B-lineage cells reconcile the competing needs of proliferation and genome stability.
Abstract
DNA double-strand breaks (DSBs) activate a canonical DNA damage response, including highly conserved cell cycle checkpoint pathways that prevent cells with DSBs from progressing through the cell cycle. In developing B cells, pre–B cell receptor (pre–BCR) signals initiate immunoglobulin light (Igl) chain gene assembly, leading to RAG-mediated DNA DSBs. The pre–BCR also promotes cell cycle entry, which could cause aberrant DSB repair and genome instability in pre–B cells. Here, we show that RAG DSBs inhibit pre–BCR signals through the ATM- and NF-κB2–dependent induction of SPIC, a hematopoietic-specific transcriptional repressor. SPIC inhibits expression of the SYK tyrosine kinase and BLNK adaptor, resulting in suppression of pre–BCR signaling. This regulatory circuit prevents the pre–BCR from inducing additional Igl chain gene rearrangements and driving pre–B cells with RAG DSBs into cycle. We propose that pre–B cells toggle between pre–BCR signals and a RAG DSB-dependent checkpoint to maintain genome stability while iteratively assembling Igl chain genes.
Developing B cells must assemble and express genes encoding the immunoglobulin heavy (Igh) and light (Igl) chains of the B cell receptor (BCR; Rajewsky, 1996). This process occurs through V(D)J recombination, a reaction that assembles the second exon of lymphocyte antigen receptor genes from component variable (V), joining (J), and, at some loci, diversity (D) gene segments (Fugmann et al., 2000). V(D)J recombination is initiated when the RAG endonuclease, composed of RAG-1 and RAG-2, introduces DNA double-strand breaks (DSBs) at a pair of recombining gene segments, generating two blunt signal DNA ends and two hairpin-sealed coding DNA ends (Fugmann et al., 2000). RAG DSBs are generated in G1 phase lymphocytes, where they activate the ataxia-telangiectasia mutated (ATM) DNA damage response (DDR) kinase (Desiderio et al., 1996; Helmink and Sleckman, 2012). In G1-phase cells, ATM inhibits S-phase entry by initiating canonical cell cycle checkpoint pathways through induction of p53 and directs DSB repair by nonhomologous end joining (NHEJ; Shiloh, 2003; Helmink and Sleckman, 2012). In addition, in response to RAG DSBs, ATM activates a genetic program that may function to regulate processes required for normal lymphocyte development (Bredemeyer et al., 2008; Bednarski et al., 2012; Helmink and Sleckman, 2012; Steinel et al., 2013).
B cell development requires the sequential assembly of Igh chain genes in pro–B cells and Igl chain genes (Igl κ [Igk] or Igl λ) in pre–B cells (Rajewsky, 1996). The ordered assembly of immunoglobulin receptor genes is directed by signals from cell surface receptors. The IL-7r signals through AKT and JAK–STAT pathways to promote survival and to regulate Igh chain gene rearrangement in pro–B cells (Bertolino et al., 2005; Clark et al., 2014). Productive assembly of an Igh chain gene leads to its expression with the surrogate light chain (λ5 and Vpre–B) and the CD79A–CD79B heterodimer (Igα and Igβ, respectively) to generate the pre–BCR (Herzog et al., 2009; Rickert, 2013). Oligomerization of the pre–BCR, through ligand-dependent or -independent mechanisms, activates the SYK tyrosine kinase, leading to phosphorylation of the adaptor protein BLNK (also known as SLP-65; Herzog et al., 2009; Rickert, 2013).
Pre–BCR signals, along with those from the IL-7r, promote the developmental transition of pro–B cells to rapidly cycling, large pre–B cells (Herzog et al., 2009; Rickert, 2013; Clark et al., 2014). Pre–BCR and IL-7r signals synergize to drive proliferation, whereas they independently regulate differentiation and survival, respectively. Activation of STAT5 by the IL-7r inhibits Igk germline transcription and activation of AKT by the IL-7r inhibits Rag1 and Rag2 gene expression, both of which prevent Igk gene assembly (Amin and Schlissel, 2008; Mandal et al., 2009, 2011; Corfe and Paige, 2012; Ochiai et al., 2012). Moreover, in cycling cells RAG-2 is degraded in S-phase (Desiderio et al., 1996). Thus, proliferative signals must be attenuated for large pre–B cells to transit to the small pre–B cell stage where Igk chain gene assembly is initiated (Rolink et al., 1991; Johnson et al., 2008; Ochiai et al., 2012; Clark et al., 2014). IL-7r signals are attenuated by the pre–BCR, which inhibits AKT, a key molecule downstream of the IL-7r (Herzog et al., 2008; Ochiai et al., 2012). Additionally, pre–BCR signals induce CXCR4, which can affect the localization of pre–B cells with respect to IL-7–producing stromal cells (Tokoyoda et al., 2004; Johnson et al., 2008). Moreover, activation of RAS by the pre–BCR in large pre–B cells promotes exit from the cell cycle (Mandal et al., 2009). Loss of IL-7r signaling leads to increased SYK and BLNK expression, which reinforces pre–BCR signaling (Ochiai et al., 2012).
Pre–BCR signals are required to initiate Igk chain gene assembly through activation of transcription factors and histone modifications that regulate Igk accessibility and RAG recruitment (Clark et al., 2014). The pre–BCR induces expression of IRF4, which, together with PU.1, binds the 3′ Igk enhancer to promote Igk germline transcription and rearrangement (Pongubala et al., 1992; Johnson et al., 2008; Clark et al., 2014). Small pre–B cells often undergo multiple sequential Igk rearrangements over several days as they attempt to generate a functional Igk chain gene (Casellas et al., 2001). Once RAG DSBs are generated, the pre–BCR must be prevented from initiating additional Igk rearrangements. Moreover, activation of SYK by the pre–BCR could drive small pre–B cells with RAG DSBs into cycle (Rolink et al., 2000; Wossning et al., 2006; Herzog et al., 2009; Rickert, 2013). In pre–B cells, RAG DSBs activate canonical cell cycle checkpoint pathways, including p53 (Guidos et al., 1996; Helmink and Sleckman, 2012). However, in other cell types these checkpoint pathways can be overridden by proliferative signals, such as those from cytokine receptors (Quelle et al., 1998; Sitko et al., 2008). Thus, unopposed pre–BCR signaling could drive pre–B cells with RAG DSBs into cycle promoting aberrant RAG DSB repair and genome instability.
We reasoned that pre–BCR signaling must be regulated to order Igk chain gene assembly and prevent these signals from driving pre–B cells with RAG DSBs into cycle. Indeed, we show here that RAG DSBs activate a cell type–specific checkpoint pathway that inhibits pre–BCR signaling. This checkpoint pathway suppresses SYK and BLNK expression, inactivating pre–BCR signals to both prevent cell cycle progression and regulate Igk chain gene assembly. We propose that pre–B cells toggle between pre–BCR signaling and this RAG DSB-dependent checkpoint pathway, allowing for iterative Igk chain gene assembly while maintaining genome stability.
RESULTS
RAG DSB signals regulate the genetic program of small pre–B cells
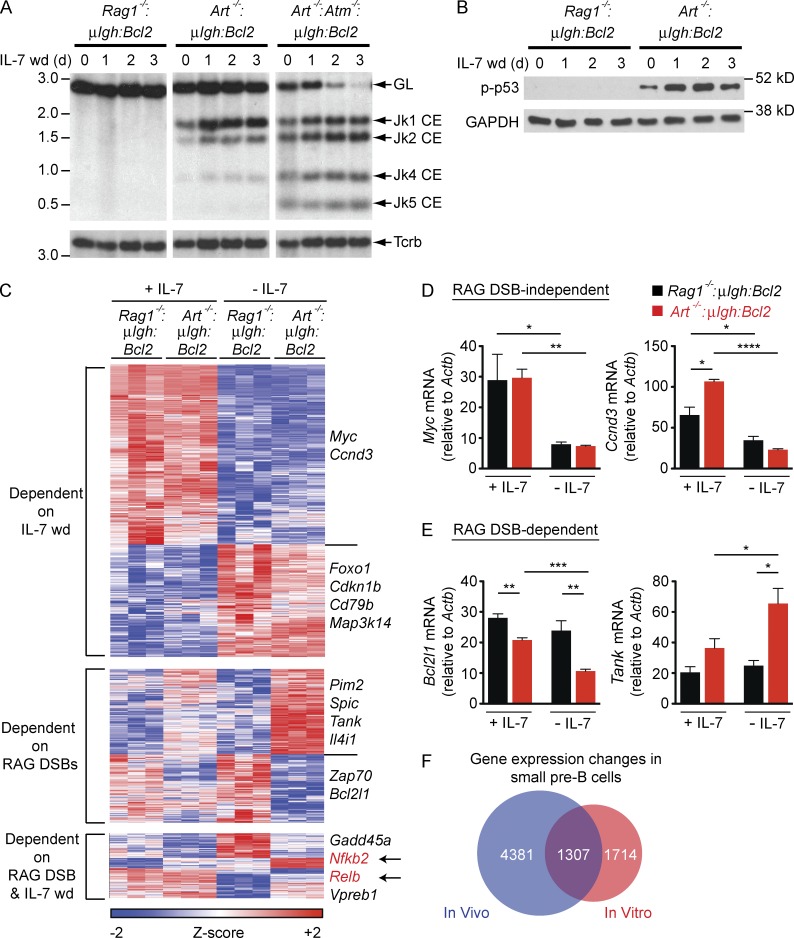
To elucidate the pre–B cell genetic program regulated by RAG DSB signals, we used mice deficient in RAG-1 or the Artemis endonuclease that express the μIgh and Bcl2 transgenes (Rag1−/−:μIgh:Bcl2 and Art−/−:μIgh:Bcl2, respectively; Bednarski et al., 2012). The μIgh transgene enables formation of a pre–BCR, allowing for pre–B cell development in these mice, and the Bcl2 transgene supports pre–B cell survival in vitro (Bednarski et al., 2012). Culturing bone marrow from these mice in the presence of IL-7 leads to the selective expansion of large pre–B cells (Rolink et al., 1991; Johnson et al., 2008; Bednarski et al., 2012). Upon withdrawal of IL-7, these cells transit to the small pre–B cell stage and induce RAG expression (Bednarski et al., 2012; Steinel et al., 2013; Fig. S1). Rag1−/−:μIgh:Bcl2 small pre–B cells do not generate RAG DSBs. Art−/−:μIgh:Bcl2 small pre–B cells generate RAG DSBs at Igk, but these DSBs are not repaired because Artemis is required to open hairpin-sealed coding DNA ends (Helmink and Sleckman, 2012; Fig. 1 A). These DSBs activate DDR, as indicated by phosphorylation of p53 (Fig. 1 B).
Figure 1.
RAG DSBs regulate the genetic program in small pre–B cells. (A) Southern blot of SacI and EcoRI digested genomic DNA from Rag1−/−:μIgH:Bcl2, Art−/−:μIgH:Bcl2 and Art−/−:Atm−/−:μIgH:Bcl2 small pre–B cells in IL-7 (0 d) and after withdrawal of IL-7 (IL-7 wd) for the indicated days. Bands reflecting the germline (GL) Igk locus and Jk coding ends (Jk1, Jk2, Jk4, and Jk5 CE) are indicated as are molecular weight markers in kilobases. (B) Western blot of phosphorylated p53 (p-p53) in pre–B cells in IL-7 (0 d) and after withdrawal of IL-7 (IL-7 wd) for the indicated days. GAPDH is shown as a protein loading control. Data in A and B are representative of at least three independent experiments. (C) Heat map of gene expression changes in Rag1−/−:μIgH:Bcl2 and Art−/−:μIgH:Bcl2 pre–B cells in the presence of IL-7 (+ IL-7) and 2 d after IL-7 withdrawal (− IL-7). Columns represent three independent cell cultures of each genotype. Representative genes are delineated to the right. (D and E) RT-PCR validation of representative RAG DSB-independent (D) or RAG DSB-dependent (E) gene expression changes from C. mRNA expression was assessed in Rag1−/−:μIgH:Bcl2 (black) and Art−/−:μIgH:Bcl2 (red) pre–B cells in the presence of IL-7 (+ IL-7) and 2 d after IL-7 withdrawal (− IL-7). Data are mean and standard error for three replicates. (F) Venn diagram comparing gene expression changes that occur during the transition of large to small pre–B cells in the bone marrow of WT mice (ImmGen; blue) and gene expression changes we observe in small pre–B cells after withdrawal of IL-7 (red). The ImmGen data represents all genes that were significantly changed (p-value < 0.05) between large and small pre–B cells. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005; ****, P ≤ 0.0001 by Student’s t test.
Gene profiling revealed that ∼3,000 genes change in expression (>1.5-fold) in Art−/−:μIgh:Bcl2 or Rag1−/−:μIgh:Bcl2 pre–B cells as they transition from large (in IL-7) to small (IL-7 withdrawal) pre–B cells (Fig. 1, C-E; and Table S1). Many of these gene expression changes are observed during B cell development in vivo (Heng and Painter, 2008; Fig. 1 F). Comparison of Art−/−:μIgh:Bcl2 and Rag1−/−:μIgh:Bcl2 small pre–B cells reveals that RAG DSB signals regulate a significant fraction (∼40%) of the gene expression changes that occur during the transition of large to small pre–B cells (RAG DSB dependent; Fig. 1, C–E; and Table S1).
ATM and NIK activate NF-κB2 in small pre–B cells
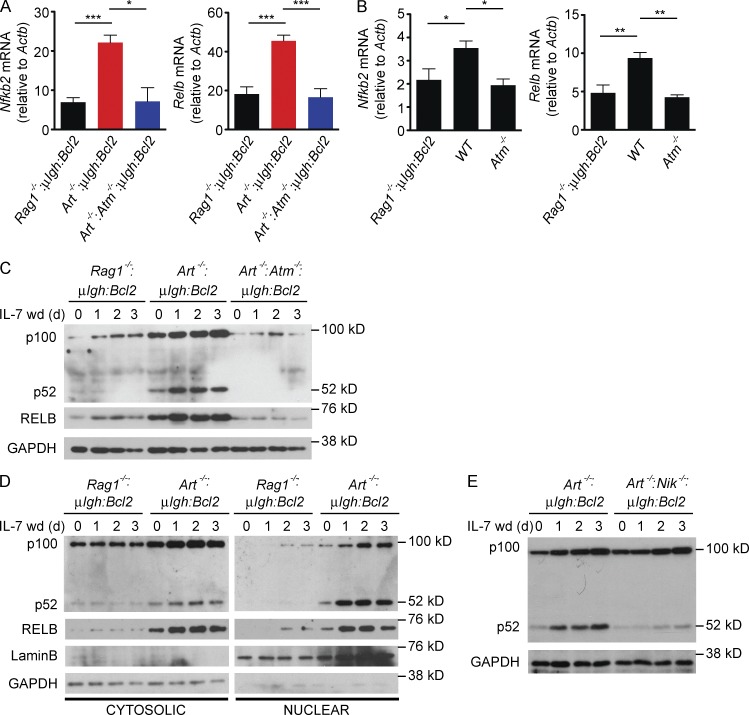
In small pre–B cells, RAG DSBs induce expression of the Relb and Nfkb2 (p100) genes, which encode the principal components of the noncanonical NF-κB (NF-κB2) pathway (Hayden and Ghosh, 2012; Sun, 2012; Fig. 2 A and Table S1). This depends on the activation of ATM as revealed by analyses of Art−/−:Atm−/−:μIgh:Bcl2 small pre–B cells in vitro and Atm−/− pre–B cells in vivo (Fig. 2, A and B). The p100 and RELB proteins are induced by RAG DSBs (compare Art−/−:μIgh:Bcl2 and Rag1−/−:μIgh:Bcl2) in small pre–B cells that express ATM and have intact DDR signals (compare Art−/−:μIgh:Bcl2 and Art−/−:Atm−/−:μIgh:Bcl2; Fig. 2 C). Formation of the transcriptionally active p52-RELB heterodimer, which translocates to the nucleus, requires cleavage of p100 to form p52 (Hayden and Ghosh, 2012; Sun, 2012). In this regard, the robust generation of p52 and the nuclear translocation of p52 and RELB are observed only in small pre–B cells with RAG DSBs (Fig. 2, C and D). In mature B cells, cleavage of p100 to form p52 depends on the NF-κB–inducing kinase (NIK; Hayden and Ghosh, 2012; Sun, 2012). Similarly, in small pre–B cells NIK is required for the generation of p52, as Art−/−:Nik−/−:μIgh:Bcl2 small pre–B cells show induction of p100 in response to RAG DSBs, but inefficient conversion of p100 to p52 (Fig. 2 E). Thus, NF-κB2 is activated in small pre–B cells through the RAG DSB-dependent induction of p100 and RELB coupled with the NIK-dependent conversion of p100 to p52.
Figure 2.
Signals from RAG DSBs activate NF-κB2. (A) Nfkb2 and Relb mRNA expression in Rag1−/−:μIgH:Bcl2 (black), Art−/−:μIgH:Bcl2 (red), and Art−/−:Atm−/−;μIgH:Bcl2 (blue) small pre–B cells 2 d after IL-7 withdrawal. Data are mean and standard error for three replicates. (B) Nfkb2 and Relb mRNA expression in sorted small pre–B cells from Rag1−/−:μIgH:Bcl2, WT, and Atm−/− mice. Data are mean and standard error from three independent mice of each genotype. (C) Western blot of NF-κB2 (p100 and p52) and RELB in pre–B cells in IL-7 (0 d) and after withdrawal of IL-7 (IL-7 wd) for the indicated days. GAPDH is shown as a protein loading control. (D) Western blot of NF-κB2 (p100 and p52) and RELB incytoplasmic andnuclear fractions from pre–B cells in IL-7 (0 d) and after withdrawal of IL-7 for the indicated days. LaminB and GAPDH are shown as protein loading controls for nuclear and cytoplasmic fractions, respectively. (E) Western blot of NF-κB2 (p100 and p52) in pre–B cells in IL-7 (0 d) and after withdrawal of IL-7 (IL-7 wd) for the indicated days. GAPDH is shown as a protein loading control. Data in all panels are representative of at least three independent experiments. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005 by Student’s t test.
NF-κB2 induces SPIC expression
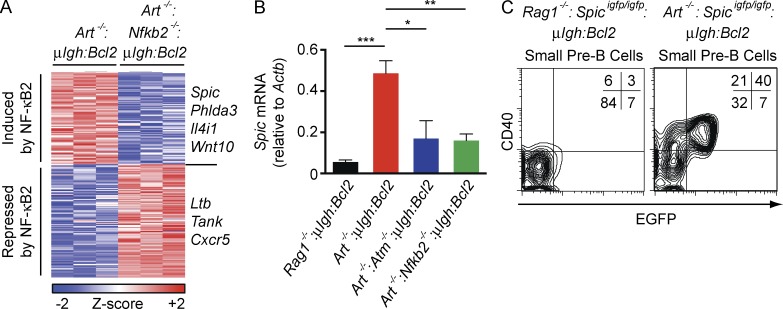
Analysis of Art−/−:μIgh:Bcl2 and Art−/−:Nfkb2−/−:μIgh:Bcl2 small pre–B cells revealed that ∼15% of the genes regulated by RAG DSBs depend on NF-κB2 (Fig. 3 A and Table S2). One of these genes encodes SPIC, an ETS family transcription factor with significant homology to PU.1 and SPIB (Bemark et al., 1999; Hashimoto et al., 1999). Induction of Spic in response to RAG DSBs depends on ATM and NF-κB2, as indicated by analysis of Art−/−:μIgh:Bcl2, Art−/−:Atm−/−:μIgh:Bcl2, and Art−/−:Nfkb2−/−:μIgh:Bcl2 small pre–B cells (Fig. 3 B). To assess SPIC expression in vivo, we used mice with a Spic reporter allele, Spicigfp, which contains an IRES-EGFP targeted to a 3′ noncoding exon of the Spic gene (Haldar et al., 2014). Approximately 40% of bone marrow small pre–B cells from Art−/−:Spicigfp/igfp:μIgh:Bcl2 mice express EGFP (Fig. 3 C). CD40 surface expression is induced by RAG DSB signals, and thus functions as a marker of small pre–B cells with RAG DSBs (Bredemeyer et al., 2008). All of the EGFP-positive small pre–B cells in Art−/−:Spicigfp/igfp:μIgh:Bcl2 mice also express CD40 (Fig. 3 C). In contrast, none of the small pre–B cells from Rag1−/−:Spicigfp/igfp:μIgh:Bcl2 mice express EGFP or CD40 (Fig. 3 C). Together, these data are consistent with the interpretation that SPIC expression in small pre–B cells is induced by RAG DSBs through the sequential activation of ATM and NF-κB2.
Figure 3.
Signals from RAG DSBs induce SPIC expression. (A) Heat map of RAG DSB-dependent gene expression changes that require NF-κB2. Art−/−:μIgH:Bcl2 pre–B cells were compared with Rag1−/−:μIgH:Bcl2 pre–B cells to identify RAG DSB-dependent gene expression changes followed by comparison of Art−/−:μIgH:Bcl2 and Art−/−:Nfkb2−/−:μIgH:Bcl2 pre–B cells to identify NF-κB2–dependent changes. Columns represent three independent cell cultures of each genotype. Representative genes are delineated to the right. (B) Spic mRNA expression in Rag1−/−:μIgH:Bcl2 (black), Art−/−:μIgH:Bcl2 (red), Art−/−:Atm−/−:μIgH:Bcl2 (blue), and Art−/−:Nfkb2−/−:μIgH:Bcl2 (green) small pre–B cells 2 d after IL-7 withdrawal. Data are mean and standard error for three replicates. (C) Flow cytometric analysis of CD40 (y-axis) and EGFP (x-axis) expression in bone marrow small pre–B cells (B220loCD43−IgM−) from Rag1−/−:Spicigfp/igfp:μIgH:Bcl2 and Art−/−:Spicigfp/igfp:μIgH:Bcl2 mice. Inset shows percent of cells in each quadrant. Data are representative of three independent experiments. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005 by Student’s t test.
RAG DSBs negatively regulate pre–BCR signaling
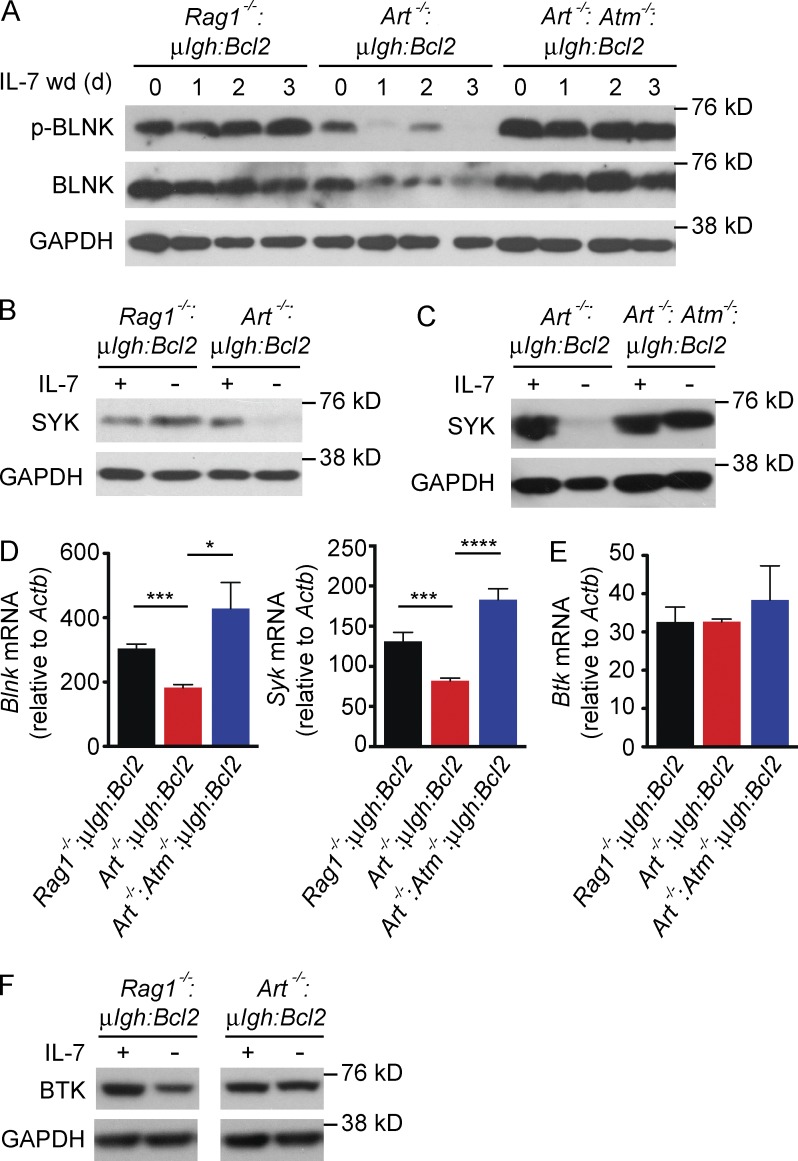
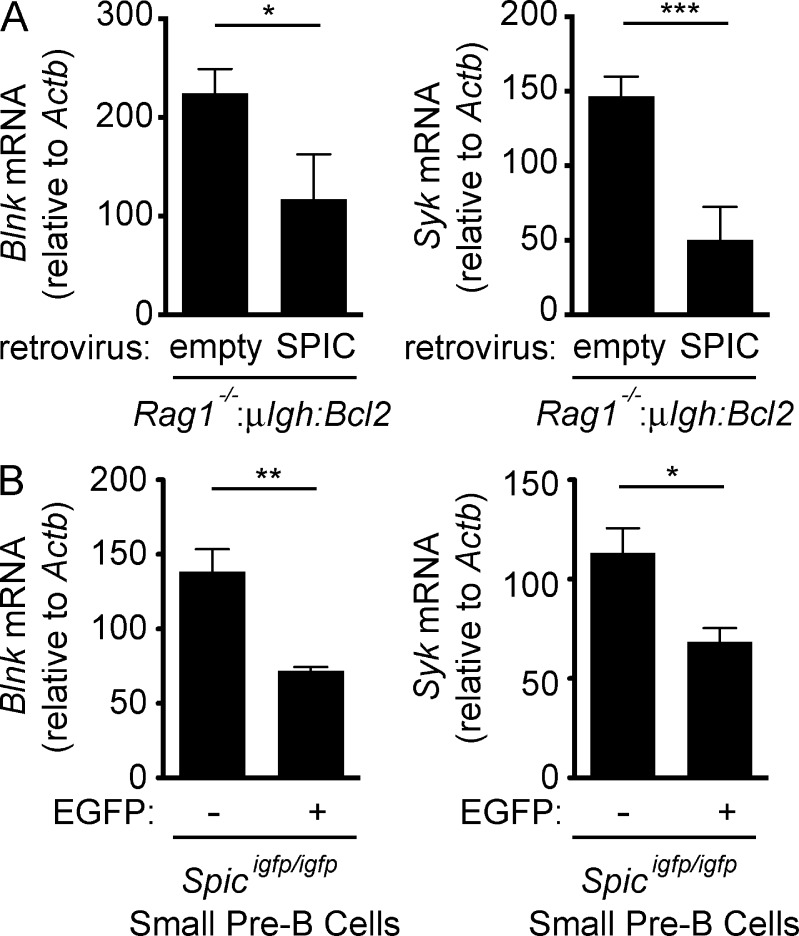
Ectopic SPIC expression in mature B cells inhibits BCR signaling, and in lymphocyte progenitors it blocks B cell development at the pre–B cell stage (Zhu et al., 2008). As RAG DSBs induce SPIC, we reasoned that they might negatively regulate pre–BCR signaling. Indeed, we find that expression of both SYK and BLNK is reduced in small pre–B cells with RAG DSBs (Art−/−:μIgh:Bcl2) but not in those without RAG DSBs (Rag1−/−:μIgh:Bcl2) or in ATM-deficient (Art−/−:Atm−/−:μIgh:Bcl2) pre–B cells with RAG DSBs (Fig. 4, A–D). Consistent with the reduction in these proteins, small pre–B cells with RAG DSBs have decreased pre–BCR signaling as evidence by reduced levels of phosphorylated BLNK (Fig. 4 A). Expression of BTK, another pre–BCR signaling protein, is not affected by RAG DSBs but is minimally decreased by loss of IL-7r signals (Fig. 4, E and F). Retroviral expression of SPIC in Rag1−/−:μIgh:Bcl2 small pre–B cells leads to inhibition of both Blnk and Syk expression (Fig. 5 A). Moreover, SPIC-expressing (EGFP-positive) small pre–B cells in Spicigfp/igfp mice have lower levels of Syk and Blnk transcripts as compared with cells that do not express SPIC (EGFP negative; Fig. 5 B). We conclude that RAG DSBs antagonize the expression of two key pre–BCR signaling proteins, SYK and BLNK, through the sequential induction of ATM, NF-κB2, and SPIC.
Figure 4.
RAG DSBs suppress pre–BCR signaling. (A) Western blot of phosphorylated BLNK (p-BLNK) and total BLNK in pre–B cells in IL-7 (0 d) and following IL-7 withdrawal (IL-7 wd) for the indicated days. GAPDH is shown as a protein loading control. (B and C) Western blot of SYK in pre–B cells cultured in IL-7 (+) and 3 d after IL-7 withdrawal (–). GAPDH is shown as a protein loading control. (D and E) Blnk, Syk and Btk mRNA expression in Rag1−/−:μIgH:Bcl2 (black), Art−/−:μIgH:Bcl2 (red), and Art−/−:Atm−/−;μIgH:Bcl2 (blue) small pre–B cells 3 d after IL-7 withdrawal. Data are mean and standard error for three replicates. (F) Western blot of BTK in pre–B cells cultured in IL-7 (+) and 3 d after IL-7 withdrawal (–). GAPDH is shown as a protein loading control. Data in A, B, C, and F are representative of at least three independent experiments. *, P ≤ 0.05; ***, P ≤ 0.005; ****, P ≤ 0.0001 by Student’s t test.
Figure 5.
SPIC inhibits expression of Syk and Blnk. (A) Rag1−/−:μIgH:Bcl2 small pre–B cells were transduced with an empty retrovirus or a retrovirus expressing SPIC. Blnk and Syk mRNA expression was assessed 2 d after IL-7 withdrawal. Data are mean and standard error for three replicates. (B) Blnk and Syk mRNA expression in EGFP-negative (–) and EGFP-expressing (+) small pre–B cells sorted from Spicigfp/igfp mice. Data are the mean and standard error from three independent mice. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005 by Student’s t test.
SPIC antagonizes PU.1 binding at Syk and Blnk
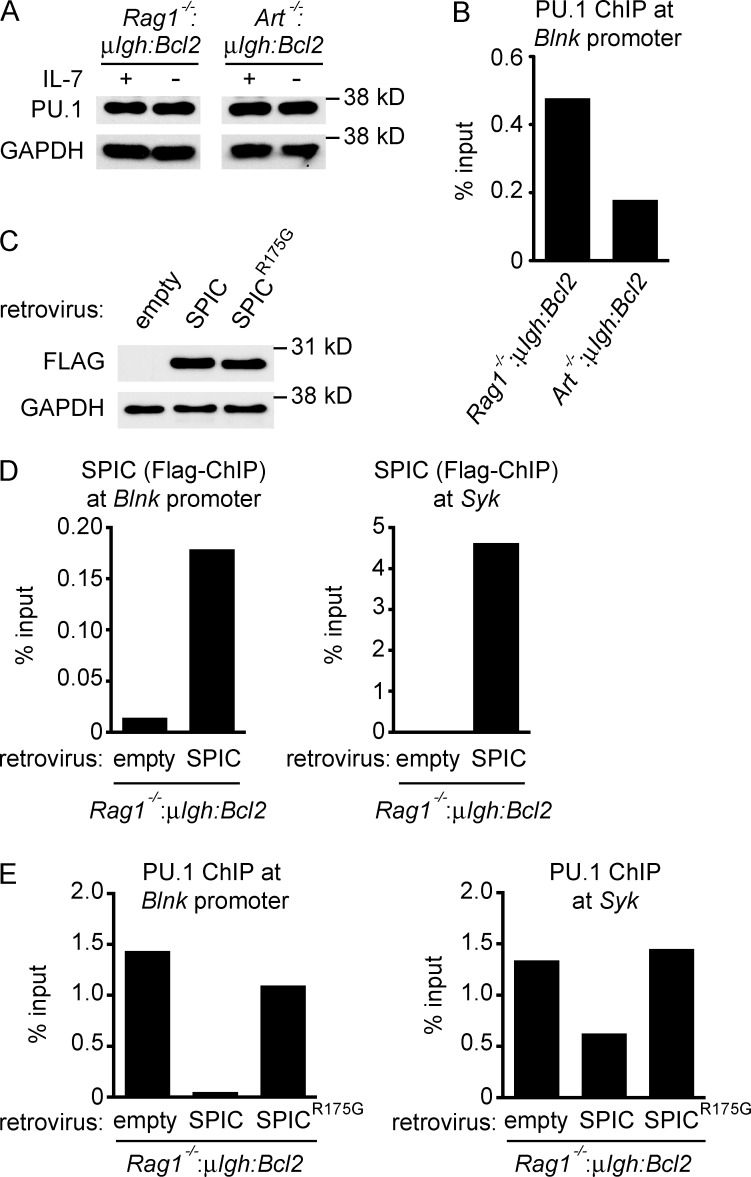
The expression of Blnk depends on PU.1 binding to its promoter (Schweitzer and DeKoter, 2004; Xu et al., 2012). SPIC functions as a transcriptional repressor that recognizes the same DNA sequence as PU.1 but, unlike PU.1, SPIC is unable to bind IRF4, which is required to initiate transcription (Bemark et al., 1999; Hashimoto et al., 1999; Carlsson et al., 2003). To determine whether SPIC induction by RAG DSBs affects PU.1 activity, we compared PU.1 binding at Blnk in Art−/−:μIgh:Bcl2 and Rag1−/−:μIgh:Bcl2 small pre–B cells. Although PU.1 levels are similar in these cells, binding of PU.1 to the Blnk promoter is markedly reduced in Art−/−:μIgh:Bcl2 small pre–B cells, which have RAG DSBs and up-regulate SPIC, as compared with Rag1−/−:μIgh:Bcl2 small pre–B cells that do not express SPIC (Fig. 6, A and B). When SPIC is retrovirally expressed in Rag1−/−:μIgh:Bcl2 small pre–B cells, we observe SPIC binding to both the Blnk promoter and a putative regulatory element upstream of the Syk gene (Zhang et al., 2012; Fig. 6, C and D). Moreover, we also observe decreased PU.1 binding at these regulatory elements in Rag1−/−:μIgh:Bcl2 small pre–B cells expressing SPIC (Fig. 6 E). Mutation of the conserved Ets domain (R175G) in SPIC (SPICR175G) ablates SPIC binding to Blnk and Syk, similar to previous findings for PU.1 (Pio et al., 1996; Suraweera et al., 2005; and unpublished data). Compared with SPIC, equivalent expression of SPICR175G in Rag1−/−:μIgh:Bcl2 small pre–B cells does not result in decreased PU.1 binding at Blnk and Syk (Fig. 6, C and E). Together, these data suggest that, after induction by RAG DSBs, SPIC directly antagonizes PU.1 at cis-acting regulatory elements in both the Blnk and Syk loci.
Figure 6.
SPIC displaces PU.1 from Syk and Blnk. (A) Western blot of PU.1 in pre–B cells cultured in IL-7 (+) and 3 d after IL-7 withdrawal (–). GAPDH is shown as a protein loading control. (B) ChIP-qPCR of PU.1 binding at the Blnk promoter in Rag1−/−:μIgH:Bcl2 and Art−/−:μIgH:Bcl2 small pre–B cells 3 d after IL-7 withdrawal. (C–E) Rag1−/−:μIgH:Bcl2 small pre–B cells were transduced with an empty retrovirus, a retrovirus expressing SPIC, or a retrovirus expressing a DNA binding–deficient SPIC (SPICR175G). (C) Western blot of FLAG was performed 2 d after IL-7 withdrawal. GAPDH is shown as a protein loading control. ChIP-qPCR of SPIC binding (D) and PU.1 binding (E) at the Blnk promoter and at a putative regulatory element located 10-kb upstream of the transcription start site for Syk was performed 2 d after IL-7 withdrawal. Data in all panels are representative of two independent experiments.
SPIC inhibits germline transcription and RAG cleavage at Igk
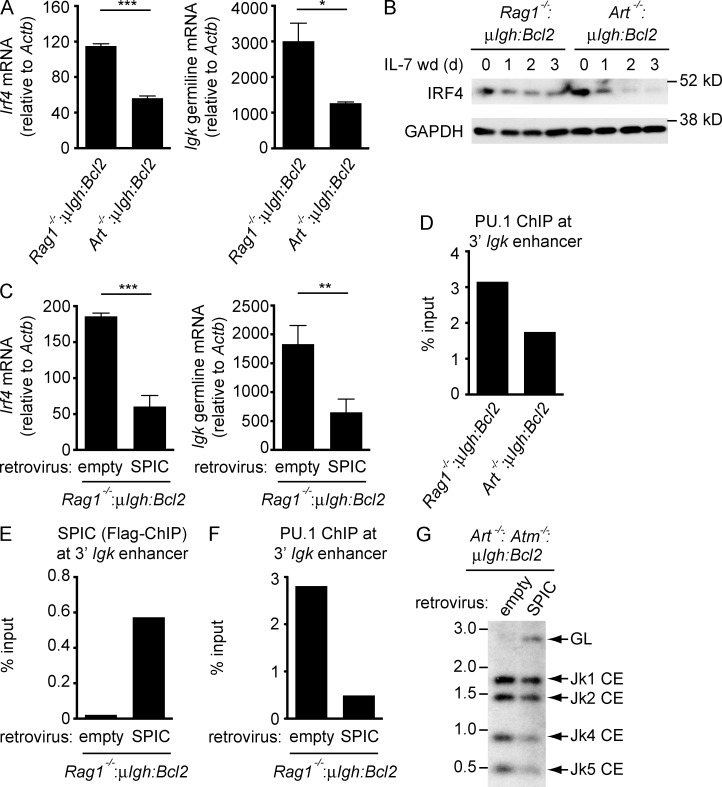
SYK and BLNK signals from the pre–BCR induce IRF4 to promote germline transcription and rearrangement of the Igk locus. Inhibition of pre–BCR signaling by RAG DSBs could feed back to suppress this process. Indeed, small pre–B cells with RAG DSBs (Art−/−:μIgh:Bcl2) exhibit decreased IRF4 expression and Igk germline transcripts as compared with Rag1−/−:μIgh:Bcl2 small pre–B cells (Fig. 7, A and B). Moreover, retroviral expression of SPIC in Rag1−/−:μIgh:Bcl2 small pre–B cells leads to a reduction in both Irf4 and germline Igk transcripts (Fig. 7 C). Germline Igk transcription requires PU.1 binding at 3′ Igk enhancer, which is significantly reduced in Art−/−:μIgh:Bcl2 small pre–B cells with RAG DSBs as compared with Rag1−/−:μIgh:Bcl2 small pre–B cells (Fig. 7 D). Moreover, when SPIC is retrovirally expressed in Rag1−/−:μIgh:Bcl2 small pre–B cells, we observe SPIC binding at the 3′ Igk enhancer and a concomitant decrease in PU.1 binding (Fig. 7, E and F). Thus, SPIC negatively regulates germline Igk transcription both by inhibiting pre–BCR signals that lead to IRF4 expression and by interfering with PU.1 binding at the 3′ Igk enhancer.
Figure 7.
SPIC inhibits Igl transcription and rearrangement. (A) Irf4 and germline Igk mRNA expression in Rag1−/−:μIgH:Bcl2 and Art−/−:μIgH:Bcl2 small pre–B cells assessed 3 d after IL-7 withdrawal. Data are mean and standard error for three replicate experiments. (B) Western blot of IRF4 in pre–B cells in IL-7 (0 d) and after IL-7 withdrawal (IL-7 wd) for the indicated days. GAPDH is shown as a protein loading control. Data are representative of three independent experiments. (C) Rag1−/−:μIgH:Bcl2 small pre–B cells were transduced with an empty retrovirus or a retrovirus expressing SPIC. Irf4 and germline Igk mRNA expression were assessed 2 d after IL-7 withdrawal. Data are mean and standard error for three replicate experiments. (D) ChIP-qPCR of PU.1 binding at the 3′ Igk enhancer in Rag1−/−:μIgH:Bcl2 and Art−/−:μIgH:Bcl2 small pre–B cells 3 d after IL-7 withdrawal. (E and F) Rag1−/−:μIgH:Bcl2 small pre–B cells were transduced with an empty retrovirus or a retrovirus expressing SPIC. ChIP-qPCR of SPIC binding (E) and PU.1 binding (F) at the 3′ Igk enhancer was performed 2 d after IL-7 withdrawal. (G) Southern blot of SacI and EcoRI digested genomic DNA from Art−/−:Atm−/−:μIgH:Bcl2 small pre–B cells transduced with an empty retrovirus or a retrovirus expressing SPIC 3 d after IL-7 withdrawal. Bands reflecting the germline (GL) Igk locus and Jk coding ends (Jk1, Jk2, Jk4, and Jk5 CE) are indicated as are molecular weight markers in kilobases. Data in D–G are representative of two independent experiments. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005 by Student’s t test.
The activation of ATM by Igk rearrangements feeds back to inhibit the generation of additional RAG DSBs as indicated by increased RAG cleavage at Igk in Art−/−:Atm−/−:μIgh:Bcl2 as compared with Art−/−:μIgh:Bcl2 small pre–B cells (Hewitt et al., 2009; Steinel et al., 2013; Fig. 1 A). Retroviral expression of SPIC in Art−/−:Atm−/−:μIgh:Bcl2 small pre–B cells results in decreased RAG cleavage at Igk (Fig. 7 G). This is indicated by the increase in Igk loci in the germline configuration and a concomitant decrease in Igk coding ends in Art−/−:Atm−/−:μIgh:Bcl2 small pre–B cells expressing SPIC as compared with Art−/−:Atm−/−:μIgh:Bcl2 small pre–B cells expressing the empty retroviral vector (Fig. 7 G). We conclude that the ATM-dependent induction of SPIC by RAG DSBs in small pre–B cells feeds back to inhibit germline transcription and the generation of additional RAG DSBs at the Igk locus.
RAG DSBs inhibit pre–BCR–driven proliferation
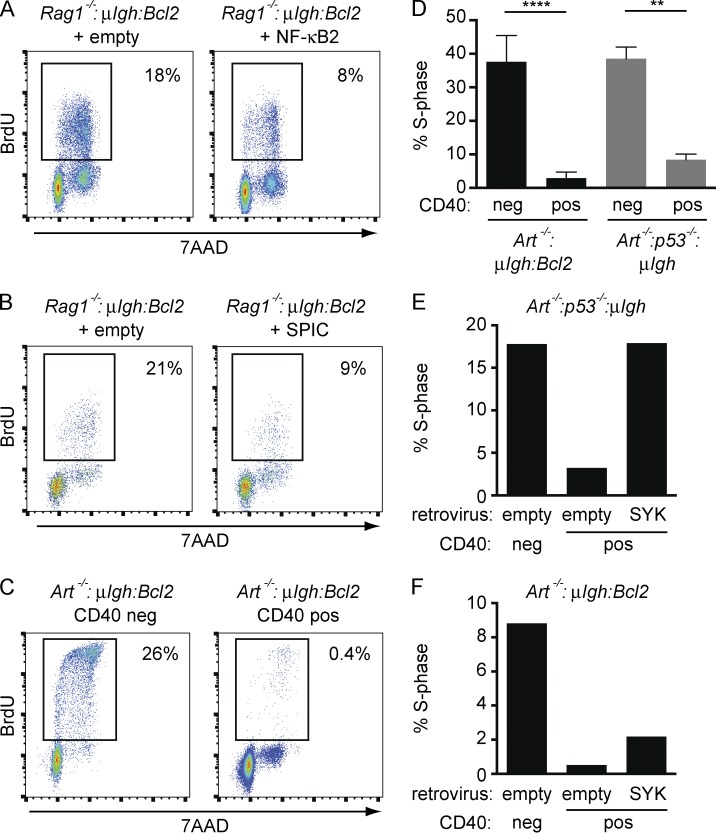
SYK activation downstream of the pre–BCR can drive small pre–B cells into cycle in the absence of IL-7r signals (Rolink et al., 2000; Wossning et al., 2006). Indeed, we find that Rag1−/−:μIgh:Bcl2 small pre–B cells proliferate after IL-7 withdrawal, and this proliferation is blocked by the SYK kinase inhibitor BAY 61–3606 (unpublished data). Persistent pre–BCR signaling, and resulting SYK activation, could drive small pre–B cells with Igk RAG DSBs into cycle. Thus, inactivation of pre–BCR signaling may be required in addition to canonical cell cycle checkpoint pathways (p53) to enforce G1 arrest in small pre–B cells with RAG DSBs. In small pre–B cells, SYK activity is suppressed through sequential activation of NF-κB2 and SPIC by RAG DSBs (Fig. 4, B–D; and Fig. 5 A). Consistent with this, expression of NF-κB2 or SPIC in Rag1−/−:μIgh:Bcl2 small pre–B cells is sufficient to reduce cell cycle entry (Fig. 8, A and B).
Figure 8.
RAG DSBs inhibit pre–BCR-driven proliferation. (A and B) Rag1−/−:μIgH:Bcl2 small pre–B cells were transduced with an empty retrovirus or a retrovirus expressing NF-κB2 (A) or SPIC (B). Flow cytometric analysis of BrdU incorporation (y axis) and DNA content (7AAD; x axis) was performed after IL-7 withdrawal. Percentage of cells that entered S-phase during BrdU labeling (box) is indicated. Data are representative of at least three independent experiments. (C) Cell cycle analysis was performed as in A on Art−/−:μIgH:Bcl2 small pre–B cells with RAG DSBs (CD40 pos) and those without RAG DSBs (CD40 neg) after IL-7 withdrawal. Percentage of cells that entered S-phase during BrdU labeling (box) is indicated. Data are representative of at least three independent experiments. (D) Cell cycle analysis of CD40 neg and CD40 pos Art−/−:μIgH:Bcl2 and Art−/−:p53−/−:μIgH small pre–B cells as in C. Graph represents the percentage of cells that entered S-phase. Data are mean and standard error for two independent experiments. (E and F) Art−/−:p53−/−:μIgH (E) and Art−/−:μIgH:Bcl2 (F) small pre–B cells were transduced with an empty retrovirus or a retrovirus expressing SYK. Cell cycle analysis of CD40 neg and CD40 pos pre–B cells was performed as in C. Graphs represent the percentage of cells that entered S-phase. Data in both panels are representative of three independent experiments. **, P ≤ 0.01; ****, P ≤ 0.0001 by Student’s t test.
Art−/−:μIgh:Bcl2 pre–B cells with RAG DSBs (CD40 pos), which both suppress SYK expression and activate p53, are arrested in the G1-phase of the cell cycle as compared with cells without RAG DSBs (CD40 neg; Fig. 8 C). Surprisingly, loss of p53 (Art−/−:p53−/−:μIgh) leads to only a modest increase in S-phase progression in cells with RAG DSBs (CD40 pos), suggesting that inhibition of pre–BCR signaling may also enforce G1 arrest in these cells (Fig. 8 D). Indeed, retroviral reconstitution of SYK significantly increases S-phase entry in Art−/−:p53−/−:μIgh small pre–B cells with RAG DSBs (CD40 pos; Fig. 8 E). Expression of SYK also increases S-phase entry in p53-sufficient Art−/−:μIgh:Bcl2 small pre–B cells with RAG DSBs (CD40 pos), but to a lesser extent than that observed in p53-deficient cells (Fig. 8, E and F). Thus, in pre–B cells, RAG DSBs must activate both the canonical checkpoint (p53) and a cell type–specific checkpoint pathway that inhibits pre–BCR signals to affect a complete G1 arrest.
DISCUSSION
Here, we show that RAG DSBs activate a cell type–specific checkpoint pathway that functions to both enforce G1 cell cycle arrest and regulate Igk chain gene rearrangement in small pre–B cell. This occurs through the sequential activation of ATM and NF-κB2, which leads to induction of SPIC, a hematopoietic-specific transcriptional repressor. SPIC inhibits pre–BCR signaling by repressing the expression of SYK and BLNK. Additionally, SPIC directly antagonizes Igk locus germline transcription and rearrangement. Thus, signals from RAG DSBs generated during Igk gene assembly are integrated into the signaling circuitry of developing pre–B cells.
In mature B cells, p100 and RELB are constitutively expressed and NF-κB2 activation occurs upon engagement of CD40 or BAFF receptors with their respective ligands, leading to stabilization of NIK and conversion of p100 to p52. In resting cells, NIK exists in a complex with the cIAP ubiquitin ligases, which mediate NIK degradation (Vallabhapurapu et al., 2008; Zarnegar et al., 2008; Hayden and Ghosh, 2012; Sun, 2012). Signals from the CD40 or BAFF receptors trigger dissociation of this complex and increased NIK levels (Vallabhapurapu et al., 2008; Zarnegar et al., 2008; Hayden and Ghosh, 2012; Sun, 2012). Similar to mature B cells, pre–B cells require NIK to promote the conversion of p100 to p52 in response to RAG DSBs. However, in contrast to mature B cells, expression of p100 and RELB must be induced in pre–B cells, and this depends on the activation of ATM by RAG DSBs generated during Igl chain gene assembly. Thus, pre–B cells require the convergence of two signals (ATM and NIK) to activate NF-κB2.
In response to RAG DSBs, NF-κB2 induces the expression of SPIC, an ETS family transcription factor with homology to PU.1 and SPIB (Bemark et al., 1999; Hashimoto et al., 1999). PU.1 and SPIB are constitutively expressed in developing B cells, where they perform critical developmental functions (Schweitzer and DeKoter, 2004). In mature B cells, SPIC regulates BCR signaling and canonical NF-κB1 activation (Zhu et al., 2008; Li et al., 2015). How SPIC is regulated and functions in early B cells has not been determined. Here, we show that in developing B cells, SPIC is induced by RAG DSBs. Consequently, SPIC is only expressed in small pre–B cells that are actively undergoing Igl chain gene rearrangement. SPIC represses transcription by interfering with PU.1 binding to critical cis-acting regulatory elements at the Igk locus and at the Blnk and Syk genes. In doing so, SPIC suppresses pre–BCR signaling, thereby enforcing a cell type–specific checkpoint in pre–B cells in response to RAG DSBs.
In large pre–B cells, signals from the pre–BCR attenuate IL-7r signals, which is required for transition to the small pre–B cell stage and for initiation of Igk chain gene assembly (Rolink et al., 1991; Johnson et al., 2008; Mandal et al., 2009; Ochiai et al., 2012; Clark et al., 2014). The pre–BCR signals through BLNK to antagonize AKT signaling downstream of the IL-7r (Herzog et al., 2008; Ochiai et al., 2012). Attenuation of IL-7r signaling leads to increased expression of SYK and BLNK, which further augments pre–BCR signaling (Ochiai et al., 2012). We show that once Igk chain gene assembly has been initiated, RAG DSB signals feedback to inhibit the pre–BCR by suppressing the expression of SYK and BLNK, which could lead to reactivation of IL-7r signals. However, pre–BCR signals may also function to reposition small pre–B cells to bone marrow niches devoid of IL-7–producing stromal cells (Tokoyoda et al., 2004; Johnson et al., 2008). Moreover, in small pre–B cells, RAG DSBs induce CD69, SWAP-70, and l-selectin, which function in lymphocyte trafficking and localization (Bredemeyer et al., 2008). This could enforce localization of pre–B cells in IL-7 poor areas of the bone marrow once Igk chain assembly has initiated. RAG DSBs also induce the PIM2 kinase, which antagonizes IL-7r signals (Bednarski et al., 2012). Thus, although pre–BCR signaling is required for the initial suppression of IL-7r signaling in small pre–B cells, continued pre–BCR signaling may not be required to maintain this suppression.
Initiation of pre–BCR signaling occurs upon the activation of SYK, which phosphorylates BLNK (Herzog et al., 2009; Rickert, 2013; Clark et al., 2014). We find that RAG DSBs inactivate pre–BCR signaling by inhibiting both SYK and BLNK. Why would it be important for RAG DSBs to inhibit both proteins? In the absence of SYK, ZAP-70 can phosphorylate BLNK, activating downstream signaling cascades (Chen et al., 2005; Fallah-Arani et al., 2008). Thus, the isolated inhibition of SYK would not attenuate pre–BCR signaling. BLNK antagonizes SYK activation by the pre–BCR (Flemming et al., 2003; Taguchi et al., 2004). Consequently, the isolated loss of BLNK increases SYK activation (Flemming et al., 2003; Taguchi et al., 2004). Indeed, both loss of BLNK and overactivation of SYK are associated with B cell transformation and leukemia (Flemming et al., 2003; Taguchi et al., 2004; Perova et al., 2014; Geng et al., 2015). Thus, complete suppression of pre–BCR signaling by RAG DSBs would rely on the inactivation of both SYK and BLNK. Here, we have defined the signaling pathway that inhibits pre–BCR signaling under physiological conditions in normal pre–B cells. Conceivably, this pathway could be therapeutically targeted in pre–B cell leukemias that depend on pre–BCR signaling.
DNA DSBs activate p53-dependent canonical checkpoint pathways to prevent cell cycle progression until the DSBs have been repaired (Shiloh, 2003). However, in some cell types proliferative signals can bypass p53-mediated checkpoints, driving cells with DSBs through cell cycle and promoting genome instability (Quelle et al., 1998; Sitko et al., 2008). RAG DSBs activate p53 in G1-phase small pre–B cells (Guidos et al., 1996). However, we find that even in the absence of p53, small pre–B cells with RAG DSBs are still arrested in G1. Furthermore, restoration of pre–BCR signaling through SYK expression overrides the p53-mediated checkpoint leading to the S-phase progression of small pre–B cells with RAG DSBs. Thus, enforcement of the G1–S checkpoint in small pre–B cells depends on activation of both the canonical checkpoint (p53) and a cell type–specific checkpoint pathway that inhibits pre–BCR proliferative signals. In the absence of ATM, these checkpoint pathways are defective. Consequently, ATM-deficient pre–B cells with RAG DSBs enter cell cycle and persist in the periphery for several weeks (Callén et al., 2007).
Small pre–B cells undergo multiple rounds of Igl chain gene rearrangement over a span of several days as they attempt to generate a productive Igl chain that can pair with the Igh chain to form a nonautoreactive BCR (Rajewsky, 1996; Casellas et al., 2001). Pre–BCR signals initiate Igl chain rearrangement through induction of IRF4 and subsequent activation of germline Igk transcription (Johnson et al., 2008; Clark et al., 2014). Once RAG DSBs are generated in the Igk locus, additional RAG cleavage events must be inhibited. In this regard, we show that RAG DSBs suppress pre–BCR signaling, leading to a reduction in Igk germline transcription and inhibition of additional DSBs at the Igk locus. This occurs through the ATM- and NF-κB2–dependent induction of SPIC, which inhibits SYK and BLNK and interferes with PU.1 binding to the Igk enhancer. This feedback circuit prevents pre–B cells with RAG DSBs from entering cycle and prevents additional RAG DSBs at the Igl locus until the initial rearrangement has been completed and ATM signaling is terminated. If this rearrangement encodes an Igk chain that forms a functional BCR, the cell transits to the immature B cell stage. However, if the Igk chain gene is not functional, loss of ATM signaling would lead to the reexpression of SYK and BLNK, leading to reactivation of pre–BCR signaling and induction of another Igk chain gene rearrangement. We propose that small pre–B cells toggle between pre–BCR signals and a RAG DSB-dependent cell-type specific checkpoint to order iterative Igl chain gene rearrangements and maintain genomic stability by preventing pre–B cells with RAG DSBs from entering cell cycle.
MATERIALS AND METHODS
Mice
All mice were bred and maintained under specific pathogen–free conditions at the Washington University School of Medicine and were handled in accordance to the guidelines set forth by the Division of Comparative Medicine of Washington University. Wild-type B6 mice (The Jackson Laboratory) were used as controls. p53−/− mice were purchased from The Jackson Laboratory. Spicigfp/igfp, Nik−/−, and Nfkb2−/− mice were generated as previously described (Yin et al., 2001; Zarnegar et al., 2004; Haldar et al., 2014). Spicigfp/igfp mice were on a B6 background. All other mice are on a mixed genetic background. In vivo studies were conducted on 4–5-wk-old mice.
Cell culture
Primary pre–B cell cultures were generated from bone marrow harvested from 4–6-wk-old mice and cultured for 7–10 d at 2 × 106 cells/ml in media containing 5 ng/ml of IL-7 (Life Technologies). For IL-7 withdrawal experiments, cells were resuspended in media without IL-7 and maintained at 2 × 106 cells/ml.
Retroviral transduction and cDNA expression
pMSCV-mCherry-SYK was purchased from Addgene (plasmid 50045; Strijbis et al., 2013). cDNA encoding SPIC with a 5′Flag tag was cloned into the pOZ retrovirus containing an IRES sequence, followed by truncated human CD25 (hCD25) cDNA as a marker of transduced cells. A DNA binding–deficient SPIC was generated by mutating arginine 175 to glycine (R175G) using QuikChange II XL (Agilent) according to the manufacturer’s protocol (Pio et al., 1996; Suraweera et al., 2005). Retrovirus was produced in platE cells by transfection of the retroviral plasmid with Lipofectamine 2000 (Life Technologies) according to the manufacturer’s protocol. Viral supernatant was collected and pooled from 24–72 h after transfection. Viral supernatant was used immediately to transduce cells or was concentrated before transduction. To concentrated viral particles, PEG-8000 (Sigma-Aldrich; final concentration 8%) was added to viral supernatant, incubation at 4°C overnight, and centrifugation at 2,500 RPM for 20 min. Precipitated virus was resuspended at 300× concentration in sterile PBS. Pre–B cells were transduced with unconcentrated virus (40 × 106 cells in 3 ml viral supernatant) or with concentrated virus (40 × 106 in 1 ml with 10× viral particles) in IL-7–containing media with polybrene (5 µg/ml; Sigma-Aldrich) by centrifugation for 2 h at 1,200 RPM at room temperature. 4 h later, fresh IL-7 media (2 mls) was added, and the cells were incubated overnight. Virus-containing media was removed and cells were cultured in fresh IL-7 media (2 × 106/ml). Cells expressing the retrovirus construct were identified by flow cytometric assessment of hCD25 or mCherry expression using a FACSCalibur (BD). Transduced cells were sorted using biotin-conjugated anti-hCD25 (BD) and anti-biotin magnetic beads (Miltenyi Biotec) on MS columns (Miltenyi Biotec) according to the manufacturer’s protocol.
Flow cytometric analyses and cell sorting
Flow cytometric analyses were performed on a FACSCalibur or BD LSRFortessa (BD). Sorting was conducted on a BD Aria (user-operated) or on a Sony Sy3200 through the Siteman Cancer Center Flow Cytometry Core Facility. FITC-conjugated anti-CD45R/B220 (clone RA3-6B2), phycoerythrin (PE)-conjugated anti-CD43 (clone S7), FITC-conjugated anti-CD43 (clone S7), PE-Cy7–conjugated anti-CD45/B220 (clone RA3-6B2), allophycocyanin (APC)-conjugated anti-IgM (clone II/41), PE-conjugated anti-CD40 (clone 1C10), and biotin-conjugated anti-CD40 (clone 3/23) were purchased from BD. APC-conjugated streptavidin was purchased from eBioscience. PE-conjugated anti-hCD25 (clone BC96) and APC-conjugated anti-hCD25 (clone BC96) were purchased from BioLegend. APC-conjugated anti-mCherry (clone 16D7) was purchased from Life Technologies. Staining for intracellular proteins was performed using Cytofix/Cytoperm solution (BD).
Cell cycle analysis
To assess pre–BCR driven proliferation, pre–B cells were resuspended in media without IL-7 and maintained at 2 × 106 cells/ml. 8 h after removal from IL-7, cells were pulsed BrdU for 2 h using the BrdU-FITC kit (BD) per the manufacturer’s instructions. DNA content was assessed by 7AAD (BD) or DAPI (Sigma-Aldrich). The SYK kinase inhibitor BAY 61–3606 (EMD Millipore) was used at 10 µM and was added 24 h before IL-7 withdrawal.
Cell fractionation and Western blot
Cell fractionation was performed as previously described (Méndez and Stillman, 2000). Western blots were done on whole-cell lysates as previously described (Bredemeyer et al., 2008). Anti–NF-κB2 (recognizes p100 and p52; polyclonal), anti-RELB (clone C1E4), anti-BLNK (clone D8R3G), anti-SYK (clone D1I5Q), anti-BTK (clone D3H5), and anti-phospho(S15)-p53 (polyclonal) antibodies were purchased from Cell Signaling Technology. Anti-TRAF2 (polyclonal), anti-TRAF3 (polyclonal), anti-IRF4 (clone E-7), anti-PU.1 (polyclonal), and anti-LaminB (clone G-1) were obtained from Santa Cruz Biotechnology. Anti-phosphorylated BLNK (clone J117-1278) was purchased from BD. Anti-GAPDH (clone GAPDH-71.1) and anti-FLAG (clone M2) were from Sigma-Aldrich. Secondary reagents were horseradish peroxidase (HRP)–conjugated anti–mouse IgG (Cell Signaling Technology) or anti–rabbit IgG (Cell Signaling Technology). Western blots were developed with ECL (Thermo Fisher Scientific) and ECL Prime (GE Healthcare).
Southern blot
Southern blot analyses of coding ends generated during rearrangement at the Igk locus were performed on genomic DNA digested with SacI and EcoRI using the JκIII probe and Tcrβ probe, as described previously (Bredemeyer et al., 2008).
RT-PCR
RNA was isolated using RNeasy (QIAGEN) and reversed transcribed using either a polyT primer or a random hexamer primer with SuperScriptII (Life Technologies) according to the manufacturers’ protocol. RT-PCR was performed using Brilliant II SYBR Green (Agilent) and acquired on an Mx3000P (Stratagene). Each reaction was run in triplicate. Primer sequences are listed in Table S3.
Gene arrays
Three independent IL-7 cultures for each genotype (Rag1−/−:μIgH:Bcl2, Art−/−:μIgH:Bcl2 and Art−/−:Nfkb2−/−:μIgH:Bcl2) were withdrawn from IL-7 for 48 h. RNA was isolated using RNeasy (QIAGEN). Gene expression profiling was performed using Illumina MouseRef-8 expression microarrays by the Washington University Genome Technology Access Center according to the manufacturer’s protocols. Data have been deposited in NCBI’s Gene Expression Omnibus under accession no. GSE67854. Differentially expressed genes were identified by error-weighted analysis of variance (ANOVA; Partek Genomics Suite v6.12). Fold changes were calculated based on the mean of three cell lines for each genotype. Only those genes with a fold change of ≥1.5 and a p-value ≤ 0.05 were considered for further analysis. ImmGen (Immunological Genome project) project gene expression data were independently analyzed to identify all genes expression changes that were significantly changed (P < 0.05) between large and small pre–B cells (datasets: preB_FrC_BM vs. preB_FrD_BM; Heng and Painter, 2008).
Chromatin immunoprecipitation (ChIP)
ChIP was performed using anti-PU.1 (Santa Cruz Biotechnology), anti-FLAG (Sigma-Aldrich), control rabbit IgG (Santa Cruz Biotechnology), and control mouse IgG antibodies (eBioscience) as previously described (Ochiai et al., 2013). In brief, DNA from 5 × 106 cells was cross-linked with 1% formaldehyde for 10 min at room temperature Reaction was stopped with 125 µM Glycine. Cells were lysed with NP-40 and nuclei were frozen in liquid nitrogen, and then lysed with SDS. DNA was sonicated with 30-s pulses for 60–90 cycles. DNA fragmentation was in the range of 0.5–1 kb and was monitored by agarose gel electrophoresis. Immunoprecipitation was performed with 1 µg of anti-PU.1 (polyclonal; Santa Cruz Biotechnology) or control rabbit IgG (sc-2027) and protein A–Sepharose beads (Thermo Fisher Scientific). For FLAG ChIP, 2 µg of anti-FLAG (clone M2; Sigma-Aldrich) or control mouse IgG (clone P3.6.2.8.1; eBioscience) was first incubated with rabbit anti-mouse IgG (EMD Millipore) and immunoprecipitation was performed with Protein A Dynabeads (Life Technologies). DNA was purified with QIAquick PCR purification kit (QIAGEN). Quantitative PCR was performed using Brilliant II SYBR Green (Agilent) and acquired on an Mx3000P (Stratagene). Primers are listed in Table S3.
Statistical analysis
For analyses other than gene array, p-values were generated via Student’s t test (unpaired, two-tailed) using Prism (GraphPad Software).
Online supplemental material
Fig. S1 shows IL-7 culture system for studying RAG DSBs. Table S1 shows genes regulated by RAG DSBs. Table S2 shows genes regulated by NF-κB2. Table S3 shows sequences of primers. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20151048/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Robert Schreiber for providing the Nik−/− mice. We thank the Siteman Flow Cytometry Core (P30 CA91842) and Genome Technology Access Center (P30 CA91842 and UL1 TR000448) at Washington University School of Medicine for their assistance with experiments.
This work was supported by the National Institutes of Health grants CA136470 (B.P. Sleckman), AI074953 (B.P. Sleckman), AI47829 (B.P. Sleckman), K08AI102946-01 (J.J. Bednarski), K08AI106953 (M. Haldar), and R01AI112621 (C.H. Bassing). J.J. Bednarski was supported by a Hyundai Hope on Wheels Scholar Award and an Alex’s Lemonade Stand Foundation Award.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- ATM
- ataxia-telangiectasia mutated
- BCR
- B cell receptor
- ChIP
- chromatin immunoprecipitation
- DDR
- DNA damage response
- DSB
- double-strand break
- NHEJ
- nonhomologous end joining
References
- Amin R.H., and Schlissel M.S.. 2008. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat. Immunol. 9:613–622. 10.1038/ni.1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarski J.J., Nickless A., Bhattacharya D., Amin R.H., Schlissel M.S., and Sleckman B.P.. 2012. RAG-induced DNA double-strand breaks signal through Pim2 to promote pre-B cell survival and limit proliferation. J. Exp. Med. 209:11–17. 10.1084/jem.20112078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemark M., Mårtensson A., Liberg D., and Leanderson T.. 1999. Spi-C, a novel Ets protein that is temporally regulated during B lymphocyte development. J. Biol. Chem. 274:10259–10267. 10.1074/jbc.274.15.10259 [DOI] [PubMed] [Google Scholar]
- Bertolino E., Reddy K., Medina K.L., Parganas E., Ihle J., and Singh H.. 2005. Regulation of interleukin 7-dependent immunoglobulin heavy-chain variable gene rearrangements by transcription factor STAT5. Nat. Immunol. 6:836–843. 10.1038/ni1226 [DOI] [PubMed] [Google Scholar]
- Bredemeyer A.L., Helmink B.A., Innes C.L., Calderon B., McGinnis L.M., Mahowald G.K., Gapud E.J., Walker L.M., Collins J.B., Weaver B.K., et al. . 2008. DNA double-strand breaks activate a multi-functional genetic program in developing lymphocytes. Nature. 456:819–823. 10.1038/nature07392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callén E., Jankovic M., Difilippantonio S., Daniel J.A., Chen H.T., Celeste A., Pellegrini M., McBride K., Wangsa D., Bredemeyer A.L., et al. . 2007. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 130:63–75. 10.1016/j.cell.2007.06.016 [DOI] [PubMed] [Google Scholar]
- Carlsson R., Persson C., and Leanderson T.. 2003. SPI-C, a PU-box binding ETS protein expressed temporarily during B-cell development and in macrophages, contains an acidic transactivation domain located to the N-terminus. Mol. Immunol. 39:1035–1043. 10.1016/S0161-5890(03)00032-4 [DOI] [PubMed] [Google Scholar]
- Casellas R., Shih T.A., Kleinewietfeld M., Rakonjac J., Nemazee D., Rajewsky K., and Nussenzweig M.C.. 2001. Contribution of receptor editing to the antibody repertoire. Science. 291:1541–1544. 10.1126/science.1056600 [DOI] [PubMed] [Google Scholar]
- Chen L., Apgar J., Huynh L., Dicker F., Giago-McGahan T., Rassenti L., Weiss A., and Kipps T.J.. 2005. ZAP-70 directly enhances IgM signaling in chronic lymphocytic leukemia. Blood. 105:2036–2041. 10.1182/blood-2004-05-1715 [DOI] [PubMed] [Google Scholar]
- Clark M.R., Mandal M., Ochiai K., and Singh H.. 2014. Orchestrating B cell lymphopoiesis through interplay of IL-7 receptor and pre-B cell receptor signalling. Nat. Rev. Immunol. 14:69–80. 10.1038/nri3570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corfe S.A., and Paige C.J.. 2012. The many roles of IL-7 in B cell development; mediator of survival, proliferation and differentiation. Semin. Immunol. 24:198–208. 10.1016/j.smim.2012.02.001 [DOI] [PubMed] [Google Scholar]
- Desiderio S., Lin W.C., and Li Z.. 1996. The cell cycle and V(D)J recombination. Curr. Top. Microbiol. Immunol. 217:45–59. [DOI] [PubMed] [Google Scholar]
- Fallah-Arani F., Schweighoffer E., Vanes L., and Tybulewicz V.L.. 2008. Redundant role for Zap70 in B cell development and activation. Eur. J. Immunol. 38:1721–1733. 10.1002/eji.200738026 [DOI] [PubMed] [Google Scholar]
- Flemming A., Brummer T., Reth M., and Jumaa H.. 2003. The adaptor protein SLP-65 acts as a tumor suppressor that limits pre-B cell expansion. Nat. Immunol. 4:38–43. 10.1038/ni862 [DOI] [PubMed] [Google Scholar]
- Fugmann S.D., Lee A.I., Shockett P.E., Villey I.J., and Schatz D.G.. 2000. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu. Rev. Immunol. 18:495–527. 10.1146/annurev.immunol.18.1.495 [DOI] [PubMed] [Google Scholar]
- Geng H., Hurtz C., Lenz K.B., Chen Z., Baumjohann D., Thompson S., Goloviznina N.A., Chen W.Y., Huan J., LaTocha D., et al. . 2015. Self-enforcing feedback activation between BCL6 and pre-B cell receptor signaling defines a distinct subtype of acute lymphoblastic leukemia. Cancer Cell. 27:409–425. 10.1016/j.ccell.2015.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidos C.J., Williams C.J., Grandal I., Knowles G., Huang M.T., and Danska J.S.. 1996. V(D)J recombination activates a p53-dependent DNA damage checkpoint in scid lymphocyte precursors. Genes Dev. 10:2038–2054. 10.1101/gad.10.16.2038 [DOI] [PubMed] [Google Scholar]
- Haldar M., Kohyama M., So A.Y.-L., Kc W., Wu X., Briseno C.G., Satpathy A.T., Kretzer N.M., Rajasekaran N.S., and Wang L., et al. . 2014. Heme-mediated BACH1 degradation induces SPI-C to promote monocyte differentiation into iron-recycling macrophages. Cell. 156:1223–1234. 10.1016/j.cell.2014.01.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto S., Nishizumi H., Hayashi R., Tsuboi A., Nagawa F., Takemori T., and Sakano H.. 1999. Prf, a novel Ets family protein that binds to the PU.1 binding motif, is specifically expressed in restricted stages of B cell development. Int. Immunol. 11:1423–1429. 10.1093/intimm/11.9.1423 [DOI] [PubMed] [Google Scholar]
- Hayden M.S., and Ghosh S.. 2012. NF-κB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 26:203–234. 10.1101/gad.183434.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmink B.A., and Sleckman B.P.. 2012. The response to and repair of RAG-mediated DNA double-strand breaks. Annu. Rev. Immunol. 30:175–202. 10.1146/annurev-immunol-030409-101320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng T.S., and Painter M.W.. Immunological Genome Project Consortium . 2008. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9:1091–1094. 10.1038/ni1008-1091 [DOI] [PubMed] [Google Scholar]
- Herzog S., Hug E., Meixlsperger S., Paik J.H., DePinho R.A., Reth M., and Jumaa H.. 2008. SLP-65 regulates immunoglobulin light chain gene recombination through the PI(3)K-PKB-Foxo pathway. Nat. Immunol. 9:623–631. 10.1038/ni.1616 [DOI] [PubMed] [Google Scholar]
- Herzog S., Reth M., and Jumaa H.. 2009. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat. Rev. Immunol. 9:195–205. 10.1038/nri2491 [DOI] [PubMed] [Google Scholar]
- Hewitt S.L., Yin B., Ji Y., Chaumeil J., Marszalek K., Tenthorey J., Salvagiotto G., Steinel N., Ramsey L.B., Ghysdael J., et al. . 2009. RAG-1 and ATM coordinate monoallelic recombination and nuclear positioning of immunoglobulin loci. Nat. Immunol. 10:655–664. 10.1038/ni.1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K., Hashimshony T., Sawai C.M., Pongubala J.M., Skok J.A., Aifantis I., and Singh H.. 2008. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity. 28:335–345. 10.1016/j.immuni.2007.12.019 [DOI] [PubMed] [Google Scholar]
- Li S.K., Solomon L.A., Fulkerson P.C., and DeKoter R.P.. 2015. Identification of a negative regulatory role for spi-C in the murine B cell lineage. J. Immunol. 194:3798–3807. 10.4049/jimmunol.1402432 [DOI] [PubMed] [Google Scholar]
- Mandal M., Powers S.E., Ochiai K., Georgopoulos K., Kee B.L., Singh H., and Clark M.R.. 2009. Ras orchestrates exit from the cell cycle and light-chain recombination during early B cell development. Nat. Immunol. 10:1110–1117. 10.1038/ni.1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M., Powers S.E., Maienschein-Cline M., Bartom E.T., Hamel K.M., Kee B.L., Dinner A.R., and Clark M.R.. 2011. Epigenetic repression of the Igk locus by STAT5-mediated recruitment of the histone methyltransferase Ezh2. Nat. Immunol. 12:1212–1220. 10.1038/ni.2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez J., and Stillman B.. 2000. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20:8602–8612. 10.1128/MCB.20.22.8602-8612.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai K., Maienschein-Cline M., Mandal M., Triggs J.R., Bertolino E., Sciammas R., Dinner A.R., Clark M.R., and Singh H.. 2012. A self-reinforcing regulatory network triggered by limiting IL-7 activates pre-BCR signaling and differentiation. Nat. Immunol. 13:300–307. 10.1038/ni.2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai K., Maienschein-Cline M., Simonetti G., Chen J., Rosenthal R., Brink R., Chong A.S., Klein U., Dinner A.R., Singh H., and Sciammas R.. 2013. Transcriptional regulation of germinal center B and plasma cell fates by dynamical control of IRF4. Immunity. 38:918–929. 10.1016/j.immuni.2013.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perova T., Grandal I., Nutter L.M., Papp E., Matei I.R., Beyene J., Kowalski P.E., Hitzler J.K., Minden M.D., Guidos C.J., and Danska J.S.. 2014. Therapeutic potential of spleen tyrosine kinase inhibition for treating high-risk precursor B cell acute lymphoblastic leukemia. Sci. Transl. Med. 6:236ra62 10.1126/scitranslmed.3008661 [DOI] [PubMed] [Google Scholar]
- Pio F., Kodandapani R., Ni C.Z., Shepard W., Klemsz M., McKercher S.R., Maki R.A., and Ely K.R.. 1996. New insights on DNA recognition by ets proteins from the crystal structure of the PU.1 ETS domain-DNA complex. J. Biol. Chem. 271:23329–23337. 10.1074/jbc.271.38.23329 [DOI] [PubMed] [Google Scholar]
- Pongubala J.M., Nagulapalli S., Klemsz M.J., McKercher S.R., Maki R.A., and Atchison M.L.. 1992. PU.1 recruits a second nuclear factor to a site important for immunoglobulin κ 3′ enhancer activity. Mol. Cell. Biol. 12:368–378. 10.1128/MCB.12.1.368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quelle F.W., Wang J., Feng J., Wang D., Cleveland J.L., Ihle J.N., and Zambetti G.P.. 1998. Cytokine rescue of p53-dependent apoptosis and cell cycle arrest is mediated by distinct Jak kinase signaling pathways. Genes Dev. 12:1099–1107. 10.1101/gad.12.8.1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewsky K. 1996. Clonal selection and learning in the antibody system. Nature. 381:751–758. 10.1038/381751a0 [DOI] [PubMed] [Google Scholar]
- Rickert R.C. 2013. New insights into pre-BCR and BCR signalling with relevance to B cell malignancies. Nat. Rev. Immunol. 13:578–591. 10.1038/nri3487 [DOI] [PubMed] [Google Scholar]
- Rolink A., Kudo A., Karasuyama H., Kikuchi Y., and Melchers F.. 1991. Long-term proliferating early pre B cell lines and clones with the potential to develop to surface Ig-positive, mitogen reactive B cells in vitro and in vivo. EMBO J. 10:327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolink A.G., Winkler T., Melchers F., and Andersson J.. 2000. Precursor B cell receptor-dependent B cell proliferation and differentiation does not require the bone marrow or fetal liver environment. J. Exp. Med. 191:23–32. 10.1084/jem.191.1.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer B.L., and DeKoter R.P.. 2004. Analysis of gene expression and Ig transcription in PU.1/Spi-B-deficient progenitor B cell lines. J. Immunol. 172:144–154. 10.4049/jimmunol.172.1.144 [DOI] [PubMed] [Google Scholar]
- Shiloh Y. 2003. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer. 3:155–168. 10.1038/nrc1011 [DOI] [PubMed] [Google Scholar]
- Sitko J.C., Yeh B., Kim M., Zhou H., Takaesu G., Yoshimura A., McBride W.H., Jewett A., Jamieson C.A., and Cacalano N.A.. 2008. SOCS3 regulates p21 expression and cell cycle arrest in response to DNA damage. Cell. Signal. 20:2221–2230. 10.1016/j.cellsig.2008.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinel N.C., Lee B.S., Tubbs A.T., Bednarski J.J., Schulte E., Yang-Iott K.S., Schatz D.G., Sleckman B.P., and Bassing C.H.. 2013. The ataxia telangiectasia mutated kinase controls Igκ allelic exclusion by inhibiting secondary Vκ-to-Jκ rearrangements. J. Exp. Med. 210:233–239. 10.1084/jem.20121605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strijbis K., Tafesse F.G., Fairn G.D., Witte M.D., Dougan S.K., Watson N., Spooner E., Esteban A., Vyas V.K., Fink G.R., et al. 2013. Bruton’s Tyrosine Kinase (BTK) and Vav1 contribute to Dectin1-dependent phagocytosis of Candida albicans in macrophages. PLoS Pathog. 9:e1003446. 10.1371/journal.ppat.1003446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S.C. 2012. The noncanonical NF-κB pathway. Immunol. Rev. 246:125–140. 10.1111/j.1600-065X.2011.01088.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraweera N., Meijne E., Moody J., Carvajal-Carmona L.G., Yoshida K., Pollard P., Fitzgibbon J., Riches A., van Laar T., Huiskamp R., et al. . 2005. Mutations of the PU.1 Ets domain are specifically associated with murine radiation-induced, but not human therapy-related, acute myeloid leukaemia. Oncogene. 24:3678–3683. 10.1038/sj.onc.1208422 [DOI] [PubMed] [Google Scholar]
- Taguchi T., Kiyokawa N., Takenouch H., Matsui J., Tang W.R., Nakajima H., Suzuki K., Shiozawa Y., Saito M., Katagiri Y.U., et al. . 2004. Deficiency of BLNK hampers PLC-γ2 phosphorylation and Ca2+ influx induced by the pre-B-cell receptor in human pre-B cells. Immunology. 112:575–582. 10.1111/j.1365-2567.2004.01918.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokoyoda K., Egawa T., Sugiyama T., Choi B.I., and Nagasawa T.. 2004. Cellular niches controlling B lymphocyte behavior within bone marrow during development. Immunity. 20:707–718. 10.1016/j.immuni.2004.05.001 [DOI] [PubMed] [Google Scholar]
- Vallabhapurapu S., Matsuzawa A., Zhang W., Tseng P.H., Keats J.J., Wang H., Vignali D.A., Bergsagel P.L., and Karin M.. 2008. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-κB signaling. Nat. Immunol. 9:1364–1370. 10.1038/ni.1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wossning T., Herzog S., Köhler F., Meixlsperger S., Kulathu Y., Mittler G., Abe A., Fuchs U., Borkhardt A., and Jumaa H.. 2006. Deregulated Syk inhibits differentiation and induces growth factor-independent proliferation of pre-B cells. J. Exp. Med. 203:2829–2840. 10.1084/jem.20060967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L.S., Sokalski K.M., Hotke K., Christie D.A., Zarnett O., Piskorz J., Thillainadesan G., Torchia J., and DeKoter R.P.. 2012. Regulation of B cell linker protein transcription by PU.1 and Spi-B in murine B cell acute lymphoblastic leukemia. J. Immunol. 189:3347–3354. 10.4049/jimmunol.1201267 [DOI] [PubMed] [Google Scholar]
- Yin L., Wu L., Wesche H., Arthur C.D., White J.M., Goeddel D.V., and Schreiber R.D.. 2001. Defective lymphotoxin-β receptor-induced NF-κB transcriptional activity in NIK-deficient mice. Science. 291:2162–2165. 10.1126/science.1058453 [DOI] [PubMed] [Google Scholar]
- Zarnegar B., He J.Q., Oganesyan G., Hoffmann A., Baltimore D., and Cheng G.. 2004. Unique CD40-mediated biological program in B cell activation requires both type 1 and type 2 NF-κB activation pathways. Proc. Natl. Acad. Sci. USA. 101:8108–8113. 10.1073/pnas.0402629101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnegar B.J., Wang Y., Mahoney D.J., Dempsey P.W., Cheung H.H., He J., Shiba T., Yang X., Yeh W.C., Mak T.W., et al. . 2008. Noncanonical NF-κB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat. Immunol. 9:1371–1378. 10.1038/ni.1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.A., Mortazavi A., Williams B.A., Wold B.J., and Rothenberg E.V.. 2012. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell. 149:467–482. 10.1016/j.cell.2012.01.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Schweitzer B.L., Romer E.J., Sulentic C.E., and DeKoter R.P.. 2008. Transgenic expression of Spi-C impairs B-cell development and function by affecting genes associated with BCR signaling. Eur. J. Immunol. 38:2587–2599. 10.1002/eji.200838323 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.