Abstract
Currently, the reliable prognostic biomarkers for WHO grade II diffuse astrocytomas (DA) are still limited. We investigated the relations between the level of 5-Hydroxymethylcytosine (5hmC), an oxidated production of 5-methylcytosine (5mC) by the ten eleven translocated (TET) enzymes, and clinicopathological features of glioma patients. With an identified anti-5hmC antibody, we performed immunohistochemistry in 287 glioma cases. We detected that 5hmC variably reduced in most gliomas and 5hmC reduction was closely associated with higher pathological grades and shortened survival of glioma patients. In multivariate analysis, 5hmC had no independent prognostic value in the entire patient cohort. However, multivariate analysis within subtypes of gliomas revealed that 5hmC was still a prognostic marker confined to DA. In addition, we detected that IDH1 mutation by DNA sequencing was associated with favorable survival within DA. Lastly, we detected that the combination of 5hmC/KI67 was a useful prognostic marker for restratification of DA.
Human gliomas are a heterogeneous group of tumors and traditionally classified into various subtypes and grades mostly based on their microscopic characteristics for therapeutic decision-making1,2. However, histopathological criteria usually unavoidablely cause subjectively diagnostic interobserver variability3,4. Moveover, classification based on microscopic characteristics rather than molecular pathogenesis of gliomas limits the adequate assessment of prognosis and appropriate planning of treatment. For these regards, “ISN-Haarlem” guidelines recently proposed to define diagnostic entities as narrowly as possible and to include applicable molecular data to come up with a more objective and reproducible “integrated diagnosis” for glioma classification5. For example, molecular biomarkers isocitrate dehydrogenase (IDH1/IDH2) mutation and 1p/19q codeletion were proposed to resolve oligoastrocytoma as either oligodendroglioma or astrocytoma6. In addition, IDH1/IDH2 mutation, 1p/19q codeletion, TP53 mutation and MGMT promoter methylation were used for prognostic modeling and stratification into molecularly determined treatment groups5,7,8,9,10. However, some questions remain ambiguous. For instance, within WHO grade II diffuse astrocytomas (DA) the prognostic relevance of the molecular markers has remained debate10,11,12,13,14,15,16,17,18 (Supplementary Tables S1 and S2). Therefore, more reliable molecular markers for predicting the course of disease and outcome of gliomas are still needed.
DNA methylation at the 5-carbon position of cytosine (5mC) is the most extensively studied epigenetic modification in human cancer19. In 2009, breakthrough studies indicated that 5mC can be converted to 5-hydroxymethylcytosine (5hmC) by the ten eleven translocated (TET) enzymes20,21. HPLC-MS analysis and immunohistochemistry revealed that 5hmC is present with highest level in central nervous system22. Subsequent studies indicated that 5hmC is not merely serving as an intermediate of DNA demethylation, but also acts as a stable epigenetic marker23. Meanwhile, abundant evidence detected that 5hmC globally decreased in most human malignancies, including gliomas24,25,26,27,28,29,30,31. Initially, 5hmC loss in gliomas was proposed to be related with IDH1/IDH2 mutations26. However, subsequently numerous trials from larger clinical samples argued against this claim25,29,30,31. It was interesting that 5hmC loss were suggested to be prognostic for malignant gliomas (World Health Organization grade III or IV)29. Due to small sample and lack detailed information about management and adjuvant treatment in this study29, much more work needs to verify the prognostic value of 5hmC in gliomas. Here, we performed immunohistochemical investigation in 287 glioma cases with a well identified homemade anti-5hmC antibody. The results showed that 5hmC was an prognostic marker confined to DA but not grade III or IV glioma patients. Moreover, we detected that IDH1 mutation by DNA sequencing and the combination of 5hmC/KI67 was associated with prognosis of DA respectively.
Results
Patient characteristics
The clinicopathological characteristics of the patients were summarized in Table 1. In total 287 patients, 143 (50%) cases were no more than age 40 with median age 41 (ranged from 16–76). The patient group consisted of 166 (58%) males and 121 (42%) females. Most gliomas (89%) located in the supratentorial areas. There were 23 (8%) grade I, 130 (45%) grade II, and 69 (24%) grade III and 64 (23%) grade IV glioma cases respectively. In the subtypes, most cases (33%) were DA. The overall follow-up durations ranged from 2 to 103 months (median, 24 months). A total of 144 (50%) patients were alive at the end of the follow-up, while 143 (50%) patients died of gliomas. The preoperative KPS scores of 179 (62%) patients were more than 80. Tumor volumes of 130 (45%) cases were less than 50 cm3. 212 (74%) cases had total tumor resection and 75 (26%) cases had subtotal tumor resection. Subsequent to surgery, 118 (41%) patients received combined radiotherapy and chemotherapy. 26 (9%) and 68 (24%) patients were treated with either radiotherapy or chemotherapy respectively. 75 (26%) patients did not receive further therapy.
Table 1. Patient Characteristics.
| Total cases | N = 287 | % |
|---|---|---|
| Age in years | ||
| Median. range | 41 (16 ∼ 76) | |
| ≤40:>40 | 143:144 | 50:50 |
| Gender | ||
| Male: Female | 166:121 | 58:42 |
| Glioma location | ||
| Supratentorial: other* | 254:33 | 89:11 |
| WHO grades | ||
| I: II: III: IV | 24:130:69:64 | 8:45:24:23 |
| Subtypes | ||
| PA(GI) | 24 | 8 |
| DA:OA:OG:PXA(GII) | 95:4:24:7 | 33:2:8:2 |
| AA:AOA:AO(GIII) | 37:2:30 | 14:1:10 |
| GBM(G IV) | 64 | 22 |
| Vital status | ||
| Alive:death | 144:143 | 50:50 |
| ≥80:<80 | 179:108 | 62:38 |
| 1Tumor size (cm) | ||
| <50 cm3: ≥ 50cm3: lack data | 130:130:27 | 45:45:10 |
| Extent of resection | ||
| Total:subtotal | 212:75 | 74:26 |
| Adjuvant treatment | ||
| RC:R:C:L | 118:26:68:75 | 41:9:24:26 |
*other: cerebellum, pons, brain stem and spinal cord.
PA, pilocytic astrocytoma; DA, diffuse astrocytoma; OA, oligoastrocytoma; OG, oligodendroglioma; PXA, pleomorphic xanthoastrocytoma; AA, anaplastic astrocytoma; AOA, anaplastic oligoastrocytoma; AO, anaplastic oligodendroglioma; GBM, glioblastoma multiforme. GI, grade I; GII, grade II; GIII, grade III; G IV, grade IV.
RC, radiation and chemotherapy; R, radiation therapy; C, chemotherapy; L, lack adjuvant treatment.
1Available data were 260 cases.
Identification of anti-5hmC antibody
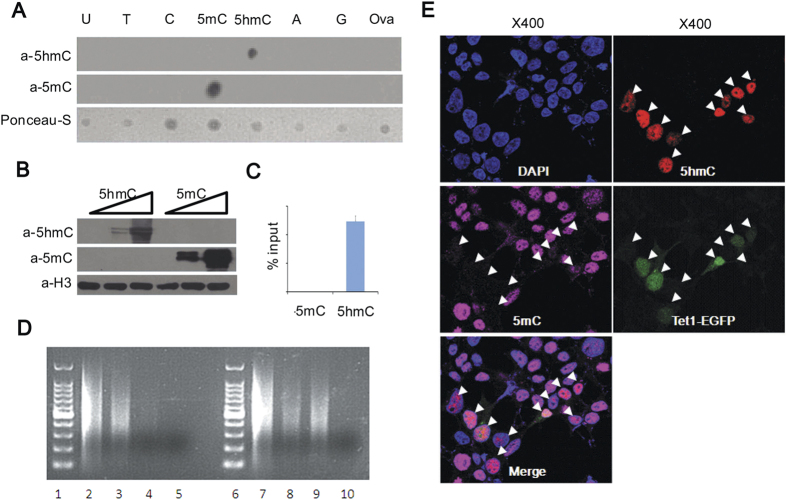
To evaluate specificity of anti-5hmC antibody generated by our lab, we firstly performed dot-blot analysis. The result showed that the rabbit polyclonal anti-5hmC antibody specifically recognized 5hmC instead of other bases (Fig. 1A). West-blot and immunoprecipitation also confirmed this result (Fig. 1B–D). Immunofluorescence analysis showed strong 5hmC staining was in the nucleus of Tet1 transfected cells. Concomitantly, the level of 5mC decreased in Tet1 transfected cells (Fig. 1E). Therefore, these results strongly demonstrated the anti-5hmC antibody had high specificity to recognize 5hmC.
Figure 1. Identification of the specificity of anti-5hmC antibody.
(A) Dotblot analysis. Equal conjugated bases were loaded and immunnoblotted with an antibody against 5hmC (top) or 5mC (middle). Loading was shown by Ponceau staining (bottom). (B) Western blotting. 0, 5, 50 pg of nucleoside-conjugated-ovalbumin was mixed with 293T cell lysate and separated by SDS-PAGE and immunnoblotted with anti-5hmC (upper), anti-5mC (middle) and anti-H3 antibodies (bottom). (C) qPCR quantifying immunoprecipitated DNA fragments containing 5mC or 5hmC by the anti-5mC (left) or anti-5hmC (right) antibody. (D) Denatured fragmented DNA from Tet1 nontransfected (Lane 2–5) or transfected (Lane 7–10) 293T cells was immunoprecipitated by an antibody for 5mC (lanes 3 and 8), 5hmC (Lanes 4 and 9), or control IgG (Lanes 5 and 10). Lanes 1 and 6 are 100-bp DNA marker and lanes 2 and 7 have 10% of DNA input. (E) Immunofluorescence analysis. 293T cells transfected with cDNA encoding the Tet1 catalytic domain-IRES-GFP (middle right, arrowheads) were stained with DAPI (top left), anti-5hmC antibody (top right), or anti-5mC antibody (middle left). Merged Image was in bottom left.
Relations between 5hmC reduction and clinicopathological features
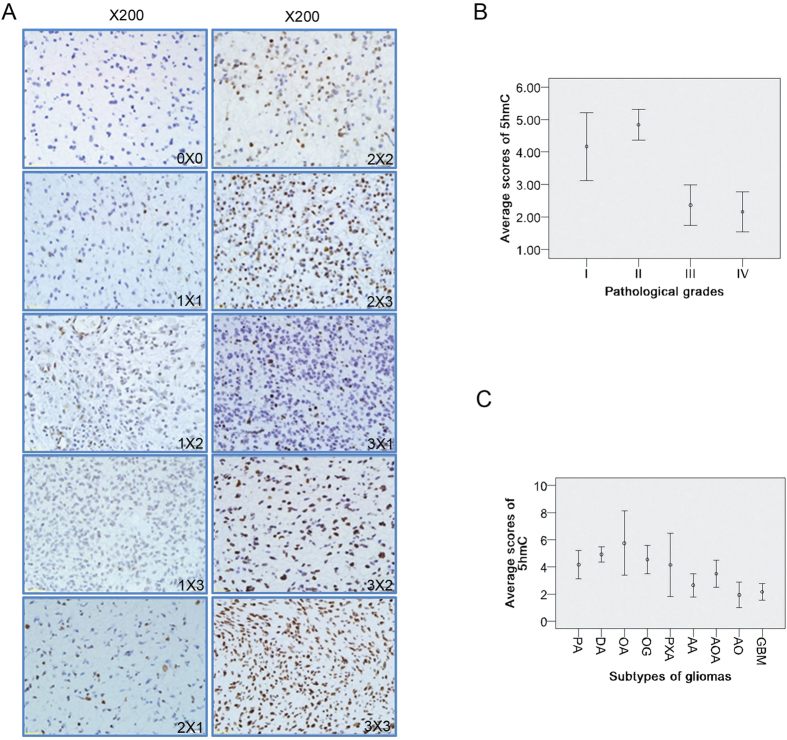
5hmC level was analyzed by immunohistochemistry (IHC) in 287 glioma cases. In glioma tissues some samples had strong staining while other had weak or no staining, and cells in the same sample also had different degrees of staining intensity (Fig. 2A). To facilitate the analysis of immunohistochemical results, specific nuclear immunoreactivity was scored using a 9-point scale on the basis of the product of staining intensity (no staining = 0, weak staining = 1, moderate staining = 2, strong staining = 3), and staining extent (% of positive cells; <5% = 0, 5%–30% = 1, 30%–60% = 2, >60% = 3)32. To facilitate statistical analysis, we divided the samples into 2 groups according to staining scores. Group 1 had no or weak staining with the scores of 0 to 3. Group 2 had moderate and strong staining with the scores from 4 to 9. The relations between 5hmC level and clinicopathological features were summarized in Table 2. On Chi square analysis, the 5hmC reduction (scores 0–3) were significantly associated with the following variables: over age 40 (P < 0.001), high pathological grades (grade III and IV, P < 0.001), vital status (death, P < 0.001), lower KPS scores (<80, P = 0.018) and larger tumor size (≥50 cm3, P = 0.015). Nonsignificant variables included gender (P = 0.422), extent of resection (P = 0.225) and adjuvant treatment (P = 0.544). The average scores in grade I, II, III, and IV for 5hmC were 4.167 ± 0.524, 4.839 ± 0.235, 2.362 ± 0.310 and 2.156 ± 0.310 respectively (Fig. 2B). Spearman correlation analysis showed there was a significantly inverse correlation between the pathological grades of gliomas and the 5hmC scores (r = −0.407, P < 0.001).
Figure 2. 5hmC level examined by immunohistochemistry, and its relations with pathological grades or subtypes of gliomas.
(A) Representative images of 5hmC immunostaining in glioma tissues. 200×. (B,C) Relations between 5hmC level and pathological grades (B) or subtypes (C) of gliomas. Error bar represents the standard error (SE).
Table 2. Relationship between the 5hmC level and clinicopathological features of patients with gliomas.
| Clinicopathological features | Total cases (n = 287) |
||
|---|---|---|---|
| 5hmC (0–3) | 5hmC (4–9) | Pvalue | |
| Gender | |||
| Male: Female | 93:62 | 73:59 | 0.422 |
| Age | |||
| ≤40:>40 | 61:94 | 82:50 | 0.000 |
| WHO grade | |||
| I∼II: III∼IV | 54:101 | 100:32 | 0.000 |
| Vital status | |||
| Alive:death | 59:96 | 85:47 | 0.000 |
| KPS | |||
| ≥80:<80 | 87:68 | 92:40 | 0.018 |
| *Tumor size (cm) | |||
| <50 cm3: ≥ 50cm3 | 60:95 | 70:62 | 0.015 |
| Extent of resection | |||
| Total:Subtotal | 119:36 | 93:39 | 0.225 |
| Adjuvant treatment | |||
| Yes:No | 113:42 | 99:33 | 0.544 |
*Available data were 260 cases.
For subtypes of gliomas, the average scores in pilocytic astrocytomas (PA); diffuse astrocytomas (DA); oligoastrocytomas (OA); oligodendrogliomas (OG); pleomorphic xanthoastrocytomas (PXA); anaplastic astrocytomas (AA); anaplastic oligoastrocytomas (AOA); anaplastic oligodendrogliomas (AO) and glioblastoma multiforme (GBM) for 5hmC were 4.167 ± 0.524, 4.926 ± 0.278, 5.750 ± 1.181, 4.542 ± 0.521, 4.143 ± 1.164, 2.649 ± 0.427, 3.500 ± 0.500, 1.933 ± 0.474 and 2.156 ± 0.310 respectively (Fig. 2C). Kruskal-Wallis test revealed significant difference between these groups (X2 = 61.678, P < 0.001).
5hmC level on patient survival in total glioma cases
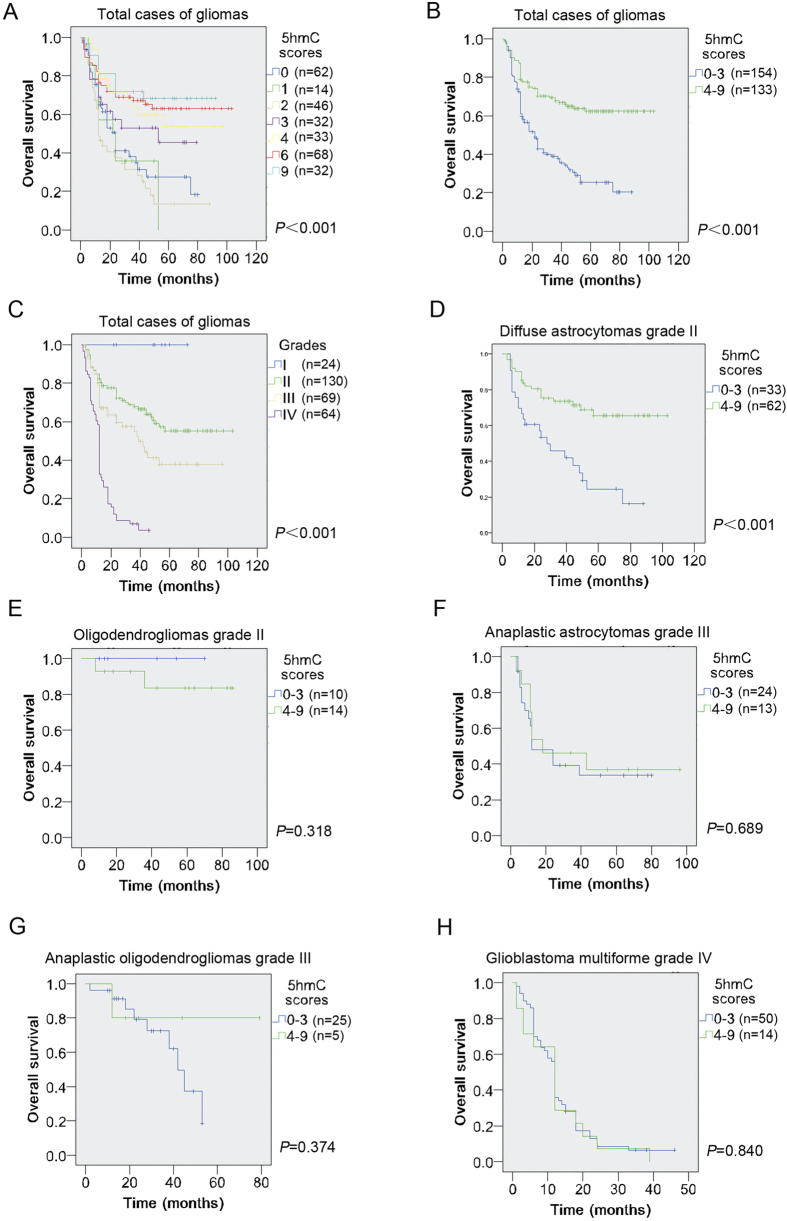
To determine the relations between 5hmC level and the survival of the glioma patients, we firstly divided the samples into seven groups according to the 5hmC score 0, 1, 2, 3, 4, 6 and 9. Kaplan-Meier survival analysis revealed low 5hmC scores strongly correlated with poor prognosis while high 5hmC scores correlated with better survival (Fig. 3A; χ2 = 40.570, P < 0.001). Next, we divided glioma patients into two groups. The cases with scores of 0 to 3 were defined as group 1, while patients with scores from 4 to 9 group 2. On Kaplan-Meier survival analysis, 5hmC reduction group statistically significantly correlated with poorer survival of patients, while 5hmC positive group with the better prognosis (Fig. 3B, χ2 = 31.109, P < 0.001). In addition, as previous reports1,2, higher pathological grade correlated with worse prognosis in total cases (Fig. 3C, χ2 = 124.243, P < 0.001).
Figure 3. Prognostic relevance between 5hmC level and glioma patients by Kaplan-Meier analysis.
(A,B) Kaplan-Meier survival analysis of glioma patients when all samples were divided into 7 groups (A) based on 5hmC scores 0, 1, 2, 3, 4, 6 and 9, or two groups (B) based on 5hmC scores 0–2 and 3–9. (C) Prognostic relevance between pathological grades and glioma patients was examined by Kaplan-Meier analysis. (D–H) Relevance between 5hmC level and patient survival within diffuse astrocytomas (D); oligodendrogliomas (E); anaplastic astrocytomas (F); anaplastic oligodendrogliomas (G) and glioblastoma multiforme (H) was tested by Kaplan-Meier analysis.
To further evaluate the prognostic quality of 5hmC level, univariate and multivariate Cox regression analyses were performed. Univariate analysis (Table 3) confirmed the association of low 5hmC level with shorter patient survival time (P < 0.001). Additionally, the following variables were also significantly associated with poorer overall survival: aged over 40 at diagnosis (P < 0.001), higher pathological grades (P < 0.001), worse KPS (P = 0.026), larger tumor size (P = 0.014), lack adjuvant treatment after surgery (P = 0.032) and subtotal resection (P = 0.049). Nonsignificant variables included gender (P = 0.068) and tumor locations (P = 0.155). However, when the data were analyzed in the multivariate Cox regression model (Table 3), low 5hmC level was not found to have independent prognostic power (P = 0.720), while higher pathological grades (P < 0.001), lack adjuvant treatment (P = 0.009) and extent of resection (P = 0.017) remained statistically significant. In an alternative multivariate model exclusive of pathological grades, a strong interaction was found between 5hmC level and pathological grades (Table 3).
Table 3. Univariate and multivariate analysis of variables associated with survival in total glioma cases (n = 287).
| Univariate analysis | Multivariate analysis | Alternative multivariate analysis | |||||||
|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value | HR | 95% CI | P-value | |
| Gender | |||||||||
| (male vs female) | 1.373 | 0.977–1.930 | 0.068 | 1.484 | 0.994–2.215 | 0.053 | 1.678 | 1.115–2.526 | 0.013 |
| Age | |||||||||
| >40 vs ≤40) | 1.956 | 1.396–2.742 | <0.001 | 1.035 | 0.675–1.588 | 0.875 | 1.704 | 1.152–2.519 | 0.008 |
| Location | |||||||||
| (supratentorial: other) | 1.536 | 0.851–2.775 | 0.155 | 1.615 | 0.778–3.350 | 0.198 | 1.169 | 0.569–2.401 | 0.671 |
| WHO grade | |||||||||
| (IV vs III. vs II vs I) | 2.641 | 2.141–3.192 | <0.001 | 2.641 | 2.064–3.380 | <0.001 | |||
| KPS score | |||||||||
| (≤80 vs >80) | 1.459 | 1.046–2.036 | 0.026 | 1.192 | 0.823–1.725 | 0.353 | 1.198 | 0.826–1.738 | 0.342 |
| 1Tumor size (cm) | |||||||||
| (>50cm3 vs ≤ 50cm3) | 1.558 | 1.094–2.217 | 0.014 | 1.270 | 0.867–1.859 | 0.220 | 1.446 | 1.006–2.078 | 0.046 |
| Extent of resection | |||||||||
| (subtotal vs total) | 1.424 | 1.000–2.027 | 0.049 | 1.660 | 1.093–2.520 | 0.017 | 1.584 | 1.059–2.369 | 0.025 |
| Adjuvant treatment | |||||||||
| (no vs yes) | 1.472 | 1.034–2.095 | 0.032 | 1.703 | 1.142–2.539 | 0.009 | 1.731 | 1.139–2.630 | 0.010 |
| 5hmC scores | |||||||||
| (0–3 vs 4–9) | 2.588 | 1.816–3.690 | <0.001 | 1.081 | 0.706–1.654 | 0.720 | 1.837 | 1.235–2.733 | 0.003 |
HR, hazard ratio. 1Available data were 260 cases.
5hmC level on patient survival in subtypes of gliomas
Given the significant interaction between pathological grades and 5hmC level, we next analyzed the relations between 5hmC level and the patient survival within subtypes of gliomas. Kaplan-Meier survival analysis revealed low 5hmC scores were strongly associated with unfavorable survival within DA (Fig. 3D, χ2 = 14.788, P < 0.001) but not OG (Fig. 3E, χ2 = 0.997, P = 0.318), AA (Fig. 3F, χ2 = 0.160, P = 0.689), AO (Fig. 3G, χ2 = 0.790, P = 0.374) and GBM (Fig. 3H, χ2 = 0.041, P = 0.840). The median survival time for DA with low 5hmC level was 37.66 (95% CI, 26.66 ∼ 48.67) months, while that with moderate and high 5hmC level was 75.25 (95% CI, 64.90 ∼ 85.62) months.
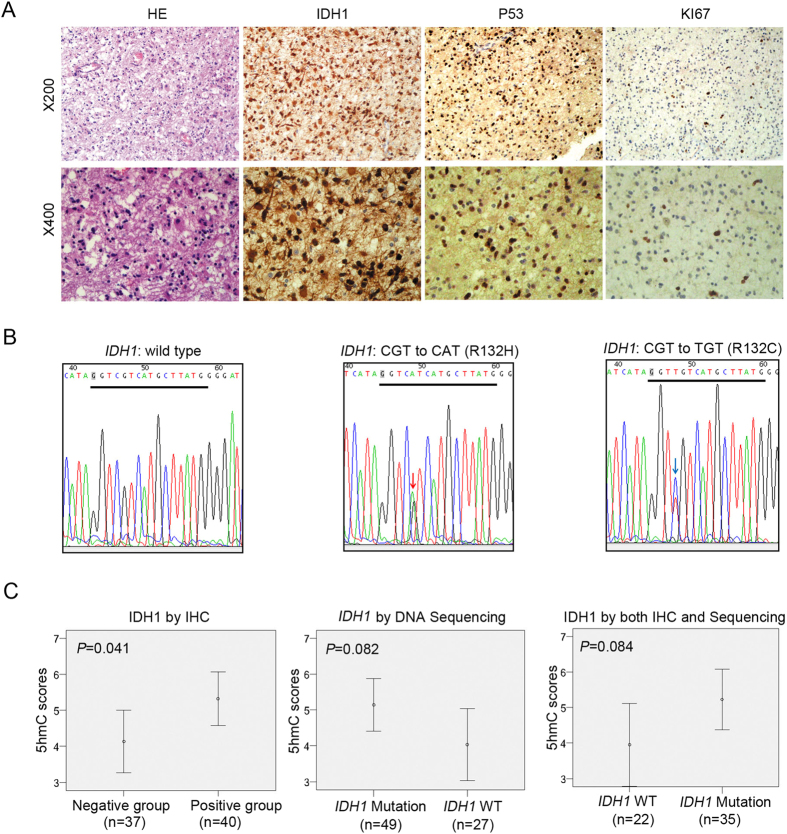
To compare the prognostic value of 5hmC with other well-known prognostic markers within DA, we examined the level of IDH1, p53 and KI67 by immunohistochemistry in these tumors (Fig. 4A). Available data for IDH1, P53 and KI67 were 77, 69 and 71 cases respectively. 52% (40 of 77) of DA cases showed IDH1-R132H positive immunostaining. Immunodetection of P53-positive or KI67-positive cell nuclei ranged up to 80% or 30% respectively. According to the 10% cutoff, P53-positive cases were detected in 46% (32 of 69) of DA patients. With cutoff as 4%, KI67-positive cases were 35% (25 of 71) of DA cases. We also examined the mutation status of IDH1/2 by direct DNA sequencing and detected 64% (49/76) of DA bear heterozygous IDH1 mutations (Fig. 4B). The predominant amino acid sequence alteration in IDH1 was R132H accounting for 98% (48/49) of the detected mutations. Only one case was found to bear IDH1-R132C mutation (Fig. 4B). We did not detect codon 172 of IDH2 mutation at present study (data not shown). Of 70 samples by both DNA sequencing and immunohistochemistry assays, 45 cases were identified as carrying IDH1 mutation by DNA sequencing. However, only 78% (35/45) DA with IDH1 mutation by DNA sequencing was positive for IDH1-R132H antibody. The three cases with positive immunoreactivity for IDH1-R132H antibody did not show IDH1/2 mutation by DNA sequencing.
Figure 4. IDH1 mutation status is not associated with 5hmC reduction.
(A) HE and representative immunostaining images of IDH1, P53 and KI67 in diffuse astrocytomas. 200× (upper), 400× (bottom). (B) Representative IDH1 wild-type (WT), IDH1 (red arrow, R132H) and IDH1 (blue arrow, R132C) mutations were analyzed by Sanger sequencing. (C) Comparison of 5hmC level in DA cases between with IDH1 mutation and wild type by immunohistochemistry (IHC), DNA sequencing, or both IHC and DNA sequencing using the unpaired Student’s t-test. Error bar represents the standard error.
The relations 5hmC level on IDH1 immunostaining signaling, IDH1 mutated status, P53 and KI67 label index and patient clinicopathological features were outlined in Table 4. Chi square analyses confirmed significant association between 5hmC level and vital status (P < 0.001). Additionally, although the significant correlation between 5hmC level and IDH1 mutation was reached (P = 0.036), most cases with low 5hmC scores were detected in the IDH1 wild type cases (61%, 30 of 49). We investigated the relationship between IDH1 mutation status and 5hmC levels. Surprisingly, we detected that 5hmC reduction seem to be associated with IDH1 wild type versus mutation cases (Fig. 4C). Thus, this result was consistent with previous conclusions that 5hmC reduction didn’t result from IDH1 mutation25,29,30.
Table 4. Relationship between the 5hmC level and clinicopathological features of patients with diffuse astrocytomas (WHO grade II).
| Molecular features | Total cases (n = 95) |
||
|---|---|---|---|
| 5hmC (0–3) | 5hmC (4–9) | P | |
| Gender | |||
| Male: Female | 17:19 | 35:24 | 0.292 |
| Age | |||
| ≤36: >36 | 16:20 | 30:29 | 0.673 |
| Location | |||
| Supratentorial: other | 31:5 | 52:7 | 0.761 |
| Vital status | |||
| Alive:death | 10:26 | 43:16 | 0.000 |
| KPS | |||
| ≥80: <80 | 28:8 | 40:19 | 0.353 |
| 1Tumor size (cm) | |||
| <50 cm3: ≥50 cm3 | 19:11 | 25:23 | 0.358 |
| Extent of resection | |||
| Subtotal :Total | 13:23 | 20:39 | 0.828 |
| Adjuvant treatment | |||
| No:Yes | 9:27 | 14:45 | 1.000 |
| 2IDH1 IHC | |||
| Positive: Negative | 10:18 | 30:19 | 0.036 |
| 3P53 label index | |||
| ≤10: >10 | 14:11 | 23:21 | 0.806 |
| 4KI67 label index | |||
| ≤4: >4 | 16:9 | 30:16 | 1.000 |
| 5IDH1 Sequencing | |||
| Mutation: Wild type | 14:11 | 35:16 | 0.315 |
1–5Available data were 78, 77, 69, 71 and 76 cases respectively.
IHC: immunohistochemistry.
To further test the 5hmC prognostic value for DA patients, we performed univariate and multivariate analyses using a Cox proportional hazards model. Consistent with Kaplan-Meier and Chi square analysis, univariate and multivariate analysis verified low 5hmC level was associated with unfavorable survival of DA patients (Table 5, P < 0.001,P = 0.013). In addition, univariate Cox regression analyses also revealed that DA patients with either positive immunoreactivity for IDH1-R132H antibody (P = 0.030) or IDH1 mutation by DNA sequencing (P = 0.001) had a better survival (Table 5). However, multivariate analyses revealed that IDH1 mutation by DNA sequencing (P = 0.021) versus IDH1 positive immunstaining (P = 0.254) had an prognostic value for DA patients (Table 5). In addition, we found that neither P53 nor KI67 harbored prognostic power by uni- or multivariate analysis (Table 5).
Table 5. Univariate and multivariate analysis of variables associated with survival within diffuse astrocytomas (WHO grade II, n = 95).
| Univariate analysis |
Multivariate analysis |
|||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value | |
| Gender | ||||||
| (male vs female) | 1.449 | 0.790–2. 660 | 0.231 | 1.538 | 0.644–3.675 | 0.333 |
| Age | ||||||
| (> 40 vs ≤40) | 1.103 | 0.601–2.028 | 0.751 | 1.358 | 0.555–3.323 | 0.503 |
| Location | ||||||
| (supratentorial: other) | 1.155 | 0.486–2.784 | 0.744 | 1.189 | 0.345–4.094 | 0.784 |
| KPS score | ||||||
| (≤80 vs >80) | 1.545 | 0.820–2.910 | 0.179 | 2.084 | 0.719–6.046 | 0.176 |
| Adjuvant treatment | ||||||
| (no vs yes) | 1.369 | 0.701–2.675 | 0.358 | 1.425 | 0.576–3.525 | 0.444 |
| Extent of resection | ||||||
| (subtotal vs total) | 1.232 | 0.640–2.370 | 0.533 | 1.225 | 0.475–3.164 | 0.674 |
| 1Tumor size (cm) | ||||||
| (>50 cm3 vs ≤50 cm3) | 1.490 | 0.781–2.843 | 0.226 | 1.709 | 0.549–5.325 | 0.355 |
| 5hmC score | ||||||
| (0–3 vs 4–9) | 3.822 | 2.038–7.168 | 0.000 | 3.343 | 1.296–8.622 | 0.013 |
| 2IDH1 IHC | ||||||
| (Negative:Positive) | 2.161 | 1.079–4.326 | 0.030 | 2.158 | 0.575–8.094 | 0.254 |
| 3P53 labeling index | ||||||
| (High:Low) | 1.175 | 0.590–2.342 | 0.646 | 1.040 | 0.407–2.660 | 0.934 |
| 4KI67 labeling index | ||||||
| (High:Low) | 1.532 | 0.778–3.017 | 0.217 | 1.527 | 0.522–2.458 | 0.439 |
| 5IDH1 Sequencing | ||||||
| (WT: Mutation) | 3.066 | 1.537–6.116 | 0.001 | 5.630 | 1.304–24.302 | 0.021 |
1–5Available data were 78, 77, 69, 71 and 76 cases respectively. HR, hazard ratio; IHC: immunohistochemistry; WT: wild type.
Molecular markers in combination for prognostic relevance within DA
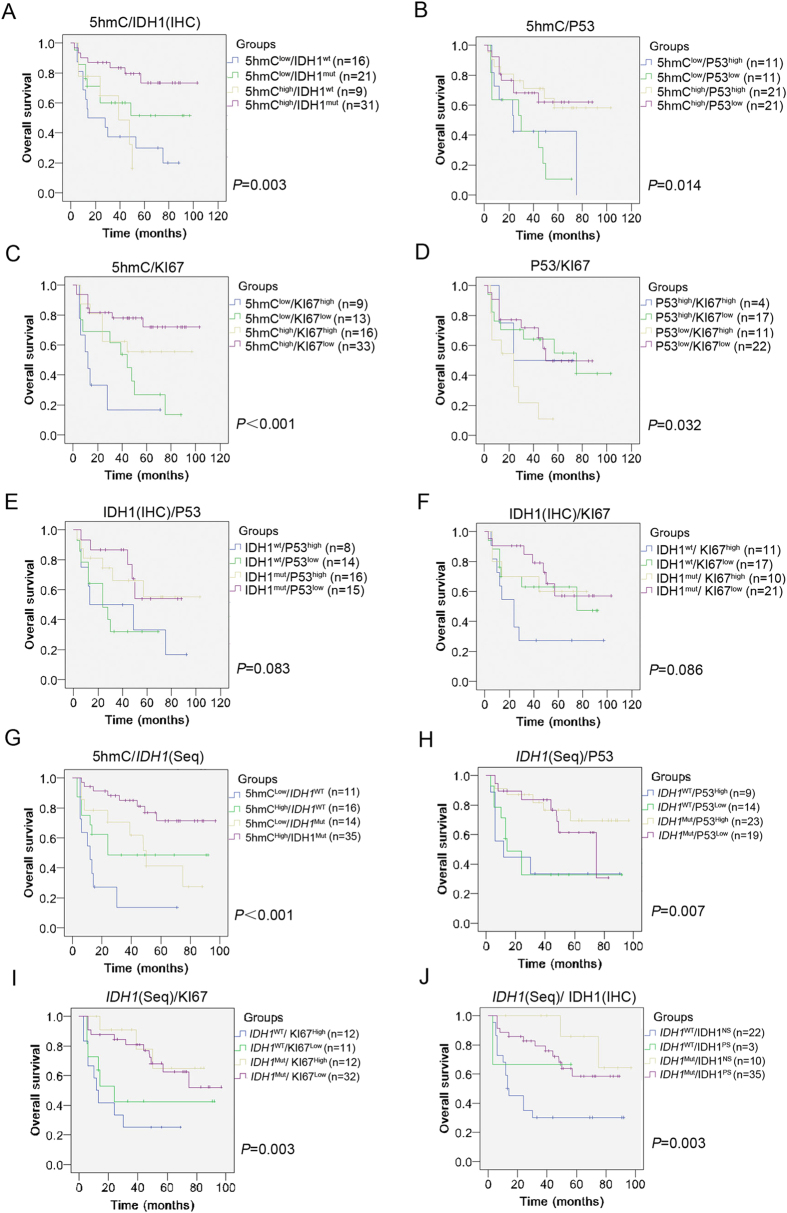
To test whether combination of 5hmC, IDH1, P53 or KI67 can be applied for assessment of prognosis and restratification of DA, we stochastically divided patients into subgroups with two molecular combinations. Kaplan-Meier survival analysis revealed combinations of 5hmC/IDH1 (IHC)(Fig. 5A, X2 = 13.661, P = 0.003), 5hmC/P53 (Fig. 5B, X2 = 10.616, P = 0.014), 5hmC/KI67 (Fig. 5C, X2 = 17.717, P = 0.000), P53/KI67 (Fig. 5D, X2 = 8.776, P = 0.032), 5hmC/IDH1 (Seq) (Fig. 5G, X2 = 14.312, P = 0.000), IDH1 (Seq)/P53 (Fig. 5H, X2 = 11.113, P = 0.007), IDH1 (Seq)/KI67 (Fig. 5I, X2 = 12.989, P = 0.003) and IDH1 (IHC)IDH1 (Seq) (Fig. 5J, X2 = 11.412, P = 0.003) were associated with OS. Nonsignificant combinations included IDH1 (IHC)/P53 (Fig. 5E, X2 = 6.667, P = 0.083) and IDH1 (IHC)/KI67 (Fig. 5F, X2 = 6.582, P = 0.086). Surprisingly, the group for best assessment of prognosis and restratification of DA was 5hmC/KI67 combinations (Table 6). Within this group, the mean OS for 5hmClow/KI67high, 5hmClow/KI67low, 5hmChigh/KI67high and 5hmChigh/KI67low groups were 22 (95% CI, 6–38), 42 (95% CI, 26–58), 63 (95% CI, 44–82) and 80 (95% CI, 67–94) months respectively. In contrast, the mean OS for 5hmClow/IDH1NS, 5hmClow/IDH1PS, 5hmChigh/IDH1NS and 5hmChigh/IDH1PS groups were 37 (95% CI, 21–53), 59 (95% CI, 41–77), 35 (95% CI, 22–48) and 83 (95% CI, 69–96) months respectively. Even for 5hmC/IDH1 (Seq) combinations, the mean OS for 5hmCLow/IDH1WT, 5hmCLow/IDH1Mut, 5hmCHigh/IDH1WT and 5hmCHigh/IDH1Mut groups were 20 (95% CI, 6–33), 51 (95% CI, 34–68), 51 (95% CI, 30–70) and 79 (95% CI, 67–89) months respectively. Therefore, the combination of 5hmC/KI67 was a useful prognostic marker for restratification of DA.
Figure 5. Overall survival by molecular markers in combination within diffuse astrocytomas.
(A–J) Relevance between patient survival and subgroups of 5hmC/IDH1 (IHC) (A), 5hmC/P53 (B), 5hmC/KI67 (C), P53/KI67 (D), IDH1 (IHC)/P53 (E), IDH1 (IHC)/KI67 (F), 5hmC/IDH1 (Seq) (G), IDH1 (Seq)/P53 (H), IDH1 (Seq)/KI67 (I) and IDH1 (Seq)/IDH1 (IHC) (J) was tested by Kaplan-Meier analysis. IHC: immunohistochemistry. Seq: sequencing.
Table 6. Molecular markers in combination for prognostic relevance within diffuse astrocytomas (WHO grade II).
| Groups | Subgroups | Mean time (months) of OS (95% CI) | P-value |
|---|---|---|---|
| 5hmC/IDH1 (IHC) | 5hmCLow/IDH1NS: 5hmCLow/IDH1PS: | 37 (95% CI, 21–53): 59 (95% CI, 41–77): | 0.003 |
| 5hmCHigh/IDH1NS: 5hmCHigh/IDH1PS | 35 (95% CI, 22–48): 83 (95% CI, 69–96) | ||
| 5hmC/P53 | 5hmCLow/P53High: 5hmCLow/P53Low: | 40 (95% CI, 19–60): 31 (95% CI, 17–44): | 0.014 |
| 5hmCHigh/P53High: 5hmCHigh/P53Low | 71 (95% CI, 53–87): 62 (95% CI, 48–76) | ||
| 5hmC/KI67 | 5hmCLow/KI67High: 5hmCLow/KI67Low: | 22 (95% CI, 6–38): 42 (95% CI, 26–58): | 0.000 |
| 5hmCHigh/KI67High: 5hmCHigh/KI67Low | 63 (95% CI, 44–82): 80 (95% CI, 67–94) | ||
| P53/KI67 | P53High/KI67High: P53High/KI67Low: | 45 (95% CI, 18–72): 62 (95% CI, 42–83): | 0.032 |
| P53Low/KI67High: P53Low/KI67Low | 22 (95% CI, 12–33): 60 (95% CI, 43–73) | ||
| IDH1 (IHC)/P53 | IDH1NS/P53High: IDH1NS/P53Low | 41 (95% CI, 16–64): 33 (95% CI, 20–47): | 0.083 |
| IDH1PS/P53High: IDH1PS/P53Low | 68 (95% CI, 47–90): 64 (95% CI, 48–81) | ||
| IDH1 (IHC)/KI67 | IDH1NS/KI67High: IDH1NS/KI67Low | 38 (95% CI, 16–60): 60 (95% CI, 42–79): | 0.086 |
| IDH1PS/KI67High: IDH1PS/KI67Low | 57 (95% CI, 36–78): 75 (95% CI, 58–91) | ||
| 5hmC/IDH1 (Seq) | 5hmCLow/IDH1WT: 5hmCLow/IDH1Mut: | 20 (95% CI, 6–33): 51 (95% CI, 34–68): | 0.000 |
| 5hmCHigh/IDH1WT: 5hmCHigh/IDH1Mut | 51 (95% CI, 30–70): 79 (95% CI, 67–89) | ||
| IDH1 (Seq)/P53 | IDH1WT/P53High: IDH1WT/P53Low | 37 (95% CI, 12–62): 38 (95% CI, 18–59): | 0.007 |
| IDH1Mut/P53High: IDH1Mut/P53Low | 76 (95% CI, 61–90): 61 (95% CI, 48–73) | ||
| IDH1 (Seq)/KI67 | IDH1WT/KI67High: IDH1WT/KI67Low | 26 (95% CI, 11–40): 45 (95% CI, 21–70): | 0.003 |
| IDH1Mut/KI67High: IDH1Mut/KI67Low | 68 (95% CI, 51–84): 70 (95% CI, 57–82) | ||
| IDH1 (Seq)/IDH1 (IHC) | IDH1WT/IDH1NS: IDH1WT/IDH1PS | 36 (95% CI, 20–52): 38 (95% CI, 10–66): | 0.003 |
| IDH1Mut/IDH1NS: IDH1Mut/IDH1PS | 85 (95% CI, 71–99): 65 (95% CI, 54–76) |
OS, overall survival. IHC: immunohistochemistry; NS: negative staining; PS: positive staining; Mut: Mutation; WT: wild type; Seq: sequencing.
Discussion
The majority of the DA exhibits a relatively good prognosis. However, some DA show unexpectedly aggressive clinical course leading to early patient death10. This creates a diagnostic dilemma for routine histopathology. The heterogeneous clinical courses of DA may be associated with different epigenetic and genetic abnormalities33. Therefore, molecular markers would be useful for the accurate restratification of these tumors and provide help for prognostication or therapeutic decision making. However, currently, available molecular markers are limited.
Epigenetic modifications play crucial roles in normal development and frequently alter during carcinogenesis33. 5hmC was a new detected DNA modification and the knowledge about its roles in gliomas remained limited. With an identified homemade anti-5hmC antibody, we detected that 5hmC variably reduced in most gliomas and the reduction was closely associated with higher pathological grades. These findings were consistent with previous reports24,25,28,29,30. In clinical practice, it still remains a challenge for pathologists to histologically define AA and DA based on morphological features of anaplasia, such as mitotic activity and microvascular proliferation, for poor interobserver agreement34. IDH1/2 mutation has been accepted as a favorable prognostic biomarker for gliomas5. However, IDH1/2 mutated status can’t be used to differentiate AA from DA because both tumors have high rates of IDH1/2 mutation. In the present study, we detected 5hmC level decreased more severely in grades III/IV versus I/II gliomas (Fig. 2B). This means 5hmC loss may be associated with anaplastic progression and can be used to stratify low-grade gliomas (I/II) from anaplastic gliomas (III/IV). In addition, we also detected that 5hmC dramatically reduced in AA and GBM compared to DA (Fig. 2C). Therefore, 5hmC loss may be a usefully marker for differentiating AA from DA.
Apart from the finding that 5hmC loss was a marker for anaplastic progression of gliomas, we also detected that 5hmC reduction was closely associated with shortened survival of glioma patients. However, in multivariate analysis, 5hmC had no independent prognostic value in the entire patient cohort. Alterative multivariate model found that pathological grade was the major interacting factor (Table 3). Further analysis within subtypes of gliomas revealed that 5hmC was still a prognostic marker confined to diffuse astrocytomas WHO II (Fig. 3D, Table 5). Inconsistent with Orr et al. report29, we didn’t find prognostic relevance between 5hmC level and AA or GBM (Fig. 3F, H). The discrepancy may be associated with sample size (only 12 cases of AA with prognostic data in Orr et al. study), evaluation of 5hmC level methods, patient races or choice of treatment. Although Orr et al. had 52 adult GBM for evaluating prognostic relevance with 5hmC level, the short survival time (mean 15 months) and most cases without detected 5hmC may limited the 5hmC prognostic value within GBM. In addition, only 10 cases with prognostic data in Orr’s study may account for their failed detection of the relevance between 5hmC level and DA.
Many studies have confirmed an association of IDH1 mutations with favorable outcome for patients with malignant gliomas (WHO III/IV)7,35,36,37. However, prognostic value of IDH1 mutations for low-grade gliomas (LGG, WHO II), especial DA, was subject to debate11,14,17,38,39 (supplementary Table S1 and S2). For example, in a largest series IDH1 was of no prognostic value for 360 patients suffering from LGG, in which 186 was oligodendroglial tumors and 174 astrocytic tumors17. A recent Cancer Genome Atlas Research showed IDH mutations had significant survival prediction in lower grade gliomas (WHO II and III), including oligodendroglioma, oligoastrocytoma and astrocytoma. However, the report didn’t particularly analyze prognostic value of IDH1 mutations within DA (WHO II)38. Ahmadi et al. performed IDH1/2 mutation assays in a series of 100 DAs patients and detected no survival benefit of IDH1 mutations in these patients11. However, by immunohistochemistry, two groups recently independently found positive IDH1 staining still hold prognostic relevance with DA patients14,39. In the present study, both univariate and multivariate Cox regression analyses confirmed that IDH1 mutation by DNA sequencing was associated with better survival for DA patients (Table 5). Therefore, our results suggested that IDH1 mutation was still a favorable prognostic marker for DA.
Currently, IDH1, TP53 and KI67 are routinely used molecular markers for assisting prognostic decision5,10,40,41,42 (supplementary Table S1). To test the prognostic value of combination of 5hmC, IDH1, P53 or KI67, we stochastically divided patients into subgrokups with two molecular combinations in DA. It is interesting that the combination of 5hmC/KI67 harbored the best value for assessment of prognosis and restratification of DA. (Fig. 5, Table 6). The reasons for 5hmC/KI67 but not other combinations in best predicting DA patient survival are unknown. One possible reason is that 5hmC/KI67 is a combination of cell differentiation (5hmC) and proliferation (KI67) markers. Several lines demonstrated that differentiated cells instead of stem cells harbor high 5hmC level24,29. In addition, an inverse relationship between 5hmC levels and cell proliferation was detected in proliferating or differentiated cells25. Functionally, double-knockout of Tet1 and Tet2 resulted in reduced 5hmC level and delayed brain development43. Therefore, the complementary combination of 5hmC and KI67 may be reason for their best assessment of prognosis and restratification of DA. It is interesting that the emerging data for superior prognosis of ATRX/IDH co-mutant diffuse astrocytomas was recently addressed in some reports38. Further work needs to compare prognostic value between 5hmC/KI67 and ATRX/IDH within DA.
Previously, some reports suggested IDH1 mutations might account for 5hmC reduction in gliomas by means of the presumed role of 2-hydroxyglutarate as an inhibitor of TET oxidases26. However, this suggestion was challenged by other observations that 5hmC reduction not associated with IDH1 mutations25,29,30. In the present study, we detected a higher versus lower level of 5hmC in IDH1 mutated DA compared to IDH1 wild type tumors (Fig. 4C). Therefore, our results didn’t support 5hmC reduction was associated with IDH1 mutations. This conclusion was further supported by the observation that 5hmC loss was associated with poorer prognosis (Fig. 3A,B,D), while IDH1 mutations correlated with better survival in DA (Table 5). Since 5hmC can be converted from 5mC by TET enzymes20,21, 5hmC loss may be associated with 5mC reduction in malignancies. However, Kraus et al. observed that 5hmC level was unrelated to 5mC values by isotope-based liquid chromatography mass spectrometry assays28. Therefore, 5hmC decrease in cancer cells may primarily result from the alterations of TET genes. Indeed, one of TET family genes, TET2, was detected with high mutation rates in some hematologic malignancies, which simultaneously had aberrant levels of 5hmC in their genomes44,45. Some other evidence showed that 5hmC loss was associated with decreased expression or nuclear exclusion of TETs proteins29,30. Much more work need to confirm the relations between TETs alteration and 5hmC reduction in gliomas.
Summarizing, our data suggested that the 5hmC level, IDH1 mutation and 5hmC/KI67 combination harbor the value for assessment of prognosis of DA. Some limitations existed in this study. These data are derived from an unselected single-center collective. The sample size for each entity was not large, the design was in retrospective, and choice of treatment was not standardized. Thus, the prognostic value of 5hmC level, IDH1 mutation and 5hmC/KI67 combination in DA needs further verify.
Methods
Sample collection and clinical follow-up
Samples collection and analysis were approved by the ethics committee of the Xijing Hospital, Fourth Military Medical University, Xi’an, P. R. China. Written informed consent was obtained from all of the patients. All specimens were handled and made anonymous according to the ethical and legal standards. To collect samples, patients who died of diseases not directly related to gliomas were excluded. After that, a total of 287 glioma samples with 2–10 years follow-up information were collected from Xijing Hospital, Fourth Military Medical University between 2003 and 2013. Patients’ clinicopathological features such as age at diagnosis, gender, tumor size, Karnofsky performance status (KPS) score, extent of resection and adjuvant treatment were collected. Three pathologists independently reviewed all histological slides and graded each glioma according to 2007 World Health Organization criteria. In case of a discrepancy, the 3 observers simultaneously reviewed the slides to achieve a consensus. All methods were carried out in accordance with the approved guidelines of Fourth Military Medical University.
Production and examination of anti-5hmC antibody
To examine 5hmC level in glioma tissues, we produced polyclonal rabbit anti-5hmC antibody. The specificity of the anti-5hmC antibody was examined by dot blot, western blot, immunoprecipitation and immunofluorescence. In brief, each nucleoside was conjugated to ovalbumin and quantified by mass spectrometrical analysis. Equal conjugated bases were spotted onto nitrocellulose filter membrane and reacted with the 5hmC (1:1000) or 5mC (1:500, Calbiochem) antibody. For western blot, 5mC or 5hmC conjugated with ovalbumin was mixed with 293T cell lysates and separated by SDS-PAGE and immunoblotted. For immunoprecipitation assay, a 367-bp DNA fragment spanning 4605333-4605699 in mouse chromosome 10 was PCR-amplified with 5mC or 5hmC in place of cytodine. 8.7 ng PCR products was mixed with 5μg sonicated genomic DNA of 293T cells. Then 2.5 μg of 5hmC antibody was added to the denatured DNA for precipitation. IP-DNA was extracted for quantitative reverse transcriptase PCR or regular PCR. To test whether the antibody could be used for immunofluorescence analysis, we transfected cDNA encoding the Tet1 catalytic domain into 293T cells by mean of that Tet1 can convert 5mC to 5hmC. Subsequently, immunofluorescence was performed with fluorescence-conjugated secondary antibody (1:500; Santa Cruz) as described.
Immunohistochemistry (IHC) analysis
Immunohistochemistry for 5hmC was performed in tissue sections of glioma samples as described previously32. In brief, Paraffin embedded tissue blocks were cut to 4μm sections and deparaffinized and rehydrated using xylene and ethanol; 3% H2O2 in phosphate buffered saline (PBS) was used to inactivate the endogenous peroxidase. DNA was denatured by immersing sections in 2 N HCl for 30 minutes then neutralizing in boric acid buffer for 10 minutes at room temperature. The slides were blocked with goat serum to reduce nonspecific binding and then incubated with primary 5hmC (1:1000 dilution) antibody overnight at 4 °C. On the following day, the peroxidase-conjugated secondary antibody (1:500 dilution; Santa Cruz) was incubated for 1 hour at room temperature. Diaminobenzidine (DAB) substrate was used for detection and hematoxylin was used for counterstaining. The samples were then mounted for visualization. The cells with brown nuclei were considered positively stained. The level of 5hmC staining was accessed independently by 3 pathologists.
Immunohistochemistries for IDH1-R132H (H09, Dianova, Hamburg, Germany; dilution 1:100), P53 (DO-7, Dako, Carpinteria,CA, USA; dilution 1:100) and KI67 (MIB-1, Dako, Glostrup, Denmark; dilution 1:50) were performed as described above exclusive the step of DNA denature by HCl. Each slide stained for IDH1-R132H, P53 and KI-67 was individually reviewed and scored by 3 independent observers. Microscopic areas with highest labeling intensity were chosen for calculation. The p53 or KI-67 labeling index (LI) was defined as the percentage of immunoreactive tumor cell nuclei. In each case either at least 1000 tumor cell nuclei were counted were examined. For statistical analysis, cutoff value of LI for P53 and KI67 was 10% and 4% respectively according to previous reports41,42,46.
PCR amplification and genes sequencing
For IDH1 and IDH2 mutations assays, DNA extraction from formalin fixed paraffin embedded tissue was used. Tumor content of at least 80% was histologically determined for each sample used for DNA extraction. Nucleic acid extraction was performed by standard procedures. 150 ng of genomic DNA was used for PCR amplification in a total volume of 50 μl. The primer sequences for IDH1 and IDH2 and PCR amplification conditions were described previously47. IDH1 codon 132 and IDH2 codon 172 were analyzed by direct sequencing. If results were ambiguous the IDH1 or IDH2 were amplified by use of a different set of primers as described previously47.
Statistical Analysis
We used SPSS 20.0 (SPSS Inc. Chicago, IL, USA) for the statistical analysis. The associations between 5hmC level and clinicopathological features were compared by using a Chi square test or Fisher’s exact test. The nonparametric Spearman correlation was used to analyze the relationship between pathological grades of glioma and 5hmC levels. Overall survival (OS) was defined as the interval between the date of diagnosis and the date of death or the last known follow-up. OS data were censored at the date of last follow-up, if the patient was still alive. Kaplan-Meier survival analysis was used to present the relationship between patient survival and 5hmC levels. Survival differences were analyzed by the log-rank test. Univariate and Multivariate analyses were performed using a Cox proportional hazards model to identify independent prognostic factors. P-values were all 2-sided and used as significance threshold less than 0.05.
Additional Information
How to cite this article: Zhang, F. et al. 5-hydroxymethylcytosine loss is associated with poor prognosis for patients with WHO grade II diffuse astrocytomas. Sci. Rep. 6, 20882; doi: 10.1038/srep20882 (2016).
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81572631 and 31000559 to FZ; 81170798 and 30671087 to QL; 81572469 to WL) and the National Natural Science Foundation of China (91219304, to CDC), National Basic Research Program of China (2010CB529705, 2011CB510103, 2014CB943100 to CDC), and the Council of Shanghai Municipal Government for Science and Technology (to CDC).
Footnotes
Author Contributions F.Z. and C.D.C. designed the project. F.Z., W.L., Z.W.Z., J.L., X.L., Y.Q.X., X.F., X.M.Z., M.Z. and Z.K.Z. collected the tissue samples, clinical data. F.Z., Y.M.W. and L.Y.Z. performed and evaluated the IHC analysis. Y.F.L. produced and identified the anti-5hmC antibody. F.Z., Y.W., J. Z. and J.Y. participated in the evaluation of the IHC data. Q.G.Y., Z.W. and Q.L. helped analyze the data. F.Z. wrote the manuscript. All authors provided editorial input. All authors read and approved the final manuscript.
References
- Louis D. N. et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 114, 97–109 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgaki H. & Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol 109, 93–108 (2005). [DOI] [PubMed] [Google Scholar]
- van den Bent M. J. Interobserver variation of the histopathological diagnosis in clinical trials on glioma: a clinician’s perspective. Acta Neuropathol 120, 297–304 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kros J. M. Grading of gliomas: the road from eminence to evidence. J Neuropathol Exp Neurol 70, 101–109 (2011). [DOI] [PubMed] [Google Scholar]
- Louis D. N. et al. International Society Of Neuropathology–Haarlem consensus guidelines for nervous system tumor classification and grading. Brain Pathol 24, 429–435 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahm F. et al. Farewell to oligoastrocytoma: in situ molecular genetics favor classification as either oligodendroglioma or astrocytoma. Acta Neuropathol 128, 551–559 (2014). [DOI] [PubMed] [Google Scholar]
- Sanson M. et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol 27, 4150–4154 (2009). [DOI] [PubMed] [Google Scholar]
- Nobusawa S., Watanabe T., Kleihues P. & Ohgaki H. IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin Cancer Res 15, 6002–6007 (2009). [DOI] [PubMed] [Google Scholar]
- Wesseling P., van den Bent M. & Perry A. Oligodendroglioma: pathology, molecular mechanisms and markers. Acta Neuropathol 129, 809–827 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura K., Narita Y. & Hawkins C. E. Diffusely infiltrating astrocytomas: pathology, molecular mechanisms and markers. Acta Neuropathol 129, 789–808 (2015). [DOI] [PubMed] [Google Scholar]
- Ahmadi R. et al. No prognostic value of IDH1 mutations in a series of 100 WHO grade II astrocytomas. J Neurooncol 109, 15–22 (2012). [DOI] [PubMed] [Google Scholar]
- Thon N. et al. IDH1 mutations in grade II astrocytomas are associated with unfavorable progression-free survival and prolonged postrecurrence survival. Cancer 118, 452–460 (2012). [DOI] [PubMed] [Google Scholar]
- Ogura R. et al. Immunohistochemical profiles of IDH1, MGMT and P53: practical significance for prognostication of patients with diffuse gliomas. Neuropathology 35, 324–335 (2015). [DOI] [PubMed] [Google Scholar]
- Waqar M. et al. Prognostic Factors in Lobar World Health Organization Grade II Astrocytomas. World Neurosurg 84, 154–162 (2015). [DOI] [PubMed] [Google Scholar]
- Li M. Y. et al. Isocitrate dehydrogenase 1 Gene Mutation Is Associated with Prognosis in Clinical Low-Grade Gliomas. PLoS One 10, e0130872 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukasa A. et al. Significance of IDH mutations varies with tumor histology, grade, and genetics in Japanese glioma patients. Cancer Sci 103, 587–592 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. H. et al. Molecular classification of low-grade diffuse gliomas. Am J Pathol 177, 2708–2714 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stander M., Peraud A., Leroch B. & Kreth F. W. Prognostic impact of TP53 mutation status for adult patients with supratentorial World Health Organization Grade II astrocytoma or oligoastrocytoma: a long-term analysis. Cancer 101, 1028–1035 (2004). [DOI] [PubMed] [Google Scholar]
- Baylin S. B. & Jones P. A. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer 11, 726–734 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S. & Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Globisch D. et al. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One 5, e15367 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Zang L., Shu Q. & Li X. From development to diseases: the role of 5hmC in brain. Genomics 104, 347–351 (2014). [DOI] [PubMed] [Google Scholar]
- Haffner M. C. et al. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget 2, 627–637 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S. G. et al. 5-Hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res 71, 7360–7365 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W. et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell 19, 17–30 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson A. R. et al. Loss of 5-hydroxymethylcytosine correlates with increasing morphologic dysplasia in melanocytic tumors. Mod Pathol 27, 936–944 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus T. F. et al. Low values of 5-hydroxymethylcytosine (5hmC), the “sixth base,” are associated with anaplasia in human brain tumors. Int J Cancer 131, 1577–1590 (2012). [DOI] [PubMed] [Google Scholar]
- Orr B. A., Haffner M. C., Nelson W. G., Yegnasubramanian S. & Eberhart C. G. Decreased 5-hydroxymethylcytosine is associated with neural progenitor phenotype in normal brain and shorter survival in malignant glioma. PLoS One 7, e41036 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller T. et al. Nuclear exclusion of TET1 is associated with loss of 5-hydroxymethylcytosine in IDH1 wild-type gliomas. Am J Pathol 181, 675–683 (2012). [DOI] [PubMed] [Google Scholar]
- Kraus T. F. et al. Loss of 5-hydroxymethylcytosine and intratumoral heterogeneity as an epigenomic hallmark of glioblastoma. Tumour Biol 36, 8439–8446 (2015). [DOI] [PubMed] [Google Scholar]
- Yang B. et al. Evaluation of global DNA hypomethylation in human prostate cancer and prostatic intraepithelial neoplasm tissues by immunohistochemistry. Urol Oncol 31, 628–634 (2013). [DOI] [PubMed] [Google Scholar]
- You J. S. & Jones P. A. Cancer genetics and epigenetics: two sides of the same coin ? Cancer Cell 22, 9–20 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olar A. et al. IDH mutation status and role of WHO grade and mitotic index in overall survival in grade II-III diffuse gliomas. Acta Neuropathol 129, 585–596 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H. et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med 360, 765–773 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller M. et al. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma Network. J Clin Oncol 27, 5743–5750 (2009). [DOI] [PubMed] [Google Scholar]
- Hartmann C. et al. Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1-mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: implications for classification of gliomas. Acta Neuropathol 120, 707–718 (2010). [DOI] [PubMed] [Google Scholar]
- Brat D. J. et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med 372, 2481–2498 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwadate Y. et al. IDH1 mutation is prognostic for diffuse astrocytoma but not low-grade oligodendrogliomas in patients not treated with early radiotherapy. J Neurooncol 124, 493–500 (2015). [DOI] [PubMed] [Google Scholar]
- Johannessen A. L. & Torp S. H. The clinical value of Ki-67/MIB-1 labeling index in human astrocytomas. Pathol Oncol Res 12, 143–147 (2006). [DOI] [PubMed] [Google Scholar]
- Takami H. et al. Revisiting TP53 Mutations and Immunohistochemistry–A Comparative Study in 157 Diffuse Gliomas. Brain Pathol 25, 256–265 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet E. et al. TP53 and p53 statuses and their clinical impact in diffuse low grade gliomas. J Neurooncol 118, 131–139 (2014). [DOI] [PubMed] [Google Scholar]
- Dawlaty M. M. et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev Cell 24, 310–323 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M. et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 468, 839–843 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefferi A. et al. Detection of mutant TET2 in myeloid malignancies other than myeloproliferative neoplasms: CMML, MDS, MDS/MPN and AML. Leukemia 23, 1343–1345 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind-Landstrom T., Habberstad A. H., Sundstrom S. & Torp S. H. Prognostic value of histological features in diffuse astrocytomas WHO grade II. Int J Clin Exp Pathol 5, 152–158 (2012). [PMC free article] [PubMed] [Google Scholar]
- Hartmann C. et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol 118, 469–474 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.