Abstract
We have recently reported that CYB5D2 plays a role in suppression of cervical cancer tumorigenesis, “CYB5D2 displays tumor suppression activities towards cervical cancer” [1]. We provide the accompany data here describing the effects of CYB5D2 overexpression and addition of recombinant CYB5D2 on HeLa cell cycle distribution. Furthermore, we will present the conditions used to specifically determine CYB5D2 expression in primary cervical and cervical cancer tissues using immunohistochemistry (IHC) and the patient cohort involved in assessing the CYB5D2 protein levels in primary cervical and cervical cancer tissues.
Specifications Table
| Subject area | Biology |
| More specific subject area | Cervical cancer tumorigenesis |
| Type of data | Figures, Table |
| How data was acquired | Western blot analysis using the Bio-Rad mini-gel apparatus; cell cycle determination using a flow cytometer (Bechman Coulter, CytomicsTM FC500) |
| Data format | Filtered and analyzed |
| Experimental factors | Cells are serum-starved for 24 h, followed by stimulation with 10% of bovine fetal serum (FBS) to examine AKT and ERK activation |
| Experimental features | Cell cycle progression and protein expression |
| Data source location | Hamilton, Ontario, Canada |
| Data accessibility | Data is within this article |
Value of the data
-
•
CYB5D2׳s effects on HeLa cell cycle distribution could be considered when investigating a role of CYB5D2 in regulating cell proliferation in other cell types.
-
•
The data on CYB5D2 in affecting ERK and AKT activation should be helpful in researching CYB5D2׳s role in regulating growth factor receptor signaling.
-
•
The data is useful for future investigations of CYB5D2-mediated cellular processes.
1. Data
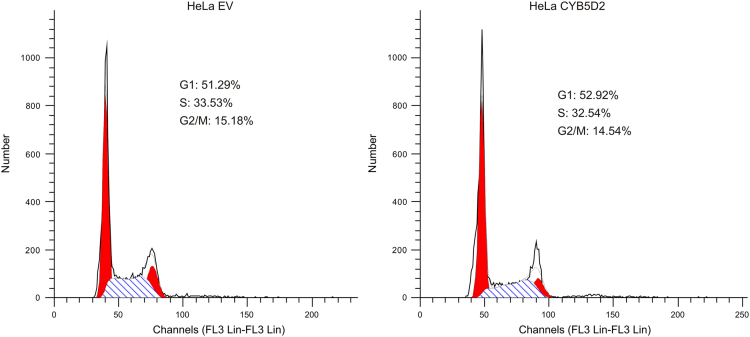
Fig. 1 examines the cell cycle distribution of HeLa cells stably expressing either an empty vector (HeLa EV) or CYB5D2 (HeLa CYB5D2).
Fig. 1.
Examination of the effects of CYB5D2 overexpression on HeLa cell cycle distribution. HeLa EV and HeLa CYB5D2 cells were seeded in 60 mm plates, and cultured for 2 days. At density of approximately 80% confluency, cell cycle distributions were determined using a flow cytometer.
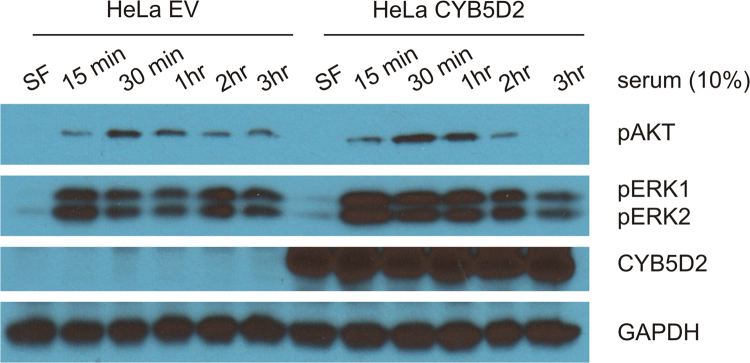
Fig. 2 shows the status of AKT and ERK1/2 activation in HeLa EV and HeLa CYB5D2 cells. Activation of AKT and ERK1/2 was indirectly determined according to the specific phosphorylation events (see Fig. 2 legend for details).
Fig. 2.
The impacts of ectopic expression of CYB5D2 on serum-induced activation of AKT and ERK kinases. HeLa EV and HeLa CYB5D2 cells at approximately 90% confluency were serum starved for 24 h, and stimulated with 10% of fetal bovine serum for the indicated periods, followed by western blot analysis for the phosphorylation of AKT at serine 473 (pAKT) and ERK at threonine 183 and tyrosine 185 (pERK1/2) as well as CYB5D2 and GAPDH. Experiments were performed twice; typical results from a single repeat are shown. SF: serum free.
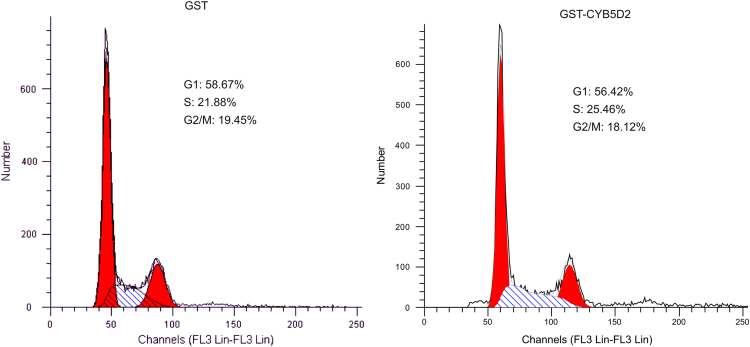
CYB5D2 can be a secretory protein [2], [3] that has been indicated to inhibit Neuro2a cell proliferation [2]. The cell cycle distribution of HeLa cells was determined in the presence of either GST or GST-CYB5D2 (Fig. 3).
Fig. 3.
Determination of the effects of recombinant CYB5D2 on HeLa cell cycle distribution. GST and GST-CYB5D2 recombinant proteins were purified from E. coli. HeLa EV and HeLa CYB5D2 cells were incubated with GST and GST-CYB5D2 at 1 mg/ml for 24 h, followed by the determination of cell cycle distribution. Cell proliferation in the presence of either protein was clearly observed. Experiments were carried out twice; typical results from a single repeat are shown.
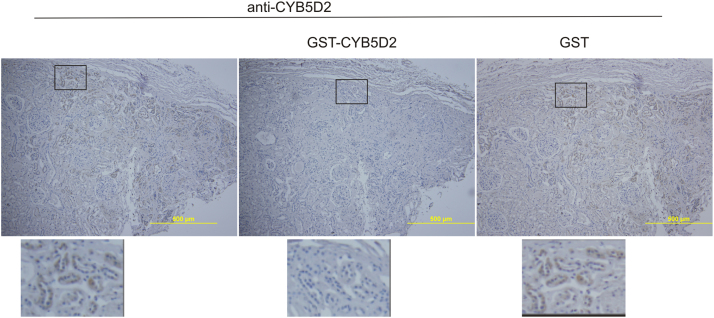
Fig. 4 shows recognition of the CYB5D2 protein in human kidney tissues by the anti-CYB5D2 antibody in the presence of GST or GST-CYB5D2 as a competitor.
Fig. 4.
Anti-CYB5D2 antibody specifically recognizes CYB5D2. Normal human kidney tissue was immunohistochemistry (IHC) stained with anti-CYB5D2 antibody without and with addition of recombinant GST-CYB5D2 or GST. The indicated regions were enlarged 3 fold and presented underneath of the individual panels. Recombinant GST-CYB5D2 and GST were produced in E. coli BL21. The recombinant protein GST-CYB5D2 was generated by N-terminal fusion of the transmembrane domain deletion mutant of CYB5D2 to GST. Anti-CYB5D2 antibody was affinity-purified by using GST-CYB5D2 as previously described [4]. For the competition experiments, GST-CYB5D2 or GST at 1 mg/ml was pre-incubated for one hour on ice with anti-CYB5D2 antibody (1:250) before applying to human kidney tissues.
Table 1 shows the tissues used to examine the CYB5D2 protein levels in normal cervical and cervical cancer tissues.
Table 1.
Patient׳s clinical information.
| Patients | Pathological diagnosis | Age | Grade |
|---|---|---|---|
| 1 | Endocervical type adenocarcinoma | 42 | 1 |
| 2 | Endocervical type adenocarcinoma | 42 | 1 |
| 3 | Endometrioid adenocarcinoma with squamous metaplasia | 48 | 1 |
| 4 | Endometrioid adenocarcinoma with squamous metaplasia | 48 | 1 |
| 5 | Endocervical type adenocarcinoma | 52 | 1–2 |
| 6 | Endocervical type adenocarcinoma | 52 | 1–2 |
| 7 | Endometrioid adenocarcinoma | 32 | 1–2 |
| 8 | Endometrioid adenocarcinoma | 32 | 1–2 |
| 9 | Instestinal type adenocarcinoma | 72 | 2 |
| 10 | Instestinal type adenocarcinoma | 72 | 2 |
| 11 | Endocervical type adenocarcinoma | 43 | 2 |
| 12 | Endocervical type adenocarcinoma | 43 | 2 |
| 13 | Clear cell adenocarcinoma | 40 | – |
| 14 | Clear cell adenocarcinoma | 40 | – |
| 15 | Instestinal type adenocarcinoma | 51 | 2 |
| 16 | Instestinal type adenocarcinoma | 51 | 2–3 |
| 17 | Endocervical type adenocarcinoma | 50 | 2–3 |
| 18 | Endocervical type adenocarcinoma | 50 | 2–3 |
| 19 | Instestinal type adenocarcinoma | 34 | 2 |
| 20 | Instestinal type adenocarcinoma | 34 | 2 |
| 21 | Adenocarcinoma | 44 | 3 |
| 22 | Adenocarcinoma | 44 | 3 |
| 23 | Adenocarcinoma | 52 | 3 |
| 24 | Adenocarcinoma | 52 | 3 |
| 25 | Adenocarcinoma | 59 | 3 |
| 26 | Adenocarcinoma | 59 | 3 |
| 27 | Endometrioid adenocarcinoma | 26 | 3 |
| 28 | Endometrioid adenocarcinoma | 26 | 3 |
| 29 | Adenocarcinoma (fibrous tissue and blood vessel) | 32 | – |
| 30 | Adenocarcinoma (fibrous tissue and blood vessel) | 32 | – |
| 31 | Adenosquamous carcinoma | 43 | – |
| 32 | Adenosquamous carcinoma | 43 | – |
| 33 | Adenosquamous carcinoma | 64 | – |
| 34 | Adenosquamous carcinoma | 64 | – |
| 35 | Adenosquamous carcinoma | 38 | – |
| 36 | Adenosquamous carcinoma | 38 | – |
| 37 | Adenosquamous carcinoma | 54 | – |
| 38 | Adenosquamous carcinoma | 54 | – |
| 39 | Adenosquamous carcinoma | 43 | – |
| 40 | Adenosquamous carcinoma | 43 | – |
| 41 | Squamous cell carcinoma | 53 | 2 |
| 42 | Squamous cell carcinoma | 53 | 2 |
| 43 | Squamous cell carcinoma | 27 | 2 |
| 44 | Squamous cell carcinoma | 27 | 2 |
| 45 | Squamous cell carcinoma | 68 | 2–3 |
| 46 | Squamous cell carcinoma | 68 | 2–3 |
| 47 | Squamous cell carcinoma | 37 | 3 |
| 48 | Squamous cell carcinoma | 37 | 3 |
| 49 | Squamous cell carcinoma | 43 | 3 |
| 50 | Squamous cell carcinoma | 43 | 3 |
| 51 | Squamous cell carcinoma | 69 | 2 |
| 52 | Squamous cell carcinoma with necrosis | 69 | 2 |
| 53 | Squamous cell carcinoma (sparse) | 48 | 2 |
| 54 | Squamous cell carcinoma | 48 | 2 |
| 55 | Squamous cell carcinoma | 36 | 3 |
| 56 | Squamous cell carcinoma | 36 | 3 |
| 57 | Squamous cell carcinoma | 63 | 2 |
| 58 | Squamous cell carcinoma | 63 | 2 |
| 59 | Squamous cell carcinoma | 47 | 2 |
| 60 | Squamous cell carcinoma | 47 | 1–2 |
| 61 | Squamous cell carcinoma | 40 | 2 |
| 62 | Squamous cell carcinoma | 40 | 2 |
| 63 | Squamous cell carcinoma | 76 | 2 |
| 64 | Squamous cell carcinoma | 76 | 2 |
| 65 | Squamous cell carcinoma | 38 | 3 |
| 66 | Squamous cell carcinoma (fibrous tissue and blood vessel) | 38 | – |
| 67 | Squamous cell carcinoma | 36 | 2–3 |
| 68 | Squamous cell carcinoma | 36 | 2–3 |
| 69 | Squamous cell carcinoma | 62 | 3 |
| 70 | Squamous cell carcinoma | 62 | 3 |
| 71 | Squamous cell carcinoma | 51 | 3 |
| 72 | Squamous cell carcinoma | 51 | 3 |
| 73 | Squamous cell carcinoma | 32 | 3 |
| 74 | Squamous cell carcinoma | 32 | 3 |
| 75 | Squamous cell carcinoma | 58 | 3 |
| 76 | Squamous cell carcinoma | 58 | 3 |
| 77 | Squamous cell carcinoma | 27 | 3 |
| 78 | Squamous cell carcinoma | 27 | 3 |
| 79 | Squamous cell carcinoma | 39 | 2 |
| 80 | Squamous cell carcinoma | 39 | 3 |
| 81 | Cancer adjacent normal cervical tissue | 45 | – |
| 82 | Cancer adjacent normal cervical tissue | 45 | – |
| 83 | Cancer adjacent normal cervical canals tissue | 62 | – |
| 84 | Cancer adjacent normal cervical canals tissue | 62 | – |
| 85 | Cancer adjacent normal cervical canals tissue | 50 | – |
| 86 | Cancer adjacent normal cervical canals tissue | 50 | – |
| 87 | Cancer adjacent normal cervical tissue of No 13 | 40 | – |
| 88 | Cancer adjacent normal cervical tissue of No 13 | 40 | – |
| 89 | Cancer adjacent normal cervical tissue (fibrous tissue and blood vessel) | 60 | – |
| 90 | Cancer adjacent normal cervical tissue | 60 | – |
| 91 | Normal cervical tissue | 18 | – |
| 92 | Normal cervical tissue | 18 | – |
| 93 | Normal cervical tissue | 15 | – |
| 94 | Normal cervical tissue | 15 | – |
| 95 | Normal cervical tissue (fibrous tissue and blood vessel) | 21 | – |
| 96 | Normal cervical tissue (fibrous tissue and blood vessel) | 21 | – |
| 97 | Normal cervical tissue (with hyperplasia of glandular epithelium) | 21 | – |
| 98 | Normal cervical tissue (with hyperplasia of glandular epithelium) | 21 | – |
| 99 | Normal cervical tissue (fibrous tissue and blood vessel) | 19 | – |
| 100 | Normal cervical tissue (fibrous tissue and blood vessel) | 19 | – |
2. Experimental design, materials and methods
2.1. Experimental design and subjects
A tissue microarray slide was selected from US Biomax that contained 40 cervical squamous cell carcinoma and 20 normal cervical tissues (Table 1). HeLa cells stably expressing EV or CYB5D2 were recently constructed [1], [4].
2.2. Cell cycle distribution determination
Cell cycle distribution was determined by individualizing cells using 0.02% EDTA in PBS. Cells were stained with a propidium iodide (PI) solution (10 mM Tris pH7.5, 150 mM NaCl, 0.05 mg/ml PI, 0.1% sodium citrate, 0.2% Triton X-100, and 0.2 mg/ml DNase-free RNase A) overnight at 4 °C in dark.Cell cycle distribution was analyzed using a fluorescent automated cell sorting (FACS) (Bechman Coulter, CytomicsTM FC500).
Acknowledgments
We like to dedicate this work to a great mother Ms. Guorui Zeng. This work was supported in part by a grant (No. 81302210) from National Natural Science Foundation of China to Y. Xie, and Heart and Stroke Foundation of Canada and CIHR (MOP-84381) to D. Tang.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.dib.2016.01.036.
Appendix A. Supplementary material
Supplementary material
References
- 1.Xie Y., Shen Y.T., Kapoor A., Ojo D., Wei F., Melo J. De, Lin X., Wong N., Yan J., Tao L., Major P., Tang D. CYB5D2 displays tumor suppression activities towards cervical cancer. BBA Mol. Basis Dis. 2016 doi: 10.1016/j.bbadis.2015.12.013. 10.1016/j.dib.2016.01.036 (in press) [DOI] [PubMed] [Google Scholar]
- 2.Kimura I., Nakayama Y., Konishi M., Kobayashi T., Mori M., Ito M., Hirasawa A., Tsujimoto G., Ohta M., Itoh N., Fujimoto M. Neuferricin, a novel extracellular heme-binding protein, promotes neurogenesis. J. Neurochem. 2010;112:1156–1167. doi: 10.1111/j.1471-4159.2009.06522.x. [DOI] [PubMed] [Google Scholar]
- 3.Bruce A., Rybak A.P. CYB5D2 requires heme-binding to regulate HeLa cell growth and confer survival from chemotherapeutic agents. PloS One. 2014;9:e86435. doi: 10.1371/journal.pone.0086435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie Y., Bruce A., He L., Wei F., Tao L., Tang D. CYB5D2 enhances HeLa cells survival of etoposide-induced cytotoxicity. Biochem. Cell Biol. 2011;89:341–350. doi: 10.1139/o11-004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material