Abstract
Transcription factors affect spatiotemporal patterns of gene expression often regulating multiple aspects of tissue morphogenesis, including cell-type specification, cell proliferation, cell death, cell polarity, cell shape, cell arrangement and cell migration. In this work, we describe a distinct role for Ribbon (Rib) in controlling cell shape/volume increases during elongation of the Drosophila salivary gland (SG). Notably, the morphogenetic changes in rib mutants occurred without effects on general SG cell attributes such as specification, proliferation and apoptosis. Moreover, the changes in cell shape/volume in rib mutants occurred without compromising epithelial-specific morphological attributes such as apicobasal polarity and junctional integrity. To identify the genes regulated by Rib, we performed ChIP-seq analysis in embryos driving expression of GFP-tagged Rib specifically in the SGs. To learn if the Rib binding sites identified in the ChIP-seq analysis were linked to changes in gene expression, we performed microarray analysis comparing RNA samples from age-matched wild-type and rib null embryos. From the superposed ChIP-seq and microarray gene expression data, we identified 60 genomic sites bound by Rib likely to regulate SG-specific gene expression. We confirmed several of the identified Rib targets by qRT-pCR and/or in situ hybridization. Our results indicate that Rib regulates cell growth and tissue shape in the Drosophila salivary gland via a diverse array of targets through both transcriptional activation and repression. Furthermore, our results suggest that autoregulation of rib expression may be a key component of the SG morphogenetic gene network.
Keywords: Drosophila, morphogenesis, Ribbon, salivary gland, size control
Introduction
Construction of a functionally specialized epithelial organ engages multiple aspects of cell structure and function. During the early stages of epithelial organ development, the regulation of general cell attributes such as specification and proliferation are critical (Inman et al., 2015; Maeda et al., 2007; Pan and Wright, 2011; Rawlins, 2011). Following specification, regulation of epithelial cell-type specific attributes such as integrity and polarity are required for tissue organization (Bryant and Mostov, 2008; Chung and Andrew, 2008; Laprise et al., 2009; Sotillos et al., 2013; Wang et al., 2012). Regulation of morphogenetic attributes such as changes in cell shape, orientation and migration are essential to define overall morphology and to achieve correct organ positioning (Andrew and Ewald, 2010).
Transcriptional regulation is the premier mechanism used by cells to control the differential gene expression that drives key steps during organogenesis. To understand the regulatory architecture at work during various stages of epithelial organogenesis, it is necessary to identify the targets of transcription factors that orchestrate organ development. Since transcription factors often impact multiple aspects of cell behavior that contribute to organogenesis, it is rare, if not impossible, to ascribe a selective role for any factor. For example, the bHLH transcription factor Ptf1a functions in multiple steps in specification and differentiation of pancreatic acinar cells (Cleveland et al., 2012). It is expressed and required in multipotent pancreatic progenitor cells, ultimately becoming restricted to the tip cells, where it is required for differentiation of the acini of the exocrine pancreas. In the differentiated acinar cells, Ptf1a directly binds and activates expression of the enzyme genes specific to the exocrine pancreas. Similarly, myogenic regulatory factors (Mrfs, including Myf5, MyoD, Mrf4) function early in myogenic precursor cells as muscle cell fate determinants in a partially redundant manner (Comai and Tajbakhsh, 2014). Different combinations of these same Mrfs are later also required for muscle differentiation where they directly regulate expression of genes directing myofiber formation and the structural proteins required for muscle function.
Development of the embryonic salivary gland in Drosophila melanogaster has been well characterized (Chung et al., 2014). After specification of the salivary gland primordia, the cells of the placode invaginate and form an incipient tube (Myat and Andrew, 2000a, b). An elaborate set of cell shape changes and rearrangements enable tube elongation, which occurs simultaneously as migration of the epithelial collective positions the organ along the anteroposterior axis by stage 15 (Chung and Andrew, 2014; Kerman et al., 2008; Myat and Andrew, 2002). Following cell specification, the number of cells remains constant; tissue growth occurs through increases in cell size rather than cell number. In addition, cell polarity is maintained throughout the duration of tube formation, elongation and migration. For these reasons, the Drosophila salivary gland is an attractive system to study organogenesis and, in particular, to track morphogenesis (Chung et al., 2014; Girdler and Roper, 2014).
Several transcription factors have been characterized for their roles in organogenesis and functional specialization of the embryonic Drosophila salivary gland (Abrams and Andrew, 2005; Abrams et al., 2006; Bradley and Andrew, 2001; Fox et al., 2010; Fox et al., 2013; Myat and Andrew, 2000a, b, 2002). Among these factors is Ribbon (Rib), a broadly expressed BTB domain protein, required for both salivary gland and tracheal tube elongation (Bradley and Andrew, 2001; Shim et al., 2001). The tube elongation defects in rib mutants have been linked to delays in apical/lumenal expansion, with the salivary gland lumen ultimately achieving only 60% the length of the WT lumen (Cheshire et al., 2008). Consistent with increases in apical domain stiffness, loss of rib is linked to reduced expression of the gene encoding the apical domain determinant Crumbs (Crb), increased levels of active, phosphorylated Moesin (Moe), an apically-localized ERM family protein known to link Crb to the actin cytoskeleton, and decreased apical accumulation of the recycling endosome factor Rab11 (Kerman et al., 2008). Whether Rib has additional affects on cell shape and tube morphogenesis has not been determined. It is unknown if Rib directly regulates crb expression or how Rib affects Moesin activity and localization of Rab11. Rib nuclear localization in the salivary gland, trachea and other ectodermal derivatives is dependent on the BTB-domain containing Lola-like (Lolal) transcription factor (Kerman et al., 2008). Rib’s nuclear location suggests that it exerts its effects on epithelial tube elongation by transcriptional regulation, perhaps through direct DNA binding with Lolal. Other than crb, genes whose expression is affected by Rib remain undiscovered. It is also unknown whether Rib functions as a transcriptional activator, repressor or both.
Here, we describe a distinct and unexpected role for Ribbon (Rib) in the regulation of morphogenetic attributes contributing to organogenesis. Rib regulates morphogenesis without affecting either the general or epithelial cell-specific characteristics essential for the construction of the salivary gland. Using both microarray gene expression analysis and SG-enriched Rib ChIP-seq data, we identified many target genes that are regulated by Rib and are likely to mediate salivary gland morphogenesis. These data suggest that the regulatory function of Rib is accomplished both by transcriptional activation and repression. Interestingly, the Rib gene regulatory network also includes auto-regulation, which may be critical for the function of Rib in SG morphogenesis.
Materials and methods
Fly strains
Oregon R embryos were used as the wild-type control in all experiments. The trans-allelic combination of rib1 (Bradley et al., 2001) and ribP7 (Shim et al., 2001) was used to analyze rib mutant phenotypes and gene expression changes by microarray, qRT-PCR and whole mount in situ hybridization. The rib alleles are EMS-induced mutations induced at different times and in different labs; ribP7 has a premature stop codon introduced at residue 22 and rib1 has a stop codon introduced at residue 283. UAS-rib-GFP was built by cloning a PCR amplification of the full-length rib ORF into the pENTR-D vector and subsequent gateway cloning into the pTWG vector, placing the entire GFP coding region downstream of and in frame with the Rib ORF. The following lines were generated and crossed together to test for rescue of rib mutant SG phenotypes: rib1 fkh-Gal4/CyO, ftz-lacZ and ribP7 UAS-rib-GFP/Cyo, ftz-lacZ. UAS-Nuclear-lacZ expression driven by fkh-Gal4 and sage-Gal4 was used to determine the full set of cell types where these drivers are active.
Immunohistochemistry and in situ hybridization
Embryo fixation and immunostaining were performed as described (Reuter and Scott, 1990). The primary antibodies used include rabbit α-SAS (D. Cavener, 1:500), guinea pig α-Sage (1:100), rabbit α-Forkhead (S. Beckendorf,1: 500)1: rat α-CrebA (1:1000), rat α-Pasilla (1:5000), rabbit α-Phospho histone H3 (Abcam, 1:500), rabbit α-CC3 (Cell Signaling, 1:100), α-PKC ζ (C-20, Santa Cruz Biotech,1:500), rat α-DE-Cadherin (DCAD2, DSHB,1:10), mouse α-Coracle (C566.9, DSHB, 1:200), mouse α-alpha-spectrin (3A9, DSHB, 1:1), rabbit α-Laminin (J. Fessler, 1:1000), mouse α-DCSP-2 (6D6, DSHB, 1:200), mouse α-Lamin (ADL84.12, DSHB, 1:200), mouse α-beta-gal (Promega,1:1000) and rabbit α-GFP (Molecular probes, 1:1000). For HRP staining, Biotin-labeled secondary antibodies were used at 1:500 dilution (Molecular Probes). The HRP staining signal was amplified using the Vectastain ABC kit (Vector Labs). For fluorescence staining, Alexa-488- or Alexa-568- or Alexa-647- labeled secondary antibodies were used at a 1:500 dilution (Molecular Probes). Confocal images were obtained using the LSM 700 confocal microscope (Carl Zeiss, Inc.). Whole-mount in situ hybridization was performed as described previously (Lehmann and Tautz, 1994). Images from in situ hybridized and HRP stained embryos were obtained with an Axiophot microscope (Carl Zeiss, Inc.).
Chromatin immunoprecipitation and deep sequencing
Chromatin extraction and immunoprecipitation were performed as described previously (Negre et al., 2006). Briefly, chromatin from three independent collections of stage 11 – 16 fkh-Gal4::UAS-rib-GFP embryos and three of sage-Gal4::UAS-rib-GFP embryos were cross-linked at room temperature in 1.8% formaldehyde in 2 ml of homogenization buffer (60 mM KCl, 15 mM NaCl, 15 mM HEPES {pH7.6}, 4 mM MgCl2, 0.5 mM DTT, 0.5% Triton X-100 and cOmplete™ protease inhibitor cocktail [1 tablet per 50 ml buffer]). The cross-linked material was resuspended in 0.1%SDS and 0.5% N-lauroylsarcosine in 0. lysis buffer (140mM NaCl, 15mM HEPES [pH 7.6], 1 mM EDTA, 0.5 mM EGTA, 0.1% sodium deoxycholate, 1% Triton X-100, 0.5mM DTT and cOmplete™ protease inhibitor cocktail [1 tablet per 50 ml buffer]). Chromatin was sonicated three times at 4°C using the Sonic Dismembrator Model 100 (Fisher Scientific) under the following conditions: power setting 3, 20s ON, 20s OFF. Immediately after sonication, the chromatin extract was stored at −80°C prior to immunoprecipitation. Immunoprecipitations were performed as described in (Negre et al., 2006), using a polyclonal goat anti-GFP antibody (a gift from Kevin White). Immunoprecipitated DNA was prepared for Illumina sequencing using the TruSeq ChIP Sample Prep Kit (Illumina) and sequenced on an Illumina HiSeq 2000 (50 bp reads) according to the manufacturer’s standard protocols. Library preparation and Illumina sequencing were performed at the University of Minnesota Genomics Center. Acquisition of GEO accession number (http://www.ncbi.nlm.nih.gov/geo/) for ChIP-seq data is pending.
ChIP-seq data processing
ChIP-seq peaks (binding sites) were called by comparing biological replicates to an input DNA control (from non-immunoprecipitated chromatin). Sequenced DNA was processed using FASTQC and FASTQ Groomer (Blankenberg et al., 2010), then mapped to the Drosophila melanogaster BDGP release 6 (dm6, August 2014) using Burrows-Wheeler Alignment (BWA) tool with default parameters (dos Santos et al., 2015; Li and Durbin, 2009; St Pierre et al., 2014). Sequencing reads from biological replicates were combined after mapping using Picard (http://broadinstitute.github.io/picard) and the MACS (v2) peak caller was used to identify and score peaks. (Zhang et al., 2008). Peak calling was carried out using the following MACS parameters (P-value: 10e-5; mfold: 10, 32), comparing ChIP DNA to matching input control samples.
Microarray gene expression analysis
Three independent collections of stage 11 – 16 rib1/ribP7 embryos and three of wild-type embryos were isolated using a COPAS Select embryo sorter (Union Biometrica). Total RNA was isolated by Trizol (Invitrogen) extraction and cleaned up with the Qiagen RNeasy kit (Qiagen). Total RNA (100ng) was labeled and amplified using standard Affymetrix protocols. Three samples for each genotype were hybridized to Drosophila Genome 2.0 Chips. Scanned intensity values were normalized using RMA (Partek software (Irizarry et al., 2003a; Irizarry et al., 2003b) and statistical analyses were performed using the Spotfire software package (TIBCO). Target genes were identified as those that were upregulated/downregulated (+ 1.5fold change cutoff, P < 0.05) in rib1/ribP7 embryos when compared with Oregon R controls. All microarray data have been deposited at GEO with accession number GSE72598.
Real-time quantitative PCR (RT-qPCR)
Total RNA from three independent samples each of wild-type and rib1/ribP7 mutant embryos (stage 11–16) was reverse transcribed using SuperScript III Reverse Transcriptase (Invitrogen) for two-step qPCR reactions with oligo(dT)18 primers (IDT). Gene-specific primers were used to amplify the corresponding cDNA by RT-qPCR on a CFX96 Real-Time PCR Detection System (BioRad) using SYBR Green Supermix (BioRad). Primer sets for each putative gene were chosen to give reaction products of 133–198 bp using PerlPrimer or Primer3Plus. The specificity of each primer set and molecular weight of the amplicon were monitored by dissociation curve analysis and verified by running the PCR products against size standards on a 1% agarose gel. A sample volume of 20 μl was used for all assays, which contained a 1X final concentration of SYBR Green mix, 8 μL RNase-free water, 1 μl of 5μM gene-specific primers, and 1 μl of 5 nM template. All samples were run in triplicate. Genes were chosen for RT-qPCR based on their fold change in the microarray analysis, presence of nearby Rib ChIP peak(s), and/or SG expression according to the BDGP in situ hybridization database.
Probability analysis
We used a hypergeometric distribution analysis (Mathematica) to calculate the probability of finding the number of overlaps in Rib binding sites and SG expressed genes by chance (p = 0.0021), as well as the number of overlaps in Rib binding sites and genes downregulated (p = 0.5537) or upregulated (p = 0.0486) in rib mutants based on microarray analysis. A total estimate of 18,000 Drosophila encoded genes was used for this analysis.
Image processing
Nuclear counts were performed on confocal image sections using Adobe Photoshop Elements 6.0. Cross-sectional nuclei were counted based on the following criteria: 1) presence of DAPI+ DNA, 2) connectivity of cells to the lumen and 3) strong Lamin signal separating individual DAPI+ nuclei. Manual surface renderings of the SG in the YZ -plane and the associated volumetric analyses were performed using Imaris ×64 version 7.7.2 (Bitplane, Oxford Instruments).
Motif analysis
The SeqPos tool from the Cistrome Galaxy-based platform (http://cistrome.org/ap/) was used to identify enriched motifs in ChIP-seq peaks (He et al., 2010; Liu et al., 2011).
DAVID analysis
Functional clustering of Rib targets from microarray and ChIP-seq data were performed to place them under gene ontological categories (GO terms) according to the Database for Annotation, Visualization and Integrated Discovery (DAVID) version 6.7 (Huang da et al., 2009a, b).
Statistical Analysis
Statistical analyses were carried out using Sigmaplot 11.0 (Systat software Inc.).
Results
Salivary gland cells are properly specified in rib mutant embryos
Embryos missing rib function (rib1/ribP7) have severe defects in overall SG morphology (Fig. 1A′,1B′). SGs are shorter, have irregular shapes as well as bulging lumens at late embryonic stages. We asked if these abnormalities could be due to changes in specification, proliferation or apoptosis of SG cells. Immunofluorescence staining of the SG with several SG-expressed nuclear proteins, including the bHLH transcription factor Sage (Fig. 1C, G), the winged-helix FoxA transcription factor Fork head (Fkh) (Fig. 1D, H), the bZip transcription factor CrebA (Fig. 1E, I), and the nuclear RNA binding protein Pasilla (Ps) (Fig. 1F, J) revealed similar staining in wild-type and rib mutant SGs. Once specified, WT SG cells do not undergo additional cell divisions: the tissue grows by increase in cell size, not cell number (Myat and Andrew, 2000a) (Sonnenblick, 1950). Staining with phospho-histone H3, a marker for dividing cells, indicated that there were no cryptic cell divisions in the rib mutant glands (Fig 1K – K‴, Fig 1L – L‴). Also, apoptosis does not occur during normal SG development (Ismat et al., 2013; Myat and Andrew, 2000a). Cleaved caspase 3 staining (Fig. 1M – M‴, Fig. 1N – N‴) showed no evidence for cell death in rib mutant or WT SGs. Furthermore, cell counts revealed no significant difference in the number of cells in the rib1/ribP7 SGs compared with WT SGs (Figure 1O). Collectively, these results indicate that the defective SG morphology in rib mutants is not caused by abnormalities in general cell attributes such as cell specification, cell division or cell death.
Figure 1.
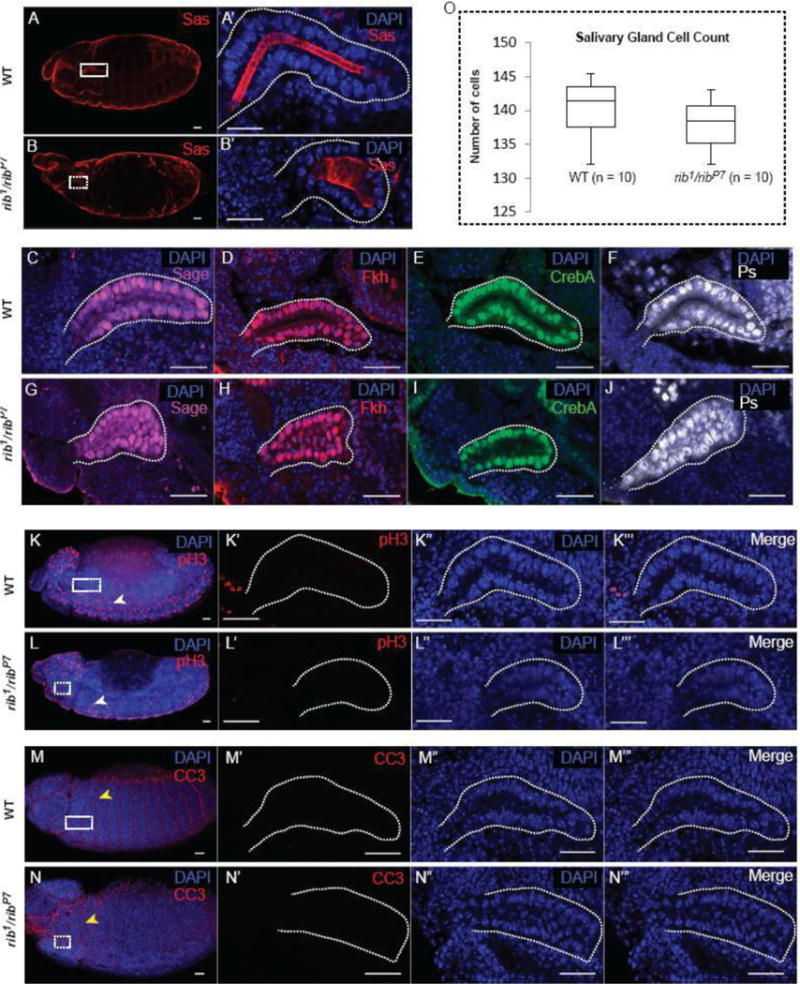
Cell specification, cell division and apoptosis are not affected in rib mutant salivary glands (SGs). (A, B) Wild-type (WT) and rib1/ribP7 embryos stained for the apical cell membrane protein SAS and DAPI show overall embryo morphology. (A′) A high magnification image of the WT SG (white box in A) reveals an elongated SG tube. (B′) A high magnification image of the rib1/ribP7 SG (white box in B) reveals a short, wide SG tube. (C – J) WT and rib mutant SGs were stained with DAPI and four different cell-specification markers: Sage (C, G), Fkh (D, H), CrebA (E, I), and Ps (F, J). (K, L) WT and rib mutant salivary glands were stained with DAPI and phospho-histone H3 (pH3) antiserum to assay for cell division. The white arrowheads indicate one of the many pH3+ neuroblasts undergoing cell division. (K′ – K‴ and L′ – L‴) High-magnification images of WT and rib mutant SGs (white boxes in K and L, respectively) are shown. (M, N) WT and rib mutant SGs were stained with DAPI and cleaved caspase3 antibody to assay for apoptosis. The yellow arrowheads indicate one of the many CC3+ intersegmental surface epithelial cells undergoing apoptosis. (M′ – M‴ and N′ – N‴) High-magnification images of WT and rib mutant SGs (white boxes in M and N, respectively) are shown. (O) Cell count from WT and rib mutants showed no statistically significant difference in the number of cells present in WT versus rib mutant SGs (n = 10 glands). Scale bars: 20 μm.
Salivary gland cell integrity, polarity and secretory potential are unaffected in rib mutants
Tissue integrity and polarity are critical determinants of epithelial organogenesis (Andrew and Ewald, 2010). As with other epithelia, tissue integrity is maintained in the developing SG epithelium by the adherens junction (AJ) and septate junction (SJ) complexes. Staining for E-Cad and other components of the AJ complex revealed their proper localization in rib mutant SGs (Fig. 2A, B; data not shown). Likewise, staining for SJ proteins, including Cora and Dlg, revealed no differences between rib mutant and WT SGs (Fig. 2C,D; data not shown). SG cells maintain apicobasal polarity throughout organogenesis with the apical surface facing the developing gland lumen (Chung and Andrew, 2014). Staining for the apical polarity marker Sas and lateral polarity marker alpha-spectrin showed that the apicolateral polarity of SG cells in rib mutants is not compromised (Fig. 2E, F). This staining also confirmed the altered cell shape in rib mutants (cuboidal vs columnar in WT) that had been reported earlier (Blake et al., 1998). Moreover, basal cell polarity is also intact based on the presence of laminin deposits observed on the basal surfaces of both WT and rib mutant SGs (Fig. 2G, H). In glandular epithelia such as the SG, cell secretion and the resulting hydrostatic pressure can affect the normal trajectory of organogenesis and hence gland morphology (Navis and Bagnat, 2015). Staining for the secretory vesicle marker CSP2 showed apical enrichment suggesting uncompromised secretory potential in rib mutant SGs (Fig. 2I, J). Taken together, these results suggest that the defective tube morphology in rib mutant SGs is not attributable to abnormal epithelial cell type-specific characteristics such as cell integrity, polarity or secretory potential.
Figure 2.
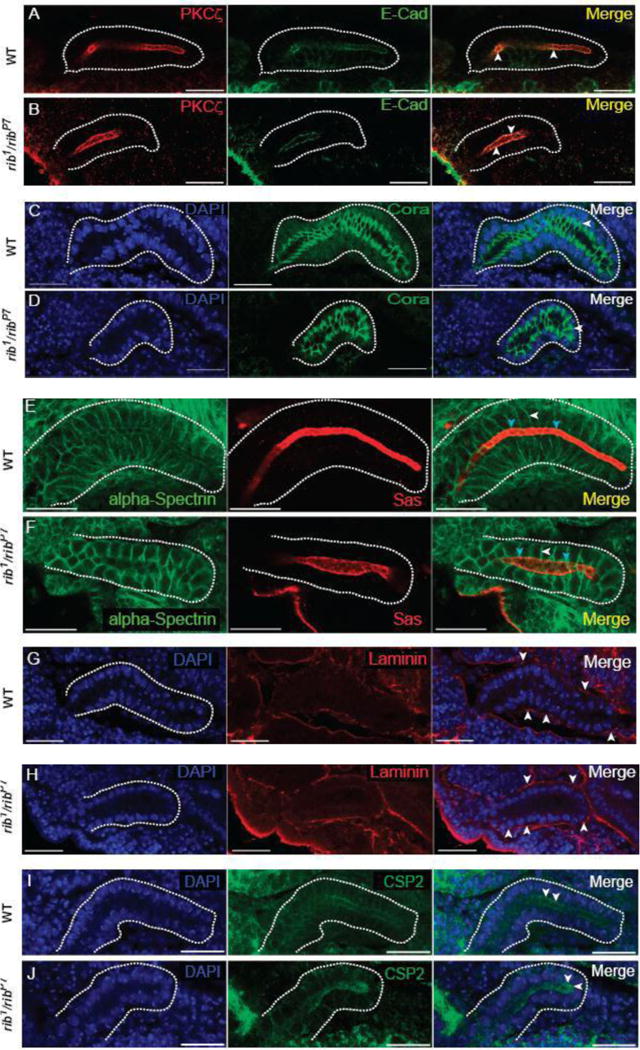
Cell junction integrity, cell polarity and secretory vesicle marker localization are not affected in rib mutant SGs. (A, B) WT and rib1/ribP7 SGs stained with aPKC (apical domain marker) and E-Cad (adherens junction (AJ) marker). Arrowheads indicate co-localization of markers and apical localization of AJs. (C, D) WT and rib1/ribP7 SGs stained with DAPI and Cora (septate junction (SJ) marker). Arrowheads indicate sub-apicolateral localization of SJs. (E, F) WT and rib1/ribP7 SGs stained with alpha-Spectrin (lateral membrane marker) and Sas (apical membrane marker). White and cyan arrowheads indicate distinct localization of the lateral and apical cell membrane domains, respectively. (G, H) WT and rib1/ribP7 SGs stained with DAPI and the basement membrane component laminin as an indirect marker for the basal cell domain. Arrowheads indicate basal deposition/localization of laminin. (I, J) WT and rib1/ribP7 SG stained with CSP2 (exocytic/secretory vesicle marker). Arrowheads indicate the apical enrichment of CSP2, implying functional secretion in rib mutant SG cells. Scale bars: 20 μm.
rib mutant SGs are significantly smaller than WT SGs
To determine if the defective morphology of rib mutant salivary glands is due to changes in cell size/shape and, consequently its volume, or is due to changes in the arrangement of cells around the lumen, we performed morphometric analyses. For volumetry, we obtained manual surface renderings of the whole SG tissue along the YZ-axis (Fig. 3A, B). The volumes obtained from luminal surface renderings (Fig. 3A′, B′) were then subtracted from the whole SG volumes to obtain the total cellular volume of the gland, termed SG volume. The mean SG volume in the rib1/ribP7 mutants was significantly decreased compared to the age-matched WT embryos (Fig. 3C). We also estimated the cell volume by dividing the individual SG volume by the total number of cells within each gland. The result of these estimates showed a significant decrease in the mean cell volume of rib1/ribP7 mutant SGs; cells in the rib mutant glands attained only 53% of the WT cell volume (Fig. 3C′). Despite the overt differences in cell and organ size, nuclear size, based on DAPI and Lamin staining, did not appear to be different between WT and rib mutant SGs (Fig. 3D,E). Since SG shape is also dependent on how the cells are arranged around the lumen (Chung and Andrew, 2014; Pirraglia et al., 2013; Xu et al., 2011), we analyzed cell arrangement by examining the cross-sectional images obtained along the luminal axis (Fig. 3D, E). We found no significant difference in cell arrangement between the rib1/ribP7 mutant and WT SGs (Fig. 3D′ – D‴ & E′ – E‴, 3F). Overall, these results suggest that a major morphological defect in rib mutant SGs is the significant decrease in average cell size/volume. Our findings also rule out a role for abnormal cell arrangement in contributing to the defective SG morphology of rib mutants.
Figure 3.
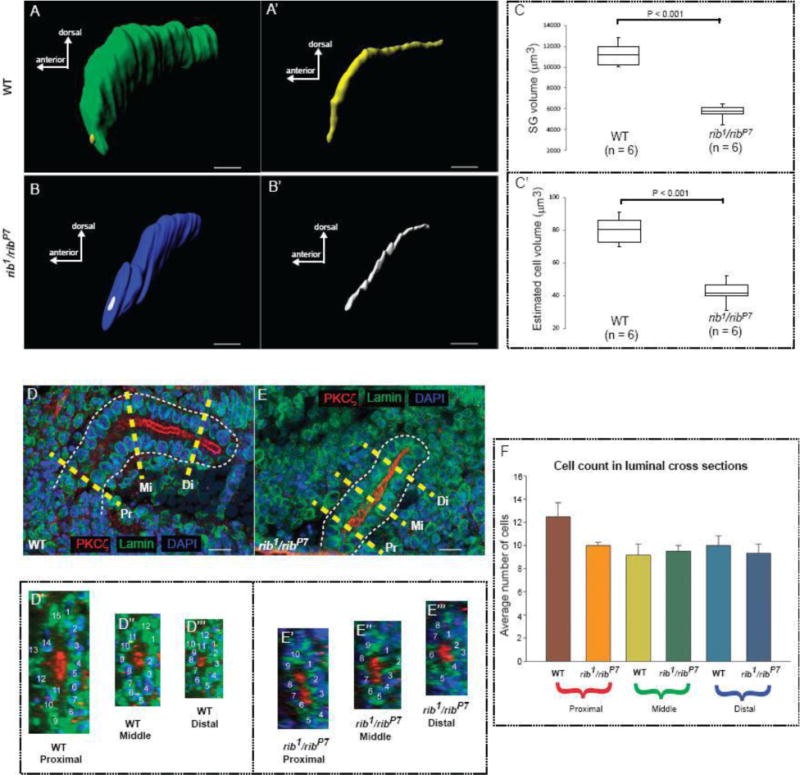
Loss of rib affects cell volume but not cell arrangement. (A) Whole surface rendering of a WT stage 12 SG. (A′) Luminal surface rendering of the gland shown in A. (B) Whole surface rendering of a rib1/ribP7 stage 12 SG. (B′) Luminal surface rendering of the gland shown in B. (C) The SG volume (difference between the whole volume and luminal volume) of the rib1/ribP7 mutants was significantly decreased (t-test, P < 0.001) compared to the WT. (C′) The estimated mean cell volume (SG volume/total cell count) of the rib1/ribP7 mutants was significantly decreased (t-test, P < 0.001) compared to the cell volume in the WT SG. (D) Representative scheme for obtaining cross-sections of the WT SG from three different regions along the length of the organ. Yellow dashed lines show Pr: proximal, Mi: middle and Di: distal sections. (D′–D‴) Cross-sections of the WT tube shown in ‘(D)’ from the proximal, middle and distal regions with nuclear count labels. (E) Representative scheme for obtaining cross-sections of the rib1/ribP7 SG from three different regions along the length of the organ. (E′ – E‴) Cross-sections of the rib1/ribP7 tube shown in ‘(E)’ from the proximal, middle and distal regions with nuclear count labels. (F) The number of cells (mean + SE) encircling the lumen was not significantly different (one-way ANOVA) between the WT and the rib1/ribP7. Scale bars: 10 μm.
ChIP-seq analysis reveals direct binding targets of Rib in the salivary gland
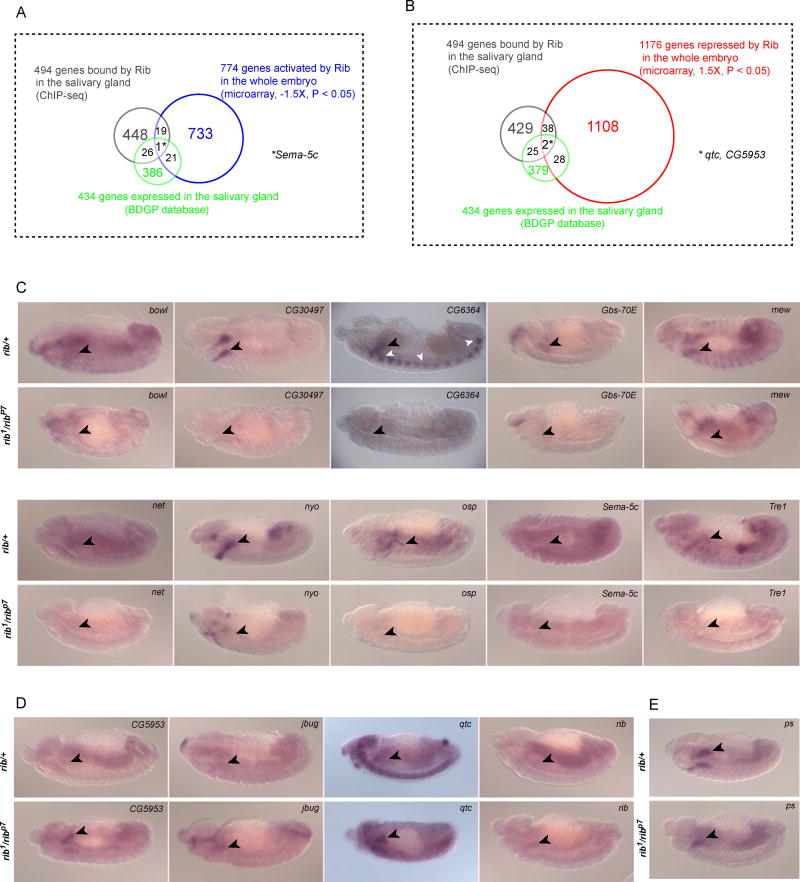
To identify direct targets of Rib in the developing SG, we performed ChIP-seq analysis. We used the fkh-GAL4 and sage-GAL4 drivers to obtain parallel ChIP-seq data sets (Fig. 4A). The C-terminal GFP-tagged UAS-Rib construct, which was driven by the two GAL4 drivers, was functional, as it rescued the SG phenotype in the rib mutant background (Figure 4B). We processed deep sequencing data from both driver conditions, and superimposed the datasets to enrich for SG-specific Rib binding sites. The fkh-GAL4 and sage-GAL4 constructs each drive strong expression of a UAS-lacZN reporter in the salivary gland as well as a variety of other tissues (Fig. 4C). Analysis of the individual expression patterns from these drivers indicates that they share high-level salivary gland expression. Hence, identification of binding events common to both data sets (fkh-GAL4::UAS-rib-GFP and sage-GAL4::UAS-rib-GFP), as shown for representative genes Hsp70Ba and Obp99b (Fig. 4D), should reveal Rib-binding sites in the SG. Using this strategy, we identified 494 genes bound and, hence, potentially directly regulated by Rib in the developing SG (Supplementary Table 1).
Figure 4.
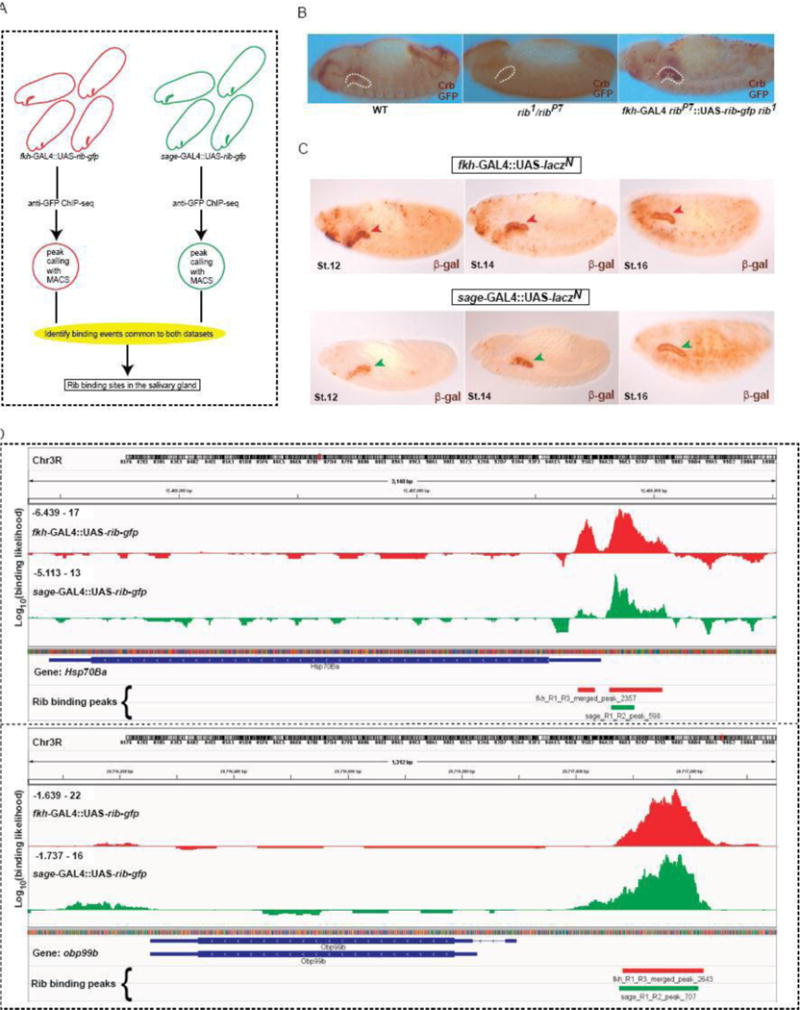
ChIP-seq analysis identifies Rib binding sites in salivary gland cells. (A) Schematic outline of the experimental approach to identify SG-specific Rib binding sites. ChIP-seq datasets were obtained from samples using two different GAL4 constructs to drive expression of UAS-rib-gfp in the SG. The overlap of binding events observed with both drivers enriches for SG-specific Rib binding. (B) Rescue of the SG phenotype in the rib1/ribP7 mutant background with fkh-Gal4::UAS-rib-GFP verified the functionality of UAS-rib-gfp construct used in the ChIP-seq experiments. (C) Tissue expression of fkh-GAL4 and sage-GAL4 drivers spanning the stages used for the ChIP-seq analyses. Arrowheads indicate the SG at different developmental stages. (D) SG-enriched ChIP-seq signals correspond to Rib binding events in the vicinity of two Rib target genes – Hsp70Ba and Obp99b.
Rib functions as both a transcriptional activator and repressor
To determine the directionality of transcriptional regulation (activation/repression) of potential targets by Rib, we performed microarray gene expression analysis in both WT and rib1/ribP7 embryos spanning the stages of SG organogenesis (Fig.5A). Microarray analysis revealed that 774 transcripts were downregulated in the rib1/ribP7 mutants compared to WT embryos (fold change < 1.5x, P < 0.05), whereas 1176 transcripts were upregulated at least 1.5x (P < 0.05) in the rib1/ribP7 mutants compared to WT embryos (Supplementary Tables 2 and 3). Intersection of the microarray gene expression results with the SG-specific ChIP-seq data sets resulted in 20 candidates for direct Rib activation and 40 candidates for direct Rib repression in the SG (Fig. 5B–D), a number consistent with chance for the activated genes (p = 0.5537) but that is higher than expected by chance for the repressed genes (p = 0.0486). A subset of these candidates was further validated by qRT-PCR analysis (Fig. 5E, F). Of the 15 candidates chosen for qRT-PCR verification, 13 showed significant change in expression levels consistent with the directionality of change suggested by the microarray gene expression analysis. Notable among the candidate targets identified by our analysis for Rib regulation is rib itself, suggesting the possibility of autoregulation. Unlike most genes represented on the Affymetrix array chips, the gene Prosap was covered by two sets of probes (one corresponding to the 5′ end near the known mapped promoter and one corresponding to the 3′ end near the coding exons), resulting in discordant results for regulation of this gene in the microarray analysis. Since the array is biased toward 3′ measures of gene expression and since Rib occupancy mapped just upstream of the 3′ probes, a 3′ region was used for the qRT-PCR analysis. The qRT-PCR verified the microarray findings for the 3′ region of the gene; Prosap expression was significantly increased in the rib1/ribP7 embryos compared to controls, consistent with Rib repression.
Figure 5.
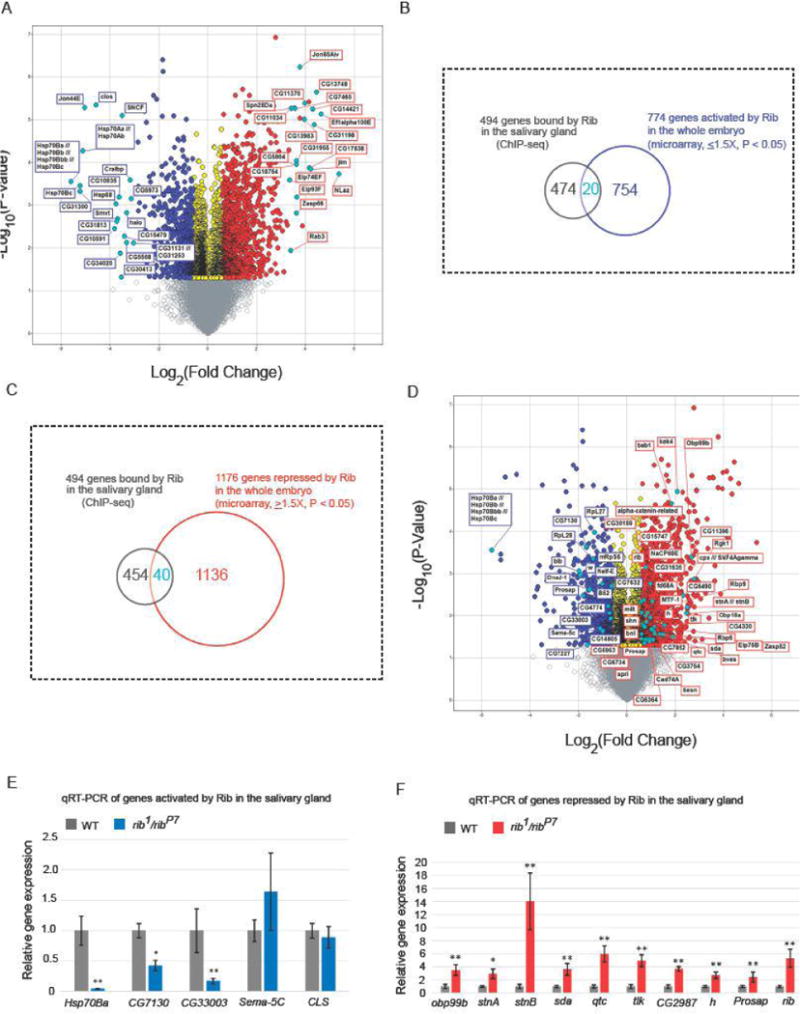
Microarray gene expression analysis indicates the direction of transcriptional control of Rib targets. (A) RNA was isolated from three individual samples each of stage 11–16 WT and rib1/ribP7 embryos. Volcano plot shows genes that were downregulated (blue) or upregulated (red) at least 1.5-fold (P < 0.05) in rib mutants compared to WT. In each direction, the top 20 annotated candidates, based on fold change, are marked (cyan). Transcripts with fold change values between −1.5 and 1.5 (P < 0.05) are shown in yellow. Fold change values that are not statistically significant (P > 0.05) are indicated by gray. (B, C) Venn diagrams representing the overlap of 494 genes from the ChIP-seq and microarray (774 targets activated and 1176 repressed by Rib, respectively) data sets are shown. (D) The set of transcripts that are downregulated (blue) or upregulated (red) at least 1.5-fold (P < 0.05, microarray analysis) in rib mutants compared to WT and testing positive for SG Rib binding (ChIP-seq analyses) are marked (cyan). (E,F) qRT-PCR results for a subset of genes obtained from the overlap of ChIP-seq and microarray data confirms significant expression change in the same direction as observed with microarray analysis for all but two examples, Sema-5C and CLS. *P < 0.01, **P < 0.001, Mann-Whitney U test.
Functional clustering of potential Rib targets by DAVID gene ontological (GO) classification was done on the following four data sets: 1) List of genes activated by Rib in the whole embryo, 2) List of genes repressed by Rib in the whole embryo, 3) List of genes activated by Rib in the whole embryo and bound by Rib in the SG, and 4) List of genes repressed by Rib in the whole embryo and bound by Rib in the SG. The results from functional annotation clusters with a significant (P < 0.05) enrichment score of at least 2.0 (Tables 1–4) showed that the targets activated by Rib in the whole embryo were generally grouped under the GO terms: DNA-related biological processes (including DNA replication and repair), nuclear chromosome part, nuclear lumen and cellular stress response. The candidate direct SG-specific targets of Rib were generally grouped under stress response, but included sub-categories of the spliceosome, endocytosis and nucleotide binding. The target genes repressed by Rib in the whole embryo included neurobiological processes and neuronal morphogenesis as well as cofactor/coenzyme binding and biosynthesis. Curiously, the GO term salivary gland morphogenesis was the predominant category for candidate direct SG-specific targets repressed by Rib. Taken together, these results indicate that the SG-specific Rib gene regulatory network is involved in the activation of processes linked to cellular stress response and repression of genes linked to morphogenesis. The finding that Rib regulates its own expression by auto-regulation – specifically, repression – suggests that Rib auto-repression may be a critical component of the regulatory architecture promoting morphogenesis during SG development.
Table 1.
Results from DAVID clustering analysis of GO terms for genes activated by Ribbon based on microarray data.
| Annotation cluster (enrichment score) | GO term | Fold enrichment | P-value |
|---|---|---|---|
| 1 (5.34) | DNA metabolic process | 3.5 | 1.0E-09 |
| DNA repair | 4.3 | 2.7E-07 | |
| Response to DNA damage stimulus | 4.0 | 3.6E-07 | |
| Cellular response to stress | 3.2 | 1.6E-06 | |
| DNA repair | 4.1 | 6.4E-03 | |
| DNA damage | 3.9 | 8.4E-03 | |
| 2 (2.42) | Nuclear chromosome part | 4.3 | 8.5E-05 |
| Nuclear chromosome | 3.8 | 2.3E-04 | |
| DNA replication | 2.9 | 2.6E-03 | |
| DNA-dependent DNA replication | 3.3 | 1.1E-02 | |
| DNA replication initiation | 6.2 | 2.4E-02 | |
| Origin recognition complex | 10.5 | 3.0E-02 | |
| Nuclear origin of replication recognition complex | 10.5 | 3.0E-02 | |
| 3 (2.37) | Nuclear chromosome part | 4.3 | 8.5E-05 |
| Nuclear chromosome | 3.8 | 2.3E-04 | |
| Chromosomal part | 2.3 | 2.8E-04 | |
| Intracellular non-membrane-bounded organelle | 1.5 | 3.5E-04 | |
| Non-membrane-bounded organelle | 1.5 | 3.5E-04 | |
| Replication fork | 7.8 | 7.6E-04 | |
| Nuclear replisome | 10.2 | 1.0E-03 | |
| Nuclear replication fork | 10.2 | 1.0E-03 | |
| Replisome | 10.2 | 1.0E-03 | |
| Chromosome | 2.0 | 1.1E-03 | |
| DNA replication | 2.9 | 2.6E-03 | |
| Homologous recombination | 5.7 | 2.7E-03 | |
| Protein-DNA complex | 4.4 | 4.4E-03 | |
| Nucleotide excision repair | 3.7 | 4.4E-03 | |
| Mismatch repair | 5.1 | 4.6E-03 | |
| DNA replication | 3.3 | 1.5E-02 | |
| DNA-directed DNA polymerase activity | 4.8 | 1.8E-02 | |
| DNA polymerase activity | 4.6 | 2.1E-02 | |
| DNA polymerase complex | 9.2 | 3.9E-02 | |
| 4 (2.09) | Nuclear lumen | 1.8 | 1.6E-03 |
| Membrane-enclosed lumen | 1.6 | 2.2E-03 | |
| Organelle lumen | 1.6 | 2.6E-03 | |
| Intracellular organelle lumen | 1.6 | 2.6E-03 | |
| Nucleoplasm | 1.6 | 6.9E-02 | |
| 5 (2.08) | Stress response | 7.1 | 7.8E-05 |
| Stress-induced protein | 12.8 | 3.4E-04 | |
| Heat shock | 12.8 | 3.4E-04 | |
| Response to heat | 3.8 | 5.9E-04 | |
| Heat shock-mediated polytene chromosome puffing | 18.6 | 7.4E-04 | |
| Polytene chromosome puffing | 18.6 | 7.4E-04 | |
| Response to hypoxia | 7.3 | 1.0E-03 | |
| Response to oxygen levels | 7.3 | 1.0E-03 | |
| Response to temperature stimulus | 3.5 | 1.0E-03 | |
| Cellular response to heat | 15.5 | 1.4E-03 | |
| Chaperone HSP70 | 8.1 | 2.2E-03 | |
| Heat shock protein 70, conserved site | 8.2 | 2.3E-03 | |
| Heat shock protein Hsp70 | 7.6 | 3.2E-03 | |
| Heat shock protein 70 | 7.6 | 3.2E-03 | |
| Molecular chaperone | 8.8 | 4.2E-02 | |
| Response to protein stimulus | 8.7 | 4.3E-02 |
Table 4.
Results from DAVID clustering analysis of GO terms for genes repressed by Ribbon based on microarray data and bound by Ribbon in the salivary gland based on ChIP-Seq.
| Annotation cluster(enrichment score) | GO term | Fold enrichment | P-value |
|---|---|---|---|
| 1 (2.21) | Gland morphogenesis | 13.3 | 2.8E-03 |
| Salivary gland morphogenesis | 13.3 | 2.8E-03 | |
| Exocrine system development | 10.9 | 4.8E-03 | |
| Salivary gland development | 10.9 | 4.8E-03 | |
| Gland development | 9.3 | 7.5E-03 | |
| Regulation of transcription | 2.8 | 4.4E-02 | |
| Gland morphogenesis | 13.3 | 2.8E-03 |
Validation/Examination of Rib targets in the salivary gland
Since genes whose expression may change in only a subset of tissues in rib mutants could be missed by microarray studies, we also examined potential regulation of known SG-expressed genes. Potential Rib target genes in the salivary gland were identified in the overlap of known SG-expressed genes from the BDGP database (Supplementary Table 4) and the Rib binding sites identified by ChIP-seq: there were twenty-seven such genes (a number higher than expected by pure chance; p = 0.0021). Three-way overlaps of the microarray gene expression dataset, ChIP-seq dataset and the BDGP-identified SG-expressed genes produced sema-5c as a target activated by Rib in the SG (Fig. 6A) and qtc and CG5953 as targets repressed by Rib in the SG (Fig. 6B). We validated the regulation of these targets and twelve others, which either had Rib binding sites or showed changes in the microarray analysis, by in situ hybridization analysis (Fig. 6C, D). Of the 15 putative targets we chose to examine by in situ hybridization, ten showed reduced SG expression in rib mutants, four showed elevated SG expression in rib mutants, and one showed no detectable change in the rib mutant SGs compared with those of the rib heterozygotes (Table 5; Supplementary Fig.1). Notably, two of the putative BDGP targets that were chosen post-hoc - net and jbug - based on a peak-like chip signal in proximity to the gene, but were not called as peaks by MACS, also showed regulation by Rib. Furthermore, our in situ analysis also confirmed that rib is subject to autoregulation, and pasilla (ps), one of the putative targets and a SG specification marker used in our study (Fig. 1F, J), was not subject to Rib regulation. Overall, the in situ hybridization analysis confirms Rib as both a transcriptional activator and repressor in the developing SG, a role also confirmed by microarray gene expression analysis of whole embryos and further corroborated by identification of Rib binding sites by ChIP-seq.
Figure 6.
Rib SG binding sites overlap with genes expressed in the SG and with genes whose expression changes in rib mutants based on microarray analysis. (A) Venn diagram representing the overlap of genes from the ChIP-seq (494 genes bound by Rib in the SG), microarray (774 genes activated by Rib in the whole embryo) and BDGP gene expression database (434 SG-enriched gene expression). (B) Venn diagram representing the overlap of genes from the ChIP-seq (494 genes bound by Rib in the SG), microarray (1176 genes repressed by Rib in the whole embryo) and BDGP gene expression database (434 SG-enriched gene expression). (C) In situ hybridization analysis of SG genes activated by Rib and with nearby Rib binding sites in rib1/ribP7 mutant and heterozygous (rib1/+ or ribP7/+) embryos. (D) In situ hybridization analysis of SG genes repressed by Rib and with nearby Rib binding sites, in rib mutant and heterozygous embryos. (E) In situ hybridization analysis of a SG gene with nearby Rib binding sites whose expression is not detectably changed in rib mutant compared with heterozygous embryos. Rib mutants were identified by morphological criteria and/or the absence of expression of lacZ from the ftz-lacZ containing balancer chromosomes. Black arrowheads indicate SGs and white arrowheads indicate lacZ expression from the balancer chromosome in C–E.
Table 5.
Candidate Rib target genes examined by in situ (or) qRT-PCR (or) both.
| CG/gene name | Microarray [fold change in ribP7/rib1 compared with WT] | qRT-PCR [direction of change in ribP7/rib1 compared with WT] | In situ hybridization [direction of change in ribP7/rib1 SGs compared with rib/+] | Protein domain(s)/Protein function |
|---|---|---|---|---|
| Hsp70Ba | −48.1* | ↓** | n.d. | Heat shock protein 70 family |
| CG7130 | −3.2* | ↓* | n.d. | DNAj domain, heat shock protein 40 family |
| CG33003 | −2.1* | ↓** | n.d. | Proline-rich predicted transmembrane protein |
| sema-5C | −1.9* | ↑ | ↓ | Semaphorin, transmembrane signaling protein |
| nyo | −1.9 | n.d. | ↓ | Zona pellucida domain, predicted transmembrane protein |
| CLS | −1.6* | ↓ | n.d. | Cardiolipin synthase |
| GBS-70E | −1.3* | n.d. | ↓ | Protein phosphatase 1 regulatory subunit |
| CG6364 | −1.3* | n.d. | ↓ | Uridine kinase |
| tre1 | −1.3 | n.d. | ↓ | G protein coupled receptor, Rhodopsin-like |
| ps | −1.1* | n.d. | n.c | K homology domain, mRNA binding protein |
| CG30497 | 1.1 | n.d. | ↓ | Armadillo-like helical domain |
| osp | 1.1 | n.d. | ↓ | Pleckstrin homology domain |
| bowl | 1.5 | n.d. | ↓ | Zn-finger transcription factor |
| mew | 1.7 | n.d. | ↓ | Integrin alpha chain |
| net | 3.3* | n.d. | ↓ | bHLH (myc-type) transcription factor |
| CG5953 | 1.5* | n.d. | ↑ | MADF domain (transcription factor?) |
| rib | 1.8* | ↑** | ↑ | BTB/POZ Psq-type DNA binding transcription factor |
| prosap | 1.9* | ↑** | n.d. | SH3 domain, PDZ domain, ankyrin repeat |
| jbug | 1.9* | n.d. | ↑ | Filamin A, Actin binding protein |
| h | 2.0* | ↑** | n.d. | bHLH transcription factor |
| CG2987 | 2.0* | ↑** | n.d. | Actin binding protein |
| qtc | 2.9* | ↑** | ↑ | Hook related protein family |
| tlk | 2.9* | ↑** | n.d. | Serine, threonine dual specificity protein kinase |
| slamdance | 3.7* | ↑** | n.d. | Alanine aminopeptidase/leukotriene A4 hydrolase |
| stnA | 3.7* | ↑* | n.d. | Synaptic vesicle recycling |
| stnB | 5.7* | ↑** | n.d. | Synaptic vesicle recycling |
| obp99b | 6.3* | ↑** | n.d. | Odorant binding protein (secreted) |
p value < 0.05 microarray, p value < 0.01 qRT-PCR
p value < 0.001 qRT-PCR n.d. = not done n.c. = no detectable change Blue, yellow, pink indicate genes that significantly went down, were unchanged, or went up, respectively, in microarrays of rib mutants compared to WT.
Determination of Rib consensus binding motifs
The ChIP-seq data revealed 494 genes directly bound by Rib in the salivary gland (Fig. 4). A SeqPos-based motif analysis of the Rib binding sites associated with these genes allowed us to identify DNA sequences that occurred most frequently, potentially revealing consensus sites recognized by Rib and/or its DNA binding partners. An analysis of the top ten sequence motifs identified in this way revealed shared sequence cores within several of the motifs. For example, motifs 1 and 4 share a CTTATCT core sequence, which is also the reverse complement of the six most conserved residues of motif 10. Likewise, motifs 2 and 5 share a GGCGGG core sequence, and motifs 3 and 9 share a GCGCGC core sequence. It was not possible to learn if the different motifs were associated with gene activation versus repression because of the relatively smaller number of genes for which the direction of gene regulation is known or could be inferred based on either in situ or microarray data. Nonetheless, these sequences will be prime targets for future in vitro DNA binding studies and in vivo enhancer analyses, and may help to identify Rib partners that may function in gene activation versus repression.
Discussion
In this study, we asked which aspects of organ morphogenesis are affected by the loss of the BTB-containing nuclear protein Rib; our studies ruled out cell specification, cell death, cell division, junctional integrity, and cell polarity, as well as defects in the cell rearrangements that normally accompany SG internalization and tube elongation. Indeed, we discovered that a major morphological defect in rib mutants is the failure of SG cells to achieve the same size and shape as WT SG cells. rib mutant SG cells are, on average, only 53% the volume of WT cells, even though nuclear size appears no different. To learn which genes are directly regulated by Rib in the SGs, we carried out ChIP-Seq analysis using a strategy to specifically enrich for SG binding sites. Combining the ChIP-Seq findings with gene expression studies, we have identified an interesting set of Rib targets that are likely to support changes in cell shape and in cell/tissue growth in the SG. We have also learned that Rib acts as both a transcriptional activator and repressor, and represses its own expression. Finally, we have identified sequence motifs through which Rib and/or its binding partners control SG gene expression.
Previous studies of rib function in both the SG and trachea led to the conclusion that the rib loss-of-function defects were primarily in epithelial migration (Bradley and Andrew, 2001; Shim et. al., 2001). The SGs of rib mutants failed to turn and migrate along the visceral mesoderm, as do WT SGs (Bradley and Andrew, 2001). Similarly, tracheal branches failed to elongate, with some branches having more severe defects than others (Bradley and Andrew, 2001; Shim et al., 2001), a phenotype that was linked by Shim et al (2001) to FGF signaling. Subsequent live imaging studies revealed compromised tube elongation attributable to reduced apical membrane length and increased apical stiffness in both the SGs and trachea of rib mutants (Cheshire et al., 2008). The delayed and limited apical elongation (SGs only ever achieved 60% of WT length) was molecularly linked to reduced expression of the apical determinant crb, increased apical levels of active, phosphorylated Moe, as well as decreased apical accumulation of Rab11, a component of the apical recycling endosome (Kerman et. al., 2008). These previous findings pointed to a role for Rib in affecting cell shape changes essential for SG morphogenesis at the apical surface. In the current study, which takes advantage of newly available image analysis programs to characterize gland morphometry, we discovered that loss of rib leads not only to a reduction in apical tube dimensions, its loss also significantly affects the volume of the entire gland and its constituent cells. Thus, the SG tube may simply be too small to fully contact the tissues upon which it normally migrates. Alternatively, the reduced cell size may limit the ability of SG cells to generate sufficient collective forces to elongate and simultaneously position the tubular organ along the anteroposterior axis.
The nuclear localization of Rib in all tissues, including the SG, and the possibility of its cooperation with another BTB domain-containing factor Lolal (a transcription factor) in epithelial cells suggests that Rib regulates gene transcription (Kerman, 2008). Thus, we sought to determine the targets of Rib in the SG by ChIP-seq and we identified 494 genes with Rib binding sites. A first pass inspection of the genes associated with Rib binding in the salivary gland revealed twenty-three functional clusters based on DAVID (Supplementary Table 5). Notable GO terms identifying these gene clusters included ribosome, microtubule-based processes, cell adhesion, cell motion, cell projection morphogenesis, imaginal disc morphogenesis, cell fate commitment, epithelial morphogenesis, plasma membrane, salivary gland, tracheal, eye and muscle development, stem-cell maintenance and epithelial-structure maintenance. These findings suggest that Rib may directly regulate cell size through translational regulation by affecting expression of genes encoding ribosomal proteins (Annotation cluster 1). The clustering analysis suggests that Rib probably also directly affects morphogenesis through effects on the cytoskeleton, cell adhesion and other morphogenetic events (Annotations clusters 2, 3, 4–6, 9–12). The David analysis also suggests that Rib could bind the same DNA sites in all tissues in which it is expressed and required, based on the finding that genes linked to the salivary gland (Annotation clusters 13, 15), trachea (Annotation cluster 19), eye (Annotation cluster 8), wing disc (Annotation clusters 4, 18), germ cells (Annotation cluster 14) and muscle cells (Annotation cluster 22) were found to be enriched in the sites bound by Rib in the SG. This final conclusion can be directly tested using a strategy similar to the one we used for the SG by doing the ChIP-seq analysis with Gal4 drivers expressed in one or more of these other tissues.
Combining the microarray gene expression analyses with the ChIP-seq results revealed that Rib functions in both transcriptional activation and repression, and provided evidence for Rib autorepression. The intersection of genes activated by Rib and Rib binding sites determined from ChIP-seq identified twenty putative targets activated by Rib in the SG. Similarly the overlap of genes repressed by Rib and ChIP-seq sites identified forty putative targets repressed by Rib in the SG. qRT-PCR on a subset of these putative targets confirmed the directionality of regulation of approximately 85% of the genes, further validating this unbiased approach to identifying Rib targets. The genes identified as targets for activation by Rib in the whole embryo spanned several GO categories such as DNA-related biological processes (including DNA replication and repair), nuclear chromosome part, nuclear lumen and cellular stress response, whereas the targets for repression by Rib in the whole embryo were categorized under neurobiological processes and neuronal morphogenesis as well as cofactor/coenzyme binding and biosynthesis. Meanwhile, the targets activated by Rib in the SG, identified by the two-way overlap, fell primarily under cellular stress response genes – most notably because this group included several heat shock protein (HSP) genes that require Rib for their high level expression. This finding is again consistent with a direct role for Rib in cell volume increases. Although HSPs were initially identified because their expression increases under conditions of high temperature and other stresses, HSPs are now known to primarily function as chaperones to facilitate protein folding and block aggregation, a potential problem associated with high levels of protein production. Alternatively, cellular stress response genes have also been shown to promote cell migration and morphogenesis (Cobreros et al., 2008; Li et al., 2012). The potential direct targets repressed by Rib in the SG, came primarily under the GO term salivary gland morphogenesis. Altogether, this finding could suggest that Rib represses morphogenesis through its targets to allow for sufficient cell size increase (growth) prior to releasing the “brake” on morphogenesis through self-repression.
We also performed a three-way superposition of gene lists obtained from 1) ChIP-seq data, 2) Microarray gene expression data, and 3) SG-expressed genes from the BDGP database. The three-way overlap to identify known SG-expressed genes activated by Rib produced Sema-5c as the single likely candidate. Similar analysis for the identification of SG-expressed genes repressed by Rib resulted in qtc and CG5953 as likely candidates. Indeed, we confirmed the directionality of regulation of all three Rib targets by in situ hybridization analyses in rib mutants. Sema-5c is a transmembrane protein belonging to the semaphorin family of growth cone guidance molecules implicated in neural morphogenesis (Khare et al., 2000). Our in situ analysis of embryos from stages 12–14, when vigorous tube elongation occurs, shows Sema-5c expression in the WT SG, which is downregulated in rib mutants. This finding is consistent with the microarray gene expression analysis, but is not consistent with the qRT-PCR results (Fig. 4E; Table 5). qtc was among the candidates repressed by Rib (three-way overlap). qtc has been implicated in the regulation of normal male sexual behavior (Gaines et al., 2000). Although its mechanism of function is unknown, portions of the predicted Qtc protein have limited sequence identity to a large family of myosin and myosin-like proteins, all of which contain α-helical coiled coil domains with common structural features (Lupas, 1996). The other candidate repressed by Rib (three-way overlap) is CG5953, which encodes a MADF domain containing protein (FlyBase). MADF (Myb/SANT-like domain in Adf-1) domain is composed of an 80-amino acid module that directs sequence specific DNA binding to sites with multiple tri-nucleotide repeats (Interpro). Based on the examples of proteins containing MADF domain (Adf-1 and Dip3) functioning as transcription factors (Cutler et al., 1998; England et al., 1992), we presume that CG5953 encodes for a transcription factor as well. Rib-dependent downstream transcription factors could be responsible for the large number of genes whose expression changes in rib mutants but that do not contain Rib binding sites. Indeed, although Rib occupancy was observed near the crb promoter in both ChIP-Seq datasets, this result fell out of our analysis due to a lack of signal overlap using the different drivers (Supplemental Figure 1Y). The lack of overlap may simply be a result of the stringent ChIP-seq peak calling thresholds we used for identifying high confidence direct Rib targets. Alternatively, this could reflect a less significant role for Rib in direct SG regulation of crb, opening up the possibility that genes such as CG5953 or hairy, a bHLH transcription factor previously shown to repress SG gene expression (Myat and Andrew, 2002), could be involved. We also found no change in crb expression by microarray. This finding suggests that the microarray of whole embryos (stages 11 – 16) may not be sensitive enough to detect the stage-specific reduction in crb transcripts previously observed in the SG and trachea by in situ and by qRT-PCR of stage 12 embryos (Kerman et al., 2008). In general, our experience indicates that in situ analysis is a much more sensitive measure of tissue-specific changes in gene expression, especially for targets expressed in multiple tissues under the control of a variety of regulators (Maruyama et al., 2011).
We also examined the expression levels of mew, which encodes for the integrin alpha subunit and functions in cell adhesion to the extracellular matrix to facilitate migration of the SG and other tissues (Bradley et al., 2003; Brown et al., 2000). Although the whole embryo mew transcript levels weren’t significantly changed in the rib mutants with microarray gene expression analysis, we chose to examine its levels in the SG because of a strong ChIP-seq Rib binding peak in the proximity of the gene. Our in situ analysis showed decreased expression of mew transcripts in the mutants compared to WT SGs. In addition to mew, we examined the SG expression of Tre1, which encodes a G-protein coupled receptor involved in cell migration (Kunwar et al., 2003). The Tre1 transcript levels were downregulated in rib mutants based on the microarray gene expression analysis, and our in situ analysis confirmed these results, suggesting that Tre1 is a direct target of Rib in the SG. The results that mew and Tre1 are activated by Rib suggest that Rib activates genes known or presumed to be involved in SG migration. Overall, the known functions of target genes bound and regulated by Rib in whole embryos and in the SG are consistent with the defects in cell volume observed in the rib mutants. They are also consistent with a more direct role for Rib in morphogenesis, including a role in cell migration.
Supplementary Material
Figure 7.
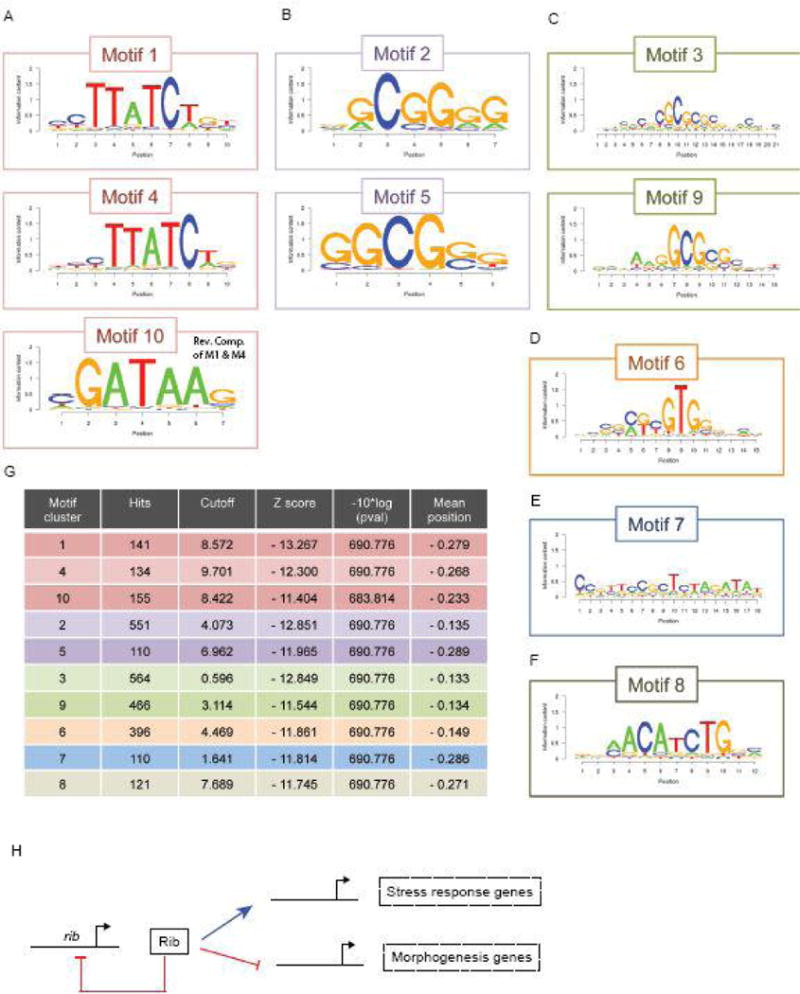
The top ten consensus sites found within Rib bound DNA fragments from the ChIP-seq analysis reveal conserved core motifs. (A) Motif 1 and Motif 4 are displaced by a single nucleotide and Motif 10 is the reverse complement of the most conserved sequence within Motifs 1 and 4. (B and C) Similarly, Motifs 2 and 5 contain a shared core sequence, as do Motifs 3 and 9. (D–lF) Motifs 6 – 8 are distinct, although the N-terminal portion of Motif 6 is related to Motifs 3 and 9. (G) Table of top 10 Motif clusters with the number of hits, the Cutoff, Z score, p value and mean position is shown. (H) A hypothetical model of the Rib gene regulatory network in the context of SG development.
Table 2.
Results from DAVID clustering analysis of GO terms for genes repressed by Ribbon based on microarray data.
| Annotation cluster (enrichment score) | GO term | Fold enrichment | P-value |
|---|---|---|---|
| 1 (3.54) | Cofactor binding | 2.3 | 1.0E-05 |
| Coenzyme binding | 2.2 | 2.7E-04 | |
| FAD binding | 2.4 | 8.7E-03 | |
| 2 (2.41) | Transmission of nerve impulse | 2.5 | 1.5E-06 |
| Synaptic transmission | 2.4 | 5.4E-06 | |
| Cell-cell signaling | 2.2 | 2.0E-05 | |
| Regulation of neurotransmitter levels | 2.7 | 5.8E-05 | |
| Synapse | 2.5 | 8.2E-05 | |
| Synaptic vesicle exocytosis | 3.5 | 2.6E-04 | |
| Secretion by cell | 2.4 | 4.5E-04 | |
| Exocytosis | 3.0 | 5.9E-04 | |
| Generation of a signal involved in cell-cell signaling | 2.5 | 6.6E-04 | |
| Secretion | 2.3 | 9.4E-04 | |
| Neurotransmitter transport | 2.3 | 1.0E-03 | |
| Synaptic vesicle transport | 2.6 | 1.2E-03 | |
| Neurotransmitter secretion | 2.4 | 1.7E-03 | |
| Neurological system process | 1.4 | 4.5E-03 | |
| Synaptic vesicle | 2.8 | 5.0E-03 | |
| Synapse part | 2.1 | 8.0E-03 | |
| Clathrin-coated vesicle | 2.5 | 9.6E-03 | |
| Coated vesicle | 2.2 | 2.1E-02 | |
| Cytoplasmic membrane-bounded vesicle | 1.9 | 2.5E-02 | |
| Membrane-bounded vesicle | 1.9 | 3.4E-02 | |
| Cytoplasmic vesicle | 1.8 | 4.1E-02 | |
| 3 (2.37) | Synapse | 2.5 | 8.2E-05 |
| Synapse | 3.0 | 1.5E-03 | |
| Synapse part | 2.1 | 8.0E-03 | |
| Cell junction | 2.1 | 2.3E-02 | |
| 4 (2.03) | Neuron differentiation | 1.7 | 3.7E-04 |
| Cell part morphogenesis | 1.7 | 2.5E-03 | |
| Cell projection morphogenesis | 1.7 | 2.6E-03 | |
| Neuron development | 1.6 | 3.2E-03 | |
| Cell morphogenesis | 1.5 | 5.6E-03 | |
| Cell projection organization | 1.5 | 1.2E-02 | |
| Axon guidance | 1.9 | 1.3E-02 | |
| Neuron projection morphogenesis | 1.6 | 1.5E-02 | |
| Neuron projection development | 1.6 | 1.5E-02 | |
| Axonogenesis | 1.7 | 1.9E-02 | |
| Cell morphogenesis involved in neuron differentiation | 1.5 | 2.7E-02 | |
| Cell morphogenesis involved in differentiation | 1.5 | 3.0E-02 | |
| Cellular component morphogenesis | 1.3 | 3.3E-02 | |
| 5 (2.00) | Amine biosynthetic process | 3.4 | 1.2E-03 |
| Organic acid biosynthetic process | 2.9 | 1.3E-03 | |
| Carboxylic acid biosynthetic process | 2.9 | 1.3E-03 | |
| Cellular amino acid biosynthetic process | 4.0 | 3.0E-03 | |
| Glutamate metabolic process | 7.2 | 3.4E-03 | |
| Glutamine family amino acid biosynthetic process | 5.5 | 9.9E-03 | |
| Glutamine family amino acid metabolic process | 3.4 | 1.5E-02 |
Table 3.
Results from DAVID clustering analysis of GO terms for genes activated by Ribbon based on microarray data and bound by Ribbon in the salivary gland based on ChIP-Seq.
| Annotation cluster (enrichment score) | GO term | Fold enrichment | P-value |
|---|---|---|---|
| 1 (2.41) | Polytene chromosome puffing | 340.2 | 2.5E-05 |
| Heat shock-mediated polytene chromosome puffing | 340.2 | 2.5E-05 | |
| Cellular response to heat | 283.5 | 3.7E-05 | |
| Chaperone HSP70 | 129.8 | 1.3E-04 | |
| Heat shock protein 70, conserved site | 149.9 | 1.5E-04 | |
| Response to heat | 33.3 | 1.6E-04 | |
| Heat shock protein 70 | 138.4 | 1.8E-04 | |
| Heat shock protein Hsp70 | 138.4 | 1.8E-04 | |
| Response to temperature stimulus | 31.1 | 2.0E-04 | |
| Stress response | 94.1 | 4.0E-04 | |
| Response to hypoxia | 89.5 | 4.2E-04 | |
| Response to oxygen levels | 89.5 | 4.2E-04 | |
| Spliceosome | 12.3 | 1.4E-03 | |
| ATP | 31.8 | 3.5E-03 | |
| Response to abiotic stimulus | 10.6 | 4.5E-03 | |
| Endocytosis | 12.8 | 1.4E-02 | |
| Nucleotide binding | 3.0 | 2.5E-02 | |
| Cellular response to stress | 9.7 | 3.2E-02 | |
| ATP-binding | 4.9 | 3.9E-02 |
Highlights.
-
-
the BTB transcription factor Ribbon controls cell shape and volume in the salivary gland
-
-
Ribbon directly activates and represses its SG target genes
-
-
Rib autoregulation may be a key aspect of its morphogenetic activities
Acknowledgments
We would like to thank the members of the Andrew lab for helpful discussions. We thank Susan Celniker for providing the comprehensive list of genes expressed in the embryonic salivary gland, based on BDGP studies. We acknowledge Flybase and the decades-long efforts of Bill Gelbart who made our genome-wide analyses possible. We thank Caitlyn Hanlon for the Tre1 RNA probe and for critiques of the manuscript. We thank the members of the Johns Hopkins School of Medicine microscopy facility for assistance with microscopy and image analysis. We thank Conover Talbot at the JHMI microarray core facility for help with microarray gene expression analysis. Our work was supported by a grant from the National Institutes of Health Grant NIH RO1 DE012873.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrams EW, Andrew DJ. CrebA regulates secretory activity in the Drosophila salivary gland and epidermis. Development. 2005;132:2743–2758. doi: 10.1242/dev.01863. [DOI] [PubMed] [Google Scholar]
- Abrams EW, Mihoulides WK, Andrew DJ. Fork head and Sage maintain a uniform and patent salivary gland lumen through regulation of two downstream target genes, PH4alphaSG1 and PH4alphaSG2. Development. 2006;133:3517–3527. doi: 10.1242/dev.02525. [DOI] [PubMed] [Google Scholar]
- Andrew DJ, Ewald AJ. Morphogenesis of epithelial tubes: Insights into tube formation, elongation, and elaboration. Dev Biol. 2010;341:34–55. doi: 10.1016/j.ydbio.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake KJ, Myette G, Jack J. The products of ribbon and raw are necessary for proper cell shape and cellular localization of nonmuscle myosin in Drosophila. Dev Biol. 1998;203:177–188. doi: 10.1006/dbio.1998.9036. [DOI] [PubMed] [Google Scholar]
- Blankenberg D, Gordon A, Von Kuster G, Coraor N, Taylor J, Nekrutenko A. Manipulation of FASTQ data with Galaxy. Bioinformatics. 2010;26:1783–1785. doi: 10.1093/bioinformatics/btq281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley PL, Andrew DJ. ribbon encodes a novel BTB/POZ protein required for directed cell migration in Drosophila melanogaster. Development. 2001;128:3001–3015. doi: 10.1242/dev.128.15.3001. [DOI] [PubMed] [Google Scholar]
- Bradley PL, Myat MM, Comeaux CA, Andrew DJ. Posterior migration of the salivary gland requires an intact visceral mesoderm and integrin function. Dev Biol. 2003;257:249–262. doi: 10.1016/s0012-1606(03)00103-9. [DOI] [PubMed] [Google Scholar]
- Brown NH, Gregory SL, Martin-Bermudo MD. Integrins as mediators of morphogenesis in Drosophila. Dev Biol. 2000;223:1–16. doi: 10.1006/dbio.2000.9711. [DOI] [PubMed] [Google Scholar]
- Bryant DM, Mostov KE. From cells to organs: building polarized tissue. Nat Rev Mol Cell Biol. 2008;9:887–901. doi: 10.1038/nrm2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheshire AM, Kerman BE, Zipfel WR, Spector AA, Andrew DJ. Kinetic and mechanical analysis of live tube morphogenesis. Dev Dyn. 2008;237:2874–2888. doi: 10.1002/dvdy.21709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Andrew DJ. The formation of epithelial tubes. J Cell Sci. 2008;121:3501–3504. doi: 10.1242/jcs.037887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Andrew DJ. Cadherin 99C regulates apical expansion and cell rearrangement during epithelial tube elongation. Development. 2014;141:1950–1960. doi: 10.1242/dev.104166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Hanlon CD, Andrew DJ. Building and specializing epithelial tubular organs: the Drosophila salivary gland as a model system for revealing how epithelial organs are specified, form and specialize. Wiley Interdiscip Rev Dev Biol. 2014;3:281–300. doi: 10.1002/wdev.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland MH, Sawyer JM, Afelik S, Jensen J, Leach SD. Exocrine ontogenies: on the development of pancreatic acinar, ductal and centroacinar cells. Semin Cell Dev Biol. 2012;23:711–719. doi: 10.1016/j.semcdb.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobreros L, Fernandez-Minan A, Luque CM, Gonzalez-Reyes A, Martin-Bermudo MD. A role for the chaperone Hsp70 in the regulation of border cell migration in the Drosophila ovary. Mech Dev. 2008;125:1048–1058. doi: 10.1016/j.mod.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Comai G, Tajbakhsh S. Molecular and cellular regulation of skeletal myogenesis. Curr Top Dev Biol. 2014;110:1–73. doi: 10.1016/B978-0-12-405943-6.00001-4. [DOI] [PubMed] [Google Scholar]
- Cutler G, Perry KM, Tjian R. Adf-1 is a nonmodular transcription factor that contains a TAF-binding Myb-like motif. Mol Cell Biol. 1998;18:2252–2261. doi: 10.1128/mcb.18.4.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos G, Schroeder AJ, Goodman JL, Strelets VB, Crosby MA, Thurmond J, Emmert DB, Gelbart WM. FlyBase: introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res. 2015;43:D690–697. doi: 10.1093/nar/gku1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England BP, Admon A, Tjian R. Cloning of Drosophila transcription factor Adf-1 reveals homology to Myb oncoproteins. Proc Natl Acad Sci U S A. 1992;89:683–687. doi: 10.1073/pnas.89.2.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox RM, Hanlon CD, Andrew DJ. The CrebA/Creb3-like transcription factors are major and direct regulators of secretory capacity. J Cell Biol. 2010;191:479–492. doi: 10.1083/jcb.201004062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox RM, Vaishnavi A, Maruyama R, Andrew DJ. Organ-specific gene expression: the bHLH protein Sage provides tissue specificity to Drosophila FoxA. Development. 2013;140:2160–2171. doi: 10.1242/dev.092924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaines P, Tompkins L, Woodard CT, Carlson JR. quick-to-court, a Drosophila mutant with elevated levels of sexual behavior, is defective in a predicted coiled-coil protein. Genetics. 2000;154:1627–1637. doi: 10.1093/genetics/154.4.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girdler GC, Roper K. Controlling cell shape changes during salivary gland tube formation in Drosophila. Semin Cell Dev Biol. 2014;31:74–81. doi: 10.1016/j.semcdb.2014.03.020. [DOI] [PubMed] [Google Scholar]
- He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, Mieczkowski P, Lieb JD, Zhao K, Brown M, Liu XS. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Inman JL, Robertson C, Mott JD, Bissell MJ. Mammary gland development: cell fate specification, stem cells and the microenvironment. Development. 2015;142:1028–1042. doi: 10.1242/dev.087643. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003a;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003b;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Ismat A, Cheshire AM, Andrew DJ. The secreted AdamTS-A metalloprotease is required for collective cell migration. Development. 2013;140:1981–1993. doi: 10.1242/dev.087908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerman BE, Cheshire AM, Myat MM, Andrew DJ. Ribbon modulates apical membrane during tube elongation through Crumbs and Moesin. Dev Biol. 2008;320:278–288. doi: 10.1016/j.ydbio.2008.05.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare N, Fascetti N, DaRocha S, Chiquet-Ehrismann R, Baumgartner S. Expression patterns of two new members of the Semaphorin family in Drosophila suggest early functions during embryogenesis. Mech Dev. 2000;91:393–397. doi: 10.1016/s0925-4773(99)00297-x. [DOI] [PubMed] [Google Scholar]
- Kunwar PS, Starz-Gaiano M, Bainton RJ, Heberlein U, Lehmann R. Tre1, a G proteincoupled receptor, directs transepithelial migration of Drosophila germ cells. PLoS Biol. 2003;1:E80. doi: 10.1371/journal.pbio.0000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laprise P, Lau KM, Harris KP, Silva-Gagliardi NF, Paul SM, Beronja S, Beitel GJ, McGlade CJ, Tepass U. Yurt, Coracle, Neurexin IV and the Na(+),K(+)-ATPase form a novel group of epithelial polarity proteins. Nature. 2009;459:1141–1145. doi: 10.1038/nature08067. [DOI] [PubMed] [Google Scholar]
- Lehmann R, Tautz D. In situ hybridization to RNA. Methods Cell Biol. 1994;44:575–598. doi: 10.1016/s0091-679x(08)60933-4. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Sun X, Wang Z, Chen L, Li D, Zhou J, Liu M. Regulation of vascular endothelial cell polarization and migration by Hsp70/Hsp90-organizing protein. PLoS One. 2012;7:e36389. doi: 10.1371/journal.pone.0036389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Ortiz JA, Taing L, Meyer CA, Lee B, Zhang Y, Shin H, Wong SS, Ma J, Lei Y, Pape UJ, Poidinger M, Chen Y, Yeung K, Brown M, Turpaz Y, Liu XS. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 2011;12:R83. doi: 10.1186/gb-2011-12-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A. Coiled coils: new structures and new functions. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- Maeda Y, Dave V, Whitsett JA. Transcriptional control of lung morphogenesis. Physiol Rev. 2007;87:219–244. doi: 10.1152/physrev.00028.2006. [DOI] [PubMed] [Google Scholar]
- Myat MM, Andrew DJ. Fork head prevents apoptosis and promotes cell shape change during formation of the Drosophila salivary glands. Development. 2000a;127:4217–4226. doi: 10.1242/dev.127.19.4217. [DOI] [PubMed] [Google Scholar]
- Myat MM, Andrew DJ. Organ shape in the Drosophila salivary gland is controlled by regulated, sequential internalization of the primordia. Development. 2000b;127:679–691. doi: 10.1242/dev.127.4.679. [DOI] [PubMed] [Google Scholar]
- Myat MM, Andrew DJ. Epithelial tube morphology is determined by the polarized growth and delivery of apical membrane. Cell. 2002;111:879–891. doi: 10.1016/s0092-8674(02)01140-6. [DOI] [PubMed] [Google Scholar]
- Navis A, Bagnat M. Developing pressures: fluid forces driving morphogenesis. Curr Opin Genet Dev. 2015;32:24–30. doi: 10.1016/j.gde.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N, Lavrov S, Hennetin J, Bellis M, Cavalli G. Mapping the distribution of chromatin proteins by ChIP on chip. Methods Enzymol. 2006;410:316–341. doi: 10.1016/S0076-6879(06)10015-4. [DOI] [PubMed] [Google Scholar]
- Pan FC, Wright C. Pancreas organogenesis: from bud to plexus to gland. Dev Dyn. 2011;240:530–565. doi: 10.1002/dvdy.22584. [DOI] [PubMed] [Google Scholar]
- Pirraglia C, Walters J, Ahn N, Myat MM. Rac1 GTPase acts downstream of alphaPS1betaPS integrin to control collective migration and lumen size in the Drosophila salivary gland. Dev Biol. 2013;377:21–32. doi: 10.1016/j.ydbio.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins EL. The building blocks of mammalian lung development. Dev Dyn. 2011;240:463–476. doi: 10.1002/dvdy.22482. [DOI] [PubMed] [Google Scholar]
- Reuter R, Scott MP. Expression and function of the homoeotic genes Antennapedia and Sex combs reduced in the embryonic midgut of Drosophila. Development. 1990;109:289–303. doi: 10.1242/dev.109.2.289. [DOI] [PubMed] [Google Scholar]
- Shim K, Blake KJ, Jack J, Krasnow MA. The Drosophila ribbon gene encodes a nuclear BTB domain protein that promotes epithelial migration and morphogenesis. Development. 2001;128:4923–4933. doi: 10.1242/dev.128.23.4923. [DOI] [PubMed] [Google Scholar]
- Sonnenblick BP. Early embryology of Drosophila melanogaster. In: Demerec M, editor. Biology of Drosophila. Hafner; New York: 1950. [Google Scholar]
- Sotillos S, Krahn M, Espinosa-Vazquez JM, Hombria JC. Src kinases mediate the interaction of the apical determinant Bazooka/PAR3 with STAT92E and increase signalling efficiency in Drosophila ectodermal cells. Development. 2013;140:1507–1516. doi: 10.1242/dev.092320. [DOI] [PubMed] [Google Scholar]
- St Pierre SE, Ponting L, Stefancsik R, McQuilton P. FlyBase 102–advanced approaches to interrogating FlyBase. Nucleic Acids Res. 2014;42:D780–788. doi: 10.1093/nar/gkt1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YC, Khan Z, Kaschube M, Wieschaus EF. Differential positioning of adherens junctions is associated with initiation of epithelial folding. Nature. 2012;484:390–393. doi: 10.1038/nature10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Bagumian G, Galiano M, Myat MM. Rho GTPase controls Drosophila salivary gland lumen size through regulation of the actin cytoskeleton and Moesin. Development. 2011;138:5415–5427. doi: 10.1242/dev.069831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.