J. Respir. Crit. Care Med. 178:1023–1032. 10.1164/rccm.200801-086OC [DOI] [PubMed] [Google Scholar]
- Wang N., Zhu M., Tsao S.W., Man K., Zhang Z., and Feng Y.. 2013. MiR-23a-mediated inhibition of topoisomerase 1 expression potentiates cell response to etoposide in human hepatocellular carcinoma. Mol. Cancer. 12:119 10.1186/1476-4598-12-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C., Calado D.P., Galler G., Thai T.H., Patterson H.C., Wang J., Rajewsky N., Bender T.P., and Rajewsky K.. 2007. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 131:146–159. 10.1016/j.cell.2007.07.021 [DOI] [PubMed] [Google Scholar]
- Zhang H., Hao Y., Yang J., Zhou Y., Li J., Yin S., Sun C., Ma M., Huang Y., and Xi J.J.. 2011. Genome-wide functional screening of miR-23b as a pleiotropic modulator suppressing cancer metastasis. Nat. Commun. 2:554 10.1038/ncomms1555 [DOI] [PubMed] [Google Scholar]
- Zheng J., Jiang H.Y., Li J., Tang H.C., Zhang X.M., Wang X.R., Du J.T., Li H.B., and Xu G.. 2012. MicroRNA-23b promotes tolerogenic properties of dendritic cells in vitro through inhibiting Notch1/NF-κB signalling pathways. Allergy. 67:362–370. 10.1111/j.1398-9995.2011.02776.x [DOI] [PubMed] [Google Scholar]
- Zhou X., Jeker L.T., Fife B.T., Zhu S., Anderson M.S., McManus M.T., and Bluestone J.A.. 2008. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J. Exp. Med. 205:1983–1991. 10.1084/jem.20080707 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.