Abstract
Sciadopitys verticillata is an evergreen conifer and an economically valuable tree used in construction, which is the only member of the family Sciadopityaceae. Acquisition of the S. verticillata chloroplast (cp) genome will be useful for understanding the evolutionary mechanism of conifers and phylogenetic relationships among gymnosperm. In this study, we have first reported the complete chloroplast genome of S. verticillata. The total genome is 138,284 bp in length, consisting of 118 unique genes. The S. verticillata cp genome has lost one copy of the canonical inverted repeats and shown distinctive genomic structure comparing with other cupressophytes. Fifty-three simple sequence repeat loci and 18 forward tandem repeats were identified in the S. verticillata cp genome. According to the rearrangement of cupressophyte cp genome, we proposed one mechanism for the formation of inverted repeat: tandem repeat occured first, then rearrangement divided the tandem repeat into inverted repeats located at different regions. Phylogenetic estimates inferred from 59-gene sequences and cpDNA organizations have both shown that S. verticillata was sister to the clade consisting of Cupressaceae, Taxaceae, and Cephalotaxaceae. Moreover, accD gene was found to be lost in the S. verticillata cp genome, and a nucleus copy was identified from two transcriptome data.
Generally, seed plant chloroplast (cp) DNAs present a conserved quadripartite structure, with a pair of inverted repeats (IR) separating the whole genome into a large single copy (LSC) and small single copy region (SSC)1. The most significant feature of coniferous cp genomes is that one copy of the canonical rRNA-containing IR has been lost. However, evolution has endowed conifers cp genomes with short novel repeats2,3,4,5. These repeats can replace the function of canonical IR to promote homologous recombination (HR), thus generating isomeric genomic forms3,5,6, such as type 1 and 3 repeats in Pinaceae3 and trnQ repeats in Cephalotaxus oliveri5 and Juniperus6. There are no common short inverted repeats (sIRs) that are larger than 100 bp found within cupressophytes per se. Nevertheless, Cupressaceae, Taxaceae, and Cephalotaxaceae share a common sIR embracing trnQ-UUG (optional expanding to 5′ end of chlB in the genus Cephalotaxus), ranging from 230 bp in Taxus mairei to 544 bp in C. oliveri. In C. oliveri, the trnQ-UUG sIR was inferred to mediate HR, and two isomeric cpDNA forms were detected5. In Taiwania cryptomerioides and Cryptomeria japonica, the trnQ-UUG sIR is only approximately 280 bp, but not any alternative form was detected5. By contrast, in four Juniperus species, the substoichiometric presence of the rearranged isomeric form caused by the trnQ-UUG sIR was identified by using both PCR and high-throughput read-pair mapping method6. Comparative genomic studies demonstrated that the predominant and substoichiometric arrangements of the sIR have shifted several times during the cupressophyte evolution6. Considering the absence of the sIR in Araucariaceae and Podocarpaceae and its presence in other cupressophytes, the sIR was deduced to originate in the common ancestor of Cupressaceae, Cephalotaxaceae, and Taxaceae6. However, the underlying evolutionary mechanisms of the formation of sIR have not been well studied. More cupressophyte cpDNAs are needed to dissect the detailed pathway. To date, the cp genomic sequences of cupressophytes have been reported for 26 species from five families, i.e. Taxaceae7, Cephalotaxaceae5,8, Cupressaceae9, Araucariaceae10, and Podocarpaceae11. Thus Sciadopityaceae becomes the only one family in cupressophyte whose chloroplast genome has not been reported. In this study, we have sequenced the entire cpDNA of Sciadopitys verticillata and performed a comparative analysis of the cupressophyte cp genomes.
Currently, it is still controversial about the phylogenetic position of Sciadopityaceae. Most of the molecular phylogenetic studies supported that Sciadopityaceae is phylogenetically isolated of all conifers, branching off firstly from the Cupressaceae-Taxaceae-Cephalotaxaceae clade12,13. Few studies also insisted that Sciadopityaceae is sister to a clade comprising Podocarpaceae and Araucariaceae14. Chloroplast genome information can serve as useful markers for resolving phylogenetic relationships between Sciadopityaceae and other cupressophytes.
Chloroplast is considered to originate from cyanobacteria through ancient endosymbiosis15. In this process, many nonessential genes were lost, and some functional genes were relocated from chloroplast to the nuclear genome16. Four genes have been reported functionally transferred from the plastid to the nucleus: infA in multiple lineages, including most rosids17; rpl22 in Fabaceae and Fagaceae18,19; rpl32 in Rhizophoraceae and Salicaceae (two families of Malpighiales) and Thalictroideae20,21,22 and accD in Campanulaceae and Fabaceae23,24. Moreover, independent loss of accD has also been documented in the cp genomes of Acoraceae25, Poaceae26,27, Geraniaceae28, and Gnetales29. In Poaceae, the prokaryotic type acetyl-CoA carboxylase (ACCase) in the plastid has been completely replaced by a nucleus-encoded eukaryotic type ancestry30. However, it remains unknown whether the accD loss has occurred in the cp genome of other gymnosperms beside Gnetales. As Sciadopityaceae is the only one family whose cp genome has not been determined in gymnosperms, sequencing the cp genome of Sciadopitys verticillata will also provide useful information on this issue.
To further understand the evolution of cupressophyte cp genomes and elucidate the phylogenetic relationships among gymnosperms, we have 1) determined the complete cp genome of S. verticillata; 2) compared the overall gene content and organization of S. verticillata cp genome with those of other cupressophytes, detailing gene loss and its substitution mechanism; 3) detected repeat sequences in S. verticillata and further investigated the evolutionary mechanism of sIR in cupressophytes; and 4) examined the phylogenetic position of Sciadopityaceae among cupressophytes.
Results
General features of S. verticillata cp genome
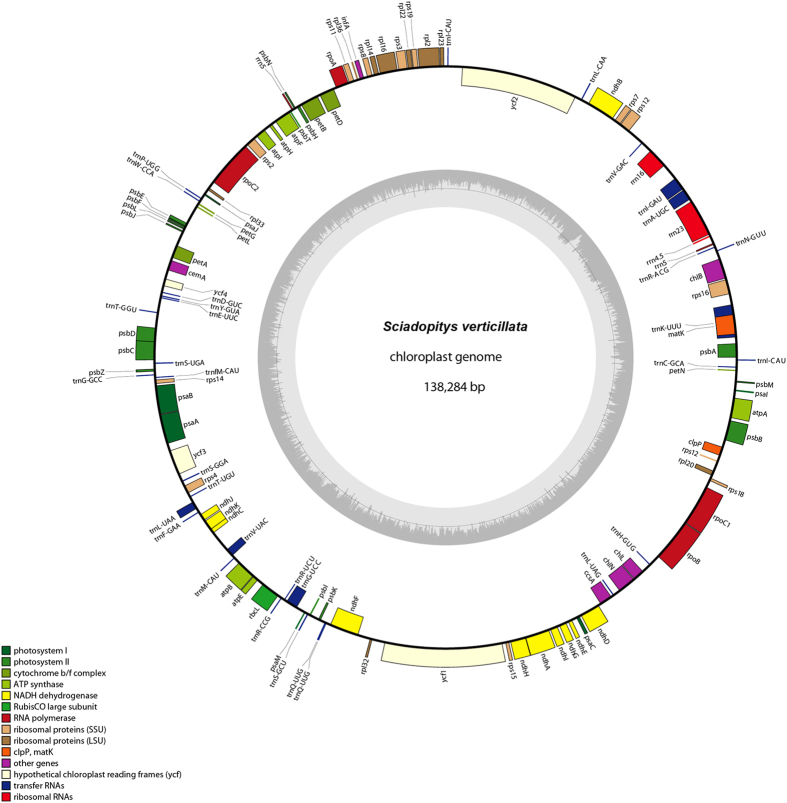
The complete cp genome of S. verticillata is a circular molecule of 138,284 bp (Fig. 1), only shorter than that of Agathis dammara (145,625 bp) in cupressophytes. Like other sequenced cp genomes of conifers, the S. verticillata cp genome does not contain one copy of canonical IR. The overall GC content is 35.42% (protein-coding genes, 36.35%; tRNA genes, 52.79%; rRNA genes, 52.45%; introns, 35.93%; intergenic spacers, 30.65%) (Table 1). In S. verticillata cp genome, 118 unique genes were identified, including 83 protein-coding genes, 31 tRNA genes and 4 rRNA genes. Among these 118 unique genes, rrn5, trnI-CAU and trnQ-UUG are duplicated. Rrn5 occurred as an inverted repeat sequence, while two copies of trnI-CAU and trnQ-UUG arranged in the same orientation. Ten protein-coding genes and six tRNA genes each have one intron, while ycf3 contains two. When comparing S. verticillata cpDNA with other conifers, the trnP-GGG and accD gene were lost. The trnP-GGG is also absent in Ephedra equisetina, whereas it is present as complete and functional in other conifers, Cycas, Ginkgo, Gnetum and Welwitschia29. The loss of accD was also found in Gnetales29. AccD gene encodes acetyl-CoA carboxylase β subunit and is essential in fatty acid synthesis26. At last, a C-to-U RNA editing site was identified at the initial codon of rps8 and verified by cDNA sequencing.
Figure 1. Gene map of the Sciadopitys verticillata plastid genome.
Genes shown inside and outside the circle are transcribed clockwise and counterclockwise, respectively.
Table 1. Comparison of chloroplast genomic characteristics between the four cupressophytes.
| Taxa | Amentotaxus argotaenia (KR780582) | Sciadopitys verticillata (KT601210) | Cephalotaxus wilsoniana (NC_016063) | Taxus mairei (NC_020321) |
|---|---|---|---|---|
| Genome size (bp) | 136657 | 138284 | 136196 | 127665 |
| Protein-coding genes (%) | 55.08 | 55.26 | 56.78 | 50.12 |
| tRNA genes (%) | 1.82 | 1.80 | 1.61 | 1.57 |
| rRNA genes (%) | 3.37 | 3.4 | 3.27 | 3.58 |
| Introns (%) | 8.78 | 9.12 | 9.15 | 10.34 |
| Spacers (%) | 30.95 | 30.4 | 29.19 | 34.37 |
| Gene density (no. of gene/Kb) | 0.88 | 0.87 | 0.85 | 0.88 |
| GC content (%) | ||||
| Genome | 35.85 | 35.42 | 35.08 | 34.72 |
| Protein-coding genes | 36.90 | 36.35 | 36.04 | 36.76 |
| tRNA genes | 53.31 | 52.79 | 52.94 | 52.36 |
| rRNA genes | 52.99 | 52.45 | 52.14 | 52.57 |
| Introns | 36.10 | 35.93 | 35.08 | 34.63 |
| Spacers | 31.03 | 30.65 | 30.29 | 29.97 |
The repeat sequence
In this study, we have analyzed the occurrence, nature, organization, and distribution of simple sequence repeat (SSR) in the S. verticillata cp genome. In total, 53 SSRs were identified (Supplementary Table S1). Of them, 34 SSR loci are located within intergenic spacers (IGS), 6 in introns and 13 in coding regions. Three regions containing multiple SSRs (3 SSRs in trnI-GAU to rrn16 IGS, 2 in rps12 intron, 3 in petA to cemA IGS) are particular valuable for population genetics studies as they are co-located in short regions31. Among these SSRs, 36 are mononucleotide, 11 are dinucleotide, 5 are trinucleotide and only one is tetranucleotide. This suggests that the most common type of cp repeat is the mononucleotide, whereas di-, tri- or tetranucleotide repeats are rare.
Tandem repeat finder was used to identify the tandem repeats in S. verticillata cp genome, with the identity and size of the repeats were limited to no less than 90% and 30 bp in unit length. In total, 18 forward tandem repeats in S. verticillata cp genome were identified (Supplementary Table S2), of which 11 are located in coding regions of ycf2 (6), trnQ-UUG (1), and ycf1 (4). The other seven tandem repeats are distributed in the IGS of trnI-GAU/rrn16 (2), rrn16/trnV-GAC (1), ycf2/trnI-CAU (1), trnM-CAU/atpB (1), rpl32/ycf1 (1), and rpoC1 intron (1). The cp genome of S. verticillata has 18 forward tandem repeats, similar to C. oliveri cp genome (17)5, but less than the cp genome of Podocarpus lambertii (28)11. The ycf1 gene in the three genomes all contains tandem repeat. Interestingly, one tandem repeat containing trnQ-UUG gene was detected in S. verticillata cp genome, which exactly forming two copies of trnQ-UUG gene concatenated.
The duplication of trnQ-UUG in S. verticillata cp genome
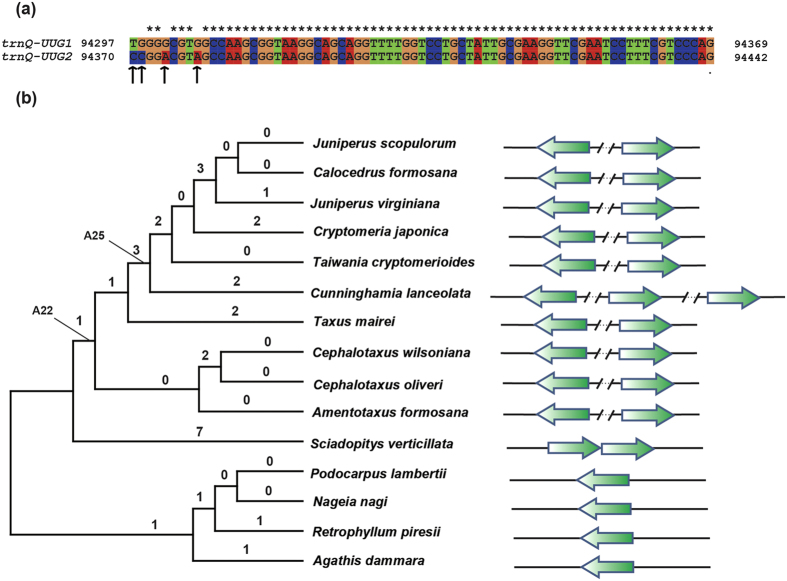
Although Sciadopityaceae cp genome does not contain trnQ-UUG sIR, two copies of trnQ-UUG were clustered as a tandem repeat (Fig. 2). Four nucleotide sites were different between the two copies of trnQ-UUG gene (Fig. 2a). Sciadopityaceae was the first species that started to contain two copies of trnQ-UUG. Podocarpaceae and Araucariaceae each have only one copy of trnQ-UUG. In order to understand the evolutionary process of trnQ-UUG in cupressophytes, the cp genome rearrangement and ancestral plastomic organization among cupressophytes were inferred. The permutation with 33 locally collinear blocks (LCBs) was generated on the basis of whole plastome alignments among the 15 cupressophyte species. The permutation was then used to construct the most parsimonious tree (Fig. 2b). The resulting tree has two major clades, one comprising Cephalotaxaceae, Taxaceae, Cupressaceae, and Sciadopityaceae which have the trnQ-UUG repeat, while the other including Podocarpaceae and Araucariaceae which does not contain trnQ-UUG repeat. The Sciadopityaceae containing the tandem repeat was located at the basal position within the first clade. So we speculate that the tandem repeat of trnQ-UUG came first, the inverted repeats may result from the evolution of rearrangement.
Figure 2. The evolution of trnQ-UUG in cupressophytes.
(a) The alignment of two trnQ-UUG copies in Sciadopitys verticillata chloroplast genome. Four black arrows under the alignment denote the different nucleotide sites. The numbers in the left and right denote the relative position in the complete chloroplast genome. The two copies of trnQ-UUG formed a tandem repeat sequence. (b) The topology in the left was the phylogenetic tree of the cupressophyte inferred from the matrices of chloroplast DNA locally collinear blocks. The number of rearrangements leading to a clade was shown above the branch. A22 indicates the common ancestor of Cephalotaxaceae, Taxaceae, and Cupressaceae, and A25 indicates the common ancestor of Cupressaceae. The green arrows in the right denote trnQ-UUG gene. The direction of the arrows denotes the relative direction of trnQ-UUG gene. The regions between the two slashes indicate the omitted gene.
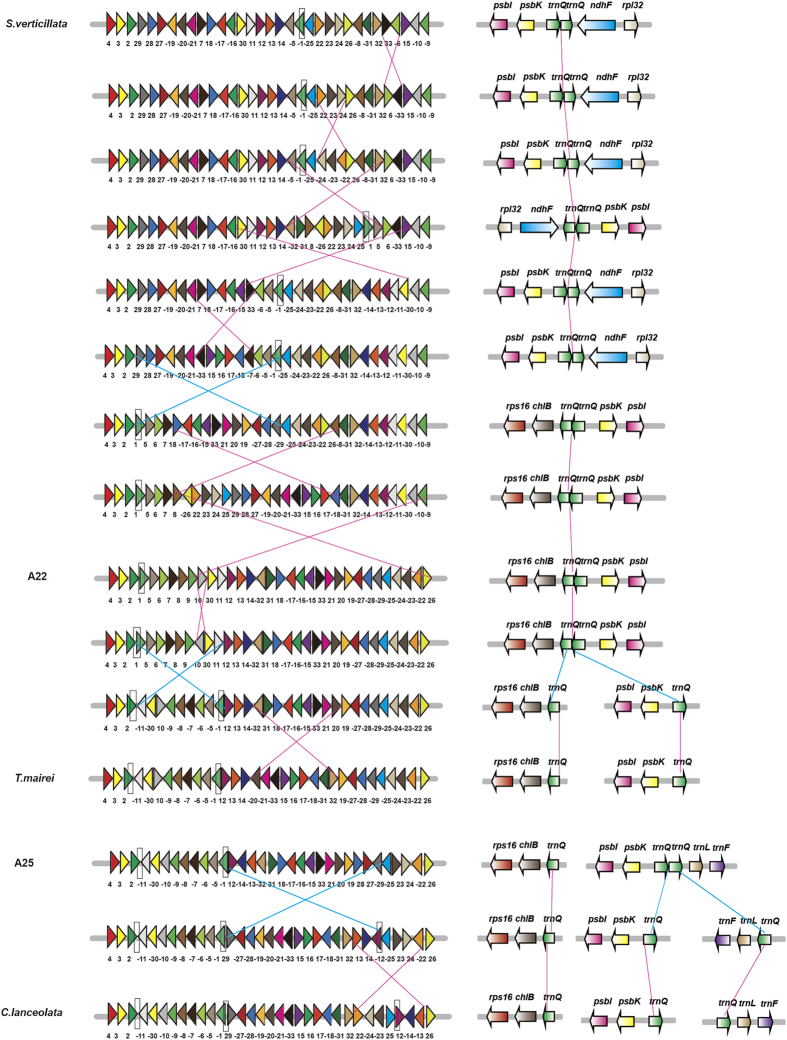
Figure 3 shows two detailed evolutionary scenarios of plastomic rearrangements: one from S. verticillata to Taxus mairei through A22 (the ancestral plastomic organization of Cupressaceae, Taxaceae, and Cephalotaxaceae), the other from A25 (the ancestral plastomic organization of Cupressaceae) to Cunninghamia lanceolata. Here, we propose an alternative mechanism for the formation of trnQ-UUG sIR following the rearrangement of cp genome. Two tandem trnQ-UUG copies are first located in the spacer region of LCB 1 and 25, which is between psbI-psbK and ndhF-rpl32 gene (Fig. 3). From S. verticillata to A22, one inversion was happened between LCB 29 and 1, making LCB 1 adjacent to LCB 2 which contains rps16 and chlB gene (Fig. 3 the blue line). During this process, the two copies of trnQ-UUG moved following the LCB 1, forming the rps16-chlB-trnQ-trnQ-psbK-psbI gene order (Fig. 3). From A22 to Taxus mairei, one inversion occurred between LCB 1 and 11 (Fig. 3 the blue line). During this inversion, one copy of trnQ-UUG gene followed LCB1, while the other followed the LCB 2. Then the tandem trnQ-UUG genes were divided into two parts: one located near rps16 and chlB genes, the other located near psbK and psbI genes. In C. lanceolata, three copies of trnQ-UUG gene were identified, with two copies of the same orientation and another with the opposite orientation (Fig. 2). Here, we also speculate one possible evolutionary process for the formation of the three copies of trnQ-UUG. We suppose that two copies of trnQ-UUG were located between LCB 1 and 12 in A25. From A25 to C. lanceolata, one inversion happened between LCB 12 and 29 (Fig. 3 the blue line). This inversion separates the two trnQ-UUG gene into two parts, making one copy of trnQ-UUG located in LCB1 and the other in LCB 12. Combined with one copy of trnQ-UUG located in LCB 2, finally, the three copies of trnQ-UUG were located in LCB 2, 1, and 12 respectively. In conclusion, the trnQ-UUG gene first formed tandem repeats, then a rearrangement divided the tandem trnQ-UUG repeat into different parts, forming inverted or forward repeats locating at different regions.
Figure 3. The formation mechanism of trnQ-UUG inverted repeat sequence along with the plastomic rearrangements in cupressophytes.
Two hypothetical evolutionary scenarios for plastomic rearrangements were shown: from Sciadopitys verticillata to Taxus mairei and from A25 to Cunninghamia lanceolata. Plastomes are circular but here are shown in gray bars. Locally collinear blocks (LCBs) with their relative orientations were indicated with color triangles. Inversions between two plastomes were linked by carmine and blue lines. Three pairs of blue lines denote the inversion occurred between LCB 29 and 1, LCB 1 and 11, LCB 12 and 29. The locations of trnQ-UUG in the LCB were shown in the black box. The arrows in the right panel denote the specific location of trnQ-UUG. The direction of arrows denotes the orientations of the gene. Carmine and blue lines between trnQ-UUG genes denote the evolutionary scenarios along with plastomic rearrangements.
All the species containing trnQ-repeats (including tandem repeats and inverted repeats) formed a monophyletic group, while Podocarpaceae and Araucariaceae composed the other (Fig. 2b). This dichotomy is exactly consistent with the phylogenetic relationship inferred from other gymnosperm phylogenetic studies13,14. Furthermore, the transition stage of trnQ-UUG also indicated S. verticillata as the basal of the Cupressaceae-Taxaceae-Cephalotaxacea clade. Therefore, trnQ-UUG repeats in cupressophytes may be considered as informative markers for conifer phylogeny.
AccD has been functionally transferred from the chloroplast to the nucleus
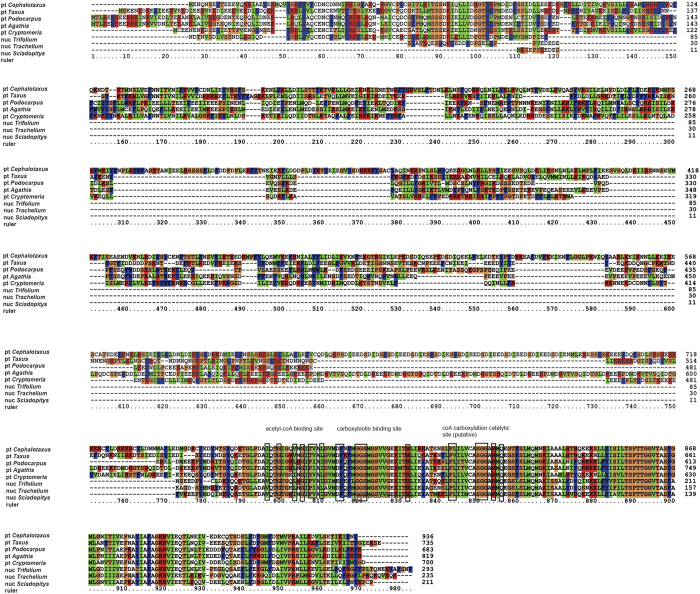
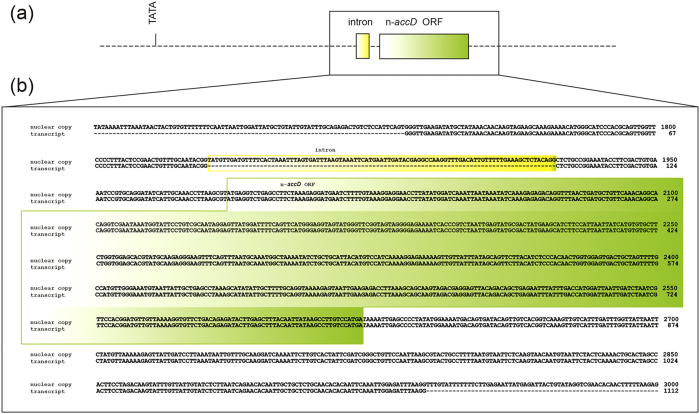
The complete cp genome sequence of S. verticillata showed that its accD gene was completely lost. A partial transcript containing high sequence identity to plastid accD was assembled using two S. verticillata transcriptome databases. Then the entire transcript of the accD sequence was obtained by using RACE. This transcript encodes a protein of 212 amino acids, compared with the approximately 680–1070 amino acids encoded by the accD gene in the chloroplast genome of other cupressophytes (Fig. 4). The 212 amino acids in this transcript are 84% identity to the accD carboxylase domain encoded by other cupressophyte plastid-accD genes. From Fig. 4, we infer that residues 25–212 (position 787–983 in the alignment) in the accD origin from plastid. The sequences located at 1985 to 1 bp upstream from the translation start site and 1 to 1066 downstream of the stop codon were obtained by anchored PCR (Fig. 5). Predicted eukaryotic TATA box were identified −1599 bp 5′ of the translation start site of the gene. The flanking region of accD sequence showed no plastid sequence similarity. In addition, blastn of this sequence to the nucleotide database in GenBank returns no match, suggesting that it is likely originated from nuclear or mitochondria. The flanking regions of the accD containing sequence also had no significant hits with blast to Cycas taitungensis mitochondria genome, the only one reported mitochondria genome in gymnosperm, which indicated that accD is more likely located in nucleus. Five different softwares (BaCelLO, MultiLoc, Predotar, Protein Prowler, and TargetP) were used to predict the target peptide-encoding sequences of nuclear accD (n-accD) sequence. None of the softwares identified any target peptide-encoding sequences to chloroplast. The BaCelLO, MultiLoc and Protein Prowler predicted target peptide-encoding sequences to cytoplasm, while Predotar and TargetP predicted to elsewhere (Supplementary Table S3). The n-accD gene was also amplified from genomic DNA. A single intron of 93 bp is present at the 5′ UTR region, which is located at −65 and −64 bp upstream of the ORF (Fig. 5). The nucleus copy and transcript of n-accD sequences showed 100% identity. Thus no RNA editing sites were found in the n-accD copy (Fig. 5).
Figure 4. Multiple alignment of the predicted nuclear accD from Trifolium repens, Trachelium caeruleum, Sciadopitys verticillata and plastidic accD from Cephalotaxus oliveri, Taxus mairei, Podocarpus lambertii, Agathis dammara and Cryptomeria japonica.
The putative acetyl-CoA binding site, the carboxybiotin-binding site and the CoA-carboxylation catalytic site were shown in black box. The T. repens and T. caeruleum accD gene sequence only contained the 5′-terminal region of the accD gene (the position 513–805 for T. repens and the position 96–330 for T. caeruleum of the entire transcript).
Figure 5. The gene structure of the nuclear accD sequence from Sciadopitys verticillata.
(a) The coding region and intron are indicated by green and yellow boxes, respectively. Dashed lines denote noncoding regions. (b) Nucleotide sequence alignment of nuclear and transcript copies. The alignment region corresponds to the black box section shown in (a). Intron in the 5′ UTR and nuclear accD open reading frame are indicated with yellow and green boxes, respectively.
Phylogenetic distribution of accD loss in gymnosperm
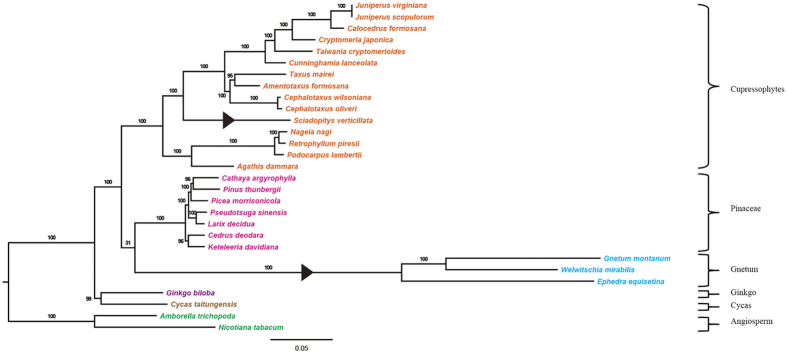
In order to determine the phylogenetic distribution of accD loss in gymnosperm, we constructed a maximum likelihood tree inferred from the concatenated 59 cpDNA genes (50555 nucleotides) with Amborella trichopoda and Nicotiana tabacum as outgroup (Fig. 6). In this topology, Cycas and Ginkgo were grouped together; then Gnetales was clustered with Pinaceae (the gnepines hypothesis). For cupressophytes, the two families, Araucariaceae and Podocarpaceae, diverged first, and then Sciadopityaceae was strongly supported to be sister to a well-supported clade containing Cupressaceae, Taxaceae and Cephalotaxaceae. At last, Taxaceae and Cephalotaxaceae formed a monophyly and then clustered with Cupressaceae. The phylogenetic position of Sciadopityaceae in our study was consistent with previous molecular phylogenetic studies12,13. A functional plastid accD still exists in Cycas, Ginkgo and other conifers. However, for Sciadopityacea and Gnetales, accD has been lost from their plastomes. This topology (Fig. 6) indicates two independent losses of accD from their plastomes. However, for Gnetales, no studies have been performed to demonstrate if there is a functional copy in the nucleus.
Figure 6. Maximum likelihood tree of 29 taxa from 59 plastid genes.
Bootstrap values are shown along branches. Triangles on nodes indicate loss of accD from plastome.
The origin of nuclear accD gene
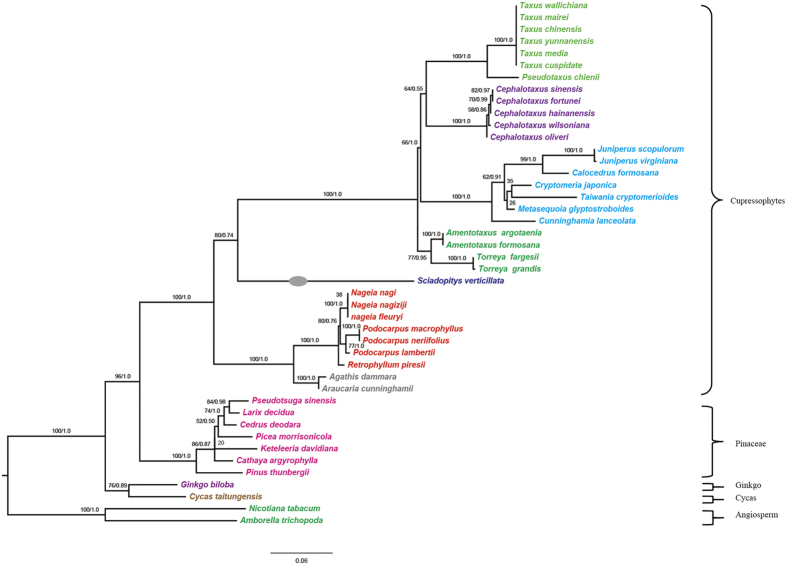
Phylogenetic analysis of the n-accD transcript in S. verticillata and plastid-encoded counterparts from 43 other plants was used to infer the origin of n-accD in S. verticillata, with A. trichopoda and N. tabacum as outgroup (Fig. 7). The results showed that n-accD transcript of S. verticillata was located in the basal clade of the Cephalotaxaceae, Taxaceae and Cupressaceae plastid copies, which was consistent with the phylogeny tree constructed by 59 cpDNA genes. Considering that other cupressophytes (Araucariaceae, Podocarpaceae, Cupressaceae, Taxaceae and Cephalotaxaceae) contain an intact copy of plastid accD gene, we suggest that accD has been functionally transferred to the nucleus independently near the time of divergence of the Cephalotaxaceae-Taxaceae-Cupressaceae and Sciadopityaceae.
Figure 7. Phylogenetic relationship of Sciadopitys verticillata nuclear accD sequences to chloroplast accD sequences from other gymnosperms.
The tree was constructed using Maximum likelihood and Bayes methods. Bootstrap and posterior probability values are shown along branches. Gray oval on node indicates the position of S. verticillata in the topology.
Disscussion
In this study, we have determined the complete chloroplast genome sequence of S. verticillata, which is the only one species of Sciadopityaceae. Like other sequenced cp genomes of cupressophytes4,5, the S. verticillata cp genome has no IRs (Fig. 1). It is most similar to the cp genome of Amentotaxus formosana, with 8 inversions involved in (Fig. 2b). When compared with C. oliveri, S. verticillata had 10 specific rearrangements (Fig. 2b). The large number of rearrangements between S. verticillata and other cupressophytes cp genome indicate that S. verticillata has a distinctive cp genome structure among cupressophytes. Previous researches have shown that the presence of IRs can stabilize cp genome organization, while their absence often leads to rearrangements4,32,33. The extensively rearranged cp genome of S. verticillata without IRs supports this suggestion.
We have proposed one mechanism of the formation of small inverted repeats. The tandem repeat of trnQ-UUG occurred first by a process of slipped-strand mispairing in S. verticilla, and the subsequent inversion resulted in the inverted repeat (Fig. 3). Tandem repeats may form totally by chance, for example, as a result of replication slippage34. For the formation of inverted repeat, Knox has proposed three mechanisms: coincidental similarities, transposed copies, and duplications at inversion endpoints35. Coincidental similarities usually lead to a half dozen of inverted repeats with moderate-to-high sequence similarity in a typical plastome35. The dispersed repeats generated by duplicative transposition of cp DNA would not be inverted relative to the source region, but subsequent inversion put these copies in inverted orientation35,36. Some inversions raise duplications that produce inverted fragments at both junctions35,37. Hence it is more difficult to form inverted repeat than tandem repeat. There are two types of genome organizations around trnQ-UUG in the clade that contains trnQ-UUG inverted repeat (Supplementary Fig. S1). One type is that one copy of trnQ-UUG is located near chlB and trnT-UGU, and the other situated between psbK and trnL-UAA. The other type shows that one copy of trnQ-UUG is located near chlB and psbK, while the other is adjacent to trnT-UGU and trnL-UAA. However, the two types can transform into each other by the occurrence of an inversion located between the two inverted copies of trnQ-UUG (Supplementary Fig. S1). Similarly, a large 36-kb inversion among four Juniperus plastomes was also suggested to be caused by an approximately 250 bp-IR containing trnQ-UUG6. The relative fixed location of the two trnQ-UUG genes indicates that the inverted repeat in Taxaceae, Cephalotaxaceae, and Cupressaceae originates from a common ancestor. We thus suggest that the inverted repeat is originated from tandem repeats in Sciadopityaceae. These inverted repeats can serve as a molecular basis of inversions, and inversions in turn promote the formation of inverted repeats6,35.
The accD gene encodes the β-carboxyl transferase subunit of ACCase. ACCase catalyzes the formation of malonyl-CoA from acetyl-CoA and is used in de novo fatty acid synthesis26. Plants have two forms of ACCase: the eukaryote form located in cytosol and the prokaryote form located in plastids26. The eukaryote-form ACCase is composed of a single multifunctional polypeptide, whereas the prokaryote-form is comprised of four subunits: the α-carboxyltransferase subunit (accA), the biotin carboxyl carrier (accB), the biotin carboxylase (accC), and the β-carboxyltransferase subunit (accD). AccA, accB, accC are all nucleus-encoded. In contrast, accD is encoded in the plastome. AccD is widely distributed in plants, even in some parasitic and non-photosynthetic plants38. In Campanulaceae and Fabaceae, accD has been functionally transferred to the nucleus23,24. In this study, we found that accD gene is lost in the plastome of Sciadopityaceae; and furthermore, the chloroplastic accD gene of Sciadopity have been transferred to the nucleus (Fig. 4).
The case of accD in Sciadopityaceae represents the third documented transfer for this gene from plastid to nucleus with the other two occuring in the unrelated angiosperm families Campanulaceae and Fabaceae. In Trachelium caeruleum (Campanulaceae) and Trifolium repens (Fabaceae), the nucleus (n)-accD transcripts encode 235 and 293 amino acids of plastid origin, respectively. Both in Campanulaceae and Fabaceae, the n-accD genes encode only the 3′-end region of the plastid gene. Consistently, the S. verticilla n-accD gene also only encodes the 3′-end region of accD gene (Fig. 4). These indicates that the C-terminus region of ACCD protein is the main functional domain. In the potato plastid accD, three functionally relevant sites have been identified: a putative acetyl-CoA binding site, a CoA-carboxylation catalytic site, and a carboxybiotin-binding site39. The three sites, clustering at the C-terminus of the protein, are present in the n-accD sequence in Campanulaceae, Fabaceae, and Sciadopityaceae (Fig. 4).
The major difference among the three transfers is that the Sciadopityaceae n-accD has no chloroplastic transit peptide sequence at the N-terminus of its transcript. In T. repens and T. caeruleum, the n-accD shares a transit peptide with LPD2 and KASI, respectively. More specifically, in T. caeruleum, a chloroplastic transit peptide was verified experimentally by showing that the product of n-accD was imported into the chloroplast24. For the acquisition of transit peptide, two different strategies were generally used. Some nucleus copies have acquired a novel transit peptide, such as the transfer of rpl32 in Thalictrum and Aquilegia22; infA in multiple lineages of rosid17, and rpoA in moss Physcomitrella patens40. In contrast, other transferred genes acquired their transit peptide by transferring into a duplicate copy of a nuclear gene that has been already targeted to the plastid. For example, in Bruguiera and Populus, rpl32 is fused to an existing nuclear gene (Cu-Zn superoxide dismutase)20,21; in Campanulaceae and Fabaceae, accD is fused to KASI and LPD2, respectively23,24. However, in Sciadopityaceae, we falied to identify the chloroplastic transit peptide sequence by using five softwares (Supplementary Table S3). Further studies are needed to explore the mechanism of function replacement of accD gene in Sciadopityaceae.
We have used two data sets to infer the phylogenetic position of Sciadopityaceae: the permutation of LCBs based on arrangements of cpDNA organization and 59-gene sequences. The two data sets yield the same result: Sciadopityaceae is sister to the clade comprising Taxaceae, Cephalotaxaceae, and Cupressaceae (Figs 2b and 6). Our result is in good agreement with previous molecular phylogenetic studies13,41. This further shows the usefulness of the cpDNA organization to resolve the phylogenetic relationships at the familial level and above. Rarrangements of cpDNA fragments have previously provided novel evidence for gnepines hypothesis9. Nonetheless, isomeric plastomes caused by sIR and homoplasious character should be treated cautiously when using genomic rearrangements in phylogenetic inferences7.
Conclusions
We have determined the complete cp genome sequence of S. verticillata. The S. verticillata cp genome is highly divergent with a distinctive genome structure comparing with other cupressophyes. Our data suggest a molecular mechanism for the formation of small inverted repeat sequences in cupressophytes; that is rearrangements divide the tandem repeat into different parts, forming inverted or forward repeats locating at different regions. One unusual feature of the S. verticilla cp genome is the loss of accD gene. Examination of transcriptome database indicates that this gene has been transferred to the nucleus. The transfer time was estimated to be in the divergence of the Sciadopityaceae and Cephaloceae-Taxaceae-Cupressaceae. Moreover, phylogenetic relationships yielded by using 59-gene sequences and cp genome organizations both strongly support the placement of Sciadopityaceae as the sister of the clade containing Cephalotaxacea, Taxaceae, and Cupressaceae.
Methods
Plant material and chloroplast genome sequencing
S. verticillata is an evergreen conifer endemic to Japan. It is monoecious and wind pollinated reaching heights of 45 m, diameters of up to 2 m42. As an economically valuable tree, S. verticillata is often used in construction. The inheritance of chloroplast and mitochondria in S. verticillata is through paternal transmission43. S. verticillata is particularly valuable as the last member of the formerly widespread and more diverse conifer family, the Sciadopityaceae.
Young leaves of S. verticillata were collected from a single individual growing in Lushan Botanical Garden, Jiangxi Province. The materials used for RNA extraction was saved in RNAfixer (Bioteke Corporation, Beijing, China).Voucher specimens were deposited at the herbarium of Wuhan Botanical Garden, Chinese Academey of Science with the accession number of Lijiachenshanshan001. The genomic DNAs were isolated from the fresh leaves using the modified CTAB method. 1 μg DNA was sheared by Covaris M220 (Covaris, USA), yielding fragments of 500 bp in length. Paired-end libraries were constructed using NEBNext UltraTM DNA Library Prep Kit for Illumina (NEB, USA) according to the manufacturer’s instructions. Genomic DNA was sequenced on a single lane using the Illumina HiSeqTM 2500 platform (Illumina Inc., San Diego, CA). Approximately 3.14 GB of 150-bp paired-end raw reads were generated. FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) was used to remove adaptor and low-quality reads. The clean reads were de novo assembled by velvet (V1.2.07)44, with the coverage cutoff value as 30 (-cov_cutoff 30). Among the 22 assembled contigs, 15 contigs were found to match the published conifers cpDNA sequence and used for complete genome finishing. The gaps were bridged by polymerase chain reaction (PCR) amplification based on the cp contig sequence. Overlapping regions of adjacent PCR products were set to at least 300 bp. PCR amplification was carried out in 50 μl volumes containing 2.5 ng of DNA template, 5 μl 10 × LA PCR Buffer II (Mg2 + Plus), 8 μl dNTP mixture (each 2.5 mM), 2.5 U of LA Taq (TaKaRa Bio Inc, Dalian, China), and 2 μl each of forward and reverse primers (10 μM). The thermo-cycling program was set as: 5 min at 95 °C (1 cycle); 30 s at 95 °C, 3 min at 62 °C (32 cycles); 20 min at 72 °C (1 cycle). Positive PCR amplicons were sequenced on ABI 3730 xl DNA Analyzer (Applied Biosystems, Foster City, CA, USA). At last, all the contigs and PCR amplification sequences were assembled into a complete chloroplast genome using Bioedit45.
Genome annotation
The cp genome of S. verticillata was initially annotated by Dual Organellar GenoMe Annotator (DOGMA)46. The exact boundaries of each gene was determined by comparisons with homologous genes in other published gymnosperm cpDNAs. tRNA genes were further verified by two programs, ARAGORN47 and tRNAscan-SE48. The circular gene map was drawn by the software OGDRAW49.
Identification and isolation of accD gene in the S. verticillata nuclear genome
Two S. verticillata transcriptome data (SRR065035, ERR364344) from 454 and illumina sequencing were downloaded from Sequence Read Archive (SRA) database (http://www.ncbi.nlm.nih.gov/Traces/sra/). Using Nageia nagi accD sequences as the query to blast against these two transcriptome dataset, 19 and 100 reads were identified for SRR065035 and ERR364344, respectively. All of these reads were then assembled into a approximately 1000 bp long sequence using Bioedit45. This sequence was first verified by PCR using appropriate primer pairs (Supplementary Table S4, naccDF and naccDR). The flanking sequence of this 1000 bp sequence was acquired by Anchored PCR50 using gene specific primers (Supplementary Table S4).
Total RNA was extracted using GREENspin Plus Plant RNA kit (ZoManBio, Beijing, China) and genomic DNA was removed using RNase-free DNase I (Takara, Dalian, China). cDNA templates were synthesized using Reverse Transcriptase M-MLV Kit (Takara Bio Inc., Dalian, China). Rapid amplification of cDNA ends (RACE) was conducted using SMARTTM RACE cDNA Amplification Kit (Clontech, Palo Alto, CA). The primers used for cloning 5′ and 3′ cDNA ends are the same as those used in Anchored PCR (Supplementary Table S4). All kits were used according to the manufacturers’ instructions.
Five softwares were used to identify transit peptides, BaCello51, MultiLoc52, Predotar53, Protein Prowler54 and TargetP55.
Computational methods and phylogenetic analysis
The Perl script MISA (http://pgrc.ipk-gatersleben.de/misa/) was used to identify simple sequence repeats (SSR) in S. verticillata cp genome. The thresholds for SSR search was defined as ten repeat units for mono-nucleotides, five repeat units for di-nucleotides, and four repeat units for tri-, tetra-, penta-, and hexa-nucleotides. Tandem repeats were analyzed using Tandem Repeats Finder (http://tandem.bu.edu/trf/trf.submit.options.html) with the basic model. All of the repeats found were manually verified, and the nested or redundant results were removed.
Phylogenetic analyses were performed on three data sets. The first contained 15 cupressophyte chloroplast sequences (Supplementary Table S5). The second data set included 15 cupressophyte species from the first data sets, 12 other gymnosperm and 2 angiosperms whose plastid genomes were completely sequenced (Supplementary Table S5). The third data set included 25 accD sequences extracted from the second data set (excluding three Gnetales species and Sciadopity whose accD gene was lost), 18 accD sequences reported in previous research and n-accD in S. verticillata (Supplementary Table S6).
For the first data set, whole-genome alignment was performed using progressive Mauve implemented in MAUVE v.2.3.156. Thirty-three LCBs were identified. MGR 2.0.1 (http://grimm.ucsd.edu/MGR/pubs.html) was used to build the phylogenetic trees and the process of rearrangement based on matrices of LCBs with the model of unichromosomal circular reversal distance.
Fifty-five common protein-coding and four rRNA genes were extracted from the cp genomes of the second data set (Supplementary Table S5). Each gene was aligned using the MUSCLE program implemented in MEGA 6. The aligned sequences were concatenated into a 59-gene data set and then used for reconstructing the gymnosperm phylogeny. Maximum likelihood (ML) tree was performed by RaxML v8.1.x57 with a GTRCAT substitution model as suggested (see RAxML manual). Clade supports were identified with 100 bootstrap replicates.
Forty-four accD sequences from the third data set were aligned using MUSCLE as implemented in MEGA 6. ML and Bayes inference (BI) were used to construct the phylogeny relationship of this forty-four accD sequences. ML trees were conducted with GTRCAT model using RaxML v8.1.x. Supports for nodes of trees were evaluated by 100 bootstrap replications. Bayesian inference (BI) trees were constructed by MrBayes v.3.1.258 with the GTR + I + G model selected by Modeltest 3.7. Four independent Markov chain Monte Carlo chains were run with 1000000 generations. The first 25% of the sampled trees were removed as burn in. The remaining trees were used to construct a 50% majority-rule consensus tree.
Data availability
All sequencing data produced in the present work have been submitted to Genbank under the accession KT601208-KT601211 and Sequence Read Archive with the accession of SRP067546.
Additional Information
How to cite this article: Li, J. et al. Evolution of short inverted repeat in cupressophytes, transfer of accD to nucleus in Sciadopitys verticillata and phylogenetic position of Sciadopityaceae. Sci. Rep. 6, 20934; doi: 10.1038/srep20934 (2016).
Supplementary Material
Acknowledgments
We thank Puxin Gao (Lushan Botanical Garden, Jiangxi Province) for collecting samples, Yuan Zhou and Zhiwei Wang for experimental assistance. This work was supported by the National Natural Science Foundation of China (Nos 30771763, 31370364, 31400207, and 31570652).
Footnotes
Author Contributions J.L., L.G. and T.W. conceived this study; J.L., S.S.C. and K.T. carried out laboratory work and data analyses; J.L., L.G. and Y.J.S. drafted the main manuscript; T.W. polished the manuscript.
References
- Jansen R. K. & Ruhlman T. A. in Genomics of chloroplasts and mitochondria. Vol. 35, (eds Bock R. & Knoop V.) 103–126 (Springer, 2012). [Google Scholar]
- Lin C. P., Huang J. P., Wu C. S., Hsu C. Y. & Chaw S. M. Comparative chloroplast genomics reveals the evolution of Pinaceae genera and subfamilies. Genome Biol. Evol. 2, 504–517 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. S., Lin C. P., Hsu C. Y., Wang R. J. & Chaw S. M. Comparative chloroplast genomes of Pinaceae: insights into the mechanism of diversified genomic organizations. Genome Biol. Evol. 3, 309–319 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao T., Watanabe A., Kurita M., Kondo T. & Takata K. Complete nucleotide sequence of the Cryptomeria japonica D. Don. chloroplast genome and comparative chloroplast genomics: diversified genomic structure of coniferous species. BMC plant biology 8, 70 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X., Gao L., Wang B., Su Y. J. & Wang T. The complete chloroplast genome sequence of Cephalotaxus oliveri (Cephalotaxaceae): evolutionary comparison of Cephalotaxus chloroplast DNAs and insights into the loss of inverted repeat copies in gymnosperms. Genome Biol. Evol. 5, 688–698 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W. et al. Predominant and substoichiometric isomers of the plastid genome coexist within Juniperus plants and have shifted multiple times during cupressophyte evolution. Genome Biol. Evol. 6, 580–590 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C. Y., Wu C. S. & Chaw S. M. Ancient nuclear plastid DNA in the yew family (Taxaceae). Genome Biol. Evol. 6, 2111–2121 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. S., Wang Y. N., Hsu C. Y., Lin C. P. & Chaw S. M. Loss of different inverted repeat copies from the chloroplast genomes of Pinaceae and cupressophytes and influence of heterotachy on the evaluation of gymnosperm phylogeny. Genome Biol. Evol. 3, 1284–1295 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. S. & Chaw S. M. Highly rearranged and size-variable chloroplast genomes in conifers II clade (cupressophytes): evolution towards shorter intergenic spacers. Plant Biotechnol. J. 12, 344–353 (2014). [DOI] [PubMed] [Google Scholar]
- Yap J. Y. et al. Complete chloroplast genome of the Wollemi pine (Wollemia nobilis): structure and evolution. PLoS One 10, e0128126 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira L. N. et al. The complete chloroplast genome sequence of Podocarpus lambertii: genome structure, evolutionary aspects, gene content and SSR detection. PLoS One 9, e90618 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. Y., Ran J. H. & Wang X. Q. Three genome-based phylogeny of Cupressaceae s.l.: further evidence for the evolution of gymnosperms and Southern Hemisphere biogeography. Mol Phylogenet. Evol. 64, 452–470 (2012). [DOI] [PubMed] [Google Scholar]
- Crisp M. D. & Cook L. G. Cenozoic extinctions account for the low diversity of extant gymnosperms compared with angiosperms. New Phytol. 192, 997–1009 (2011). [DOI] [PubMed] [Google Scholar]
- Lu Y., Ran J. H., Guo D. M., Yang Z. Y. & Wang X. Q. Phylogeny and divergence times of gymnosperms inferred from single-copy nuclear genes. PLoS One 9, e107679 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T. et al. Genomes of Stigonematalean cyanobacteria (subsection V) and the evolution of oxygenic photosynthesis from prokaryotes to plastids. Genome Biol. Evol. 5, 31–44 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis J. N., Ayliffe M. A., Huang C. Y. & Martin W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5, 123–135 (2004). [DOI] [PubMed] [Google Scholar]
- Millen R. S. et al. Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell 13, 645–658 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantt J. S., Baldauf S. L., Calie P. J. & Weeden N. F. Transfer of rp122 to the nucleus greatly preceded its loss from the chloroplast and involved the gain of an intron. EMBO J. 10, 3073–3078 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R. K., Saski C., Lee S. B., Hansen A. K. & Daniell H. Complete plastid genome sequences of three Rosids (Castanea, Prunus, Theobroma): evidence for at least two independent transfers of rpl22 to the nucleus. Mol. Biol. Evol. 28, 835–847 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusack B. P. & Wolfe K. H. When gene marriages don’t work out: divorce by subfunctionalization. Trends Genet. 23, 270–272 (2007). [DOI] [PubMed] [Google Scholar]
- Ueda M. et al. Loss of the rpl32 gene from the chloroplast genome and subsequent acquisition of a preexisting transit peptide within the nuclear gene in Populus. Gene 402, 51–56 (2007). [DOI] [PubMed] [Google Scholar]
- Park S., Jansen R. K. & Park S. Complete plastome sequence of Thalictrum coreanum (Ranunculaceae) and transfer of the rpl32 gene to the nucleus in the ancestor of the subfamily Thalictroideae. BMC Plant Biol. 15, 40 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee A. M. et al. Localized hypermutation and associated gene losses in legume chloroplast genomes. Genome Res. 20, 1700–1710 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau-Gueutin M. et al. Potential functional replacement of the plastidic acetyl-CoA carboxylase subunit (accD) gene by recent transfers to the nucleus in some angiosperm lineages. Plant Physiol. 161, 1918–1929 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goremykin V. V., Holland B., Hirsch-Ernst K. I. & Hellwig F. H. Analysis of Acorus calamus chloroplast genome and its phylogenetic implications. Mol. Biol. Evol. 22, 1813–1822 (2005). [DOI] [PubMed] [Google Scholar]
- Konishi T. & Sasaki Y. Compartmentalization of two forms of acetyl-CoA carboxylase in plants and the origin of their tolerance toward herbicides. Proc. Natl. Acad. Sci. 91, 3598–3601 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris M. E., Meyer G., Vandergon T. & Vandergon V. O. Loss of the acetyl-CoA carboxylase (accD) gene in poales. Plant Mol. Biol. Rep. 31, 21–31 (2012). [Google Scholar]
- Guisinger M. M., Kuehl J. V., Boore J. L. & Jansen R. K. Genome-wide analyses of Geraniaceae plastid DNA reveal unprecedented patterns of increased nucleotide substitutions. Proc. Natl. Acad. Sci. 105, 18424–18429 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. S., Lai Y. T., Lin C. P., Wang Y. N. & Chaw S. M. Evolution of reduced and compact chloroplast genomes (cpDNAs) in gnetophytes: selection toward a lower-cost strategy. Mol Phylogenet. Evol. 52, 115–124 (2009). [DOI] [PubMed] [Google Scholar]
- Konishi T., Shinohara K., Yamada K. & Sasaki Y. Acetyl-CoA carboxylase in higher plants: most plants other than gramineae have both the prokaryotic and the eukaryotic forms of this enzyme. Plant Cell Physiol. 37, 5 (1996) [DOI] [PubMed] [Google Scholar]
- Nock C. J., Baten A. & King G. J. Complete chloroplast genome of Macadamia integrifolia confirms the position of the Gondwanan early-diverging eudicot family Proteaceae. BMC genomics 15 (Suppl 9), S13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S., Schneeweiss G. M., dePamphilis C. W., Muller K. F. & Quandt D. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol. 76, 273–297 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J. D. & Thompson W. F. Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell 29, 537–550 (1982). [DOI] [PubMed] [Google Scholar]
- Gemayel R., Vinces M. D., Legendre M. & Verstrepen K. J. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu. Rev. Genet. 44, 445–477 (2010). [DOI] [PubMed] [Google Scholar]
- Knox E. B. The dynamic history of plastid genomes in the Campanulaceae sensu lato is unique among angiosperms. Proc. Natl. Acad. Sci. 111, 11097–11102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle R. C., Fourcade H. M., Boore J. L. & Jansen R. K. Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J. Mol. Evol. 66, 350–361 (2008). [DOI] [PubMed] [Google Scholar]
- Knox E. B. & Palmer J. D. The chloroplast genome arrangement of Lobelia thuliniana (Lobeliaceae): expansion of the inverted repeat in an ancestor of the Campanulales. Pl. Syst. Evol. 214, 49–64 (1999). [Google Scholar]
- Molina J. et al. Possible loss of the chloroplast genome in the parasitic flowering plant Rafflesia lagascae (Rafflesiaceae). Mol. Biol. Evol. 31, 793–803 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. S. et al. Characterization of the plastid-encoded carboxyltransferase subunit (accD) Gene of Potato. Mol. cells 15, 422–429 (2004). [PubMed] [Google Scholar]
- Sugiura C., Kobayashi Y., Aoki S., Sugita C. & Sugita M. Complete chloroplast DNA sequence of the moss Physcomitrella patens: evidence for the loss and relocation of rpoA from the chloroplast to the nucleus. Nucleic Acids Res. 31, 5324–5331 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran J. H., Gao H. & Wang X. Q. Fast evolution of the retroprocessed mitochondrial rps3 gene in Conifer II and further evidence for the phylogeny of gymnosperms. Mol. Phylogen. Evol. 54, 136–149 (2010). [DOI] [PubMed] [Google Scholar]
- Eckenwalder J. E. in Conifers of the world (Timber Press, London, 2009). [Google Scholar]
- Worth J. R., Yokogawa M. & Isagi Y. Outcrossing rates and organelle inheritance estimated from two natural populations of the Japanese endemic conifer Sciadopitys verticillata. J. Plant Res. 127, 617–626 (2014). [DOI] [PubMed] [Google Scholar]
- Zerbino D. R. & Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. A. BioEdit: a user friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser. 41, 95–98 (1999). [Google Scholar]
- Wyman S. K., Jansen R. K. & Boore J. L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20, 3252–3255 (2004). [DOI] [PubMed] [Google Scholar]
- Laslett D. & Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32, 11–16 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T. M. & Eddy S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M., Drechsel O. & Bock R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 52, 267–274 (2007). [DOI] [PubMed] [Google Scholar]
- Chen B. J., Sun C., Wang Y., Hu Y. L. & Lin Z. P. Anchored PCR (A-PCR): A new method for chromosome walking. Chin. Sci. Bull. 49, 1772–1774 (2004). [Google Scholar]
- Pierleoni A., Martelli P. L., Fariselli P. & Casadio R. BaCelLo: a balanced subcellular localization predictor. Bioinformatics 22, e408–416 (2006). [DOI] [PubMed] [Google Scholar]
- Hoglund A., Donnes P., Blum T., Adolph H. W. & Kohlbacher O. MultiLoc: prediction of protein subcellular localization using N-terminal targeting sequences, sequence motifs and amino acid composition. Bioinformatics 22, 1158–1165 (2006). [DOI] [PubMed] [Google Scholar]
- Small I., Peeters N., Legeai F. & Lurin C. Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4, 1581–1590 (2004). [DOI] [PubMed] [Google Scholar]
- Hawkins J. & Boden M. Detecting and sorting targeting peptides with neural networks and support vector meahines. J. Bioinf. Comput. Biol. 4, 1–18 (2006). [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., Brunak S. & Heijne G. v. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016 (2000). [DOI] [PubMed] [Google Scholar]
- Darling A. E., Mau B. & Perna N. T. ProgressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5, e11147 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck J. P. & Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data produced in the present work have been submitted to Genbank under the accession KT601208-KT601211 and Sequence Read Archive with the accession of SRP067546.