Abstract
This study is one of the few investigations which analyze albumen prints, perhaps the most important photographic heritage of the late 19th and early 20th centuries. The chemical composition of photographic samples was assessed using Fourier-transform infrared spectroscopy and X-ray fluorescence. These two non-invasive techniques revealed the complex nature of albumen prints, which are composed of a mixture of proteins, cellulose and salts. Microbial sampling was performed using cellulose nitrate membranes which also permitted the trapped microflora to be observed with a scanning electron microscope. Microbial analysis was performed using the combination of culture-dependent (cultivation in different media, including one 3% NaCl) and culture-independent (bacterial and fungal cloning and sequencing) approaches. The isolated microorganisms were screened for their lipolytic, proteolytic, cellulolytic, catalase and peroxidase activities. The combination of the culture-dependent and -independent techniques together with enzymatic assays revealed a substantial microbial diversity with several deteriogen microorganisms from the genera Bacillus, Kocuria, Streptomyces and Geobacillus and the fungal strains Acrostalagmus luteoalbus, Bjerkandera adusta, Pleurotus pulmonarius and Trichothecium roseum.
In the digital era it is very easy take a picture with a modern digital camera and it is equally easy to print it with ever newer and more rapid printing methods. But until the last decade of the 20th century, it was not so simple, and photographs were printed on special photographic paper, composed of a light-sensitive emulsion containing of silver halide salts suspended in a colloidal material, usually gelatin, coated onto a paper, a resin coated paper or a polyester support1.
From the mid-19th century to the beginning of the 20th century, the most commonly used colloidal material was albumen. Albumen photographs were invented around 1850 by a French photographer, Louis Désiré Blanquart-Evrard. In these photographs, a paper sheet was covered by a layer of albumen, a type of protein found in egg whites, in order to bind the light-sensitive halide salts to the paper; they became the dominant form of photographic positives from 1855 to the turn of the 20th century, peaking during the years 1860–90. The use of albumen paper greatly diminished around the year 19202.
Many albumen-based photographs must exist in archives, galleries and museums around the world, both on display and in storage, and several of them are part of important historical collections; like the other historical objects in these collections, they too need to be safeguarded against aging and biodeterioration processes. It is already known how many microorganisms, mainly fungi, attack various types of archival materials, including books, parchments and photographic materials3,4, but, though many studies have been carried out to investigate the microflora responsible for the deterioration of different materials preserved in indoor environments3,4,5, few have examined the biodeterioration of photographic materials, and most of those that do have focused only on gelatin-based photographs6,7,8,9,10.
A methodological strategy for assessing the biodeterioration of albumen photographs, one of the most valuable “collections of memories” from the 19th and 20th centuries, is sorely lacking.
In this study we analyzed the material composition of and microflora present in two albumen photographs. Fourier-transform infrared spectroscopy (FTIR) and X-ray fluorescence (XRF), used to investigate the material, were combined with scanning electron microscopy (SEM), culture-dependent and culture-independent microbial analyses and different biodegradative agar assays in order to assess the nature of the microbial contaminants.
Results
Microscopic observation and chemical analysis
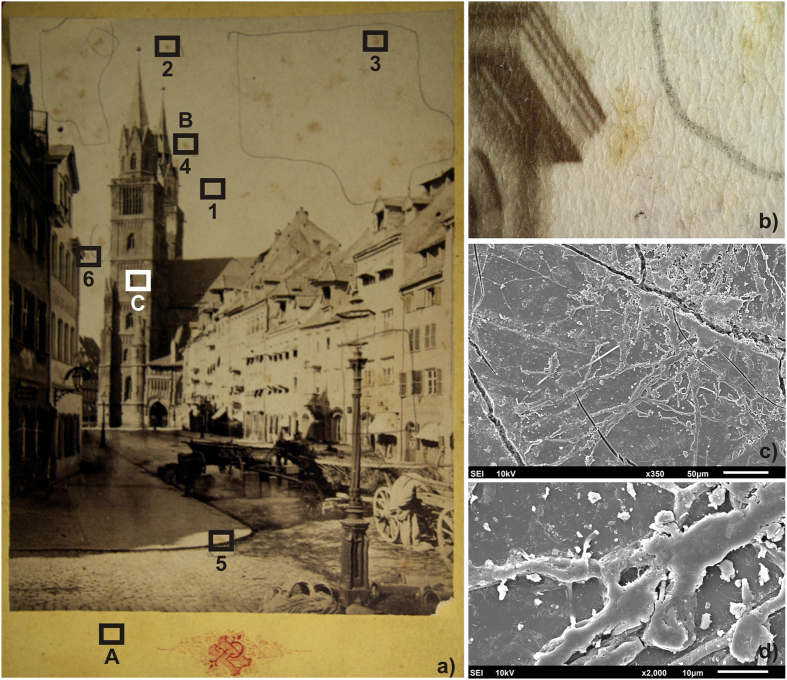
A preliminary observation of various parts of the two albumen prints was made using a stereo microscope. On the Gyula photograph (Fig. 1a) the presence of fungal contamination and different impurities on its surface near the sampling areas were evident (Fig. 1b,c). Cellulose nitrate membranes were used to lift microbial and (predominantly) fungal contaminants from the indicated areas of the photograph (red circles, Fig. 1a), and their presence was confirmed by the SEM analysis of a small portion of membrane (Fig. 1d,e). No microbial contamination could be clearly seen on the antiquarian print (Fig. 2a) using the stereo microscope (Fig. 2b), but SEM observation did reveal the presence of hyphae on the photograph’s surface (Fig. 2c,d).
Figure 1. Gyula photograph.
(a) The albumen surface is 9.7 × 13.9 cm; the size of the complete photograph including the cardboard is 10.8 × 16.2 cm. The microbial sampling sites are marked with two red circles. The sites F0–F4 were subjected to FTIR analysis, while the letters XA–XC indicate the sites where XRF measurements were performed. (b,c) Stereo microscope images with a magnification of 40× (site F4) and 20× (area near to site F4) respectively. (d,e) SEM photographs showing the fungal hyphae and spores recovered by a cellulose nitrate membrane. The photograph was supplied by the Slovak National Archive and is used with permission.
Figure 2. Antiquarian photograph.
(a) The albumen surface measures 10.1 × 13.9 cm; the size of complete photograph including the cardboard is 10.9 × 16.6 cm. Numbers 1–6 indicate the sites where FTIR measurements were taken, while sites A–C were subjected to XRF examination. (b) A 20× stereo microscope image of site 6 (c,d) SEM photographs of site 2 where the presence of fungal hyphae and possible mineral structures is evident.
FTIR–ATR spectroscopy confirmed that the two photographs are, indeed, albumen prints. The typical absorption peaks for an albumen photograph appear between 1632 cm−1 and 1161 cm−1 and can be seen in Fig. 3.
Figure 3. ATR-FTIR spectra of the two albumen prints, Gyula and antiquarian.
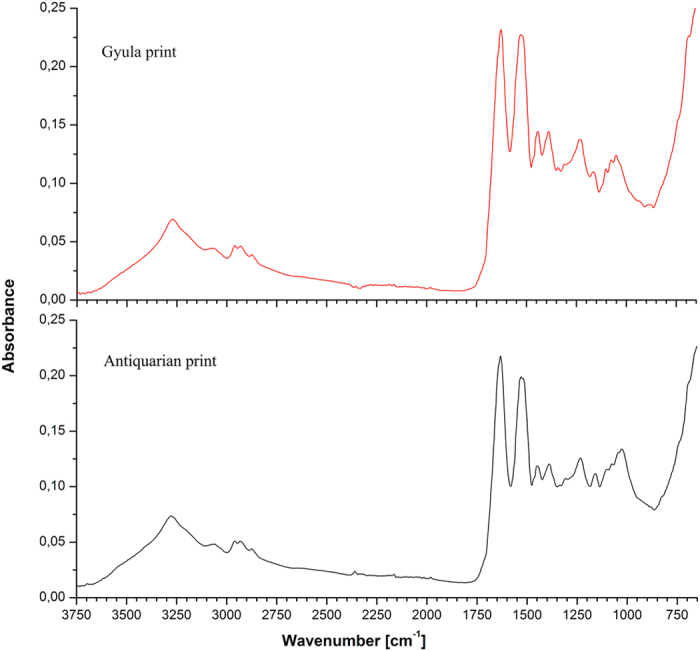
Figure 4 shows two different spectra from the Gyula print. The most intense peak in the F0 spectrum (the control sample) is at 1632 cm−1, corresponding to protein Amide I, the C = O and C-N stretching vibrations of (mainly) the peptide linkage. The peak at 1530 cm−1 belongs to Amide II, arising from the N-H bending and C-N stretching vibrations of the peptide bond Amide III is represented by the series of peaks in the 1400–1200 cm−1 11 range. Because the albumin layers are deposited on paper supports in these kinds of photographs, the peaks in the range 1250–850 cm−1 indicate the presence of cellulose.
Figure 4. Comparison of μ-FTIR spectra between control site (F0) and the foxing site F4 of the Gyula print.
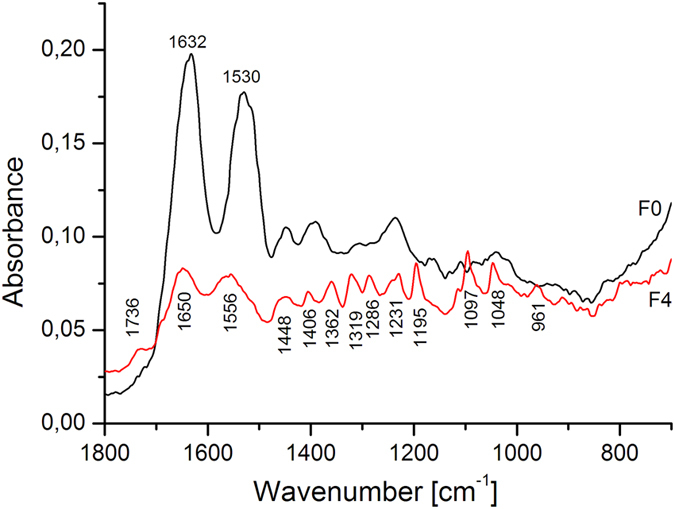
The μ-FTIR spectrum of the foxing stain (F4) showed shifts of the Amide I and Amide II bands to higher wavenumbers compared to F0 (Fig. 4). The foxing stain spectrum showed the greatest number of changes in the number of peaks and their shapes in the 1422–960 cm −1 region, and a number of new peaks appeared (1736, 1406, 1362, 1319, 1286, 1231, 1195, 1097, 1048 and 961 cm−1). The functional groups corresponding to these peaks are described in Supplementary Table S1.
An XRF analysis of the Gyula photograph (Fig. 1a; XA–XC) detected high values of Ba and Ti in all three analyzed sites (Table 1), suggesting the presence of barium sulfate and titanium dioxide. The dark foxing site (Fig. 1a; XC) contained a small amount of silver which had been used to create the photographic image. Other elements detected in the two foxing stains included Ca, Fe and Sr. The cardboard of the antiquarian photograph (Fig. 2a; A) contained the elements Ag, Al, Fe, Cu, Mg and Zn (Table 1). Elevated concentrations of sulfur and silicon were also detected in all analyzed sites of this photograph. Bromine was present in both foxing sites (B and C), while the dark foxing site (C) also contained Ag and Al.
Table 1. XRF analysis of the Gyula and antiquarian photographs performed at three different sites: the cardboard frame (XA; A), pale foxing sites (XB; B) and dark foxing sites (XC; C).
| Elements | Gyula photograph |
Antiquarian photograph |
||||
|---|---|---|---|---|---|---|
| Cardboard frame (XA) % | Foxing PS (XB) % | Foxing DS (XC) % | Cardboard frame (A) % | Foxing PS (B) % | Foxing DS (C) % | |
| Ag | n.d. | n.d. | 0.1 ± 0.0 | 0.5 ± 0.1 | n.d. | 1.1 ± 0.2 |
| Al | n.d. | n.d. | n.d. | 0.9 ± 0.1 | n.d. | 0.2 ± 0.0 |
| Ba | 9.76 ± 0.1 | 8.81 ± 0.1 | 11.83 ± 0.1 | n.d. | n.d. | n.d. |
| Br | n.d. | n.d. | n.d. | n.d. | 0.3 ± 0.0 | 0.4 ± 0.1 |
| Ca | 0.2 ± 0.0 | 0.3 ± 0.1 | 0.4 ± 0.0 | 0.4 ± 0.1 | 0.3 ± 0.1 | 0.6 ± 0.0 |
| Cu | n.d. | n.d. | n.d. | 0.5 ± 0.0 | n.d. | n.d. |
| Fe | 0.1 ± 0.0 | 0.1 ± 0.0 | 0.2 ± 0.0 | 0.3 ± 0.0 | 0.2 ± 0.0 | 0.4 ± 0.0 |
| Mg | n.d. | n.d. | n.d. | 0.9 ± 0.1 | n.d. | n.d. |
| S | n.d. | n.d. | n.d. | 2.5 ± 0.2 | 2.8 ± 0.1 | 3.2 ± 0.1 |
| Si | n.d. | n.d. | n.d. | 2.5 ± 0.1 | 0.8 ± 0.0 | 0.6 ± 0.0 |
| Sr | 0.6 ± 0.0 | 0.5 ± 0.0 | 0.6 ± 0.0 | n.d. | n.d. | n.d. |
| Ti | 11.4 ± 0.1 | 5.9 ± 0.0 | 6.4 ± 0.04 | n.d. | n.d. | n.d. |
| Zn | n.d. | n.d. | n.d. | n.d. | 0.3 ± 0.0 | n.d. |
n.d.: not detected.
Cultivation strategy and isolated microflora
Two different sampling procedures (direct inoculation and decimal dilution) and different medial compositions (some containing 3% NaCl) were used for the cultivation of microflora. Direct inoculation allowed only a few fungal strains to be isolated from the Gyula photograph (Penicillium chrysogenum P1_18_Fu, Chaetomium elatum P1_13_Fu and Pleurotus pulmonarius P1_15_Fu); the remaining fungal and bacterial strains were recovered by the inoculation of 10× diluted samples. Differences in the isolation power of the standard microbiological media used in this study (R2A and LB10; MEA and DRBC) were noted. In particular, only the two Bacillus strains (P1_1_5 and P1_2_8) isolated from Gyula photo grew on R2A and 59% of fungi were recovered on DRBC. MEA was only important for the isolation of the penicilli and aspergillli recovered (mainly) from the antiquarian photograph, but the sampling strategy for this photo also included the analysis of a piece of albumen print. It is possible to say therefore that the bacterial diversity of the albumen photograph samples was expressed better on LB10 and the fungal diversity on DRBC. The medium with 3% NaCl allowed different fungal strains to be isolated, only one of which, Trichothecium roseum (PF1_2_Fu), was also isolated on DRBC. The others, Mucor plumbeus (P1_Na_18_Fu), Pleosporales sp. (P1_Na_21_Fu), Cephalosporium sp. (PF1_Na_8_Fu) and Cladosporium macrocarpum (PF1_Na_30_Fu), were isolated only on salt medium, attesting to their slightly halophilic character. Only two halotolerant Actinobacteria were isolated, Kocuria sp. (PF1_Na_14) and Streptomyces albidoflavus (PF1_Na_10). All of the slightly halophilic microorganisms were isolated from the Gyula photograph samples (Table 2).
Table 2. Bacterial and fungal strains isolated from albumen photographs and album paper frame.
| Sample of isolation | Strain | Medium of isolation | Microorganisms identified based on the highest 16S rDNA and ITS similarity scores | Accession Number |
|---|---|---|---|---|
| Gyula albumen photograph | P1_2_8 | R2A | KF844070 Bacillus simplex 561/561 (100%) | KT200239 |
| P1_1_5 | R2A | KF995632 Bacillus sp. 613/614 (99%) | KT200240 | |
| P1_1_Fu | DRBC | GU731546 Bjerkandera adusta 551/551 (100%) | KT200251 | |
| P1_2_Fu | DRBC | KJ831970 Phlebia sp. 639/654 (98%) | KT200252 | |
| P1_3_Fu | MEA | KJ583240 Penicillium sp. 475/477 (98%) | KT200253 | |
| P1_4_Fu | MEA | KJ653464 Penicillium sp. 501/501 (100%) | KT200254 | |
| P1_6_Fu | DRBC | KP233879 Bjerkandera adusta 451/451 (100%) | KT200255 | |
| P1_13_Fu | DRBC | KC109758 Chaetomium elatum 511/511 (100%) | KT200256 | |
| P1_2_3_Fu | DRBC | KJ831970 Phlebia sp. 639/655 (98%) | KT200257 | |
| P1_1_20_Fu | DRBC | KC109758 Chaetomium elatum 511/512 (99%) | KT200258 | |
| P1_10_Fu | DRBC | KJ653464 Penicillium sp. 520/521 (99%) | KT200259 | |
| P1_15_Fu | DRBC | KM985673 Pleurotus pulmonarius 399/401 (99%) | KT200260 | |
| P1_Na_18_Fu | R2A-Gel-NaCl | JX537955 Mucor plumbeus 401/401 (100%) | KT200261 | |
| P1_Na_21_Fu | R2A-Gel-NaCl | HQ207056 Pleosporales sp. 481/496 (97%) | KT200262 | |
| P1_18_Fu | DRBC | JX139710 Penicillium chrysogenum 530/530 (100%) | KT200263 | |
| Gyula album paper frame | PF1_1 | LB10 | KC429605 Kocuria rhizophila 620/621 (99%) | KT200241 |
| PF1_2 | LB10 | KJ139433 Streptomyces sp. 611/611 (100%) | KT200242 | |
| PF1_5 | LB10 | LN774467 Staphylococcus epidermidis 639/641 (99%) | KT200243 | |
| PF1_7 | LB10 | KP128847 Streptomyces sp. 608/609 (99%) | KT200244 | |
| PF1_30 | LB10 | KP128847 Streptomyces sp. 622/624 (99%) | KT200245 | |
| PF1_Na_14 | R2A-Gel-NaCl | KM874399 Kocuria sp. 619/621 (99%) | KT200246 | |
| PF1_Na_10 | R2A-Gel-NaCl | KP122209 Streptomyces albidoflavus 505/509 (99%) | KT200247 | |
| PF1_Na_8_Fu | R2A-Gel-NaCl | HQ630979 Cephalosporium sp. 517/526 (98%) | KT200264 | |
| PF1_2_Fu | DRBC | KJ466977 Trichothecium roseum 371/375 (99%) | KT200265 | |
| PF1_3_Fu | DRBC | KM853014 Acrostalagmus luteoalbus 311/311 (100%) | KT200266 | |
| PF1_2_2_Fu | DRBC | KM853014 Acrostalagmus luteoalbus 410/411 (99%) | KT200267 | |
| PF1_4_Fu | MEA | KJ653464 Penicillium sp. 521/522 (99%) | KT200268 | |
| PF1_6_Fu | MEA | KJ653464 Penicillium sp. 471/471 (100%) | KT200269 | |
| PF1_5_Fu | MEA | KJ653464 Penicillium sp. 521/521 (100%) | KT200270 | |
| PF1_Na_12_Fu | R2A-Gel-NaCl | EF589898 Trichothecium roseum 528/541 (98%) | KT200271 | |
| PF1_Na_30_Fu | R2A-Gel-NaCl | KM396371 Cladosporium macrocarpum 497/499 (99%) | KT200272 | |
| Antiquarian photograph | PA2_1_1 | LB10 | KP178217 Bacillus subtilis 631/631 (100%) | KT200248 |
| PA2_1_2 | LB10 | KP178217 Bacillus subtilis 630/631 (99%) | KT200249 | |
| PA2_1_3 | LB10 | KP178217 Bacillus subtilis 636/636 (100%) | KT200250 | |
| PA2_4_Fu | DRBC | JN368454 Penicillium chrysogenum 533/534 (99%) | KT200273 | |
| PA2_6_Fu | MEA | JX156356 Aspergillus versicolor 513/514 (99%) | KT200274 | |
| PA2_3_Fu | MEA | LN809060 Aspergillus versicolor 501/502 (99%) | KT200275 | |
| PA2_5_Fu | MEA | JN624909 Penicillium thomii 517/522 (99%) | KT200276 | |
| PA2_2_Fu | MEA | EU910586 Penicillium thomii 525/526 (99%) | KT200277 |
MEA: Malt Extract Agar; DRBC: Dichloran Rose Bengal Chloramphenicol; LB10: 10x diluted Luria Bertani; R2A-Gel-NaCl: R2A supplemented with gelatin and 3% of NaCl.
Only a few bacterial strains were isolated from the three samples (2 isolates from the Gyula photo, 7 from the album paper frame and 3 from the antiquarian photograph). Most species were members of the Bacillus genus, and they were isolated from the albumen substrate of both photographs. The bacterial microflora isolated from the paper frame contained Actinobacteria belonged to the genera Kocuria and Streptomyces (Table 2).
The fungal community was more diverse than its bacterial counterpart. Most isolated fungi belonged to the phylum Ascomycota; members of the phylum Basidiomycota, together with a single member of the subphylum Mucoromycotina, were isolated only from the albumen substrate of the Gyula photograph. The most common genus was Penicillium, and from the antiquarian photograph only penicillia and aspergilla were isolated. The fungal diversity of the album paper frame also included members of the genus Cephalosporium, Trichothecium and Acrostalagmus (Table 2).
Culture-independent investigation
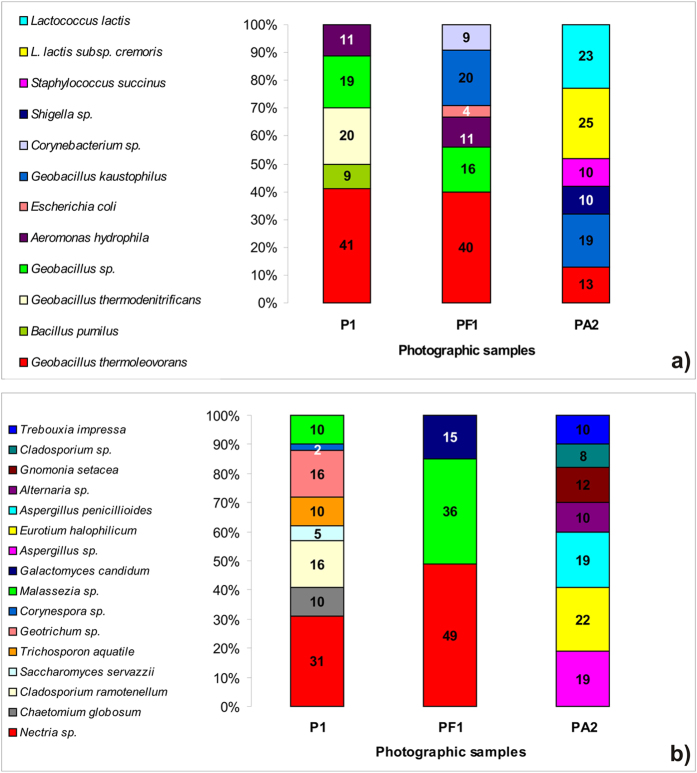
Because not every microorganism in the microbial community is able to be isolated and cultivated, we also extracted and sequenced the communities’ total DNA to identify those organisms which resisted cultivation. The extracted bacterial DNA belonged mainly to Bacilli and Gammaproteobacteria; several Corynebacterium sequences were detected from the album paper frame sample, representing the only link with the Actinobacteria class. Specifically, the most common bacteria found in the samples from the Gyula photograph belonged to the genus Geobacillus which were 80% and 76%, respectively, of the population recovered from the albumen surface (P1) and the album paper frame (PF1). Some Geobacillus species were found only on the albumen surface (G. thermodenitrificans) while others were present only on the paper frame (G. kaustophilus). The third most common bacterium was Aeromonas hydrophila, found on both the albumen and the album, while Bacillus pumilus DNA was detected only on the albumen surface (Fig. 5a).
Figure 5. Microbial communities detected in photographic samples.
Bacterial (a) and eukaryotic (b) diversity on photographic materials detected using the DGGE and clone library approach. P1: Gyula photo from Slovak National Archive; PF1: album paper frame of Gyula photo; PA2: antiquarian photograph.
The bacterial community of the antiquarian print (PA2) was markedly different from the Gyula photograph: here Lactococcus lactis OTUs were the most abundant, reaching 48%, followed by Geobacillus OTUs (G. kaustophilus and G. thermoleovorans), with 32%, and Staphylococcus succinus and Shigella sp., 10% each (Fig. 5a; Supplementary Table S2).
Differences between the fungal communities of the Gyula photo and the album paper frame were noted: the DNA of Nectria and Malassezia members occurred in both samples, but Galactomyces candidum OTUs were detected only on the paper frame. The albumen material showed the greatest fungal divesrsity, with Chaetomium, Cladosporium, Saccharomyces, Trichosporon, Geotrichum and Corynespora OTUs all detected (Fig. 5b; Supplementary Table S2).
The photograph PA2 clone library showed yet another kind of fungal community, composed mainly of Aspergillus OTUs (38%) together with Eurotium halophilicum (22%), Gnomonia setacea (12%), Alternaria sp. (10%) and Cladosporium (8%). The DNA of the green algae Trebouxia impressa was also detected (Fig. 5b; Supplementary Table S2).
It should be noted that these cloning and sequencing approaches allowed the composition of the microbial communities colonizing the two photographs to be determined, but does not by itself indicate which of their members are responsible for the biodegradation.
Biodegadative, catalase and peroxidase activities
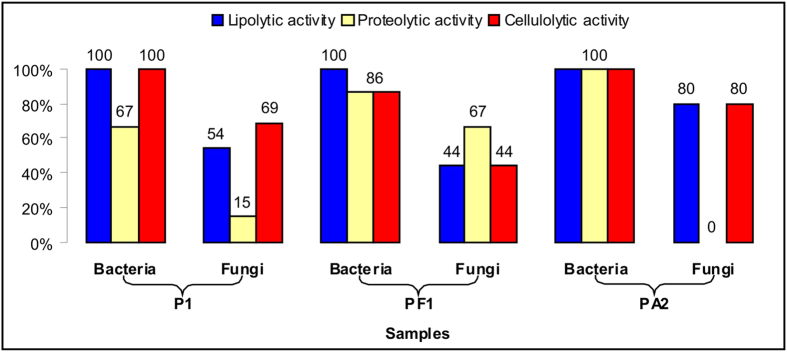
All isolated bacteria displayed significant biodegradation activities. The two Bacillus strains contaminating the Gyula photograph quickly produced a very intense halo around the colonies when cultivated on Spirit Blue (lipase activity) and Congo red (cellulase activity). The Actinobacteria from the album paper frame, members of the genus Kocuria and Streptomyces, exhibited strong lipolytic and proteolytic properties, (with the exception of Kocuria rhizophila PF1_1, which was positive only to the lipolytic assay). The three Bacillus sp. isolated from the antiquarian print, meanwhile, showed very similar biodegradative profiles, with weaker protelytic and lipolytic activities compared to those strains recovered from the other samples. The distribution of hydrolytic activities of the bacterial isolates is shown in Table 3 and Fig. 6. The Bacillus strains from both albumen photographs exhibited similar peroxidase activities (0.07–0.1 U/ml) and very high catalase activities, from the 1257 U/ml of PA2_1_3 to the 1340 U/ml of strain P1_1_5. The Actinobacteria from the album paper frame possessed weaker catalase activity than the Bacillus strains, with values between 436 U/ml for Streptomyces PF1_7 and 560 U/ml for Kocuria rhizophila PF1_1. The other Kocuria isolate (PF1_Na_14) exhibited 0.2 U/ml peroxidase activity, the highest from the bacteria (Table 3).
Table 3. Hydrolytic abilities and catalase and peroxidase activities of microorganisms isolated from different samples.
| Sample of isolation | Isolates | Lipolytic activity Sprit Blue Agar | Proteolytic activity Gelatin Agar | Cellulolytic activity Congo Red | Catalase activity U/ml | Peroxidase activity U/ml |
|---|---|---|---|---|---|---|
| Gyula albumen photograph | P1_2_8 Bacillus simplex | +++ | ++ | +++ | 1270 | 0.07 |
| P1_1_5 Bacillus sp. | +++ | ++ | +++ | 1340 | 0.08 | |
| P1_1_Fu Bjerkandera adusta | +++ | – | +++ | 343 | 0.2 | |
| P1_ 2_Fu Phlebia sp. | +++ | – | ++ | 240 | 0.2 | |
| P1_2_3_Fu Phlebia sp. | +++ | – | ++ | 237 | 0.2 | |
| P1_4_Fu Penicillium sp. | – | – | + | 270 | – | |
| P1_6_Fu Bjerkandera adusta | +++ | – | +++ | 350 | 0.2 | |
| P1_13_Fu Chaetomium elatum | + | + | ++ | 450 | 0.15 | |
| P1_15_Fu Pleurotus pulmonarius | – | +++ | ++ | 200 | 0.4 | |
| P1_Na_18_Fu Mucor plumbeus | ++ | – | ++ | 600 | 0.2 | |
| P1_Na_21_Fu Pleosporales sp. | ++ | – | ++ | 500 | 0.4 | |
| Gyula album paper frame | PF1_1 Kocuria rhizophila | +++ | – | ++ | 560 | 0.1 |
| PF1_2 Streptomyces sp. | +++ | +++ | ++ | 550 | 0.07 | |
| PF1_5 Staphylococcus epidermidis | +++ | +++ | – | 92 | – | |
| PF1_7 Streptomyces sp. | +++ | +++ | ++ | 436 | 0.09 | |
| PF1_30 Streptomyces sp. | +++ | +++ | ++ | 441 | 0.09 | |
| PF1_Na_10 Streptomyces albidoflavus | +++ | +++ | ++ | 471 | 0.1 | |
| PF1_Na_14 Kocuria sp. | +++ | +++ | ++ | 440 | 0.2 | |
| PF1_ 2_Fu Trichothecium roseum | +++ | +++ | ++ | 220 | 0.2 | |
| PF1_3_Fu Acrostalagmus luteoalbus | +++ | +++ | ++ | 257 | 0.2 | |
| PF1_2_2_Fu Acrostalagmus luteoalbus | +++ | +++ | ++ | 250 | 0.2 | |
| PF1_6_Fu Penicillium sp. | – | + | – | 350 | 0.12 | |
| PF1_Na_8_Fu Cephalosporium sp. | – | + | – | 340 | 0.2 | |
| PF1_Na_12_Fu Trichothecium roseum | +++ | +++ | ++ | 220 | 0.2 | |
| Antiquarian photograph | PA2_1_1 Bacillus subtilis | ++ | ++ | + | 1260 | 0.1 |
| PA2_1_2 Bacillus subtilis | ++ | ++ | + | 1262 | 0.1 | |
| PA2_1_3 Bacillus subtilis | ++ | ++ | + | 1257 | 0.1 | |
| PA2_6_Fu Aspergillus versicolor | ++ | – | ++ | 290 | 0.1 | |
| PA2_3_Fu Aspergillus versicolor | + | – | ++ | 300 | 0.05 | |
| PA2_5_Fu Penicillium thomii | + | – | ++ | 252 | 0.1 | |
| PA2_2_Fu Penicillium thomii | + | – | ++ | 255 | 0.1 |
+++: quick and extensive positive reaction; ++: strong positive reaction; +: weak p 838 ositive reaction; -: no reaction.
Figure 6. Percentage of hydrolytic abilities of isolated microflora.
Distribution of hydrolytic activities of microorganisms isolated from different samples. P1: Gyula photo from Slovak National Archive; PF1: album paper frame of Gyula photo; PA2: photograph from antiquarian.
Nineteen fungal strains from a total of 27 isolates demonstrated at least one biodegradative property. The fungi isolated from the two albumen photographs showed feeble activities with respect to the bacteria. Figure 6 shows that only 15% of the Gyula print isolates possessed proteolytic activity, and that none of the fungi isolated from the antiquarian photograph did. The most active fungal isolates were P1_6_Fu Bjerkandera adusta (with marked lipolytic and cellulolytic activities), P1_15_Fu Pleurotus pulmonarius (characterized by both proteolytic and cellullytic abilities) and PA2_6_Fu Aspergillus versicolor (medium lipolytic and cellulolytic activities).
Sixty-seven percent of the fungi isolated from the album paper frame displayed proteolytic activity, but only 44% were able to produce lipases and cellulases (Fig. 6). The fungi Trichothecium roseum (PF1_ 2_Fu and PF1_Na_12_Fu) and Acrostalagmus luteoalbus (PF1_3_Fu and PF1_2_2_Fu) had the best hydrolytic properties, producing a positive reaction in all three biodegradative assays (Table 3).
The fungi showed stronger peroxidase activities than the bacteria, many of them equalling the best bacterial result (0.2 U/ml). The P1_15_Fu Pleurotus pulmonarius and P1_Na_21_Fu Pleosporales sp. isolates went higher, reaching 0.4 U/ml. The fungi with the highest catalase activities were isolated from the Gyula photograph: P1_6_Fu Bjerkandera adusta, P1_13_Fu Chaetomium elatum, P1_Na_18_Fu Mucor plumbeus and P1_Na_21_Fu Pleosporales sp. exhibited 350, 450, 600 and 500 U/ml, respectively (Table 3).
Discussion
Albumen prints have important historical and cultural value, they represent the images of an epoch, the late 19th and early 20th centuries, when albumen was the most popular photographic binder. Albumen photographs are stored and exhibited in many galleries, museums, libraries and archives; it is therefore crucial to know their composition and the causes of their bio-deterioration.
The albumen paper prints were be prepared in different ways, but generally required the use of a very pure, metal-free paper support (metals may have caused black stains to appear in the final photographs). The albumen, derived from the clear white of fresh eggs, was mixed with NaCl or NH4Cl. The paper substrate was then coated by floating it in this salted albumin solution. After drying, the albumen paper print was sensitized by brushing it with a 10% AgNO3 solution, which readted with the salt to form the insoluble, light-sensitive AgCl. When this sensitized paper was exposed to sunlight, an image was formed by the reduction of the silver on surface of these AgCl crystals, which were embedded in the albumen layer. The albumen photograph was then washed in water, stabilized in a 15% sodium thiosulfate solution, and again washed2.
The chemical composition of the photographs examined here is not especially different from those analyzed in previous works12,13, and this investigation confirmed the utility of FTIR and XRF for noninvasively probing such sensitive materials.
The FTIR showed differences between the control sample (F0) and foxing areas (F1–F4) in the range 1736–960 cm1; these differences could arise from three sources: microbial contamination, impurities and degradation products of the albumen print. The interpretation of these results is difficult because the peaks from the albumen layer, paper support and biological contamination overlap. On the basis of the findings of Oberle-Kilic et al.14,15,16 it is reasonable to assume that the spectral differences arise from microbiological contaminations. Zotti et al.17 showed that the presence of active fungi on paper can be easily detected by FTIR analysis on the basis of the amide I and amide II bands around 1635 and 1540 cm−1 and a plateau between 1500 and 1200 cm−1. Our FTIR results showed that the albumen produces a FTIR spectrum very similar to that of microfilamentous fungi (Fig. 4, spectrum F0), making it impossible to clearly attest to the presence of fungi on albumen surface using FTIR alone18.
XRF analysis permitted the identification of the different inorganic elements from the various compounds used during the production of the albumen prints. Barium sulfate and titanium dioxide (detected on the Gyula photograph) were used for several purposes in photographic materials, including the creation of decorative elements on cardboard frames2. The presence of Si and Al, detected on the antiquarian photograph, suggests that kaolin [Si2Al2O5(OH)4] together with calcium carbonate (CaCO3) was used for coating the paper19. Bromine indicates that bromide salts (AgBr) were used for the production of the photograph. The other elements, such as sulfur and iron, were also detected in other photographic materials examined in a previous study6, and the increased amount of sulfur here may also indicate the presence of sulfur-containing degradation products from albumen or paper20. Cu, Mg and Zn are likely to be impurities. The absence of Ag in the pale foxing places (XB and B) indicates that it had been washed out during photographic processing. The lack of metals such as Au, Pt, and Se suggests that the two photographs were not toned2.
FTIR and XFR both showed that the albumen photographs are a mixture of proteins, cellulose and salts, which can be a suitable substrate for microbial colonization. Such biological contamination can sometimes be observed using a normal light microscope, as in the case of the Gyula photograph. Other studies have previously shown the advantages of using membranes for sampling cultural heritage objects6,21,22. The SEM analysis of a cellulose nitrate membrane performed here revealed fungal colonization, and can be considered a noninvasive alternative to SEM instruments with specialized sample chambers6.
Comparing the isolated microflora from these two albumen prints with those from similar items, for example gelatin-based photographs6,7,8,9,10, shows that there are some common members from the Bacillus, Aspergillus and Penicillium genera. Several fungi, such as Pleurotus pulmonarius, Phlebia sp., Pleosporales sp., Mucor plumbeus, Bjerkandera adusta, Chaetomium elatum, had not previously been isolated on photographic materials, although some of them had been found on other items and in other museum and archival environments23,24,25,26. In particular, Bjerkandera adusta had already been isolated from a foxing stain on a 19th century book23.
There is a physical connection between the Gyula albumen print and album paper frame and the sampling membrane lifted microorganisms and spores colonizing both materials, but there were evident microbial differences between these two samples; the album paper frame had a diverse bacterial microflora, characterized by Kocuria and Streptomyces isolates, confirming the findings of previous studies related to cinematographic films, historical documents and archival atmospheres4,5,7,9.
To our knowledge, Trichothecium roseum and Acrostalagmus luteoalbus had not been previously found in this kind of habitat. The members of these two fungi displayed significant hydrolytic properties and catalase and peroxidase activities, indicating their ability to degrade composite substrates, but unfortunately no information about their enzymatic characteristics could be found in the literature. The hydrolytic and oxidoreductive abilities of other fungi, such as Bjerkandera adusta, Pleurotus pulmonarius and Mucor plumbeus, were confirmed by our investigation, and, in fact, the different biodegradation properties of these fungi have already found application27,28,29. Once again, the bacterial cohort exhibited dangerous lipolytic, proteolytic and cellulolytic activities against cultural heritage items30,31,32, though these activities could also be exploited for biotechnological applications33.
The hydrolytic properties of the isolates in this study were complemented by catalase and peroxidase activities. In this kind of dry envrionment, oxidoreductive enzymes may have a dual role. First, they may protect the bacterial cells against oxidative stress by removing the hydrogen peroxide produced as a byproduct of oxygen metabolism; in fact previous work showed an increase in catalase and peroxidase production in different kinds of microorganisms when cultivated in water-deficient conditions34,35,36. Second, it has recently been established that oxidoreductases are present during the biodegradation of cellulose materials by fungi37,38; indeed this recent finding allowed different oxidoreductive enzymes to be included in the carbohydrate-active enzymes database (CAZy)39. It is therefore possible to colclude that oxidoreductases are also involved in biodeterioration, and bio-degradation processes in support of the hydrolytic enzymes. Both roles suggest that the high catalase and peroxidase activities of the isolates could aid these microorganisms in better colonizing and utilizing the contaminated substrate.
Several previous studies26,32,40 have shown that a combination of culture-dependent and -independent methods provides better microbiological characterization than either method alone. Indeed, only the culture-independent approach was able to detect Geobacillus members in all analyzed samples. The presence of these thermophilic organisms was surprising because they normally require a growth temperature between 45–70 °C41. The most likely hypothesis is that the Geobacillus strains contaminated the albumen supports during their preparation. Different methods were used to harden the albumen layers, including steaming and storing the albumen-coated papers inside a warm hayloft for half a year42. The temperature of a hayloft can sometimes reach 50 °C or more43, providing an optimum environment for the growth of Geobacillus strains. These bacteria were also detected on the paper frame of the photo album which was in contact with the Gyula print. If Geobacillus contamination was intrinsic in the Gyula print, the album paper frame could have become contaminated when the photo album was stored in conditions allowing the growth of these microorganisms. Geobacillus members were also detected in the albumen layer of the antiquarian photograph, making it seem likely that Geobacillus tended to colonize these types of photographs during their manufacturing. These microorganisms can cause serious deterioration problems due to to their known hydrolytic properties41.
Lactococci were the most prevalent group on the antiquarian print, and their presence too may be connected with the hardening of the albumen layer of this photo, perhaps inside a hayloft where lactic acid bacteria are known to participate in silage fermentation43.
The fungal microflorae detected in the three samples by the DGGE-cloning approach are complementary to those isolated by cultivation. Only a few links were found, namely the Aspergillus and Chaetomium strains, which were detected by both microbial investigation methods in, respectively, the antiquarian print and the Gyula print.
The paper frame of the photo album showed the lowest fungal diversity; the same fungal members (Nectria sp. and Malassezia sp.) were detected in both the album and the Gyula print, although the fungal community of the albumen print was richer, containing many other species, several of which, Trichosporon, Geotrichum/Galactomyces, Chaetomium and Cladosporium, have already been found in photographic materials or archival documents6,10,44,45. Fungal species of the genera Nectria, Corynespora and Saccharomyces were also seen, which have not frequently been encountered in archival environments. Different members of Nectria and Corynespora are plant pathogens, so they possess significant lignocellulolytic abilities46,47. Malassezia strains are normally isolated from epidermises or skin scalp48, so their presence on these samples probably is likely due to handling of the photo-book over time.
Three particular eukaryotic organisms were only detected in the antiquarian albumen photograph: Eurotium halophilicum, a xerophilic species previously found in archival items21,49, Trebouxia impressa and Gnomonia setacea. Algae of the Trebouxia genus are generally photobiont lichens and it is not surprising to find them on surfaces exposed to other kinds of environments32,50; they were very recently identified on da Vinci’s self-portrait21. Trebouxia species are heavy-metal tolerant, and their presence here may be due to the traces of different metals found in the photograph (this property has also lead to their use in detoxification applications51,52). Gnomonia setacea is an endophytic species frequently isolated from Betula trees53, but little is currently known about their hydrolytic potential; to our knowledge members of this species have never before been detected in archival objects and environments.
Conclusion
Only few studies have analyzed albumen photographs and, to our knowledge, none of them treated the biodeterioration of this important part of our heritage. Safeguarding our cultural heritage requires methods able to identify the construction materials of historical objects and to diagnose the potential biodeteriogens in order to develop targeted conservation and restoration strategies. This study has illustrated a possible armamentarium of techniques and investigative approaches for the analysis of albumen prints.
FTIR and XRF were useful in analyzing the photographic materials and the composition of foxing stains. The use of cellulose nitrate membranes for sampling the microflora on the surface of the photographs and their subsequent observation by SEM provides an easy, noninvasive procedure for acquiring a preliminary idea of the extent of microbial contamination. Exploiting different kinds of microbiological media allowed a diverse microbial community (especially the fungal) to be isolated. The cloning and sequencing approach together with DGGE screening of the clones revealed different species from those present in the cultivation trial. This combination of culture-dependent and culture-independent methods revealed complex microbial communities in all analyzed samples. Finally, enzyme assays allowed the most deteriogenic microorganisms to be identified.
Methods
Samples
Two albumen photographs were the subject of this investigation. The first photograph (Gyula; P1) is dated 1889; it was produced by an important photographer, Max Stern (1836–1901), and is one of 218 photographs contained in the official photo album of the district of Trenčin. It has been stored in the Slovak National Archives since 1991. The Gyula photograph (Fig. 1) is surrounded by the paper frame of the photographic album (Gyula; PF1) and it has clearly been affected by an easily visible microbial contamination; several different foxing stains can be seen on its surface. The Gyula photograph was offered for investigation, before various conservation treatments were undertaken, in order to determine potential microbial contaminants. Only noninvasive methods were applied to this photo.
The second albumen photograph was bought by a local antiquarian and brought to the laboratory inside a sterile bag. On this antiquarian photo (PA2) (Fig. 2) only foxing stains were visible to the naked eye. This photograph “was sacrificed”, using invasive investigation methods, in order to better understand the microbial contamination of albumen photos and to compare it with the Gyula photograph.
Microscopic observation
Both photographs were examined with a stereo microscope (Olympus SZX9; Tokyo, Japan) at 20–40× magnification and light incident at a 45° angle.
A portion of the 25 mm cellulose nitrate membrane used for the microbiological sampling of the Gyula photograph surface (F4, area outlined by red circle, Fig. 1a) and a foxing stain (point 2 on Fig. 2a) cut from the antiquarian photograph were examined by SEM (Jeol JSM 6610, Tokyo, Japan) using the facilities offered by the Institute of Materials and Machine Mechanics of the Slovak Academy of Sciences. Prior to SEM observation, the samples were sputtered with gold ions.
μ-FTIR and ATR-FTIR spectroscopy
μ-FT-IR spectra were measured using the attenuated total reflection technique using a μ-FTIR-ATR Varian 610-IR (Agilent Technology, Santa Clara, USA), equipped with a Ge/KBr beamsplitter and extended with a Continuum Microscope and a liquid nitrogen cooled MCT detector. Measurements were performed at a frequency of 20 kHz in the 4000–600 cm−1 range at a resolution of 4 cm−1, averaging 400 acquisitions per sample. The spectra were processed by the Varian Resolution Pro program (Agilent Technology). Four places in the Gyula print with foxing stains (F1–F4; Fig. 1a) were analyzed by μ-FTIR. The place F0, which lacked apparent damage, was treated as the control in order to determine the photographic technique. A similar strategy was applied to the antiquarian photograph where points 2–6 evidenced foxing alterations and point 1 (Fig. 2a) was used to identify the photographic technique.
The reflectance spectra of albumen prints in the infrared region were also measured on an Excalibur FTS 3000MX (Digilab, Randolph, USA) spectrometer with an ATR adapter containing a diamond crystal. Measurements were taken at a frequency of 20 kHz in the 4000–600 cm−1 range at a resolution of 4 cm−1, averaging 60 acquisitions per sample. The ATR spectra were processed by the Varian Resolution Pro (Agilent Technology) program.
XRF measurement
XRF measurements were performed at three different points (the cardboard around the photographs and foxing stains from pale and dark sites; Fig. 1a XA-XC; Fig. 2a A–C). A portable XRF spectrometer was used for both photographs (X-MET5100; Oxford Instruments, Abingdon, UK). The X-ray tube was operated at a voltage of 45 kV and a current of 40 mA, the scanning time was 60 s and the spectral resolution was 150 eV.
Sampling and microflora isolation
A sterile, 25 mm cellulose nitrate membrane (Sartorius, Goettingen, Germany) was pressed onto a contaminated section of the albumen part of the Gyula photograph, and a second membrane was pressed onto the album paper frame (Fig. 1a red circles).
The sampling of the antiquarian albumen print (PA2) was performed by cutting off a piece of the photograph (Fig. 2a, point 3). This piece of albumen print was suspended in 2 ml of physiological solution; after vortexing, 200 μl of the resultant suspension were plated in the same type of agar media as in the membrane approach described below. The remainder of the suspension, including the photograph fragments, was used for DNA extraction.
Each membrane was cut in five pieces: one piece was used for SEM observation (see above), 3 pieces were used for cultivation and one piece was used for DNA extraction (see below).
Cultivation was done in two different ways: (i) in direct inoculation, two pieces of membrane from each sample were put on 10× diluted Luria Bertani (LB10) agar plates for bacteria and on Malt Extract Agar (MEA) for fungi; (ii) in the membrane approach, a different piece of membrane for each sample was suspended in physiological solution, diluted 10× and plated onto agar dishes specific for the growth of bacteria or fungi. For bacterial isolation, the membrane approach used R2A (Oxoid, Basingstoke, UK), LB10 and gelatin agar with 3% NaCl (R2A-Gel-NaCl); for fungal isolation, the method used Dichloran Rose Bengal Chloramphenicol (DRBC; Hi-Media, Bombay, India), MEA (Hi-Media) and R2A-Gel-NaCl. The gelatin medium was prepared using R2A agar (Oxoid, Basingstoke, UK) plus 3% NaCl. This medium was autoclaved separately and then 0.4% of sterilized gelatin was added (Sigma-Aldrich, Germany) along with the amount of 20% MgSO4 (autoclaved separately) needed to reach a final MgSO4 concentration of 2%. All bacterial and fungal plates were incubated at room temperature (22–26 °C) for five days to 2 weeks. The agar media were supplemented with either actidione (50 mg l−1; Fluka, Seelze, Germany) or chloramphenicol (50 mg l−1; Sigma–Aldrich, Seelze, Germany) in order to avoid the growth of fungi and bacteria respectively.
DNA extraction from isolated microflora and identification
Microorganisms were selected on the basis of differing morphology and transferred to new agar plates to obtain pure colonies. The purified fungal isolates were maintained on Sabouraud (SAB) slants, the bacteria on plates of Tryptone-Soya Agar (TSA; Oxoid). For DNA extraction from fungal strains, the isolates were inoculated in SAB broth at 28 °C until growth; they were then separated from the broth by filtration through sterile filter paper, followed by DNA extraction with Ron’s fungal DNA mini kit (Bioron, Ludwigshafen, Germany), according to the instructions of the manufacturer. The ITS region of rDNA was amplified with the primers ITS1 (5′-TCC GTA GGT GAA CCT GCG G-3′) and ITS4 (5′-TCC TCC GCT TAT TGA TAT GC-3′)54. The 25 μl PCR mixture contained 50 pmol of each primer, 200 μmol l−1 of dNTP (Life Technologies, Gaithersburg, Maryland, USA), 1.5 U HotStar Taq plus DNA polymerase (Qiagen, Hilden, Germany), 1× PCR buffer and 3 μl of the extracted DNA (the PCR template). The PCR program consisted of an initial denaturation at 94 °C for 5 min, followed by 30 cycles (denaturation at 94 °C for 30 s, annealing at 54 °C for 45 s, extension at 72 °C for 1 min) and a final polymerization step at 72 °C for 10 min.
For DNA extraction from bacteria, fresh bacterial colonies were collected from the TSA plates and their DNA was isolated using InstaGene Matrix (Biorad, Hercules, CA, USA) following the instructions of the manufacturer; this extracted DNA was then used as a PCR template. The bacterial isolates were identified by partial sequencing of the 16S rDNA using PCR with the primers 27f (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 685r (5′-TCT ACG CAT TTC ACC GCT AC-3′)55; the PCR conditions and program were the same as for the ITS procedure.
The resulting PCR products from both fungal and bacterial strains were purified using ExoSAP-IT (Affymetrix, Cleveland, Ohio, USA) and sequenced at a commercial facility (GATC-Biotech, Konstanz, Germany). The resulting sequences were directly compared with those in GenBank using blast program (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and were subsequently deposited in GenBank under the accession numbers KT200239–KT200250 (bacterial isolates) and KT200251–KT200277 (fungal isolates).
Total DNA extraction and 16S rRNA, 28S rRNA gene amplification
The total microbial DNA was extracted by chaotropic solid-phase extraction (DNeasy Blood & Tissue Kit; Qiagen) from the remaining cellulose nitrate membranes (from the Gyula photo and its surroundeding paper frame) and from the suspension containing the antiquarian photograph fragments (point 3 on Fig. 2a) by the protocol described by Bučková et al.6
The bacterial 16S rDNA and eukaryotic 28S rDNA fragments were amplified in two steps. The first step involved primers 27f and 685r oriented towards the bacterial 16S rRNA gene. For the amplification of the eukaryotic 28S rRNA gene, the universal primers NL1 (5′-GCA TAT CAA TAA GCG GAG GAA AAG-3′) and NL4 (5′-GGT CCG TGT TTC AAG ACG G-3′)56 were used. The 25 μl PCR mixtures contained 50 pmol of each primer, 2.5 mmol l−1 MgCl2, 200 μmol l−1 of dNTP, 1.5 U HotStar Taq plus DNA polymerase (Qiagen), 1 × PCR buffer and 3 μl (4.2–8.45 ng μl−1) of extracted DNA. The PCR was performed with an initial 5 min denaturation step at 95 °C followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 54 °C (amplification of 16S rRNA gene) and at 56 °C (amplification of 28S rRNA gene) for 1 min, and extension at 72 °C for 1 min, subsequently followed by a 10 min extension step at 72 °C. For each DNA target, four reactions of 25 μl (100 μl altogether) were produced. The four reactions of each DNA target were mixed together and the specificity of amplification was checked by agarose gel electrophoresis. A part of these PCR products were set aside for the construction of clone libraries (see below).
Denaturing gradient gel electrophoresis fingerprint analysis
The remainder of the PCR product from the first step (1 μl) was used as a template in the second amplification step, a semi-nested PCR for each DNA target. The 16S rRNA gene was re-amplified with primers 518f (5′-CCA GCA GCC GCG GTA AT-3′) and 685r-GC (5′-CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GGG GTC TAC GCA TTT CAC CGC TAC-3′); the primers NL1f-GC (5′- CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GGG GGC ATA TCA ATA AGC GGA GGA AAA G-3′) and LS2 (5′- ATT CCC AAA CAA CTC GAC TC-3′) were used for the semi-nested amplification of the 28S rRNA gene. The PCR conditions were the same as above. Denaturing gradient gel electrophoresis (DGGE) for bacterial and eukaryotic amplicons was performed as described by Pangallo et al.57 to produce DGGE fingerprinting profiles of the bacterial and eukaryotic communities.
Construction of clone libraries and screening of clones
Clone libraries were produced from a portion of the PCR products from the first PCR amplification step following a previously described protocol57. Briefly, the PCR products were purified with the QIAquick PCR purification kit (Qiagen), ligated into a pGEM-T Easy vector (Promega,Madison, Wisconsin, USA), transformed into E. coli XLI-Blue, and spread on Luria Bertani plates with 100 μg ml−1 ampicillin 0.1 mM, X-Gal and 0.2 mM IPTG. To confirm the presence of the desired inserts, 100 white colonies of each clone library were picked and directly PCR screened for the presence of the inserts using the vector specific primers SP6 and T757. Positive clones from each library were analyzed using the by semi-nested PCR DGGE procedure described above. The profiles of individual clones were compared with each other and with that of the whole community. A diagram schematically outlining the complete cloning and DGGE screening strategy is shown in Supplementary Figure S1. The PCR products of clones with different profiles were sequenced in order to identify the microorganisms colonizing the albumen prints. To sequence these products, they were first purified using ExoSAP-IT (Affymetrix, Cleveland, USA) and sequenced using the SP6 primer by a commercial facility (GATC Biotech, Konstanz, Germany). For microorganism identification, the sequences were compared directly with those in GenBank using blast program (http://blast.ncbi.nlm.nih.gov/Blast.cgi). These sequences were deposited in GenBank under accession numbers KT200278–KT200296 (bacterial clones) and KT200297–KT200316 (eukaryotic clones).
Hydrolytic assays, catalase and peroxidase activities
Several agar plate tests were applied to assess the proteolytic (R2A-Gel agar), cellulolytic (Congo red agar) and lypolitic (Spirit blue agar) abilities of isolated strains as described by Šimonovičová et al.58. R2A-Gel agar plates were prepared by mixing autoclaved R2A Agar (Oxoid) with 0.4% of sterilized gelatin (Sigma–Aldrich, St. Louis, USA). The Congo-Red agar contained 0.5 g KH2PO4, 0.25 g MgSO4, 2 g cellulose, 0.2 g Congo-Red, 2 g gelatin, and 15 g agar in 1 l distilled water at pH 6.8–7.2. Spirit Blue agar was prepared by suspending 32.15 g of Spirit Blue agar (Himedia) in 1000 ml distilled water, autoclaved, cooled down to 50 °C and supplemented with 30 ml of lipase substrate (1 ml of Tween 80, 400 ml of warm distilled water and 100 ml of olive oil; sterilized by autoclaving). This was slow mixed and poured into Petri dishes. All chemicals, if not otherwise specified, were bought from Sigma–Aldrich. All assays were performed in triplicate using 60 mm plates incubated at room temperature (22–26 °C) generally for 3–7 days.
Catalase (CAT; EC 1.11.1.6) activity was determined spectrophotometrically at pH 7.0 by monitoring the decomposition of H2O2 at 240 nm with an extinction coefficient of 43.6 M−1 cm−1. One unit (1 U) of catalase activity was defined as the amount of enzyme that catalyzes the decomposition of 1 μmol of H2O2 per min59.
Peroxidase (POX; E.C.1.11.1.7) activity was assayed spectrophotometrically at pH 5.5 by monitoring the oxidation of o-dianisidine dihydrochloride at 460 nm (ε460 nm = 11.3 × 103 M−1). One unit of peroxide activity (U) was defined as the amount of activity that produces 1 μmol of oxidized o-dianisidine per minute60.
Additional Information
How to cite this article: Puškárová, A. et al. Microbial communities affecting albumen photography heritage: a methodological survey. Sci. Rep. 6, 20810; doi: 10.1038/srep20810 (2016).
Supplementary Material
Acknowledgments
This work was financed by the Slovak VEGA Agency, project number: 2/0103/14 “Protecting our memories: investigation into the biodeterioration of photographic and cinematographic materials”. We are very grateful to Dr. Jacob Bauer for the English revision of the text.
Footnotes
Author Contributions A.P., M.B., L.K. and D.P. performed sampling, microorganism isolation and identification, molecular analysis, hydrolytic and catalase – peroxidase assays. B.H. and A.M. were responsible for FTIR and XRF analyses and microscopic observation. D.P. wrote the article.
References
- Marien M. W. Photography: A cultural history. Laurence King Publishing (2006). [Google Scholar]
- Reilly J. M. & McCabe C. Care and identification of 19th century photographic prints. Eastman Kodak Company (1986). [Google Scholar]
- Cappitelli F. & Sorlini C. From papyrus to compact disc: the microbial deterioration of documentary heritage. Crit. Rev. Microbiol. 31, 1–10 (2005). [DOI] [PubMed] [Google Scholar]
- Sterflinger K. & Pinzari F. The revenge of time: fungal deterioration of cultural heritage with particular reference to books, paper and parchment. Environ. Microbiol. 14, 559–566 (2012). [DOI] [PubMed] [Google Scholar]
- Sterflinger K. & Piñar G. Microbial deterioration of cultural heritage and works of art-tilting at windmills? Appl. Microbiol. Biotechnol. 97, 9637–9646 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bučková M. et al. Co-occurrence of bacteria and fungi and spatial partitioning during photographic materials biodeterioration. Polym. Degrad. Stab. 108, 1–11 (2014). [Google Scholar]
- Abrusci C. et al. Isolation and identification of bacteria and fungi from cinematographic films. Int. Biodeterior. Biodegradation 56, 58–68 (2005). [Google Scholar]
- Borrego S. et al. The quality of air at archives and the biodeterioration of photographs. Int. Biodeterior. Biodegradation 64, 139–145 (2010). [Google Scholar]
- Guiamet P., Borrego S., Lavin P., Perdomo I. & de Saravia S. G. Biofouling and biodeterioration in materials stored at the Historical Archive of the Museum of La Plata, Argentine and at the National Archive of the Republic of Cuba. Colloids Surf. B Biointerfaces 85, 229–234 (2011). [DOI] [PubMed] [Google Scholar]
- Sclocchi M. C., Damiano E., Matè D., Colaizzi P. & Pinzari F. Fungal biosorption of silver particles on 20th-century photographic documents. Int. Biodeterior. Biodegradation 84, 367–371 (2013). [Google Scholar]
- Barth A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta (BBA)-Bioenergetics 1767, 1073–1101 (2007). [DOI] [PubMed] [Google Scholar]
- Ricci C., Bloxham S. & Kazarian S. G. ATR-FTIR imaging of albumen photographic prints. J. Cult. Herit. 8, 387–395 (2007). [Google Scholar]
- Čechák T., Kopecká I., Trojek T., Štanzel T. & Bártová H. Application of X-ray fluorescence in an investigation of photographic heritage. Radiat. Phys. Chem. 116, 8–13 (2015). [Google Scholar]
- Oberle-Kilic J., Dighton J. & Arbuckle-Keil G. Chemical Characterization of Starch and Starch: Lignin Films Using Micro-attenuated Total Reflectance Fourier Transform Infrared Spectroscopy (micro-ATR FTIR). Trends Biomater. Artif. Organs 26, 107–109 (2012). [Google Scholar]
- Oberle-Kilic J., Dighton J. & Arbuckle-Keil G. Atomic force microscopy and micro-ATR-FT-IR imaging reveals fungal enzyme activity at the hyphal scale of resolution. Mycology 4, 44–53 (2013). [Google Scholar]
- Oberle J., Dighton J. & Arbuckle-Keil G. Comparison of methodologies for separation of fungal isolates using Fourier transform infrared (FTIR) spectroscopy and Fourier transform infrared-attenuated total reflectance (FTIR-ATR) microspectroscopy. Fungal Biol. 119, 1100–1114 (2015). [DOI] [PubMed] [Google Scholar]
- Zotti M., Ferroni A. & Calvini P. Mycological and FTIR analysis of biotic foxing on paper substrates. Int. Biodeterior. Biodegradation 65, 569–578 (2011). [Google Scholar]
- Corsaro C. et al. Molecular degradation of ancient documents revealed by 1H HR-MAS NMR spectroscopy. Sci. Rep. 3, 2896 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak P. S. & Singh B. K. Instrumental characterization of clay by XRF, XRD and FTIR. Bull. Mater. Sci. 30, 235–238 (2007). [Google Scholar]
- Cattaneo B. et al. Physico-chemical characterization and conservation issues of photographs dated between 1890 and 1910. J. Cult. Herit. 9, 277–284 (2008). [Google Scholar]
- Piñar G., Tafer H., Sterflinger K. & Pinzari F. Amid the possible causes of a very famous foxing: molecular and microscopic insight into Leonardo da Vinci’s self-portrait. Environ. Microbiol. Rep. (in press), 10.1111/1758-2229.12313 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquarella C. et al. A multidisciplinary approach to the study of cultural heritage environments: experience at the Palatina Library in Parma. Sci. Total Environ. 536, 557–567 (2015). [DOI] [PubMed] [Google Scholar]
- Rakotonirainy M. S., Heude E. & Lavédrine B. Isolation and attempts of biomolecular characterization of fungal strains associated to foxing on a 19th century book. J. Cult. Herit. 8, 126–133 (2007). [Google Scholar]
- Skóra J. et al. Assessment of microbiological contamination in the work environments of museums, archives and libraries. Aerobiologia (in press), 10.1007/s10453-015-9372-8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraková L., Chovanová K., Puškarová A., Bučková M. & Pangallo D. A novel PCR-based approach for the detection and classification of potential cellulolytic fungal strains isolated from museum items and surrounding indoor environment. Lett. Appl. Microbiol. 54, 433–440 (2012). [DOI] [PubMed] [Google Scholar]
- Michaelsen A., Pinar G. & Pinzari F. Molecular and microscopical investigation of the microflora inhabiting a deteriorated Italian manuscript dated from the thirteenth century. Microb. Ecol. 60, 69–80 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina R. et al. Differences in the secretion pattern of oxidoreductases from Bjerkandera adusta induced by a phenolic olive mill extract. Fungal Genet. Biol. 72, 99–105 (2014). [DOI] [PubMed] [Google Scholar]
- Knop D., Yarden O. & Hadar Y. The ligninolytic peroxidases in the genus Pleurotus: divergence in activities, expression, and potential applications. Appl. Microbiol. Biotechnol. 99, 1025–1038 (2015). [DOI] [PubMed] [Google Scholar]
- Carvalho M. B. et al. Degradation pathway of pentachlorophenol by Mucor plumbeus involves phase II conjugation and oxidation–reduction reactions. J. Hazard. Mater. 198, 133–142 (2011). [DOI] [PubMed] [Google Scholar]
- Kraková L. et al. A multiphasic approach for investigation of the microbial diversity and its biodegradative abilities in historical paper and parchment documents. Int. Biodeterior. Biodegradation 70, 117–125 (2012). [Google Scholar]
- Pangallo D. et al. Disclosing a crypt: microbial diversity and degradation activity of the microflora isolated from funeral clothes of Cardinal Peter Pázmány. Microbiol. Res. 168, 289–299 (2013). [DOI] [PubMed] [Google Scholar]
- Pangallo D. et al. Biodeterioration of epoxy resin: a microbial survey through culture-independent and culture-dependent approaches. Environ. Microbiol. 17, 462–479 (2015). [DOI] [PubMed] [Google Scholar]
- Ventorino V. et al. Exploring the microbiota dynamics related to vegetable biomasses degradation and study of lignocellulose-degrading bacteria for industrial biotechnological application. Sci. Rep. 5, 8161 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaleel C. A. et al. Pseudomonas fluorescens enhances biomass yield and ajmalicine production in Catharanthus roseus under water deficit stress. Colloids Surf. B Biointerfaces 60, 7–11 (2007). [DOI] [PubMed] [Google Scholar]
- Khan A. L. et al. Co-synergism of endophyte Penicillium resedanum LK6 with salicylic acid helped Capsicum annuum in biomass recovery and osmotic stress mitigation. BMC Microbiol. 13, 51 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Xu X. & Feng L. Responses of antioxidant defenses and membrane damage to drought stress in fruit bodies of Auricularia auricula-judae. World J. Microbiol. Biotechnol. 30, 119–124 (2014). [DOI] [PubMed] [Google Scholar]
- Adav S. S., Li A. A., Manavalan A., Punt P. & Sze S. K. Quantitative iTRAQ secretome analysis of Aspergillus niger reveals novel hydrolytic enzymes. J. Proteome Res. 9, 3932–3940 (2010). [DOI] [PubMed] [Google Scholar]
- Takasuka T. E., Book A. J., Lewin G. R., Currie C. R. & Fox B. G. Aerobic deconstruction of cellulosic biomass by an insect-associated Streptomyces. Sci. Rep. 3, 1030 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levasseur A., Drula E., Lombard V., Coutinho P. M. & Henrissat B. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol. Biofuels 6, 41 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepe O. et al. Characterization in the archaeological excavation site of heterotrophic bacteria and fungi of deteriorated wall painting of Herculaneum in Italy. J. Environ. Biol. 32, 241–250 (2011). [PubMed] [Google Scholar]
- Zeigler D. R. The Geobacillus paradox: why is a thermophilic bacterial genus so prevalent on a mesophilic planet? Microbiology 160, 1–11 (2014). [DOI] [PubMed] [Google Scholar]
- James C. The book of alternative photographic processes. Cengage Learning (2015). [Google Scholar]
- Collins M. & Owens V. Preservation of forages as hay and silage; p. 443–472. In Barnes R. F. et al. (ed.) Forages. Vol. 1: An introduction to grassland agriculture. Iowa State Press, Ames (2003). [Google Scholar]
- da Silva M. et al. Inactivation of fungi from deteriorated paper materials by radiation. Int. Biodeterior. Biodegradation 57, 163–167 (2006). [Google Scholar]
- Pinheiro A. C. et al. Mould and yeast identification in archival settings: Preliminary results on the use of traditional methods and molecular biology options in Portuguese archives. Int. Biodeterior. Biodegradation 65, 619–627 (2011). [Google Scholar]
- Liaud N. et al. Exploring fungal biodiversity: organic acid production by 66 strains of filamentous fungi. Fungal Biol. Biotechnol. 1, 1 (2014).26457194 [Google Scholar]
- Shimomoto Y. et al. Pathogenic and genetic variation among isolates of Corynespora cassiicola in Japan. Plant Pathol. 60, 253–260 (2011). [Google Scholar]
- Gupta A. K., Batra R., Bluhm R., Boekhout T. & Dawson T. L. Skin diseases associated with Malassezia species. J. Am. Acad. Dermatol. 51, 785–798 (2004). [DOI] [PubMed] [Google Scholar]
- Micheluz A. et al. The extreme environment of a library: Xerophilic fungi inhabiting indoor niches. Int. Biodeterior. Biodegradation 99, 1–7 (2015). [Google Scholar]
- Urzì C., De Leo F., Bruno L. & Albertano P. Microbial diversity in paleolithic caves: a study case on the phototrophic biofilms of the Cave of Bats (Zuheros, Spain). Microb. Ecol. 60, 116–129 (2010). [DOI] [PubMed] [Google Scholar]
- Pawlik-Skowrońska B. & Bačkor M. Zn/Pb-tolerant lichens with higher content of secondary metabolites produce less phytochelatins than specimens living in unpolluted habitats. Environ. Exp. Bot. 72, 64–70 (2011). [Google Scholar]
- Sanita’ di Toppi L. et al. First and second line mechanisms of cadmium detoxification in the lichen photobiont Trebouxia impressa (Chlorophyta). Environ. Pollut. 151, 280–286 (2008). [DOI] [PubMed] [Google Scholar]
- Osono T. & Masuya H. Endophytic fungi associated with leaves of Betulaceae in Japan. Can. J. Microbiol. 58, 507–515 (2012). [DOI] [PubMed] [Google Scholar]
- White T. J., Bruns T., Lee S. & Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics; p. 315–321. In PCR Protocols: A Guide to Methods and Applications. Innis M. A., Gelfand D. H., Sninsky J. J. & White T. J., (eds.). New York: Academic Press (1990). [Google Scholar]
- Lane D. J. 16S/23S rRNA sequencing; p. 115–148. In Nucleic Acid Techniques in Bacterial Systematics. Stackenbrandt E. & Goodfellow M. (eds.) John Wiley & Sons, New York (1991). [Google Scholar]
- Kurtzman C. P. & Robnett C. J. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie van Leeuwenhoek 73, 331–371 (1998). [DOI] [PubMed] [Google Scholar]
- Pangallo D. et al. Microbial diversity and dynamics during the production of May bryndza cheese. Int. J. Food Microbiol. 170, 38–43 (2014). [DOI] [PubMed] [Google Scholar]
- Šimonovičová A. et al. Fungi on mummified human remains and in the indoor air in the Kuffner family crypt in Sládkovičovo (Slovakia). Int. Biodeterior. Biodegradation 99, 157–164 (2015). [Google Scholar]
- Roggenkamp R., Sahm H. & Wagner F. Microbial assimilation of methanol induction and function of catalase in Candida boidinii. FEBS Lett. 41, 283–286 (1974). [DOI] [PubMed] [Google Scholar]
- Claiborne A. & Fridovich I. Purification of the o-dianisidine peroxidase from Escherichia coli B. Physicochemical characterization and analysis of its dual catalatic and peroxidatic activities. J. Biol. Chem. 254, 4245–4252 (1979). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.