Abstract
In living cells, membrane morphology is regulated by various proteins. Many membrane reshaping proteins contain a Bin/Amphiphysin/Rvs (BAR) domain, which consists of a banana-shaped rod. The BAR domain bends the biomembrane along the rod axis and the features of this anisotropic bending have recently been studied. Here, we report on the role of the BAR protein rods in inducing membrane tubulation, using large-scale coarse-grained simulations. We reveal that a small spontaneous side curvature perpendicular to the rod can drastically alter the tubulation dynamics at high protein density, whereas no significant difference is obtained at low density. A percolated network is intermediately formed depending on the side curvature. This network suppresses tubule protrusion, leading to the slow formation of fewer tubules. Thus, the side curvature, which is generated by protein–protein and membrane–protein interactions, plays a significant role in tubulation dynamics. We also find that positive surface tensions and the vesicle membrane curvature can stabilize this network structure by suppressing the tubulation.
The Bin/Amphiphysin/Rvs (BAR) superfamily proteins regulate the membrane shape of cell organella as well as membrane fusion and fission; therefore, BAR protein dysfunction is implicated in neurodegenerative, cardiovascular, and neoplastic diseases1,2,3,4,5,6,7. However, the manner in which these proteins assemble on the biomembrane and cooperate to reshape the membranes is not well understood. The extension of membrane tubes from liposomes and specific adsorption of the BAR superfamily proteins onto tube regions have been observed in in vitro experiments1,2,3,8,9,10,11,12,13,14,15. Frost et al. have experimentally determined that F-BAR proteins are adsorbed on flat regions of lipid membranes using electron microscopy9. Although the assembly seems to constitute the nucleus of the tubule formation, the tubule protrusion process has not been experimentally observed. Recently, Tanaka-Takiguchi et al. reported that the formation dynamics of tubules from a liposome can differ significantly for different F-BAR proteins13. That is, FBP17 and CIP4 simultaneously generate many tubule protrusions over the entire liposome surface, while PSTPIP1 and Pacsin2 generate only a few protrusions from a narrow region of the surface. In particular, the tubules induced by CIP4 and PSTPIP1 have the same radius. Thus, the tubule nucleation process depends on the protein type. However, it is not known what causes this difference in tubule nucleation behaviour. Tanaka-Takiguchi et al. also reported that the full length of Pacsin2 induces tubulation, but its F-BAR domain region alone does not13. In contrast, Wang et al. reported the tubulation is induced by F-BAR domain of Pacsin1 more than by the full-length protein10.
In the last decade, interactions between laterally isotropic objects on biomembrane, such as transmembrane proteins and adsorbed spherical colloids, have been intensively investigated16,17,18,19,20,21,22. In contrast to such studies, however, the interactions between anisotropic adhesives have not yet been explored so far. The BAR domains are banana shaped and generate an anisotropic curvature different from the isotropic spontaneous curvature C023. This anisotropic nature has recently been receiving increasing theoretical interest. The classical Canham–Helfrich curvature free energy24,25 has been extended to anisotropic curvatures26,27,28. Dommersnes and Fournier have derived a many-body potential of long-range interactions between point-like anisotropic inclusions and found linear assemblies and egg-carton membrane structures using Monte Carlo simulations29,30. In addition, the adsorption and assembly of BAR domains have been investigated using atomic and coarse-grained molecular simulations31,32,33,34. For example, Simunovic et al. have simulated a linear aggregation of N-BAR domains parallel to the domain axis33,34. However, the relationship between this aggregation and tubulation remains unclear. Further, tubular formation has been simulated using a dynamically triangulated membrane model35,36 and, also, meshless membrane models37,38. Despite these numerous advancements, the present understanding of the physics of membrane shape deformation due to anisotropic curvature is still far from complete.
In this paper, we focus on the effects of the spontaneous (side) curvature Cside of a protein rod perpendicular to its longest axis on the assembly behaviour. The side curvature has not been focused upon in previous studies, but here we reveal that it strikingly changes the assembly dynamics. The excluded volume or van der Waals attraction between proteins and the membrane can effectively generate positive or negative Cside. We simulate flat membranes and vesicles using an implicit-solvent meshless membrane model38,39,40,41,42, which allows a large-scale simulation. A BAR domain is modelled as a banana-shaped rod, which is assumed to be strongly adsorbed onto the membrane. The rod length corresponds to 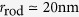 (the BAR domain lengths range from 13 to 27 nm2). To investigate the membrane-curvature-mediated interactions, no direct attractive interaction is considered between the rods. Our previous studies showed that parallel and perpendicular assemblies occur separately through membrane-mediated attractive interactions at low protein density38, and that polyhedral shapes are formed at high protein density42 for vesicles and membrane tubes.
(the BAR domain lengths range from 13 to 27 nm2). To investigate the membrane-curvature-mediated interactions, no direct attractive interaction is considered between the rods. Our previous studies showed that parallel and perpendicular assemblies occur separately through membrane-mediated attractive interactions at low protein density38, and that polyhedral shapes are formed at high protein density42 for vesicles and membrane tubes.
Results
Tubulation from Flat Membrane
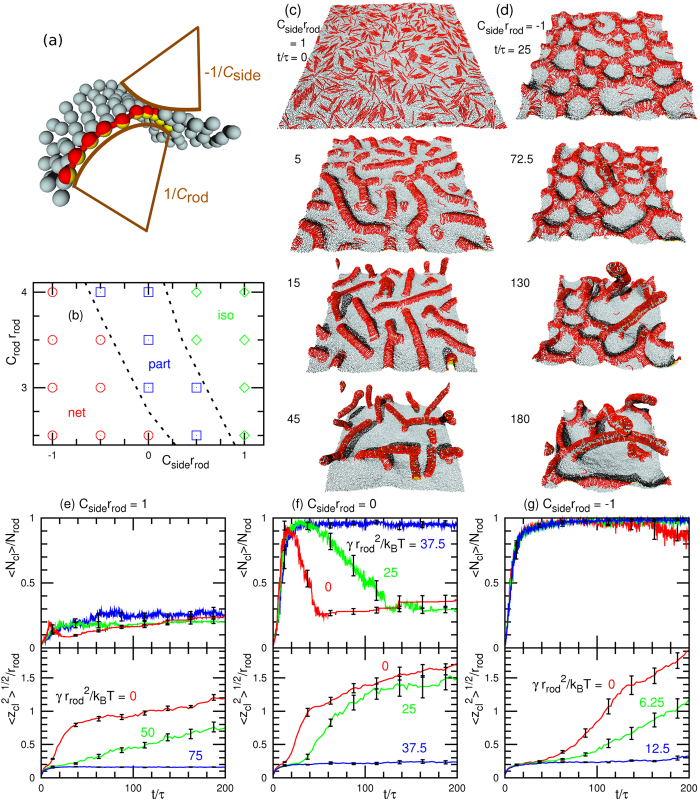
First, we investigate the tubulation from a tensionless flat membrane (surface tension γ = 0) at a high rod density, ϕrod = 0.4 (see Fig. 1). The protein rods are initially equilibrated with the rod curvature Crod = 0 and Cside = 0. Once the spontaneous curvatures are altered at t = 0, the rods begin to assemble perpendicularly to the rod axis. For a positive spontaneous curvature of Csiderrod = 1 (CsideCrod > 0), many tubules simultaneously protrude via the bending of straight rod assemblies (see Fig. 1(c) and Supplemental Movie 1). Branches of the rod network are formed on the membrane for a short time only. When the tubulation is initiated, neighbouring branches are broken through lateral shrinkage of the rod assembly.
Figure 1. Tubulation dynamics from flat membrane for high rod density, ϕrod = 0.4 (Nrod = 1,024).
(a) Protein rod with spontaneous rod and side curvatures, Crod and Cside, respectively. The protein rod is displayed as a chain of spheres, the halves of which are coloured red and yellow. The orientation vector lies along the line of the yellow to red hemispheres. The light blue spheres represent membrane particles. (b) Dynamic phase diagram of tubulation from a tensionless flat membrane. The red circles represent percolated network formation before tubulation. The green diamonds indicate that the tubules are formed from isolated clusters. The blue squares represent partial network formation, and the dashed lines are guides for the eye. (c,d) Sequential snapshots of tubulation from tensionless flat membrane for Csiderrod = (c) 1 and (d) −1 at Crodrrod = 4. (e–g) Time evolution of mean cluster size 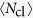 and mean cluster height
and mean cluster height 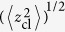 for Csiderrod = (e) 1, (f) 0, and (g) −1 at Crodrrod = 4. Error bars calculated from eight independent runs are displayed at several data points.
for Csiderrod = (e) 1, (f) 0, and (g) −1 at Crodrrod = 4. Error bars calculated from eight independent runs are displayed at several data points.
For a negative curvature Csiderrod = −1, the rods form a percolated network covering the entire membrane area [see the top snapshot in Fig. 1(d)] and a tubule protrudes under membrane undulation (see the second snapshot in Fig. 1(d) and Supplemental Movie 2). Subsequently, tubule growth occurs along the network. Thus, the tubulation dynamics is altered remarkably by a relatively small Cside. Negative and positive Cside values stabilize and destabilize the network branches, respectively. The tubulation at Csiderrod = −1 is significantly slower than that at Csiderrod = 1 and much fewer tubules protrude: the average protrusion time of the first tubule are 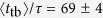 and
and 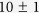 for Csiderrod = −1 and 1 at Crodrrod = 4, respectively.
for Csiderrod = −1 and 1 at Crodrrod = 4, respectively.
Such characteristic dynamics is distinguishable from the time evolution of the mean cluster size 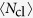 and the root mean square cluster height
and the root mean square cluster height 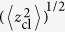 , as shown in Fig. 1(e–g). For Csiderrod = −1, the majority of the rods belong to one large percolated cluster during the tubulation. In contrast, for Csiderrod = 1,
, as shown in Fig. 1(e–g). For Csiderrod = −1, the majority of the rods belong to one large percolated cluster during the tubulation. In contrast, for Csiderrod = 1, 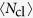 decreases as the tubules are formed and the rod assemblies are divided; subsequently,
decreases as the tubules are formed and the rod assemblies are divided; subsequently, 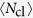 slowly increases owing to tubule fusion. Branched tubules are formed by this fusion [see the bottom snapshot in Fig. 1(c) and the late stage of Supplemental Movie 1]. Based on the evolution of
slowly increases owing to tubule fusion. Branched tubules are formed by this fusion [see the bottom snapshot in Fig. 1(c) and the late stage of Supplemental Movie 1]. Based on the evolution of 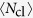 , the tubulation pathways are categorized into three groups [see Fig. 1(b)]: tubulation via percolated-network formation (net), via partial-percolated-network formation (part), and without percolation (iso). When a percolated network does not cover the entire membrane surface or a large cluster of
, the tubulation pathways are categorized into three groups [see Fig. 1(b)]: tubulation via percolated-network formation (net), via partial-percolated-network formation (part), and without percolation (iso). When a percolated network does not cover the entire membrane surface or a large cluster of 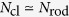 is maintained for a period shorter than 20τ, we categorize the tubulation pathway as part. (A typical dynamics is shown in Supplemental Movie 3). For the entire parameter range explored in Fig. 1(b), the final structures are tubules. As Crod decreases, the tubulation decelerates and a smaller number of large tubules are formed. The tubules are nucleated and grow from the network vertices at Crodrrod = 2.5 or 0.3 and Csiderrod = −1 (see Fig. 2). The tubule radius Rtb is roughly determined by Crod as Rtb ~ 1/Crod. At Crodrrod = 4, the tubule with circumference
is maintained for a period shorter than 20τ, we categorize the tubulation pathway as part. (A typical dynamics is shown in Supplemental Movie 3). For the entire parameter range explored in Fig. 1(b), the final structures are tubules. As Crod decreases, the tubulation decelerates and a smaller number of large tubules are formed. The tubules are nucleated and grow from the network vertices at Crodrrod = 2.5 or 0.3 and Csiderrod = −1 (see Fig. 2). The tubule radius Rtb is roughly determined by Crod as Rtb ~ 1/Crod. At Crodrrod = 4, the tubule with circumference 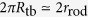 consists of two hemicylinders of the rod assembly.
consists of two hemicylinders of the rod assembly.
Figure 2. Snapshot of tubulation from tensionless flat membrane at Crodrrod = 2.5, Csiderrod = −1, and ϕrod = 0.4.

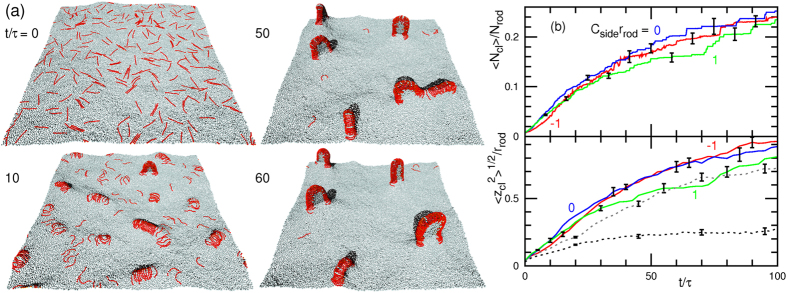
Our simulation results show that the network formation suppresses the tubulation. To confirm this more clearly, the effects of Cside on the rod–membrane interaction are investigated. A percolated network is not formed during the tubulation at a low ϕrod of 0.1. Rather, the rods assemble into linear clusters and, subsequently, the large clusters 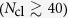 transform into tubules (see Fig. 3 and Supplemental Movie 4). Although the initial cluster formation is slightly slower for negative curvature, i.e., Csiderrod = −1, no qualitative difference is detected in the tubulation dynamics [see Fig. 3(b)]. Thus, we conclude that the suppression of the tubulation at high rod density is caused by the mesoscale network formation.
transform into tubules (see Fig. 3 and Supplemental Movie 4). Although the initial cluster formation is slightly slower for negative curvature, i.e., Csiderrod = −1, no qualitative difference is detected in the tubulation dynamics [see Fig. 3(b)]. Thus, we conclude that the suppression of the tubulation at high rod density is caused by the mesoscale network formation.
Figure 3. Membrane tubulation from flat membrane for low ϕrod = 0.1 (Nrod = 256), at Crodrrod = 4.
(a) Sequential snapshots at Cside = 0 and γ = 0. (b) Time evolution of 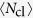 and
and 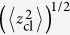 . The solid lines represent the data for Csiderrod = −1, 0, and 1 at γ = 0. The gray and black dashed lines represent the data for
. The solid lines represent the data for Csiderrod = −1, 0, and 1 at γ = 0. The gray and black dashed lines represent the data for 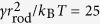 and 37.5 at Cside = 0, respectively. Error bars calculated from eight independent runs are displayed at several data points.
and 37.5 at Cside = 0, respectively. Error bars calculated from eight independent runs are displayed at several data points.
Recent experiments have demonstrated that positive surface tension can suppress tubulation by BAR proteins15 and budding by clathrin coats43. In our simulation, the positive tension and network formation cooperate to suppress tubulation [see Figs 1(e–g) and 3(b)]. At ϕrod = 0.4, the critical tension decreases with increasing Cside: 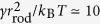 and 70 for Csiderrod = −1 and 1, respectively, where kBT denotes the thermal energy. These are experimentally measurable magnitudes (
and 70 for Csiderrod = −1 and 1, respectively, where kBT denotes the thermal energy. These are experimentally measurable magnitudes (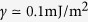 and 0.7 mJ/m2, respectively). The assembly of rods into a clustered network is not suppressed by the applied tension. In contrast, network structure breaking does not occur at higher tensions [see Fig. 1(f)]. Thus, the network formation is stabilized by the positive tensions.
and 0.7 mJ/m2, respectively). The assembly of rods into a clustered network is not suppressed by the applied tension. In contrast, network structure breaking does not occur at higher tensions [see Fig. 1(f)]. Thus, the network formation is stabilized by the positive tensions.
When a positive tension γ is imposed for the coexisting states of network and tubules as shown in the second snapshot of Fig. 1(d), the tubules continue to grow at 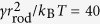 . However, the tubules shrink at
. However, the tubules shrink at 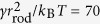 . Thus, at the critical tension
. Thus, at the critical tension 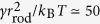 , the tubule elongation force ftb by the rod assembly is balanced with the expansion of the projected membrane area by the surface tension as
, the tubule elongation force ftb by the rod assembly is balanced with the expansion of the projected membrane area by the surface tension as 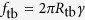 . This tension is higher than that required to suppress the tubule protrusion from the flat membranes
. This tension is higher than that required to suppress the tubule protrusion from the flat membranes 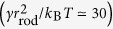 , because a nucleation barrier exists for the protrusion.
, because a nucleation barrier exists for the protrusion.
Geometrical Analysis
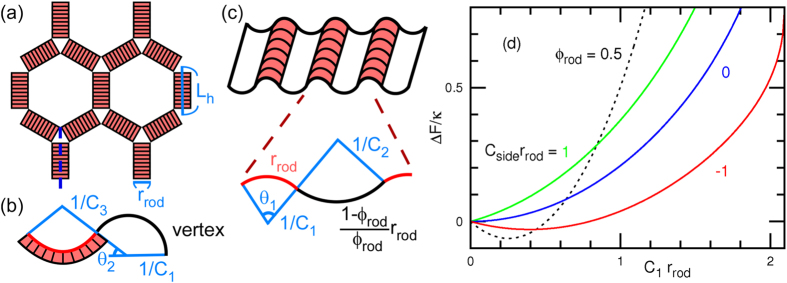
To clarify the effects of Cside on the network formation, the difference between the free energy of a hexagonal array of the rod assembly and that of a striped array is estimated using a simple geometric model (see Fig. 4). Here, a percolated network is modelled as a hexagonal array with side length Lh [Fig. 4(a)], while an unbranched rod assembly is modelled as a striped array [Fig. 4(c)]. The rod assemblies have rectangular shapes with widths equal to the rod length rrod. Our analysis shows that the hexagonal network can have lower energy for Cside < 0. This explains why the membranes are trapped in the branched network as a local free-energy minimum.
Figure 4. Energy analysis of network structure using simple geometric model.
(a–c) Schematic representation of geometric model. (a) Top view of hexagonal array at C1 = 0. (b) Side view of hexagonal array along the dashed line in (a). (c) Bird’s-eye and front views of striped array. (d) Free-energy difference 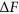 between hexagonal and striped arrays of rod assembly. The solid lines represent the data for Csiderrod = −1, 0, and 1 at ϕrod = 0.4. The dashed line represents the data for Csiderrod = −1 and ϕrod = 0.5.
between hexagonal and striped arrays of rod assembly. The solid lines represent the data for Csiderrod = −1, 0, and 1 at ϕrod = 0.4. The dashed line represents the data for Csiderrod = −1 and ϕrod = 0.5.
In the striped array, the rod assemblies are aligned in parallel with intervals of (1 − ϕrod)rrod/ϕrod. The rod assemblies are curved upwards and the other regions are curved downwards, as shown in Fig. 4(c). To maintain the continuity of the normal vector of the membrane, the curvatures have the relation C2 = ϕrodC1/(1 − ϕrod). The curvature energy Fst of the striped array per the area 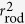 is given by
is given by
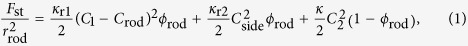 |
where κr1 and κr2 are the bending rigidities of the rod assembly parallel and perpendicular to the rod axis, respectively. In our simulation, κ = 15kBT, κr1 = 40kBT, and κr2 = κ42.
In the hexagonal array case, the membrane surface is divided into the following three regions: Region I: A rectangular rod assembly with length 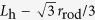 and width rrod; Region II: A triangular membrane with side length rrod at the vertex; and Region III: A hexagonal membrane with side length
and width rrod; Region II: A triangular membrane with side length rrod at the vertex; and Region III: A hexagonal membrane with side length 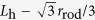 . On a flat membrane, the areas of these regions are given by
. On a flat membrane, the areas of these regions are given by 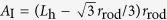 ,
, 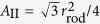 , and
, and 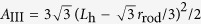 , respectively. To simplify the calculation, it is assumed that each region has constant curvatures and its area is independent of these curvatures. Region I has curvatures C1 along the rod axis and –C3 perpendicular to the rod axis [along the dashed line in Fig. 4(a)], such that it has a saddle shape. Regions II and III are triangular and hexagonal spherical caps with radii 1/C1 and 1/C4, respectively. To maintain the continuity of the normal vector of the membrane,
, respectively. To simplify the calculation, it is assumed that each region has constant curvatures and its area is independent of these curvatures. Region I has curvatures C1 along the rod axis and –C3 perpendicular to the rod axis [along the dashed line in Fig. 4(a)], such that it has a saddle shape. Regions II and III are triangular and hexagonal spherical caps with radii 1/C1 and 1/C4, respectively. To maintain the continuity of the normal vector of the membrane, 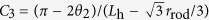 and
and 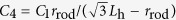 . The angle θ2 is given by
. The angle θ2 is given by 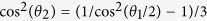 , where
, where 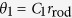 , since the vertices of three rod assemblies make contact with each other on the spherical cap and maintain three-fold rotational symmetry. At θ2 = 0, C1 has a maximum value of 2π/3rrod. As the area fraction of region I is
, since the vertices of three rod assemblies make contact with each other on the spherical cap and maintain three-fold rotational symmetry. At θ2 = 0, C1 has a maximum value of 2π/3rrod. As the area fraction of region I is 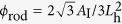 , Lh is obtained as
, Lh is obtained as
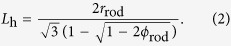 |
The curvature energy Fhex of the hexagonal array per 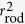 is given by
is given by
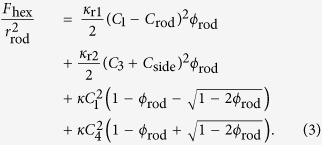 |
The energy difference 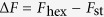 is shown in Fig. 4(d). The first terms in equations (1) and (3) cancel since both arrays have the same curvature C1 along the rods. Hence,
is shown in Fig. 4(d). The first terms in equations (1) and (3) cancel since both arrays have the same curvature C1 along the rods. Hence, 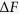 is independent of Crod and κr1. In the hexagonal array, the curvature energy of the rod assembly is reduced for Cside < 0, and Fhex is smaller than Fst for small C1. Thus, the branched network can be stabilized by a negative Cside during the formation of the rod assembly.
is independent of Crod and κr1. In the hexagonal array, the curvature energy of the rod assembly is reduced for Cside < 0, and Fhex is smaller than Fst for small C1. Thus, the branched network can be stabilized by a negative Cside during the formation of the rod assembly.
In our simulation, networks are formed in a wider range of Cside for smaller values of Crod [see Fig. 1(b)]. This dependence can be explained by an effective increase in the area fraction ϕrod. The rod assembly region contains more membrane particles with decreasing Crod (see light blue particles in rod network and tubules in Fig. 2). Thus, the area of region I increases. At ϕrod = 0.5, the minimum of 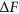 is twice that at ϕrod = 0.4, although the area fraction is only 25% larger [see the dashed line in Fig. 4(d)]. Thus, the effective increase in the rod region enhances the network formation.
is twice that at ϕrod = 0.4, although the area fraction is only 25% larger [see the dashed line in Fig. 4(d)]. Thus, the effective increase in the rod region enhances the network formation.
As ϕrod decreases, Lh of the hexagonal array increases [see equation (2)]. However, it is difficult for the hexagonal network with long Lh to form spontaneously, since the tubule formation begins before the rod assembly reaches Lh. The simulation results for ϕrod = 0.1 indicate tubulation from clusters with 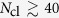 at
at 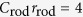 . Thus, the formation threshold of the percolated network is
. Thus, the formation threshold of the percolated network is 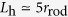 , where
, where 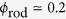 . In the simulation at ϕrod = 0.2, the clusters are typically percolated only in one direction at Crodrrod = 4 and Csiderrod = −1, which supports this estimation of the critical density.
. In the simulation at ϕrod = 0.2, the clusters are typically percolated only in one direction at Crodrrod = 4 and Csiderrod = −1, which supports this estimation of the critical density.
Shape Transformation of Small Vesicles
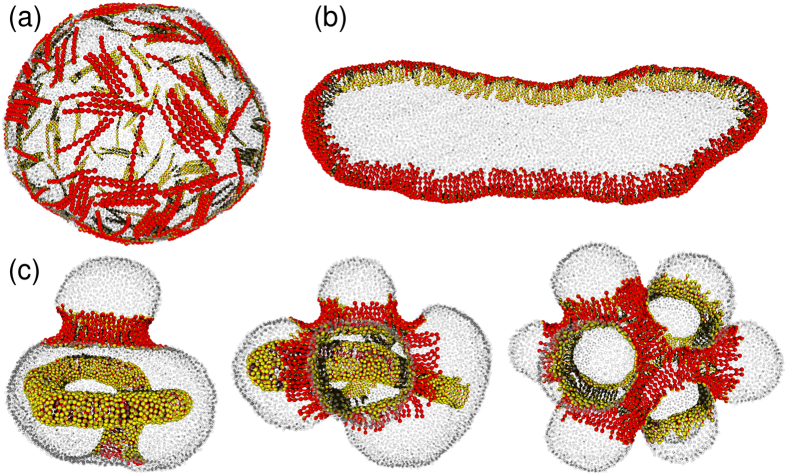
Next, we investigate the tubulation from a vesicle of radius Rves = 3.07 rrod at ϕrod = 0.3 (see Fig. 5) and demonstrate that the original membrane curvature Cves = 1/Rves = 0.33/rrod changes the tubulation dynamics. For the positive curvature Crodrrod = 4, no tubulation is obtained for Csiderrod = −1, 0, and 1. Instead, the vesicle deforms to an elliptic disk and the rods surround the disk rim [see Fig. 5(b)]. A discoidal bud is often transiently formed, but the rearrangement of the rod assemblies results in disk formation even for Csiderrod = −1 (see Supplemental Movie 5). Thus, outward tubulation and network formation are suppressed in small vesicles.
Figure 5. Snapshots of vesicles at ϕrod = 0.3 and Nrod = 288.
(a) Spherical shape in thermal equilibrium at Crod = 0 and Cside = 0. (b) Elongated discoidal shape in thermal equilibrium at Crodrrod = 4 and Csiderrod = −1. (c) Three metastable shapes at Crodrrod = −4 and Csiderrod = 1. The membrane particles are displayed as small transparent spheres for clarity.
In contrast, for the negative curvature Crodrrod = −4, tubulation into the inside of the vesicle is obtained (see Fig. 5(c) and Supplemental Movie 6). A percolated network with CsideCrod < 0 has a significantly longer lifetime than in the flat membrane case [see the middle and right snapshots in Fig. 5(c)]. The coexistence of tubules and a ring is also obtained [see the left snapshot in Fig. 5(c)]. The ring stabilizes an outward bud. For Crod < 0, the rods bend the membrane towards the interior (opposite to the original membrane curve), such that the rods locally form a saddle shape in which the two principal curvatures have opposite signs. The network and ring structures are stabilized by the positive (opposite) Cside > 0, but not by Cside < 0.
Discussion
We have revealed that, in addition to the spontaneous curvature along the protein rods Crod, the perpendicular spontaneous curvature Cside significantly influences the protrusion of membrane tubules. The percolated-network structure of the rod assembly has a long lifetime for Cside < 0, because the saddle membrane shape at branches of the rod network is stabilized by the opposite curvature of Cside with respect to Crod. Thus, the network formation decelerates the tubulation significantly, despite having a minor effect on the equilibrium property. Both positive surface tensions and membranes originally bending in the same direction as Cside < 0 can stabilize the network structure. Our findings provide new insights into the regulation of biomembrane shapes by curvature-inducing proteins.
Here, we employ Langevin dynamics, in which hydrodynamic interactions are neglected. Since the static stability of the network branch is the key factor, we do not expect the obtained Cside dependence to be qualitatively changed by the hydrodynamic interactions. However, the network formation condition may be modified. The diffusion coefficient of the proteins on the membrane depends on the protein size, and fast protein diffusion compared to the membrane deformation speed likely enhances the network formation.
The F-BAR domain of Pascin is considered to have a nonzero side curvature, since it has an S-shape on the membrane surface in the addition to the curvature perpendicular to the membrane10. Pascin induces membrane tubes in a wide range of diameter10,14. In the present simulations, we did not obtain such a behaviour. It may require a larger side curvature or attractive interactions between the rods. The rod assembly with large side curvatures is an interesting problem for further studies.
An assembly of F-BAR proteins, Cdc15, has been observed along the contractile ring of cell division44. Their adsorption to the inner leaflet of the plasma membrane is considered to yield a ring structure similar to that shown in Fig. 5(c). Our study suggests that the side curvature may play an important role in the formation of neck-like structures during cell division and membrane budding in endo/exocytosis.
Methods
We employ one of the meshless membrane models41, in which a fluid membrane is represented by a self-assembled one-layer sheet of membrane particles. A membrane particle has an excluded volume with diameter σ and an orientational degree of freedom. The solvent is implicitly accounted for by an effective potential between the membrane particles. The mechanical properties of the fluid membrane can be varied over a wide range. The details of the meshless membrane model and protein rods are described in refs 38,41, respectively. In this study, we employ the parameter set used in ref. 38 for a membrane with isotropic spontaneous curvature C0 = 0. The membrane has mechanical properties typical for lipid membranes: Bending rigidity κ/kBT = 15 ± 1, tensionless membrane area per particle  , area compression modulus
, area compression modulus 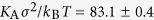 , and edge line tension
, and edge line tension 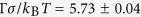 .
.
A BAR protein is modelled as a curved rod consisting of a chain of Nsg membrane particles with rrod = 10σ and Nsg = 10. The rod has anisotropic spontaneous curvature Crod along its length and spontaneous curvature Cside perpendicular to its length. When two protein rods come into contact, Cside is applied between the rods. When the rod is surrounded by membrane particles, the spontaneous curvature Cside/2 is applied between the rod and neighbouring membrane particles. A molecular dynamics with a Langevin thermostat is employed41,45. The simulation results are displayed with a time unit of 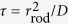 , where D is the diffusion coefficient of the membrane particles in the tensionless membranes. We use total particle numbers N = 25,600 and 9,600 for flat membranes and vesicles, respectively. The rod density is defined as ϕrod = NrodNsg/N.
, where D is the diffusion coefficient of the membrane particles in the tensionless membranes. We use total particle numbers N = 25,600 and 9,600 for flat membranes and vesicles, respectively. The rod density is defined as ϕrod = NrodNsg/N.
The mean square cluster height 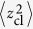 is calculated as follows. A rod is considered to belong to a cluster when the distance between the centres of mass of the rod and one of the rods in the cluster is less than rrod/2. The height variance of each cluster is calculated as
is calculated as follows. A rod is considered to belong to a cluster when the distance between the centres of mass of the rod and one of the rods in the cluster is less than rrod/2. The height variance of each cluster is calculated as 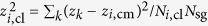 , where Ni,cl is the number of rods belonging to the i-th cluster, zi,cm is the z component of the centre of mass of the cluster, and the summation is taken over all rod segments in the cluster. Finally,
, where Ni,cl is the number of rods belonging to the i-th cluster, zi,cm is the z component of the centre of mass of the cluster, and the summation is taken over all rod segments in the cluster. Finally, 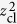 is calculated as the average of
is calculated as the average of 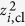 for all clusters.
for all clusters.
Additional Information
How to cite this article: Noguchi, H. Membrane tubule formation by banana-shaped proteins with or without transient network structure. Sci. Rep. 6, 20935; doi: 10.1038/srep20935 (2016).
Supplementary Material
Acknowledgments
This work was partially supported by a Grant-in-Aid for Scientific Research on Innovative Areas “Fluctuation & Structure” (No. 25103010) from MEXT, Japan.
Footnotes
Author Contributions H.N. performed the research and wrote the manuscript.
References
- Itoh T. & De Camilli P. BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochim. Biophys. Acta 1761, 897–912 (2006). [DOI] [PubMed] [Google Scholar]
- Masuda M. & Mochizuki N. Structural characteristics of BAR domain superfamily to sculpt the membrane. Semin. Cell Dev. Biol. 21, 391–398 (2010). [DOI] [PubMed] [Google Scholar]
- Mim C. & Unger M. V. Membrane curvature and its generation by BAR proteins. Trends Biochem. Sci. 37, 526–533 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerberg J. & Kozlov M. M. How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell Biol. 7, 9–19 (2006). [DOI] [PubMed] [Google Scholar]
- Baumgart T., Capraro B. R., Zhu C. & Das S. L. Thermodynamics and Mechanics of Membrane Curvature Generation and Sensing by Proteins and Lipids. Annu. Rev. Phys. Chem. 62, 483–507 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes L., Wunder C. & Bassereau P. Bending “On the Rocks”–A Cocktail of Biophysical Modules to Build Endocytic Pathways. Cold Spring Harbor Perspect. Biol. 6, a016741 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez A. et al. T. Bin/Amphiphysin/Rvs (BAR) family members bend membranes in cells. Sci. Rep. 4, 4693 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila P. K. et al. Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J. Cell Biol. 176, 953–964 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost A. et al. Structural basis of membrane invagination by F-BAR domains. Cell 132, 807–817 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. et al. Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein Pacsin/Syndapin. Proc. Natl. Acad. Sci. USA 106, 12700–12705 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorre B. et al. Nature of curvature coupling of amphiphysin with membranes depends on its bound density. Proc. Natl. Acad. Sci. USA 109, 173–178 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Das S. L. & Baumgart T. Nonlinear Sorting, Curvature Generation, and Crowding of Endophilin N-BAR on Tubular Membranes. Biophys. J. 102, 1837–1845 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka-Takiguchi Y. et al. Physicochemical Analysis from Real-Time Imaging of Liposome Tubulation Reveals the Characteristics of Individual F-BAR Domain Proteins. Langmuir 29, 328–336 (2013). [DOI] [PubMed] [Google Scholar]
- Ramesh P. et al. FBAR Syndapin 1 recognizes and stabilizes highly curved tubular membranes in a concentration dependent manner. Sci. Rep. 3, 1565 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z. & Baumbart T. Membrane tension and peripheral protein density mediate membrane shape transitions. Nature Comm. 6, 5974 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips R., Ursell T., Wiggins P. & Sens P. Emerging roles for lipids in shaping membrane-protein function. Nature 459, 379–385 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynwar B. J. et al. Aggregation and vesiculation of membrane proteins by curvature-mediated interactions. Nature 447, 461–464 (2007). [DOI] [PubMed] [Google Scholar]
- Atilgan E. & Sun S. X. Shape transitions in lipid membranes and protein mediated vesicle fusion and fission. J. Chem. Phys. 126, 095102 (2007). [DOI] [PubMed] [Google Scholar]
- de Meyer F. J., Venturoli M. & Smit B. Molecular simulations of lipid-mediated protein-protein interactions. Biophys. J. 95, 1851–1865 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auth T. & Gompper G. Budding and vesiculation induced by conical membrane inclusions. Phys. Rev. E 80, 031901 (2009). [DOI] [PubMed] [Google Scholar]
- Šarić A. & Cacciuto A. Fluid Membranes Can Drive Linear Aggregation of Adsorbed Spherical Nanoparticles. Phys. Rev. Lett. 108, 118101 (2012). [DOI] [PubMed] [Google Scholar]
- Aimon S. et al. Membrane Shape Modulates Transmembrane Protein Distribution. Dev. Cell 28, 212–218 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipowsky R. Spontaneous tubulation of membranes and vesicles reveals membrane tension generated by spontaneous curvature. Faraday Discuss. 161, 305–331 (2013). [DOI] [PubMed] [Google Scholar]
- Canham P. B. The minimum energy of bending as a possible explanation of the biconcave shape of the human red blood cell. J. Theor. Biol. 26, 61–81 (1970). [DOI] [PubMed] [Google Scholar]
- Helfrich W. Elastic properties of lipid bilayers: theory and possible experiments. Z. Naturforsch 28c, 693–703 (1973). [DOI] [PubMed] [Google Scholar]
- Fournier J. B. Nontopological saddle-splay and curvature instabilities from anisotropic membrane inclusions. Phys. Rev. Lett. 76, 4436–4439 (1996). [DOI] [PubMed] [Google Scholar]
- Kabaso D. et al. Attachment of Rod-Like (BAR) Proteins and Membrane Shape. Mini Rev. Med. Chem. 11, 272–282 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglič A. et al. Curvature-induced accumulation of anisotropic membrane components and raft formation in cylindrical membrane protrusions. J. Theor. Biol. 240, 368–373 (2006). [DOI] [PubMed] [Google Scholar]
- Dommersnes P. G. & Fournier J. B. N-body study of anisotropic membrane inclusions: Membrane mediated interactions and ordered aggregation. Eur. Phys. J. B 12, 9–12 (1999). [Google Scholar]
- Dommersnes P. G. & Fournier J. B. The many-body problem for anisotropic membrane inclusions and the self-assembly of “saddle” defects into an “egg carton”. Biophys. J. 83, 2898–2905 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkhipov A., Yin Y. & Schulten K. Four-scale description of membrane sculpting by BAR domains. Biophys. J. 95, 2806–2821 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. & Schulten K. Membrane Sculpting by F-BAR Domains Studied by Molecular Dynamics Simulations. PLoS Comput. Biol. 9, e1002892 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simunovic M., Srivastava A. & Voth G. A. Linear aggregation of proteins on the membrane as a prelude to membrane remodeling. Proc. Natl. Acad. Sci. USA 110, 20396–20401 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simunovic M. & Voth G. A. Membrane tension controls the assembly of curvature-generating proteins. Nature Comm. 6, 7219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan N., Ipsen J. H. & Kumar S. P. B. Role of disclinatons in determining the morphology of deformable fluid interfaces. Soft Matter 8, 3058–3061 (2012). [Google Scholar]
- Ramakrishnan N., Kumar S. P. B. & Ipsen J. H. Membrane-mediated aggregation of curvature-inducing nematogens and membrane tubulation. Biophys. J. 104, 1018–1028 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayton G. S. et al. New Insights into BAR Domain-Induced Membrane Remodeling. Biophys. J. 97, 1616–1625 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi H. Two- or three-step assembly of banana-shaped proteins coupled with shape transformation of lipid membranes. EPL 108, 48001 (2014). [Google Scholar]
- Noguchi H. Membrane simulation models from nanometer to micrometer scale. J. Phys. Soc. Jpn. 78, 041007 (2009). [Google Scholar]
- Noguchi H. & Gompper G. Meshless membrane model based on the moving least-squares method. Phys. Rev. E 73, 021903 (2006). [DOI] [PubMed] [Google Scholar]
- Shiba H. & Noguchi H. Estimation of the bending rigidity and spontaneous curvature of fluid membranes in simulations. Phys. Rev. E 84, 031926 (2011). [DOI] [PubMed] [Google Scholar]
- Noguchi H. Formation of polyhedral vesicles and polygonal membrane tubes induced by banana-shaped proteins. J. Chem. Phys. 143, 243109 (2015). [DOI] [PubMed] [Google Scholar]
- Saleem M. et al. A balance between membrane elasticity and polymerization energy sets the shape of spherical clathrin coats. Nature Comm. 6, 6249 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D., Coffman V. C., Lee I. J. & Wu J. Q. Assembly and architecture of precursor nodes during fission yeast cytokinesis. J. Cell Biol. 192, 1005–1012 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi H. Solvent-free coarse-grained lipid model for large-scale simulations. J. Chem. Phys. 134, 055101 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.