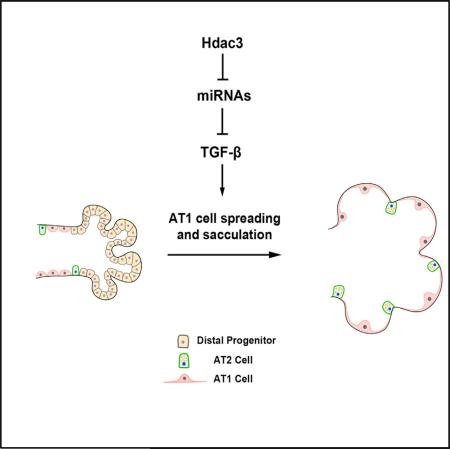
SUMMARY
The terminal stages of pulmonary development, called sacculation and alveologenesis, involve both differentiation of distal lung endoderm progenitors and extensive cellular remodeling of the resultant epithelial lineages. These processes are coupled with dramatic expansion of distal airspace and surface area. Despite the importance of these late developmental processes and their relation to neonatal respiratory diseases, little is understood about the molecular and cellular pathways critical for their successful completion. We show that a histone deacetylase 3 (Hdac3)-mediated epigenetic pathway is critical for the proper remodeling and expansion of the distal lung saccules into primitive alveoli. Loss of Hdac3 in the developing lung epithelium leads to a reduction of alveolar type 1 cell spreading and a disruption of lung sacculation. Hdac3 represses miR-17-92 expression, a micro-RNA cluster that regulates transforming growth factor β (TGF-β) signaling. De-repression of miR-17-92 in Hdac3-deficient lung epithelium results in decreased TGF-β signaling activity. Importantly, inhibition of TGF-β signaling and overexpression of miR-17-92 can phenocopy the defects observed in Hdac3 null lungs. Conversely, loss of miR-17-92 expression rescues many of the defects caused by loss of Hdac3 in the lung. These studies reveal an intricate epigenetic pathway where Hdac3 is required to repress miR-17-92 expression to allow for proper TGF-β signaling during lung sacculation.
Graphical Abstract
INTRODUCTION
The saccular stage of lung development, which extends from approximately embryonic day 16.5 (E16.5) to E18.5 of mouse gestation, is a pivotal step when the distal airspace saccules are generated as a first step toward alveologenesis. Disruption of this process can lead to serious diseases such as bronchopulmonary dysplasia in neonates. Lung sacculation and alveologenesis involve dramatic changes in the architecture and cellular composition of the distal airways. Prior to sacculation, the narrowed distal airway tubules are lined with epithelial progenitor cells that are cuboidal in shape and express markers such as Sox9 and Id2 (Rawlins et al., 2009). By E17.5 a wave of airspace expansion and alveolar epithelial differentiation occurs at the bronchoalveolar junction, which then progresses toward the distal airway tip at E18.5 (Desai et al., 2014; Treutlein et al., 2014). This results in the differentiation of two major alveolar epithelial cell lineages; the flat squamous alveolar type I (AT1) cells and the small cuboidal alveolar type II (AT2) cells. After specification, AT1 cells spread extensively and cover approximately 95% of the luminal surface of alveoli. While earlier stages of lung development including branching morphogenesis have become relatively well understood in recent studies, far less is known about sacculation and alveologenesis in the lung. In particular, how AT1 cells remodel and form the extensive surface area to mediate efficient oxygen diffusion is unclear.
Recent evidence has begun to shed light on the role of histone deacetylases (Hdacs) during lung endoderm progenitor specification (Wang et al., 2013). The class I Hdacs, Hdac1 and Hdac2, are required for development of early Sox2+ proximal lung endoderm progenitors, through regulation of Bmp4 and cell-cycle regulators including Rb1 (Wang et al., 2013). However, what roles other class I Hdacs including Hdac3 play in lung development and homeostasis has remained unclear. Importantly, Hdac3 associates with the NCoR/SMRT complex whereas Hdacs 1 and 2 associate with complexes such as NuRD/Sin3a (Guenther et al., 2000, 2001; Li et al., 2000; Zhang et al., 1997), suggesting potentially different roles for these Hdacs and chromatin remodeling complexes during lung development.
In this report, we show that Hdac3-mediated transcriptional regulation is required for the formation of distal alveolar saccules and early lung alveologenesis. Hdac3 acts in a cell-autonomous manner to regulate AT1 cell spreading, a process required for formation of the distal alveoli, without affecting specification or early differentiation of this lineage. Loss of Hdac3 results in de-repression of two major microRNA (miRNA) clusters including miR-17-92, a cluster of miRNAs that has been previously reported to be important for lung sacculation (Lu et al., 2007). miR-17-92 targets and inhibits the transforming growth factor β (TGF-β) pathway (Dews et al., 2010; Mestdagh et al., 2010), which is known to regulate cell spreading, adhesion, and tissue morphogenesis (Edlund et al., 2002; Heino et al., 1989; Ignotz et al., 1989; Massague, 2012). Overexpression of this miRNA cluster in the developing lung epithelium leads to decreased TGF-β signaling and inhibition of sacculation, whereas epithelial loss of miR-17-92 rescues much of the phenotype caused by epithelial loss of Hdac3 expression, including AT1 cell spreading and TGF-β signaling. These data reveal a molecular program regulated by Hdac3 that is essential for the spreading of AT1 cells during late lung development, a process critical for sacculation and formation of the large alveolar surface area in the lung required for postnatal gas exchange.
RESULTS
Loss of Hdac3 in the Lung Epithelium Leads to Defects in Sacculation
Lung sacculation and alveologenesis results in the extensive dilation and expansion of the distal lung region after initial specification and differentiation of AT1 and AT2 cells (Figure 1A). Examination of Hdac3 expression during lung development showed that it is broadly expressed in both epithelial and mesenchymal lineages (Figures S1A and S1C). Given the importance of Hdac1 and 2 in early lung development (Wang et al., 2013), we sought to determine whether Hdac3 is also essential for lung development through genetic inactivation of Hdac3 using the Shhcre line that drives cre-dependent recombination in the lung endoderm starting at E8.75 (Goss et al., 2009; Sun et al., 2011). Hdac3 was efficiently deleted in the developing lung epithelium of Shhcre:Hdac3f/f mutants as shown by immunostaining (Figures S1B and S1D). Approximately two-thirds of the newborn Shhcre:Hdac3f/f mutant pups died within postnatal day 2 (P2) and the rest died by P10.
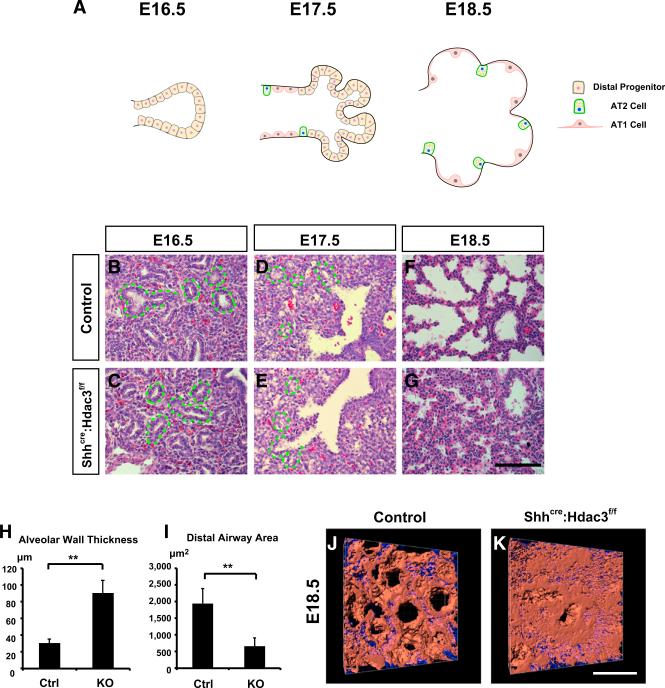
Figure 1. Loss of Epithelial Hdac3 Leads to Defects in Lung Sacculation.
(A) A model diagram describes the process of lung sacculation. Mouse lung sacculation starts at approximately E16.5 when the small distal epithelial tubules are composed of cuboidal distal progenitor cells. At E17.5, the differentiation of these progenitor cells into AT1 and AT2 cells occurs first at the bronchoalveolar junction, which then progresses toward the distal airway tip. This is accompanied by the expansion of distal airway space at the same time. At E18.5, the distal saccules are further enlarged and AT1 cells expand substantially to cover the alveolar surface.
(B–I) The Shhcre:Hdac3f/f mutants show no obvious defects in lung development at E16.5 or E17.5 by H&E staining (B–E). Green dotted lines outline the distal airways that have not undergone sacculation. At E18.5, the Shhcre:Hdac3f/f mutant lungs display disrupted lung sacculation (F and G) as evidenced by an increase in the alveolar wall thickness (H) and smaller distal airspace (I) compared with control lungs.
(J and K) The sacculation defects are further demonstrated by whole-mount staining of Aqp5 at E18.5 and a subsequent 3D reconstruction of airway surface (red indicates Aqp5 staining and blue indicates nuclei).
Ctrl, control; KO, Shhcre:Hdac3f/f. Two-tailed Student's t test: **p < 0.01. Quantification data are represented as mean ± SD. Scale bars: (G) 100 μm; (K) 50 μm.
Shhcre:Hdac3f/f mutants developed normally and showed no obvious lung defects up to E17.5, a time point that marks early sacculation (Figures 1B–1E). However, histological analysis of E18.5 Shhcre:Hdac3f/f mutant lungs revealed significantly compacted distal airspaces and increased thickness of the alveolar wall (Figures 1F–1I). These defects were further illustrated by whole-mount staining of the AT1 cell marker Aqp5 and subsequent 3D reconstruction of the saccular surfaces, which showed a compressed distal lung region with little airspace dilation in the Shhcre:Hdac3f/f mutants (Figures 1J and 1K, and Movies S1 and S2).
Previous studies have shown that one common function of Hdac proteins is to regulate cell proliferation and survival through direct de-repression of cell-cycle inhibitors (Montgomery et al., 2007; Wilson et al., 2006; Yamaguchi et al., 2010). Bromodeoxyuridine labeling or phospho-histone H3 immunostaining did not reveal any detectable changes in cell proliferation at E16.5 and E18.5 (Figures S1E–S1J). Moreover, there were no obvious changes in cell apoptosis as noted by cleaved caspase-3 immunostaining at these time points (Figures S1K–S1N). These data suggest that Hdac3 does not control lung sacculation through modulation of cell proliferation or survival.
Hdac3 Is Required for the Proper Spreading of AT1 Cells during Lung Sacculation
AT1 and AT2 cells are the major epithelial cell lineages lining the alveolar sacs and are thought to derive from a common precursor (Rawlins et al., 2009; Treutlein et al., 2014). Their differentiation and maturation from the Sox9+ distal progenitor pool is an important process occurring at sacculation, and decreased AT1 cell differentiation in particular is associated with sacculation and alveolarization defects during lung development (Becker et al., 2011; Kalin et al., 2008; Wongtrakool et al., 2003). To assess whether the alveolar epithelial cell-fate commitment occurs properly in the Shhcre:Hdac3f/f mutants, we examined the expression of several markers for AT2 and AT1 cells. AT2 cells appeared to differentiate normally in Shhcre:Hdac3f/f lungs, as the expression levels of Sftpc, Sftpb, and Abca3 were not significantly altered (Figures 2A–2F and S2A–S2C). The AT1 cell marker Aqp5 was unchanged as shown by immunostaining and qPCR (Figures 2A–2F and S2A). Expression of Pdpn, which is present in both AT1 cells and lymphatic endothelial cells, was modestly reduced by qPCR but was unaltered using immunostaining (Figures S2A, S2D, and S2E). We quantified the percentage of AT2 and AT1 cells in the distal region and showed no significant changes in the numbers of these epithelial lineages in Shhcre:Hdac3f/f mutant lungs (Figures S2H and S2I). In addition, expression of markers for proximal airway epithelial lineages and lung mesenchymal lineages were not significantly altered in the Shhcre:Hdac3f/f mutant lungs (Figures S2A and S2J–S2Q). These data suggest that the lineage commitment of multiple cell types, including that of AT1 and AT2 cells, is largely unaffected by loss of Hdac3. This is in contrast to a previous mouse model in which a mutation of SMRT that abrogated its interaction with nuclear receptors resulted in inhibition of AT1 cell differentiation via regulation of transcription factor Klf2. However, Klf2 expression in our Shhcre:Hdac3f/f lungs was not altered (Figure S2R), suggesting that Hdac3 and SMRT may have non-overlapping roles in lung development.
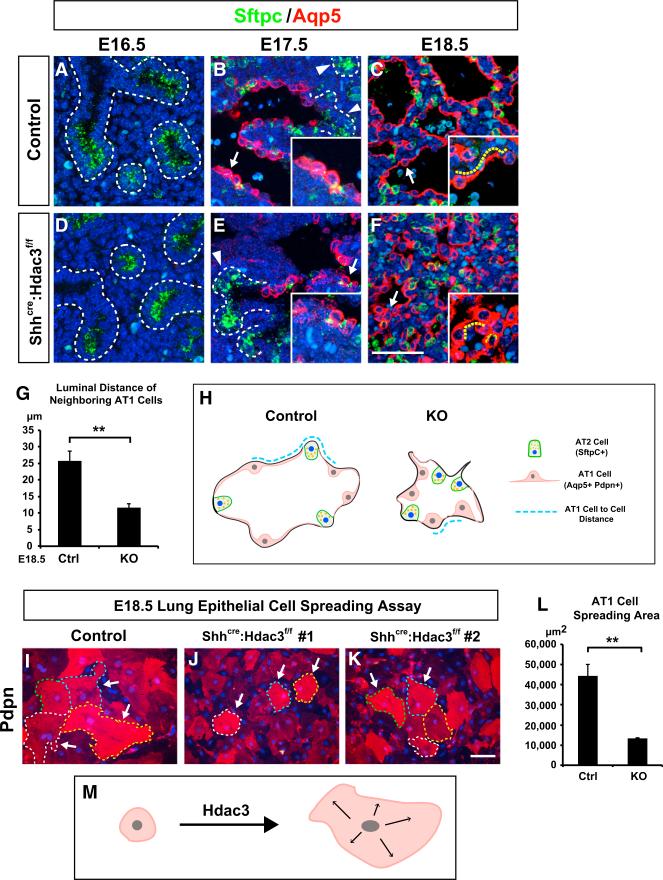
Figure 2. Hdac3 Is Required for AT1 Cell Spreading during Lung Sacculation.
(A–G) Prior to lung sacculation, Sftpc (green) is expressed in the distal airway progenitor cells at E16.5 (A) and becomes restricted to the AT2 cells as sacculation proceeds (B and C). The AT1 cell marker Aqp5 (red) is initially not expressed in the E16.5 distal airways (A). Aqp5 expression is first observed near the bronchoalveolar junction at around E17.5 and marks the nascent AT1 cells that are still cuboidal at this time point (B, arrow). Aqp5 expression is restricted to the junction region and is not found in the distal portion of the primitive saccules (B, arrowheads). At E18.5, Aqp5 expression spatially extends to the distal tip of the airways, and at the same time the AT1 cells undergo substantial cell spreading to achieve a sheet-like morphology (C, arrow). In the Shhcre:Hdac3f/f mutant lungs, the expression level and pattern of Sftpc and Aqp5 are not affected during sacculation (D–F). At 17.5, Aqp5 expression is found in the nascent AT1 cells (E, arrow) and not in the distal portion of primitive saccules (E, arrowhead) of the Shhcre:Hdac3f/f mutant lungs, similar to controls at this stage. However, the mutant AT1 cells exhibit a disruption in cell spreading as shown by Aqp5 staining at E18.5 (F, arrow). The luminal distance between two neighboring AT1 cells (yellow dotted lines in C and F) is significantly decreased in the Shhcre:Hdac3f/f mutant lungs (G). Dotted lines in (A), (B), (D), and (E) outline the distal airways where Aqp5 is not expressed.
(H) A model diagram illustrates the defects in AT1 cell spreading and distal airway expansion in the Shhcre:Hdac3f/f mutant lungs at E18.5 and how the neighboring AT1 cell-cell distances are measured.
(I–L) The E18.5 primary lung epithelial cell culture shows that the Shhcre:Hdac3f/f mutant AT1 cells have a cell-intrinsic defect in spreading as marked by the significantly decreased area of Pdpn+ cells in the culture (I–K, arrows and dotted lines). The spreading ability of AT1 cells is quantified by the mean areas of Pdpn+ cells (L).
(M) A diagram shows Hdac3 promotes the spreading of nascent AT1 cells during lung sacculation in a cell-autonomous manner.
Ctrl, control; KO, Shhcre:Hdac3f/f. Two-tailed Student's t test: **p < 0.01; n = 3. Quantification data are represented as mean ± SD. Scale bars: (F) 50 μm; (K) 100 μm.
See also Figure S2.
After their appearance at E17.5 as noted by Aqp5 expression, AT1 cells gradually spread and covered 95% of the alveolar surface to form the thin gas-diffusible interface between the airspace and pulmonary vasculature. This morphological change started to occur rapidly after AT1 cell commitment at E17.5, and the flattened morphology of AT1 cells was evident in the E18.5 wild-type lungs (Figures 2A–2C). In contrast, the majority of Shhcre:Hdac3f/f mutant AT1 cells appeared densely packed and failed to acquire a flat squamous cell shape at this stage (Figure 2F). Using ImageJ to quantitate the luminal distance between neighboring AT1 cells, we found that this distance was greatly reduced in the Shhcre:Hdac3f/f mutant lungs (Figures 2G–2H). To further examine whether this disruption of AT1 cell morphology was due to a cell-intrinsic defect in spreading or merely an indirect consequence of unexpanded distal airways, we isolated and cultured E18.5 control and Shhcre:Hdac3f/f mutant lung primary epithelial cells and examined their ability to spread in vitro without the influence of mesenchymal cells. Immunostaining for Pdpn showed that AT1 cells from Shhcre:Hdac3f/f mutant lungs were unable to spread as extensively as those from their control littermate lungs (Figures 2I–2L). These data indicate that while epithelial Hdac3 is dispensable for alveolar AT1 and AT2 cell lineage commitment, it is essential for promoting AT1 cell spreading during lung sacculation (Figure 2M).
miR-17-92 and the miRNAs in the Dlk1-Dio3 Locus Are De-repressed upon Loss of Hdac3 Expression in the Lung Epithelium
To further define the molecular changes that occur upon loss of Hdac3 expression in the lung epithelium, we examined the changes in the transcriptome of Shhcre:Hdac3f/f mutant lungs at E18.5. 1,070 genes were differentially expressed upon loss of Hdac3 expression, with 569 genes upregulated and 501 downregulated (Table S1). Strikingly, expression of 62 miRNA primary transcripts was increased in the Shhcre:Hdac3f/f mutant lungs (Figure 3A). Analysis of these miRNAs revealed that the majority of them belonged to two genetic loci: the large imprinted Dlk1-Dio3 locus located on chromosome 12 and the miR-17-92 cluster located on chromosome 14. To determine whether these miRNAs were de-repressed in a cell-autonomous manner by loss of Hdac3, we performed mature miRNA qPCR on the isolated EpCAM+ epithelial cells and EpCAM mesenchymal cells from E18.5 lungs (Figures 3B and S3A). These experiments confirmed that the miR-17-92 cluster was significantly de-repressed in lung epithelial cells but not in mesenchymal cells after an epithelial cell-specific loss of Hdac3 (Figures 3C and S3B), whereas fewer of the mature miRNAs in the Dlk1-Dio3 locus were validated to be de-repressed, and the ones that were exhibited a smaller fold change compared with the miR-17-92 cluster (Figure 3C).
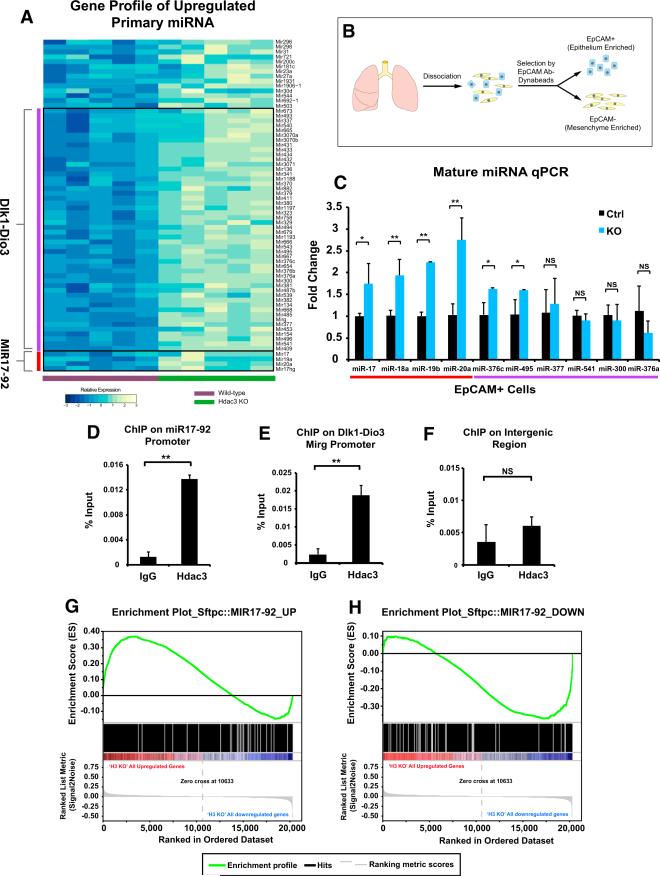
Figure 3. Loss of Epithelial Hdac3 Leads to De-repression of miRNAs, Including miR-17-92 and Dlk1-Dio3 Locus, in a Cell-Autonomous Fashion.
(A) Microarray analysis reveals differential upregulation of 62 primary miRNA transcripts in the E18.5 Shhcre:Hdac3f/f mutant lungs. Genomic location analysis of these miRNAs shows that the majority of them belong to two miRNA clusters: miR-17-92 and those in the Dlk1-Dio3 locus.
(B) Diagram showing how E18.5 lung epithelial cells are isolated by EpCAM antibody-conjugated Dynabeads and subjected to downstream experiments such as qPCR analysis.
(C) qPCR analysis on the mature form of miRNAs confirms the upregulation of most members of miR-17-92 cluster and some members of Dlk1-Dio3 locus in the Shhcre:Hdac3f/f mutant epithelial cells.
(D–F) ChIP assays show that Hdac3 can directly associate with the promoters of both miR-17-92 (D) and Mirg (E), which is the host gene for most miRNAs in the Dlk1-Dio3 locus. In contrast, Hdac3 does not associate with an unrelated intergenic region (F).
(G and H) The transcriptomes of E18.5 Sftpc::MIR17-92 and Shhcre:Hdac3f/f lungs are compared by GSEA. Enrichment plots show that the “upregulated gene set” from the Sftpc::MIR17-92 microarray were enriched in the upregulated genes in the Shhcre:Hdac3f/f microarray (G). The “downregulated gene set” from the Sftpc::MIR17-92 microarray were enriched in the downregulated genes in the Shhcre:Hdac3f/f microarray (H).
Ctrl, control; KO, Shhcre:Hdac3f/f. IgG, immunoglobulin G. Two-tailed Student's t test: *p < 0.05; **p < 0.01; NS, not significant; n = 3. qPCR data are represented as mean ± SD.
To investigate whether Hdac3 can directly repress the expression of these miRNAs, we examined whether Hdac3 could associated with the promoters of the miR-17-92 cluster and Mirg, which hosts most of the Dlk1-Dio3 miRNAs, using chromatin immunoprecipitation (ChIP) assays on E18.5 lungs. These data showed that Hdac3 associates with the miR-17-92 and Mirg promoters but not with an unrelated intergenic region (Figures 3D–3F), suggesting that Hdac3 can directly regulate the expression of these miRNAs.
Previous studies have reported that miR-17-92 is highly expressed at early stages of lung development but declines as development proceeds (Bhaskaran et al., 2009; Lu et al., 2007). Transgenic overexpression of miR-17-92 in the developing lung epithelium using the mouse Sftpc promoter (Sftpc:MIR17-92) inhibited epithelial differentiation and resulted in immature sacculation, similar to but more severe than occurs in the Shhcre:Hdac3f/f mutant lungs (Lu et al., 2007). This suggests that Hdac3 may play a role in repressing the expression of miR-17-92 to allow proper lung sacculation to proceed. To determine whether the transcriptome changes caused by loss of Hdac3 were similar to those caused by overexpression of miR-17-92, we compared the differentially upregulated and downregulated genes from the previously reported Sftpc:MIR17-92 microarray (Lu et al., 2007) with the transcriptome analysis we performed on the Shhcre:Hdac3f/f mutant lungs. Using gene set enrichment analysis (GSEA), we found that the upregulated gene set from the Sftpc:MIR17-92 microarray were enriched in the genes upregulated in the Shhcre:Hdac3f/f transcriptome data (Figure 3G). Conversely, the downregulated gene set from the Sftpc:MIR17-92 microarray were enriched in the genes downregulated in the Shhcre:Hdac3f/f transcriptome data (Figure 3H). These data demonstrate that loss of Hdac3 in the developing lung epithelium results in gene expression changes similar to those caused by overexpression of miR-17-92, suggesting that de-repressed miR-17-92 expression may underlie some aspects of the phenotype in Shhcre:Hdac3f/f mutants.
Elevated miRNA Expression Represses TGF-β Signaling and AT1 Cell Spreading
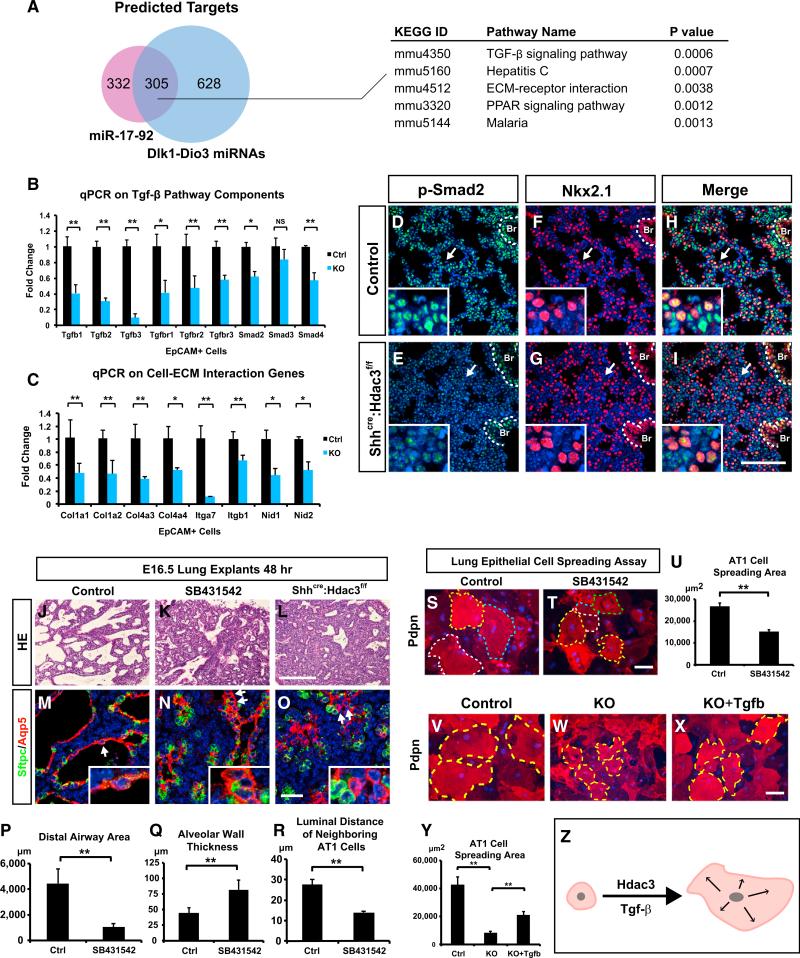
Since both the miR-17-92 and Dlk1-Dio3 locus miRNAs were de-repressed upon loss of Hdac3 in the developing lung epithelium, we compared the predicted target gene lists of these two miRNA clusters to identify common downstream molecular pathways. A total of 305 common target genes were differentially expressed in the Shhcre:Hdac3f/f mutant lungs, and signaling pathway impact analysis (SPIA) on those genes revealed TGF-β signaling and extracellular matrix (ECM)-receptor interaction as top candidate pathways targeted by these two miRNA clusters (Figure 4A and Table S2). We then performed SPIA on the predicated targets of all the upregulated miRNAs (Table S3), as well as on all the differentially downregulated genes in our microarray data (Table S4). We found that the TGF-β pathway was also among the top candidates in these analyses. Consistent with our results, previous studies have shown that miR-17-92 cluster is a potent inhibitor of TGF-β signaling through targeting key components of the TGF-β pathway as well as TGF-β target genes (Dews et al., 2010; Mestdagh et al., 2010). To verify that these TGF-β signaling components and cell-ECM genes were downregulated in the Shhcre:Hdac3f/f mutant lungs, we performed qPCR analysis, which revealed that most of these genes were significantly downregulated in the lung epithelium but not mesenchyme of Shhcre:Hdac3f/f mutants (Figures 4B, 4C, and S4A). Immunostaining showed that one of the ECM genes, Col4a3, a basement membrane component preferentially expressed in AT1 cells (Treutlein et al., 2014), was significantly downregulated in the Shhcre:Hdac3f/f mutants (Figures S4B and S4C). To determine whether the changes in TGF-β signaling components caused a decrease in TGF-β signaling activity, we performed immunostaining for p-Smad2 at E18.5. p-Smad2 level was significantly decreased in the distal epithelium and mesenchyme including the bronchoalveolar duct junction region, as noted by co-staining for the lung epithelial marker Nkx2.1, consistent with a decrease in TGF-β signaling components including TGF-β ligands (Figures 4D–4I and S4D–S4I). However, the p-Smad2 level in the proximal airway epithelium of Shhcre:Hdac3f/f mutants was relatively unaffected (Figures 4D–4I, dotted outline).
Figure 4. Elevation of miRNAs Leads to Decreased TGF-β Signaling, which in Turn Regulates Lung Sacculation and AT1 Cell Spreading.
(A) Comparison of predicted targets of miR-17-92 and miRNAs in Dlk1-Dio3 locus shows a total of 305 common genes targeted by both loci that are also differentially expressed in the Shhcre:Hdac3f/f mutant lungs. SPIA on those genes identified several top candidate pathways including TGF-β signaling and ECM-receptor interaction.
(B and C) qPCR analysis shows that multiple components of TGF-β pathway and genes involved in cell-ECM interaction are downregulated in the epithelial cells of Shhcre:Hdac3f/f mutant lungs.
(D–I) p-Smad2 is downregulated in the distal portion of Shhcre:Hdac3f/f mutant lungs (arrows) while remaining relatively unaffected in the proximal epithelium (white dotted lines). Co-staining with epithelial marker Nkx2.1 shows that p-Smad2 is downregulated in both distal epithelium and mesenchyme. Br, bronchiole.
(J–R) Treatment of TgfβRI inhibitor SB431542 on E16.5 lung explants for 48 hr disrupts lung sacculation (J and K), which mimics the Shhcre:Hdac3f/f mutant explants cultured in parallel (L). TGF-β inhibition also leads to a defect in AT1 cell spreading as shown by Aqp5 staining (M–O, arrows). The defects in saccular formation and AT1 cell spreading are quantified, which shows significant alteration under SB431542 treatment (P–R).
(S–U) Treatment of TgfβRI inhibitor SB431542 reduces AT1 cell spreading in the E18.5 lung epithelial cell culture. Dotted lines in (S) and (T) outline the Pdpn+ AT1 cells.
(V–Y) Treatment of Shhcre:Hdac3f/f epithelial culture with recombinant TGF-β1/2/3 partially restores the ability of AT1 cells to spread in vitro. Dotted lines in V–X outline the boundaries of AT1 cells in culture.
(Z) A diagram shows that TGF-β signaling promotes the spreading and expansion of AT1 cells during lung sacculation.
Ctrl, Control; KO, Shhcre:Hdac3f/f. Two-tailed Student's t test: *p < 0.05; **p < 0.01; NS, not significant; n = 3–5. qPCR and quantification data are represented as mean ± SD. Scale bars: (I, L, T, X) 100 μm; (O) 25 μm.
See also Figure S4.
TGF-β signaling is widely recognized as a crucial player in cell spreading, adhesion, and tissue morphogenesis (Edlund et al., 2002; Heino et al., 1989; Ignotz et al., 1989; Massague, 2012). Global loss of either TGF-β2 or TGF-β3 led to perinatal lung defects (Kaartinen et al., 1995; Sanford et al., 1997). TGF-β3 null lungs, in particular, were characterized by alveolar hypoplasia and mesenchymal thickening that is reminiscent of Shhcre:Hdac3f/f mutant lungs (Kaartinen et al., 1995). Moreover, lung epithelial-specific loss of TGF-β receptor type I (TgfβRI) (Alk5) also led to collapsed distal airways before birth, although detailed defects in the distal epithelial development were not explored in this study (Xing et al., 2010). To further investigate whether decreased TGF-β signaling could lead to improper remodeling of AT1 cells including reduced cell spreading, we cultured E16.5 wild-type lung explants at the air-liquid interface in the absence or presence of the TgfβRI inhibitor SB431542. The development of wild-type lung explants partially mimicked lung sacculation in vivo, as evidenced by an expansion of distal airways and the specification and early differentiation of AT1 cells, which is noted by Aqp5 expression and their flattened appearance (Figures 4J and 4M). Treatment with SB431542 reduced p-Smad2 level in the lung explants (Figures S4K and S4L), resulting in greatly narrowed distal airway lumen, and disrupted the morphology and remodeling of AT1 cells, including reduced spreading that resembled Shhcre:Hdac3f/f mutant lung explants cultured in parallel (Figures 4J–4R). This was accompanied by the downregulation of several cell-ECM genes which were also decreased in Shhcre:Hdac3f/f mutant lung epithelium (Figure S4J), suggesting that these cell-ECM genes can be regulated by TGF-β signaling. Similarly, treatment of wild-type lung epithelial cell culture with SB431542 also reduced p-Smad2 level and the spreading of AT1 cells in vitro (Figures S4M, S4N, and 4S–4U). Importantly, addition of recombinant TGF-β ligands to the Shhcre:Hdac3f/f mutant epithelial cell culture increased p-Smad2 expression and partially restored the ability of AT1 cells to spread (Figures S4O, S4P, and 4V–4Y). Together, these data suggest that epithelial-specific loss of Hdac3 in the lung suppresses TGF-β signaling and genes related to cell-ECM interaction, through upregulation of the miR-17-92 and the Dlk1-Dio3 miRNA loci. Moreover, these data demonstrate that loss of TGF-β signaling can lead to reduced AT1 cell spreading and defective lung sacculation (Figure 4Z).
Overexpression of miR-17-92 Can Phenocopy the Loss of Hdac3 and Lead to Defective AT1 Cell Spreading and Lung Sacculation
A previous study showed that transgenic overexpression of miR-17-92 led to a block in lung epithelial differentiation with an increase in Sox9+ progenitors (Lu et al., 2007). However, our Shhcre:Hdac3f/f mutant lungs did not display a change in Sox9 expression or alveolar epithelial differentiation (Figures S5A–S5D and 2A–2F). Such differences in the overall phenotype observed between these two models could be due to the differing levels of increased miR-17-92 expression, given that the level of miR-17-92 upregulation in Shhcre:Hdac3f/f mutant lungs was more modest than when using the powerful mouse Sftpc promoter (Figure 3C and Lu et al., 2007). Since our data suggest significant molecular similarities between miR-17-92 overexpression and loss of Hdac3 expression during lung epithelial development, we overexpressed miR-17-92 by utilizing the Shhcre line and a Rosa26-MIR17-92 knockin allele (R26MIR-17-92) that has been previously described to deliver a moderate level of miR-17-92 expression upon cre recombination (Xiao et al., 2008). We confirmed that the epithelial miR-17-92 expression level in these Shhcre:R26MIR-17-92 mutants was upregulated by approximately 2- to 4-fold, which was similar to that in Shhcre:Hdac3f/f mutant lungs (Figure S5E). Histological analysis showed that E18.5 Shhcre:R26MIR17-92 mutant lungs exhibited a decrease in the distal airway luminal size and an increase in the alveolar wall thickness, similar to but less severe than in the Shhcre:Hdac3f/f mutant lungs (Figures 5A–5H). Aqp5 staining revealed that a large proportion of AT1 cells in Shhcre:R26MIR17-92 lungs were densely packed and had not spread properly (Figures 5I–5K). Multiple components of the TGF-β signaling pathway, along with genes related to cell-ECM interaction, were decreased in Shhcre:R26MIR17-92 lung epithelial cells (Figures 5L and 5M), as well as in the MLE12 mouse lung epithelial cells overexpressing miR-17-92 (Figure S5F). These data suggest that elevated levels of this miRNA cluster can directly suppress TGF-β signaling in lung epithelial cells. Similarly to the Shhcre:Hdac3f/f lungs, we did not observe an increase in Sox9+ or Sox2+ epithelial progenitor cells in the Shhcre:R26MIR-17-92 lungs at E18.5 (Figures S5A–S5D and S5G–S5L), suggesting that a higher expression level of miR-17-92 may be required to trigger such a block in distal epithelial differentiation. The phenotype observed in Shhcre:R26MIR17-92 lungs including the extent of TGF-β signaling downregulation was generally milder than in Shhcre:Hdac3f/f lungs, suggesting that additional mechanisms, such as the upregulation of Dlk1-Dio3 miRNAs, might also play a role.
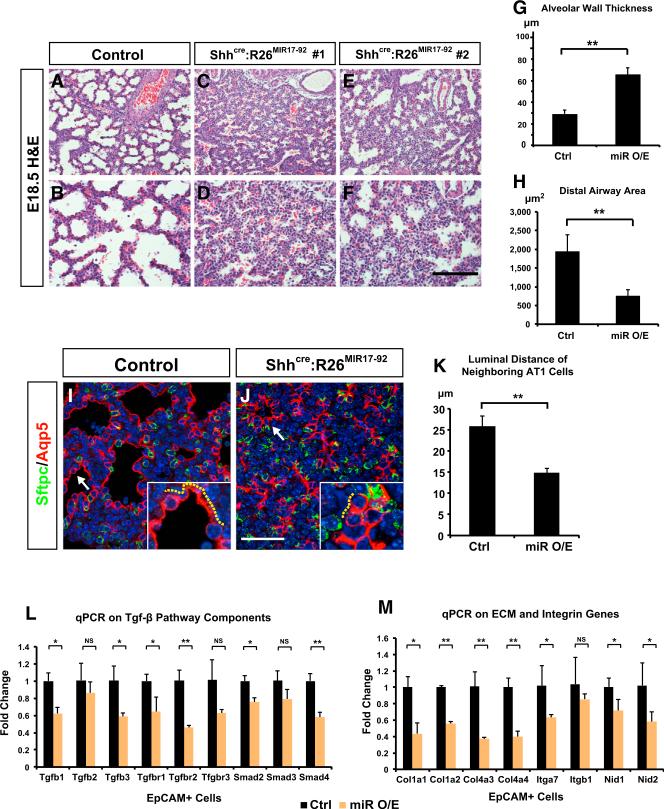
Figure 5. Overexpression of miR-17-92 Leads to Defective AT1 Cell Spreading and Lung Sacculation Similar to Loss of Hdac3.
(A–F) E18.5 Shhcre:R26MIR17-92 lungs display defective sacculation that is characterized by increased alveolar wall thickness and narrowed distal airway lumens, similar to the Shhcre:Hdac3f/f mutant lungs.
(G and H) Quantification of alveolar wall thickness and distal airway size in control and Shhcre:R26MIR17-92 lungs.
(I–K) Double staining of Sftpc and Aqp5 shows that while AT2 cells are not altered, AT1 cells demonstrate reduced spreading in Shhcre:R26MIR17-92 lungs at E18.5 (I and J, arrows and yellow dotted lines). This defect is evidenced by a decrease in the luminal distance between two neighboring AT1 cells (K).
(L and M) qPCR analysis indicates that multiple components of TGF-β pathway as well as genes involved in cell-ECM interaction are decreased specifically in the epithelial cells of Shhcre:R26MIR17-92 lungs at E18.5.
Ctrl, control; miR O/E, Shhcre:R26MIR17-92. Two-tailed Student's t test: *p < 0.05; **p < 0.01; NS, not significant; n = 3. qPCR and quantification data are represented as mean ± SD. Scale bars: (F) 100 μm; (J) 50 μm.
See also Figure S5.
Lung Sacculation Requires Hdac3-Mediated Repression of miR-17-92
To determine whether the sacculation defects in Shhcre:Hdac3f/f lungs are directly mediated by increased expression of miR-17-92, we generated mice lacking both Hdac3 and miR-17-92 expression (Shhcre:Hdac3f/f:MIR17-92f/f). In contrast to a previous report of a lung hypoplasia phenotype in the miR-17-92 global knockout mice (Ventura et al., 2008), Shhcre:MIR17-92f/f mutants were viable with no obvious lung phenotype (data not shown). Next, we generated Shhcre:Hdac3f/f:MIR17-92f/f mutants and compared these with Shhcre:Hdac3f/f mutants at E18.5. The sacculation defects observed in Shhcre:Hdac3f/f mutant lungs, including thickened alveolar wall and compacted distal saccular lumen, were significantly alleviated in Shhcre:Hdac3f/f:MIR17-92f/f mutant lungs (Figures 6A–6J and Figure S6). AT1 cells in Shhcre:Hdac3f/f:MIR17-92f/f mutant lungs also exhibited greater spreading with a more flattened appearance compared with those in the Shhcre:Hdac3f/f mutant lungs (Figures 6K–6N). Together, these studies revealed that loss of miR-17-92 significantly restores the AT1 cell remodeling and sacculation defects in Shhcre:Hdac3f/f lungs (Figure 6O).
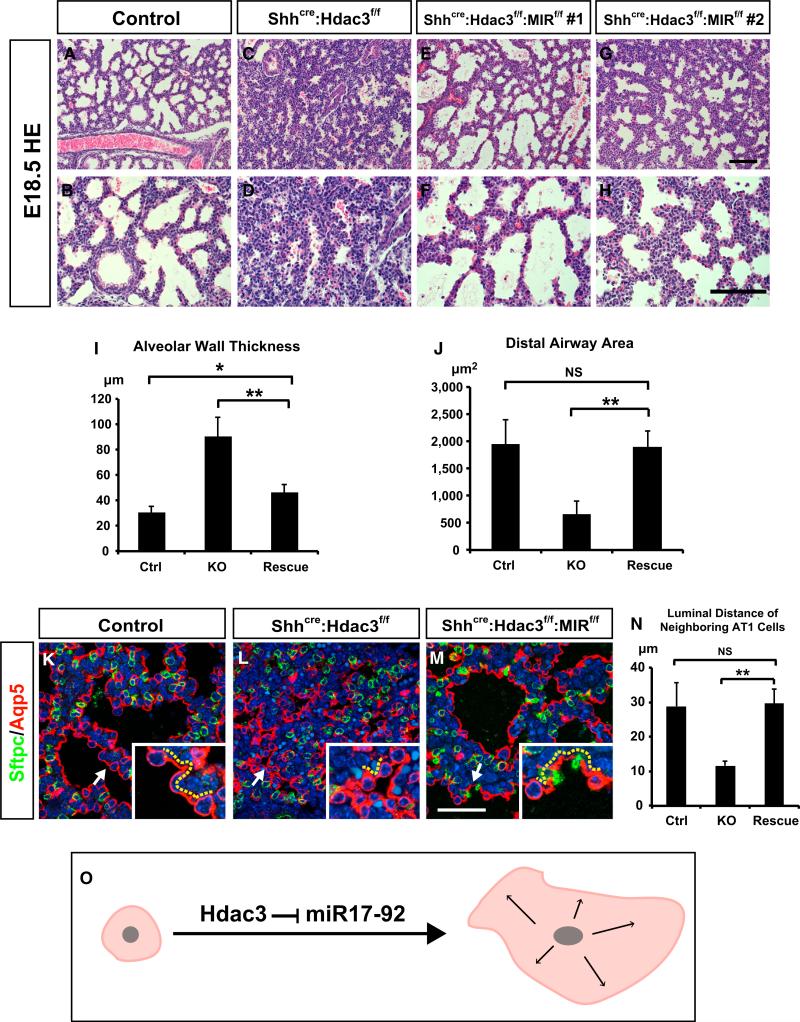
Figure 6. Loss of Epithelial miR-17-92 Alleviates the Defects of Lung Sacculation and AT1 Cell Spreading in the Shhcre:Hdac3f/f Mutant Lungs.
(A–H) H&E staining shows that the sacculation defects at E18.5 are alleviated in Shhcre:Hdac3f/f:MIR17-92f/f lungs compared with Shhcre:Hdac3f/f lungs.
(I and J) Quantification of alveolar wall thickness and distal airway area indicates that these indices are significantly restored in Shhcre:Hdac3f/f:MIR17-92f/f lungs at E18.5.
(K–N) Aqp5 immunostaining (red) and quantification of luminal distance (arrows and yellow dotted lines) between two neighboring AT1 cells show that the defect in AT1 cell spreading is significantly improved in Shhcre:Hdac3f/f:MIR17-92f/f lungs compared with Shhcre:Hdac3f/f lungs at E18.5.
(O) Diagram showing that repression of miR-17-92 expression by Hdac3 is required for AT1 cell spreading.
Ctrl, control; KO, Shhcre:Hdac3f/f; Rescue, Shhcre:Hdac3f/f:MIR17-92f/f. Two-tailed Student's t test: *p < 0.05; **p < 0.01; NS, not significant; n = 3–4. Quantification data are represented as mean ± SD. Scale bars: (H) 100 μm; (M) 50 μm.
See also Figure S6.
To test whether loss of miR-17-92 expression would normalize TGF-β signaling as well as the gene expression changes observed in Shhcre:Hdac3f/f mutants, we isolated EpCAM+ cells from Shhcre:Hdac3f/f:MIR17-92f/f mutant lungs and performed qPCR analysis. Most of the gene expression changes were completely or partially normalized in Shhcre:Hdac3f/f:MIR17-92f/f mutant lungs compared with Shhcre:Hdac3f/f mutant lungs (Figures 7A and 7B). Moreover, loss of miR-17-92 expression also increased p-Smad2 level in the distal alveolar region of Shhcre:Hdac3f/f:MIR17-92f/f mutant lungs (Figures 7C–7K). These data suggest that miR-17-92, through targeting the TGF-β pathway, is a critical modulator of Hdac3-mediated regulation of AT1 cell spreading and remodeling of the distal saccules and alveolus required for postnatal lung function (Figure 7L).
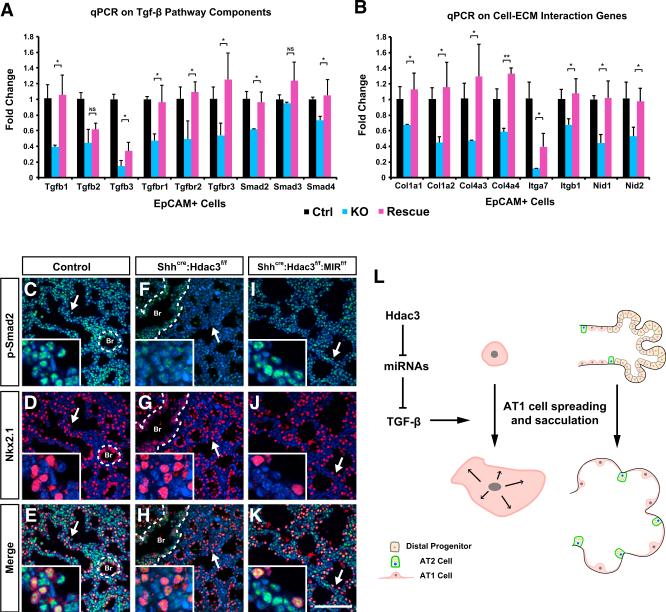
Figure 7. Loss of Epithelial miR-17-92 Significantly Restores TGF-β Pathway in the Shhcre:Hdac3f/f Mutant Lungs.
(A and B) qPCR analysis shows that multiple components of TGF-β pathway and genes involved in cell-ECM interaction are significantly restored in the E18.5 Shhcre:Hdac3f/f:MIR17-92f/f lungs compared with Shhcre:Hdac3f/f lungs.
(C–K) p-Smad2 staining level is improved in the distal region of E18.5 Shhcre:Hdac3f/f:MIR17-92f/f lungs compared with Shhcre:Hdac3f/f lungs. Nkx2.1 co-staining shows that p-Smad2 level is improved in both distal epithelium and mesenchyme (C–K, arrows). Dotted lines mark the proximal airways. Br, bronchiole.
(L) A model diagram summarizes the molecular and cellular mechanisms by which epithelial Hdac3 regulates mouse lung sacculation. Hdac3 directs lung sacculation and AT1 cell spreading through suppressing miRNAs including miR-17-92, which in turn allow for proper level of TGF-β signaling and expression of cell-ECM genes that are crucial for cell spreading and tissue morphogenesis.
Ctrl, control; KO, Shhcre:Hdac3f/f; Rescue, Shhcre:Hdac3f/f:MIR17-92f/f. Two-tailed Student's t test: *p < 0.05; **p < 0.01; NS, not significant; n = 3–4. qPCR data are represented as mean ± SD. Scale bar in (K), 100 μm.
DISCUSSION
Lung sacculation and alveologenesis remain poorly understood developmental processes that comprise elegant and complicated events of cellular differentiation and remodeling. Defects in these developmental processes can lead to serious lung diseases in the newborn, including bronchopulmonary dysplasia. How alveolar epithelial cells differentiate and remodel to form the intricate gas exchange interface required for postnatal respiration is poorly understood. In particular, how AT1 cells acquire their unique morphology during late lung development that allows them to cover 95% of alveolar luminal surface and facilitate the rapid diffusion of oxygen to the pulmonary vascular plexus is unknown. Our study demonstrates that an epigenetic pathway directed by Hdac3 regulates alveolar epithelial cell remodeling and saccular development in the mouse lung. Hdac3 is essential for AT1 cells to spread and acquire a sheet-like morphology during sacculation. Hdac3 accomplishes this developmental role through repression of the miR-17-92 and the Dlk1-Dio3 miRNA clusters, which in turn allows for appropriate levels of TGF-β signaling.
Hdac3 is a class I Hdac that interacts with the co-repressive complexes NCoR and SMRT (Guenther et al., 2000, 2001; Li et al., 2000). A previous study reported that an SMRT knockin allele that abrogated its interaction with nuclear receptors produced defects in lung sacculation and inhibited AT1 cell differentiation (Pei et al., 2011). This phenotype was partially attributed to the downstream target Klf2 and its role in promoting the AT1 cell differentiation program. In contrast to these results, loss of Hdac3 does not significantly perturb the specification of the AT1 cell lineage, and Klf2 expression levels appeared to be unchanged. One possibility for this discrepancy could be that our model examined the epithelial cell-specific function of Hdac3 while the SMRT knockin mutation is a globally expressed point mutation. Moreover, the SMRT knockin mutation used in these studies only abrogated its interaction with nuclear receptors but retained its ability to bind Hdac3. Recent studies have also implicated that SMRT has Hdac3-independent functions in some contexts (Adikesavan et al., 2014), and that SMRT and NCoR have non-redundant in vivo functions (Mottis et al., 2013; Sun et al., 2013), suggesting that despite the co-existence of Hdac3 and SMRT in the same complex, these two proteins are not completely interchangeable in their molecular functions.
Previous studies have shown that the miR-17-92 cluster is highly expressed in the early stages of lung development and declines as development proceeds (Bhaskaran et al., 2009; Lu et al., 2007). The downregulation of this cluster is important for late lung development, as transgenic epithelial-specific overexpression of miR-17-92 impeded differentiation and saccular development (Lu et al., 2007). Our study demonstrates that Hdac3 plays a regulatory role in repressing this cluster of miRNAs in the lung epithelium, and that elevated levels of miR-17-92 disrupt formation of distal saccules and primitive alveoli by inhibiting AT1 cell spreading. miR-17-92 can directly target and inhibit multiple components of TGF-β signaling including TGF-β responsive genes (Dews et al., 2010; Mestdagh et al., 2010). Consistent with this observation, we found a downregulation of genes involved in TGF-β signaling specifically in the Hdac3-deficient lung epithelium that can be partially restored by epithelial-specific loss of miR-17-92. TGF-β signaling is known as a critical regulator of cell spreading, adhesion, and migration. Previous studies have suggested that it plays a role in lung sacculation and alveologenesis, but the cellular and molecular processes it regulates during this stage of development have remained unclear (Kaartinen et al., 1995; Sanford et al., 1997; Xing et al., 2010). We found that TGF-β signaling promotes the spreading of AT1 cells during lung sacculation, supporting a model in which the morphogenetic changes of AT1 cells are directed by an Hdac3-miRNA-TGF-β molecular cascade. This process of AT1 cell spreading is likely to be important for the rapid expansion of distal saccular surface area that occurs during late lung development in preparation for alveologenesis. We also observed increased expression of miRNAs in the maternally imprinted Dlk1-Dio3 locus, which is regulated by histone acetylation in induced pluripotent stem cells (Stadtfeld et al., 2010). Several miRNAs in this locus, including miR376c, are reported or predicted to directly target components of the TGF-β pathway (Fu et al., 2013; Liu et al., 2014), suggesting that this cluster of miRNAs also plays an important role in repressing TGF-β signaling.
The generation of the thin gas-diffusible interface between the external environment and the vasculature remains a unique characteristic of the mammalian respiratory system. Our data suggest that Hdac3 plays a critical role in the initial establishment of alveolar structure through the promotion of AT1 cell spreading by inhibiting miR-17-92-mediated repression of TGF-β signaling. Such information will be beneficial to our understanding of how neonatal respiratory diseases occur and how an alveolus reestablishes its structure after postnatal injury.
EXPERIMENTAL PROCEDURES
Animals
The generation and genotyping of Hdac3flox, Shhcre, R26MIR17-92, and MIR17-92flox alleles have been previously described (Goss et al., 2009; Sun et al., 2011; Ventura et al., 2008; Xiao et al., 2008). All animal work was performed in accordance to the guidance of the University of Pennsylvania Institutional Animal Care and Use Committee.
qPCR
For mRNA qPCR, total RNA was isolated from lungs or cells at indicated time points by an RNeasy Mini Kit (Qiagen) or RNeasy Micro Kit (Qiagen). First-strand cDNA synthesis was performed using SuperScript III Reverse Transcriptase (Life Technologies). qPCR was performed using the SYBR green system (Applied Biosystems). GAPDH expression level was used as an internal control. See Supplemental Experimental Procedures for qPCR primer sequences. For mature miRNA qPCR, a MirVana miRNA Isolation Kit (Life Technologies) was used to isolate total RNA that contains small RNAs. The TaqMan MicroRNA Assays (Life Technologies) was used to quantitate various miRNA levels. Sno202 was used as an internal control for miRNA quantity.
Histology
Tissues were fixed in 4% paraformaldehyde overnight and subject to paraffin sectioning. Immunohistochemistry was performed by using the following antibodies and concentrations: Hdac3 (Santa Cruz Biotechnology, 1:10), Nkx2.1 (Santa Cruz, 1:50), p-Smad2 (Millipore, 1:1000), Sox2 (Seven Hills, 1:500), Sox9 (Santa Cruz, 1:100), Sftpc (Santa Cruz, 1:50), Sftpb (Chemicon, 1:100), Aqp5 (Abcam, 1:100), Pdpn (Hybridoma Bank, 1:50), Scgb1a1 (Santa Cruz, 1:20), TubbIV (BioGenex, 1:20), Pdgfrα (Cell Signaling, 1:25), Pecam (Pharmingen, 1:500), Col4a3 (Santa Cruz, 1:50), RAGE (R&D Systems, 1:50) c-Caspase 3 (Cell Signaling, 1:50), BrdU (Abcam, 1:100), and phospho-histone H3 (Cell Signaling, 1:200). Whole-mount staining was performed as previously described (Metzger et al., 2008).
Quantification of Alveolar Wall Thickness and Airway Lumen Area
For each sample, three pictures were taken under a 40× objective lens. The method for quantification of alveolar wall thickness was previously described (Herriges et al., 2014). The areas for distal airways was measured by using the area measurement function in ImageJ and calculated for mean value and SD.
Transcriptome and Bioinformatics Analysis
RNA was isolated from E18.5 lungs from Shhcre control and Shhcre:Hdac3f/f mutant embryos. Five lungs were collected for either control or Shhcre:Hdac3f/f groups. The RNA was then used to generate a biotinylated cRNA probe library for Affymatrix Mouse Gene 2.0ST array. Microarray data were analyzed using the Oligo package available at the Bioconductor website (www.bioconductor.org). The raw data were background-corrected by the Robust Multichip Average method and then normalized by an invariant set method. Differential gene expression between the control and mutant mice was analyzed by the Limma package available at the Bioconductor website. p Values obtained from the multiple comparison tests were corrected by false-discovery rates. Heatmap displays were created using the heatmap.2 function in the R package gplots. See Table S1 for the list of genes that were differentially expressed. The Gene Expression Omnibus accession number for the microarray data is GEO: GSE70684.
Functional enrichment analysis of pathways and gene ontology terms was performed using the GSEA software package (PMID: 16199517).
For the functional analysis of genes altered by miR-17-92 overexpression, custom “gene sets” of up- and downregulated genes were created from a previous microarray study on the E18.5 miR-17-92-overexpressing lungs (Lu et al., 2007). Functional enrichment of miR-17-92 altered gene sets in the Hdac3 mutant transcriptome was formally tested using the GSEA software package (PMID: 16199517). Predicted miRNA-gene targets were obtained from the microRNA.org website, and only target predictions that were conserved and annotated with a “good” mirSVR score were considered for analysis. Pathway analysis of genes targeted by miR-17-92 cluster and Dlk-Dio3 locus was performed using the SPIA R software package (PMID: 18990722) using the KEGG pathway annotations.
ChIP Assays
E18.5 WT lungs were dissected and cross-linked by 3.7% formaldehyde and sonicated to obtain genomic DNAs sized between 200 and 400 bp. ChIP assays were performed using the ChIP Assay Kit (Millipore) and Hdac3 antibody (Abcam). Rabbit immunoglobulin G was used as a negative control in these assays. See Supplemental Experimental Procedures for ChIP primer sequences.
Lung Explant Culture
Lung explant culture was performed as previously reported (Geng et al., 2011). In brief, E16.5 lungs were dissected and diced into 0.5- to 1-mm thick pieces and cultured on an air-liquid interface with DMEM media for 48 hr. The explants were treated with 10 μM SB431542 or vehicle.
Lung Epithelial Cell Isolation and Culture
E18.5 lungs were dissected and digested by collagenase and Dispase to obtain single-cell solution. A Dynabeads Flow Comp Flexi Kit (Life Technologies) and EpCAM antibody (eBioscience) were used to isolate primary lung epithelial cells. The epithelial cells were cultured on the surface coated by fibronectin for 8 days to achieve a well-spread epithelial sheet (Demaio et al., 2009). The epithelial cells were treated with 10 μM SB431542 from the first day of culture. For TGF-β ligand treatment, Tgfβ1, Tgfβ2, and Tgfβ3 (R&D) were added to the epithelial culture at 5 ng/ml each. Before immunostaining, cells were fixed in 4% paraformaldehyde for 10 min and permeabilized with 0.1% Tween 20.
Lentivirus
Mouse MIR17-92 overexpression construct (Lu et al., 2007) was cloned into pLenti 7.3/V5-DEST through the Gateway cloning system. Lentivirus containing empty vector or MIR17-92 overexpression construct was packaged and produced as previously reported (Herriges et al., 2014). MLE12 Cells were infected with lentivirus and allowed to grow for 48 hr before RNA was extracted for qPCR.
Supplementary Material
Highlights.
Hdac3 is important for AT1 cell spreading and remodeling during lung sacculation
Hdac3 represses miR-17-92, which in turn targets components of TGF-β signaling
Proper levels of TGF-β signaling are crucial for lung AT1 cell remodeling
ACKNOWLEDGMENTS
The authors gratefully acknowledge the Penn CVI Histology Core for histology services and Andrea Stout for help with confocal microscopy. These studies were supported by grants from the NIH to E.E.M. (HL110942) and M.A.L. (NIH R37 DK43806) and from the Penn Diabetes Center Transgenic Core (NIH P30 DK19525) and AHA fellowship to Y.W. (11PRE7580096).
Footnotes
AUTHOR CONTRIBUTIONS
Y.W., D.B.F., M.A.L., and E.E.M. designed experiments and wrote the manuscript. Y.W., D.B.F., S.Z., X.W., and M.M.L. performed experiments and analyzed data. M.P.M. analyzed bioinformatics data.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, six figures, four tables, and two movies and can be found with this article online at http://dx.doi.org/10.1016/j.devcel.2015.12.031.
REFERENCES
- Adikesavan AK, Karmakar S, Pardo P, Wang L, Liu S, Li W, Smith CL. Activation of p53 transcriptional activity by SMRT: a histone deacetylase 3-independent function of a transcriptional corepressor. Mol. Cell. Biol. 2014;34:1246–1261. doi: 10.1128/MCB.01216-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker PM, Tran TS, Delannoy MJ, He C, Shannon JM, McGrath-Morrow S. Semaphorin 3A contributes to distal pulmonary epithelial cell differentiation and lung morphogenesis. PLoS One. 2011;6:e27449. doi: 10.1371/journal.pone.0027449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskaran M, Wang Y, Zhang H, Weng T, Baviskar P, Guo Y, Gou D, Liu L. MicroRNA-127 modulates fetal lung development. Physiol. Genomics. 2009;37:268–278. doi: 10.1152/physiolgenomics.90268.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaio L, Tseng W, Balverde Z, Alvarez JR, Kim KJ, Kelley DG, Senior RM, Crandall ED, Borok Z. Characterization of mouse alveolar epithelial cell monolayers. Am. J. Physiol. Lung Cell. Mol. Physiol. 2009;296:L1051–L1058. doi: 10.1152/ajplung.00021.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai TJ, Brownfield DG, Krasnow MA. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature. 2014;507:190–194. doi: 10.1038/nature12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dews M, Fox JL, Hultine S, Sundaram P, Wang W, Liu YY, Furth E, Enders GH, El-Deiry W, Schelter JM, et al. The myc-miR-17~92 axis blunts TGF{beta} signaling and production of multiple TGF{beta}-dependent antiangiogenic factors. Cancer Res. 2010;70:8233–8246. doi: 10.1158/0008-5472.CAN-10-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund S, Landstrom M, Heldin CH, Aspenstrom P. Transforming growth factor-beta-induced mobilization of actin cytoskeleton requires signaling by small GTPases Cdc42 and RhoA. Mol. Biol. Cell. 2002;13:902–914. doi: 10.1091/mbc.01-08-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu G, Ye G, Nadeem L, Ji L, Manchanda T, Wang Y, Zhao Y, Qiao J, Wang YL, Lye S, et al. MicroRNA-376c impairs transforming growth factor-beta and nodal signaling to promote trophoblast cell proliferation and invasion. Hypertension. 2013;61:864–872. doi: 10.1161/HYPERTENSIONAHA.111.203489. [DOI] [PubMed] [Google Scholar]
- Geng Y, Dong Y, Yu M, Zhang L, Yan X, Sun J, Qiao L, Geng H, Nakajima M, Furuichi T, et al. Follistatin-like 1 (Fstl1) is a bone morphogenetic protein (BMP) 4 signaling antagonist in controlling mouse lung development. Proc. Natl. Acad. Sci. USA. 2011;108:7058–7063. doi: 10.1073/pnas.1007293108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss AM, Tian Y, Tsukiyama T, Cohen ED, Zhou D, Lu MM, Yamaguchi TP, Morrisey EE. Wnt2/2b and beta-catenin signaling are necessary and sufficient to specify lung progenitors in the fore-gut. Dev. Cell. 2009;17:290–298. doi: 10.1016/j.devcel.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Lane WS, Fischle W, Verdin E, Lazar MA, Shiekhattar R. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 2000;14:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol. 2001;21:6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heino J, Ignotz RA, Hemler ME, Crouse C, Massague J. Regulation of cell adhesion receptors by transforming growth factor-beta. Concomitant regulation of integrins that share a common beta 1 subunit. J. Biol. Chem. 1989;264:380–388. [PubMed] [Google Scholar]
- Herriges MJ, Swarr DT, Morley MP, Rathi KS, Peng T, Stewart KM, Morrisey EE. Long noncoding RNAs are spatially correlated with transcription factors and regulate lung development. Genes Dev. 2014;28:1363–1379. doi: 10.1101/gad.238782.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignotz RA, Heino J, Massague J. Regulation of cell adhesion receptors by transforming growth factor-beta. Regulation of vitronectin receptor and LFA-1. J. Biol. Chem. 1989;264:389–392. [PubMed] [Google Scholar]
- Kaartinen V, Voncken JW, Shuler C, Warburton D, Bu D, Heisterkamp N, Groffen J. Abnormal lung development and cleft palate in mice lacking TGF-beta 3 indicates defects of epithelial-mesenchymal interaction. Nat. Genet. 1995;11:415–421. doi: 10.1038/ng1295-415. [DOI] [PubMed] [Google Scholar]
- Kalin TV, Wang IC, Meliton L, Zhang Y, Wert SE, Ren X, Snyder J, Bell SM, Graf L, Jr., Whitsett JA, Kalinichenko VV. Forkhead Box m1 transcription factor is required for perinatal lung function. Proc. Natl. Acad. Sci. USA. 2008;105:19330–19335. doi: 10.1073/pnas.0806748105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang J, Wang J, Nawaz Z, Liu JM, Qin J, Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wang L, Su Z, Wu W, Cai X, Li D, Hou J, Pei D, Pan G. A reciprocal antagonism between miR-376c and TGF-beta signaling regulates neural differentiation of human pluripotent stem cells. FASEB J. 2014;28:4642–4656. doi: 10.1096/fj.13-249342. [DOI] [PubMed] [Google Scholar]
- Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev. Biol. 2007;310:442–453. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J. TGFbeta signalling in context. Nat. Rev. Mol. Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestdagh P, Bostrom AK, Impens F, Fredlund E, Van Peer G, De Antonellis P, von Stedingk K, Ghesquiere B, Schulte S, Dews M, et al. The miR-17-92 microRNA cluster regulates multiple components of the TGF-beta pathway in neuroblastoma. Mol. Cell. 2010;40:762–773. doi: 10.1016/j.molcel.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger RJ, Klein OD, Martin GR, Krasnow MA. The branching programme of mouse lung development. Nature. 2008;453:745–750. doi: 10.1038/nature07005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery RL, Davis CA, Potthoff MJ, Haberland M, Fielitz J, Qi X, Hill JA, Richardson JA, Olson EN. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottis A, Mouchiroud L, Auwerx J. Emerging roles of the core-pressors NCoR1 and SMRT in homeostasis. Genes Dev. 2013;27:819–835. doi: 10.1101/gad.214023.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei L, Leblanc M, Barish G, Atkins A, Nofsinger R, Whyte J, Gold D, He M, Kawamura K, Li HR, et al. Thyroid hormone receptor repression is linked to type I pneumocyte-associated respiratory distress syndrome. Nat. Med. 2011;17:1466–1472. doi: 10.1038/nm.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins EL, Clark CP, Xue Y, Hogan BL. The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells. Development. 2009;136:3741–3745. doi: 10.1242/dev.037317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford LP, Ormsby I, Gittenberger-de Groot AC, Sariola H, Friedman R, Boivin GP, Cardell EL, Doetschman T. TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development. 1997;124:2659–2670. doi: 10.1242/dev.124.13.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Singh N, Mullican SE, Everett LJ, Li L, Yuan L, Liu X, Epstein JA, Lazar MA. Diet-induced lethality due to deletion of the Hdac3 gene in heart and skeletal muscle. J. Biol. Chem. 2011;286:33301–33309. doi: 10.1074/jbc.M111.277707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Feng D, Fang B, Mullican SE, You SH, Lim HW, Everett LJ, Nabel CS, Li Y, Selvakumaran V, et al. Deacetylase-independent function of HDAC3 in transcription and metabolism requires nuclear receptor corepressor. Mol. Cell. 2013;52:769–782. doi: 10.1016/j.molcel.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein B, Brownfield DG, Wu AR, Neff NF, Mantalas GL, Espinoza FH, Desai TJ, Krasnow MA, Quake SR. Reconstructing line-age hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tian Y, Morley MP, Lu MM, Demayo FJ, Olson EN, Morrisey EE. Development and regeneration of Sox2+ endoderm progenitors are regulated by a Hdac1/2-Bmp4/Rb1 regulatory pathway. Dev. Cell. 2013;24:345–358. doi: 10.1016/j.devcel.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AJ, Byun DS, Popova N, Murray LB, L'Italien K, Sowa Y, Arango D, Velcich A, Augenlicht LH, Mariadason JM. Histone deacetylase 3 (HDAC3) and other class I HDACs regulate colon cell maturation and p21 expression and are deregulated in human colon cancer. J. Biol. Chem. 2006;281:13548–13558. doi: 10.1074/jbc.M510023200. [DOI] [PubMed] [Google Scholar]
- Wongtrakool C, Malpel S, Gorenstein J, Sedita J, Ramirez MI, Underhill TM, Cardoso WV. Down-regulation of retinoic acid receptor alpha signaling is required for sacculation and type I cell formation in the developing lung. J. Biol. Chem. 2003;278:46911–46918. doi: 10.1074/jbc.M307977200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat. Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Li C, Li A, Sridurongrit S, Tiozzo C, Bellusci S, Borok Z, Kaartinen V, Minoo P. Signaling via Alk5 controls the ontogeny of lung Clara cells. Development. 2010;137:825–833. doi: 10.1242/dev.040535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Cubizolles F, Zhang Y, Reichert N, Kohler H, Seiser C, Matthias P. Histone deacetylases 1 and 2 act in concert to promote the G1-to-S progression. Genes Dev. 2010;24:455–469. doi: 10.1101/gad.552310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.