Abstract
Most shotgun proteomics data analysis workflows are based on the assumption that each fragment ion spectrum is explained by a single species of peptide ion isolated by the mass spectrometer; however, in reality mass spectrometers often isolate more than one peptide ion within the window of isolation that contributes to additional peptide fragment peaks in many spectra. We present a new tool called reSpect, implemented in the Trans-Proteomic Pipeline (TPP), that enables an iterative workflow whereby fragment ion peaks explained by a peptide ion identified in one round of sequence searching or spectral library search are attenuated based on the confidence of the identification, and then the altered spectrum is subjected to further rounds of searching. The reSpect tool is not implemented as a search engine, but rather as a post search engine processing step where only fragment ion intensities are altered. This enables the application of any search engine combination in the following iterations. Thus, reSpect is compatible with all other protein sequence database search engines as well as peptide spectral library search engines that are supported by the TPP. We show that while some datasets are highly amenable to chimeric spectrum identification and lead to additional peptide identification boosts of over 30% with as many as four different peptide ions identified per spectrum, datasets with narrow precursor ion selection only benefit from such processing at the level of a few percent. We demonstrate a technique that facilitates the determination of the degree to which a dataset would benefit from chimeric spectrum analysis. The reSpect tool is free and open source, provided within the TPP and available at the TPP website.
Introduction
Tandem mass spectrometry (MS2) is currently the most widely used technique to identify proteins and quantify their abundances in complex biological samples [1]. In a typical workflow (sometimes termed shotgun proteomics) proteins extracted from a sample are either proteolytically or chemically cleaved into peptides (e.g. with an enzyme such as trypsin) which are then fractionated, further separated via liquid chromatography to reduce the complexity for analysis, ionized via electrospray, and introduced into a mass spectrometer (MS) [2]. The instrument acquires mass spectra of all precursor ions at frequent intervals to determine the m/z values of the ions entering the MS at a given moment. These precursor ion scans are commonly referred to as MS1 spectra. The instrument then sequentially opens a series of isolation windows centered at the most intense precursor ion peaks using a predefined set of rules provided in the instrument method. The ions selected by these isolation windows are fragmented and product ion spectra of the fragments are collected. In modern instruments, tens of thousands of product ion spectra are collected in each analysis. As instruments increase in speed and sensitivity, it becomes possible to reduce the number of fractions that must be collected prior to MS [3]. It has recently been reported that a majority of yeast proteins can be detected in a single run [4] and there is a need to provide comprehensive MS analysis in a single run of more complex proteomes such as human.
The subsequent interpretation of these MS2 spectra requires an informatics workflow of significant sophistication to account for the myriad of analysis approaches and hence complexity [5]. Many techniques and software tools used to identify the ions that yielded each spectrum have emerged over the past 20 years since the initial implementation of an automated tool called SEQUEST [6]. The Comet search engine [7] was recently introduced to the proteomics community and constitutes an open-source implementation of the SEQUEST algorithm. It was used in lieu of SEQUEST to process much of the data in this article as described below. Through the TPP’s support for other protein sequence search engine results such as Mascot [8], the reSpect algorithm will also work with these workflows. In general, the approach is to match each of the acquired spectra either with theoretical spectra that are generated on-the-fly from a set of candidate peptides with similar mass as the detected precursor or with spectra that have been previously observed and stored in spectral libraries [9], having been selected from a list of proteins that may be present in the sample. Programs for searching sequence databases and spectral libraries are termed sequence search engines and spectral library search engines, respectively [10].
There are dozens of search engines available to users, with new ones emerging each year. Curiously, the most recently developed search engines are not vastly better than the ones developed 20 years ago (and subsequently maintained). Yet, while most search engines yield broadly similar results, the variety in scoring functions of different engines leads to the observation that intelligently combining the results of several search engines run on the same dataset will yield an improved result over any of the search engines alone [11]. This seems to arise from the fact that different scoring functions are better at scoring different subsets of correct PSMs more highly than others.
The Trans-Proteomic Pipeline (TPP; [12–14]) is a widely used suite of open-source software tools for processing shotgun proteomics data. It includes raw data converters, both spectral library and sequence search engines, search result validation tools, quantification tools, and data exploration and visualization tools. Search engines typically yield a PSM for nearly every spectrum in a file, but many are incorrect, and many methods have been proposed to help statistically validate the search results and help separate correct from incorrect identifications. Although a common approach is to use search engine scores to specify thresholds by which to filter the search results and using decoy counting methods to estimate the false positive rate, post processing all unfiltered search results with validation software such as the TPP will typically significantly increase the number of correct PSMs (and distinct peptide sequences) that can be mined from each dataset.
There are several TPP tools that assist with this. PeptideProphet [15] models search engine output scores in conjunction with mass differences and other attributes of each PSM to assign a probability of being correct to each PSM. As of the writing of this paper, PeptideProphet can model the results of the following established search engines: SEQUEST, Comet, X!Tandem, MyriMatch, MSGF+, Mascot, Inspect, ProbID, SpectraST, Crux, Phenyx, and OMSSA.
The iProphet tool [16] further refines the probabilities of each PSM with potentially corroborating information from other PSMs, and can also combine the results of multiple search engines when applicable. ProteinProphet [17] then infers which proteins have been detected, and assigns to each a statistically robust probability based on the derived peptides. In all, the TPP provides a complete set of software tools underpinned by several XML data formats [12] that support the interoperability of all the tools.
One aspect of the shotgun workflow that is often overlooked is that several species of peptide ions can often be fragmented together and represented within the same MS2 spectrum. Even for highly fractionated samples, there are times when peptides of similar masses will occur in the same fraction and at overlapping retention times; however, for minimally fractionated samples, or otherwise very complex samples, it becomes rather common to observe several different peptide ion species contained within the isolation window along with the instrument-targeted precursor peptide. The ions that are isolated within the defined isolation window are all fragmented together in the ion-trap or collision cell and the resulting fragment ion spectrum is a composite of all the ions initially isolated. When precursor ions of similar intensities are fragmented together, the resulting chimeric spectrum may be difficult to identify. But in many other cases the intended precursor ion dominates the signal and can still be easily identified. The other, lower intensity precursor ions contribute many lower intensity fragment ion peaks in the single composite product ion spectrum.
There are previous efforts to develop software to identify the contributing peptides to chimeric spectra. The first search engine to try to identify multiple ions per spectrum was ProbIDTree [18], which would remove all identified peaks from a spectrum and immediately try another round of identification with the remaining peaks. The output supported multiple identifications for each spectrum. The M-SPLIT tool [19] attempts to model input spectra as the composite of several spectra taken from a spectral library. The MixDB tool [20] instead uses a sequence database search strategy to model each spectrum as the composite of a pair of ions of differing abundance. A recently described approach implemented in the DeMix algorithm [21] instead clones spectra that may be chimeric based on the detection of multiple precursors in the isolation window, and each of the clones are analyzed separately using a very narrow tolerance at each detected precursor m/z. A limitation in the widespread adoption of these software solutions is that they typically replace the search engine in the data analysis, potentially disrupting pipelines already established and relied upon in laboratories.
An alternate acquisition method, termed Data-Independent Acquisition (DIA), or SWATH-MS [22], or other implementations such as the MSE approach [23], attempts to generate chimeric spectra with much wider isolation windows containing many co-eluting peptides by design. Because the isolation windows are typically large enough to include many, perhaps dozens, of peptide ions, traditional search engines such as SEQUEST and Mascot are not suitable for analysis of such DIA data in their native form. Different software solutions have been developed for analyzing DIA type data [24, 25] to try to overcome this difficulty in extreme multiplexed fragmentation spectra interpretation.
Here we present a new software tool, called reSpect, which assists in the effort to identify additional peptide ions contributing to chimeric spectra in Data-Dependent Acquisition (DDA). It has the distinct advantage over other software tools for the identification of chimeric spectra in that it is not implemented as yet another search engine, but functions as a post processing step that is compatible with other sequence database search engines as well as spectral library search engines. To illustrate this point, reSpect is included with the TPP, and can be seamlessly integrated into existing pipelines utilizing any of the TPP search tools. In the following sections we describe the implementation of reSpect, select some test datasets, and then demonstrate the usefulness of the tool by examining the results of processing these test datasets with a workflow that includes reSpect.
Methods
Implementation of the software
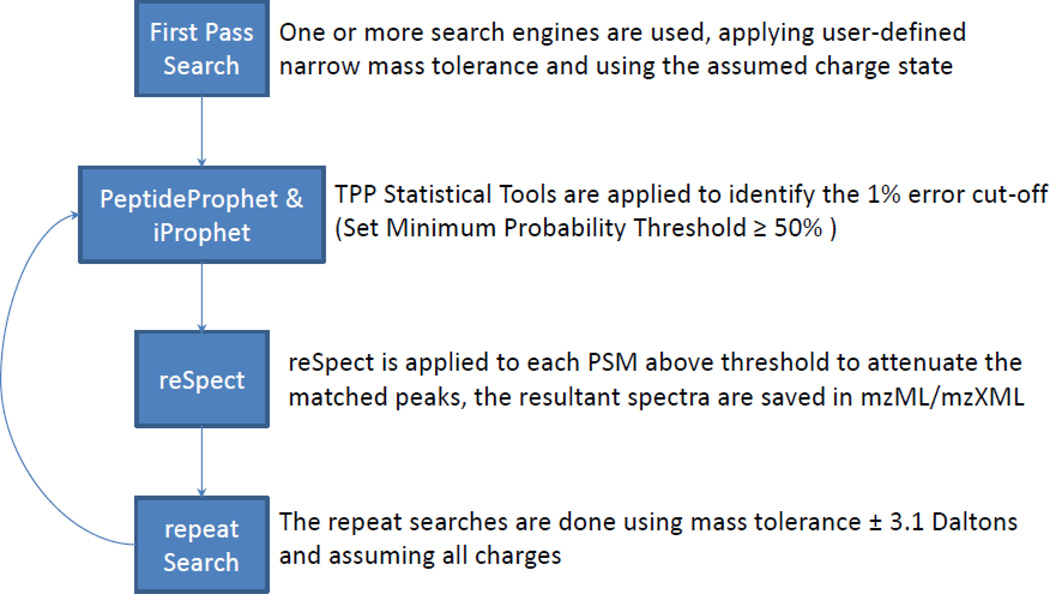
In order to enable the identification of multiple peptide ions in conglomerate MS2 spectra we have developed an iterative workflow that can be applied to most search engines and analysis environments. The workflow, as depicted in Figure 1, begins with a first pass search using any search engine(s) supported by the TPP followed by processing with PeptideProphet and iProphet to produce a pepXML file with probabilities that for each spectrum, the matched peptide ion is responsible for the major ion peaks therein. The next step is to process the result with reSpect to produce a new set of mzML files with modified MS2 spectra as described below. The process continues with a second pass search with more relaxed search parameters, opening up the mass tolerance to match the isolation window and allowing for different charge states, with the goal of identifying the remaining fragment ion peaks in the spectrum. The second pass search is followed by PeptideProphet and iProphet modeling on the new search result. Because the first and second pass peptide match statistics are likely to differ, they are modeled separately and are not combined until ProteinProphet analysis. The method may be followed by additional rounds of analysis with reSpect and re-search, each time attenuating each of the identified fragment ion peaks. At some point enough peaks will be attenuated that the remaining noise will fail to produce additional high-scoring matches, at this point the process should be halted. In this analysis we applied at most three rounds of reSpect analysis and search.
Figure 1. Overview of a workflow that includes the reSpect tool.
MS2 data are first searched by a database search engine, followed by post-processing with the PeptideProphet and iProphet tools. Then reSpect is used to reprocess each PSM from the search engine output to create a new mzML file with a subset of spectra that are modified to attenuate the peaks explained by the originally identified peptide ions. This is followed by another round of database searching, typically with a larger precursor m/z tolerance. These search results are processed by PeptideProphet and iProphet, followed by additional iterations if warranted.
Alternatively to sequence searching, spectral library searching with the SpectraST tool [26] may be used in any of the search passes as desired by the user. Spectral library search is typically faster, more sensitive, and more specific than sequence searching, partly on account of the smaller search space. However, since spectral libraries are generally incomplete relative to sequence references, the degree to which identifications are missed because they are not in the reference is much greater.
The reSpect tool takes as input a pepXML file with PSMs and probabilities based on PeptideProphet and iProphet modeling plus the original mzML or mzXML files. For each PSM with a probability greater than the set threshold (P>0.5 by default), reSpect evaluates all possible b and y ions (c and z in the case of ETD), neutral losses, and the component isotopes of the assigned peptide ion fragments. The peaks in the original spectrum that match the expected mass of the peptide ion fragments, within a user-defined mass tolerance (±0.5 by default) are deemed explained and their intensities are attenuated, with the attenuated intensity being:
Iatt = (1−P)*Iorig, where: Iatt is the attenuated intensity, Iorig is the original intensity, and P is the iProphet probability (or PeptideProphet probability if iProphet was not used)
For example, when P = 0.5, peaks are reduced by half, and when P = 1 the corresponding peaks are removed completely. The modified spectra are written out as new mzML files containing only the modified spectra. The spectrum identifiers are modified by appending “_rs” to the end so as to differentiate them from the original spectra. The following search then uses these new, reSpect-created mzML files as input.
The reSpect algorithm attenuates the matching peaks in each spectrum assuming the correctness of the match. In the case of PTM containing peptides and the possibility of false localization of the PTM by the search algorithm we suggest the use of the TPP tool PTMProphet to first help correctly localize the modifications within the peptides. This will help ensure that the correct peaks in each spectrum can be identified and thus attenuated by reSpect.
We note that each probability metric is not indicative that the assigned peptide fragment ion is the only ion that contributes to a spectrum, but rather that the assigned peptide fragment ion does contribute to the peaks in a spectrum. The reSpect tool is written in C++, and the source code is available at SourceForge under an open-source license, along with the entire implementation of the TPP. Most users will find it easiest to use the tool simply by installing the TPP package as a whole.
Demonstration datasets
To demonstrate the effectiveness of this workflow, we apply the procedure to 7 different datasets of varying complexity (Table 1) and examine the results. Second pass searching of the reSpect generated spectra was done using a ±3.1 Dalton precursor tolerance and allowing for possible charge states of 1+ to 5+. Selection of the wide mass tolerance allowed identifying non-monoisotopic peptides present in the isolation window.
Table 1.
Attributes and statistics for the datasets used to validate reSpect.
| Dataset Number |
Dataset Description | Krönik MS1 Features |
First Pass Peptides |
First Pass and reSpect Peptides |
First Pass Peptide to MS1 Feature Ratio |
% Boost in Peptide IDs with reSpect |
First Pass Peptides with Detected Feature by Krönik |
First Pass and reSpect Peptides with Detected Feature by Krönik |
Max Delta PPM |
Max Delta RT +- mins |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Yeast S288c (Moritz Lab) | 23992 | 5298 | 6903 | 0.22 | 30.32 | 4775 | 5111 | 10 | 5 |
| 2 | HeLa (Set 1 Yates Lab) | 231766 | 167776 | 178056 | 0.72 | 6.13 | 150085 | 153371 | 10 | 5 |
| 3 | HeLa (Set 2 Yates Lab) | 206855 | 193898 | 202059 | 0.94 | 4.21 | 173276 | 175852 | 10 | 5 |
| 4 | Hs_hESC_NSC_phospho | 128698 | 94509 | 100435 | 0.73 | 6.27 | 86218 | 86467 | 10 | 5 |
| 5 | HeLa (Mann Lab) | 267550 | 119327 | 134539 | 0.45 | 12.89 | 112916 | 118903 | 10 | 5 |
| 6 | Yeast (Coon Lab) | 57592 | 44793 | 45953 | 0.78 | 2.59 | 36270 | 36656 | 10 | 5 |
| 7 | iPRG2013 | 97880 | 39669 | 42893 | 0.41 | 8.13 | 38891 | 40672 | 20 | 10 |
Dataset 1 raw files (this laboratory) are stored in PeptideAtlas [27, 28] (accession # PASS00665) and a detailed description of the sample can be found in the Supplementary Material. Dataset 1 was searched with the Comet database search engine [29], using 25 ppm precursor tolerance with isotopic error enabled and using semi-tryptic enzymatic rules in the first pass search The search database utilized was UniProt [30] yeast (2014-01) with an included set of randomized decoys. The search results were processed with PeptideProphet and iProphet versions bundled with TPP version 4.7.1. PeptideProphet was run with the ACCMASS option enabled (for high mass accuracy precursor modeling), using NONPARAM option (for using the exact shape of the decoy distribution as the negative distribution) and specifying the DECOY=Random and DECOYPROBS decoy PSM handling options. All reSpect results, including the third and fourth round search results were processed along with the second-round search results so that there were sufficient data points for PeptideProphet and iProphet to model. The processing of reSpect results with PeptideProphet was done using the same options as with the first pass, but without the ACCMASS model.
Datasets 2 and 3 are provided by Dr. John R Yates III from a HEK293T cell study (PeptideAtlas accessions: PAe004080 and PAe004083). There are a total of 395 datafiles divided into two subsets. The first subset labeled Dataset 2 contains 156 datafiles and the second subset, Dataset 3, contains 239 additional datafiles. Both datasets were searched with Comet. Precursor mass tolerance of 1.1 Da was used. The search database utilized was UniProt human complete proteome (2012-10) plus alternative sequences with added peptides which contain the amino acid variants annotated by UniProt. The common contaminants and randomized decoys were added to the search database. PeptideProphet was run with the ACCMASS model enabled, using NONPARAM option and specifying the DECOY=DECOY and DECOYPROBS (for reporting the modeled probabilities of decoy hits rather than forcing them always to 0 as known false positives). The Comet PeptideProphet results were then processed with iProphet to improve the classification of correct and incorrect PSMs. The processing of reSpect results with PeptideProphet was done using the same options as with the first pass, but without the ACCMASS model and with EXPECTSCORE option enabled (using Comet expectation scores for PSM classification); iProphet was used to process the PeptideProphet validated reSpect results.
Dataset 4 is provided by Dr. Laurence Brill (Sanford-Burnham Medical Research Institute), stored in the PeptideAtlas (accession: PASS00233), and available once the dataset is published by the owner. It consists of 738 ETD and 738 CID mzML files generated on LTQ-Velos Orbitrap (Thermo-Fisher Scientific). The data contain 27 SCX fractions of comparative proteomes and total phosphoproteomes from human embryonic stem cells (hESCs) and their virtually pure neural stem cell (NSC) derivatives. The data were searched with Comet against the database used also to search Datasets 2 and 3 described above. Precursor tolerance of 50 ppm was specified with isotope error flag enabled. The search was semi-tryptic and allowed for 2 missed cleavages. Variable mods of n-terminal acetylation, methionine oxidation, and serine, tyrosine and threonine phosphorylation were used in the search. The search results were processed using PeptideProphet with ACCMASS enabled and using the semi-parametric model (NONPARAM). Further validation was done by iProphet (version 4.6.3) with all default settings. PTM site localization was modeled by PTMProphet (version 4.8.0) to indicate the most probable site of attachment.
Dataset 5 is derived from a 48-fraction HeLa cell lysate dataset [31] from the Dr. Matthias Mann Lab (Max-Plank Institut für Biochemie, Martinsried, Germany), stored in the PeptideAtlas (accession: PAe003653), collected on an LTQ-Velos Orbitrap instrument (Thermo Fisher-Scientific). Data were searched with the Comet algorithm using high resolution search setting, 20ppm precursor tolerance with isotope error enabled and using semi-tryptic enzymatic rules. The search database was generated same the same as dataset 2 but with newer version (2014-01). The search results were processed with PeptideProphet and iProphet versions bundled with TPP version 4.7. PeptideProphet was run with the ACCMASS model enabled, using NONPARAM option and specifying the DECOY=DECOY and DECOYPROBS decoy PSM handling options
Dataset 6 is derived from the One Hour Yeast Proteome dataset [4] from the Dr. Joshua Coon Lab (U. Wisconsin, WI), stored in the PeptideAtlas (accessions: PAe005216, PAe005217, PAe005218). Briefly, spectra were acquired using an Orbitrap Fusion Tribrid instrument (Thermo-Fisher Scientific). MS2 spectra were acquired with an isolation window of 0.7 m/z, using HCD with normalized collision energy of 30. Dynamic exclusion was set to use ppm accuracy around the precursor, and the exclusion duration was 45 seconds. The dataset was searched with Comet using 20ppm precursor tolerance with isotopic error disabled and using semi-tryptic enzymatic rules. The search database utilized was downloaded from http://downloads.yeastgenome.org/sequence/S288C_reference/orf_protein/orf_trans_all.fasta.gz with included set of common contaminants and randomized decoys. The search results were processed in the same way as dataset 3.
Dataset 7 analyzed for this article was the iPRG2013 study containing data derived from personal omics whole cell lysate profiling of human peripheral blood mononuclear cells [32] collected on a LTQ-Velos Orbitrap instrument (Thermo-Fisher Scientific), stored in the PeptideAtlas (accession: PAe005219). Peaks selected for fragmentation more than once within 30s were excluded from selection (10 ppm window) for 60s. The peptide digest was separated by a 2-dimensional workflow where 14 fractions were obtained in the first dimension by high pH reverse phase chromatography and each fraction was analyzed by LC-MS2 using a 240-minute low pH reversed phase separation in the second dimension. Six-plex tandem mass tag (TMT) reagents were employed for labeling these samples and cysteines were carbamidomethylated. The data were searched using Comet and X!Tandem against databases derived from RNA-Seq transcriptome analysis, novel sequences and UniProt SwissProt human databases [33].
Results and Discussion
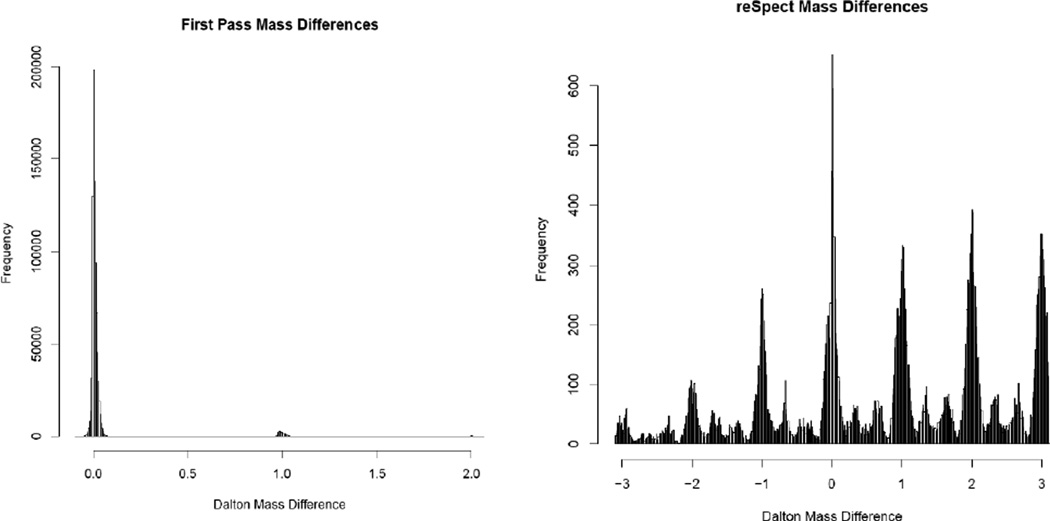
For the first pass searches we used a mass tolerance of 20 to 50 ppm, centered around the primary precursor and several neighboring isotopes. However, for subsequent post-reSpect searches, we used a much wider ±3.1 Dalton precursor tolerance, because the isolation window could contain the +1, +2 and +3 charge isotope peaks (in addition to the monoisotopic ions) of co-eluting peptides. The selection of the wide mass tolerance in the reSpect rounds of searching allows identifying the chimeric peptides that are co-eluting yet not necessarily targeted by the instrument. While the precursor ion mass of the target ion is often predicted accurately, the masses of co-eluting ions are unknown and may differ by several m/z from the target ion precursor mass. Figure 2 shows the observed m/z differences between each selected precursor ion m/z and the m/z of each putative identification for the first search (with narrow precursor mass tolerance) on the left panel and the second pass search (with a wide precursor mass tolerance) on the right panel. The pattern of peaks in the mass difference distribution of the secondary matches is likely related to whether the charge state of the original measured precursor matches that of the secondary peptide; when the charges are different, the mass differences will tend to fall between the integer offsets. In this experiment the majority of secondary matches were of charge 2+ and some were 3+; identifications containing a 2+ primary and 2+ secondary peptides charge states tend toward integer mass offsets, identifications containing a 2+ primary and 3+ secondary peptides charge states, or vice versa, tend toward mass differences with a decimal value near whole thirds (e.g. x.333 or x.666).
Figure 2. Histograms of mass differences between measured precursor m/z values and theoretical m/z values for the PSM assignments.
Representative histograms of mass differences between measured precursor m/z values and theoretical m/z values for the PSM assignments by the first pass search on the left, and PSM assignments after reSpect and re-search of a 48 fraction HeLa cell lysate dataset on right. In the left panel, the strong feature at 0 corresponds to the narrow m/z tolerance of the search, while the features at +1 and +2 Daltons are due to misassigned primary isotopes.
The performance of the reSpect algorithm was evaluated using iterative re-analysis of MS2 spectra over multiple rounds. All counts are distinct peptides at a defined peptide-level FDR of 1% or less based on decoy count estimates with PTM variants of each peptide being counted independently. If PTM variants are co-eluting and present in the same chimeric spectrum, they would have to be identified in separate iterations of reSpect processing, passing the FDR of 1% threshold each time.
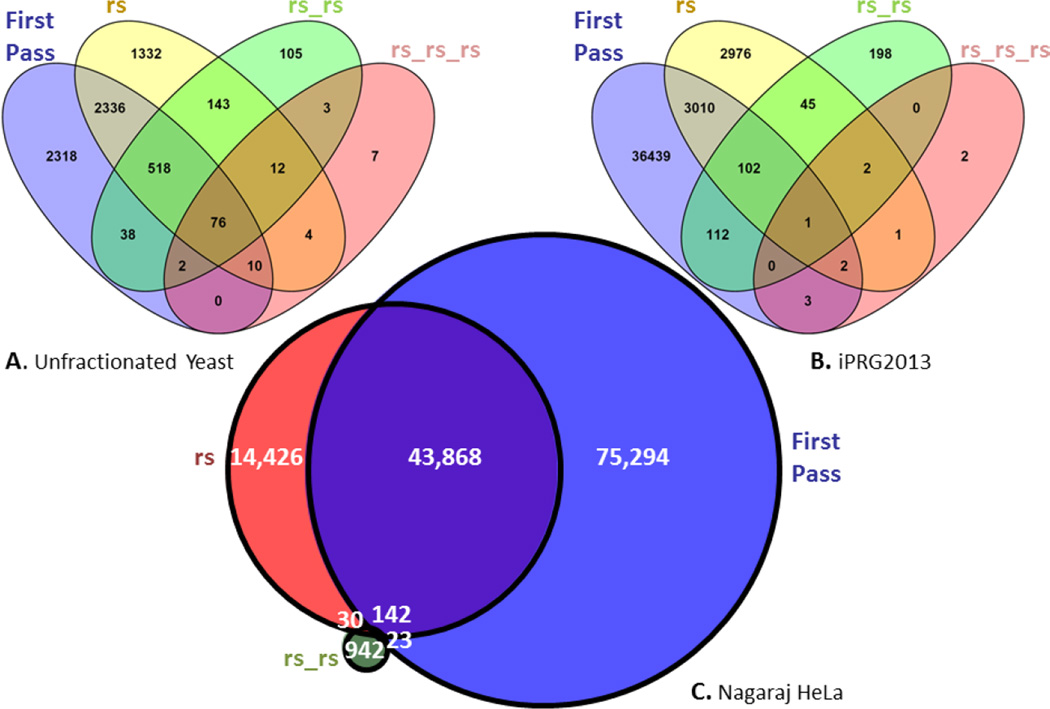
The degree of overlap in the four rounds of searching for Dataset 1 is depicted in a non-proportional Venn diagram in Figure 3A. Based only on the first round of searching 5298 distinct peptides were identified. The second round search revealed 2940 peptides that had been seen before in the first pass search, but also 1491 new peptides, missed in the first pass search. The third round search yielded an additional 108 novel peptides, and the final fourth round yielded yet an additional 7 not previously identified peptides. However in both cases, instances of previously identified peptides were also found, lending confidence that the method is working as intended. In all, the increase in the total number of distinct peptide sequences was 30.3% at the same decoy-based peptide-level FDR.
Figure 3. Peptide identification after repetitive reSpect analysis.
Venn diagrams for the overlap in identified distinct peptides after multiple rounds of reSpect, irrespective of any modifications, for datasets 1 (Panel A), 7 (Panel B), and 5 (Panel C), arranged in decreasing order of sample complexity. Datasets 1 and 7 were searched four times, with very few novel distinct peptides identified in the last round. Dataset 5 was searched three times.
The overlap in the four rounds of searching for Dataset 6 is depicted in a non-proportional Venn diagram in Figure 3B. There are 39,669 distinct peptides identified in the first round of searching. After running reSpect on these results a second round of searching resulted in 3115 distinct peptides that had been seen before, and 3024 distinct peptides not previously identified. The third round of search yielded 198 novel peptides, and the final fourth round still yielded an additional 2 peptide matches. In both cases, additional PSMs corresponding to previously identified peptides were found. Thus, the distinct newly identified peptide count increase in this fractionated dataset totaled 8.1%.
The overlap in three rounds of searching for Dataset 5 is depicted in a proportional Venn diagram in Figure 3C. In this dataset 119,327 distinct peptides were identified in the first round of searching. After reSpect analysis of these spectra, a second round of searching yielded nearly 44,010 distinct peptides that had been seen before, and 14,456 distinct peptides not previously identified in the initial database search. The second application of reSpect followed by the third round of searching yielded 942 new peptides. This analysis demonstrated a 12.8% increase of distinct peptide sequences in two reSpect rounds.
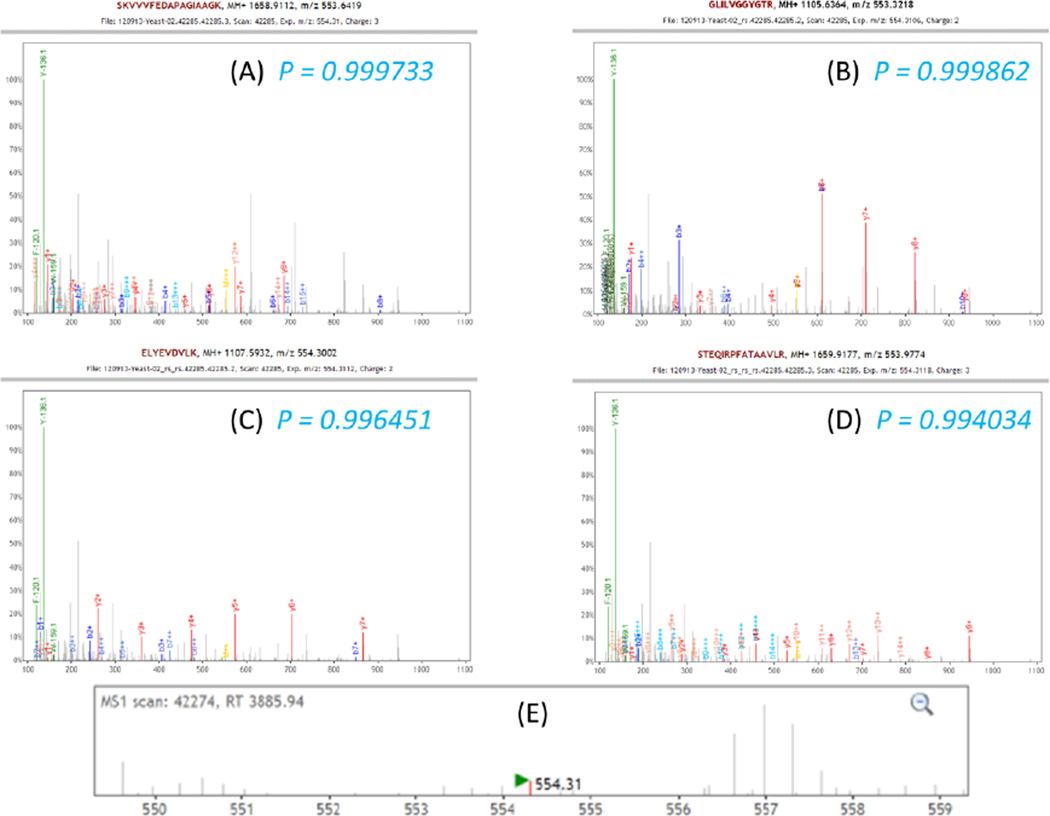
Figure 4 depicts an example of four identifications of different peptides contained within a single MS2 spectrum from the Dataset 1; all peptides were identified with probabilities greater than 0.99. Figure 4A shows the original spectrum overlaid with the primary identification, which was 3+ charge ion SKVVVFEDAPAGIAAGK with precursor m/z delta 2.0043 Daltons, or less than 3 ppm from the +2 charge isotopic peak. Although many peaks are identified, there are clearly many unidentified peaks present. Additional peaks in the precursor spectrum that preceded the fragmentation of the selected peptide ions Figure 4E suggest the presence of additional ion species within the isolation window of ±3 Daltons. All of the explained peaks were then highly attenuated, and the resulting spectrum searched again, this time with a search window matching the broadness of the isolation window. The second search yielded the second confident peptide ion identification, with a probability of 0.999 and precursor m/z delta of 1.9776 Daltons, or about 20 ppm from the original MS1 precursor. The y series of peaks from the second peptide identified in this spectrum is clearly visible in Figure 4B. After attenuation of the matched peaks from the second round identification, the third round of database searching identified another peptide shown in Figure 4C. As shown in Figure 4D, after the third round of reSpect, and the fourth round of searching, nearly all peaks in the original spectrum are explained by at least one of the matching peptides.
Figure 4. Iterative reSpect analysis of a single MS2 spectrum.
Shown above is a single spectrum from the Moritz Lab yeast dataset that yielded 4 different high confidence PSMs, all having probabilities greater than 99%. Panel A depicts the original spectrum overlaid with the primary identification, which was 3+ charge ion identified with a probability of 0.9997. Identified peaks for this peptide are attenuated by reSpect, and the second round of searching (panel B) yields a different peptide with a probability 0.9999, clearly matching to some unidentified peaks from panel A. The results of search rounds three and four are displayed in panels C and D, at which point nearly all peaks are identified. The peaks at 120.1 and 136.1 m/z are immonium ions (predominantly Y, and F), which are neither removed by reSpect, nor used for scoring by the search engines, but are labeled in the plots for further confirmation of identified peptide composition. Panel E shows the MS1 scan which triggered the original CID.
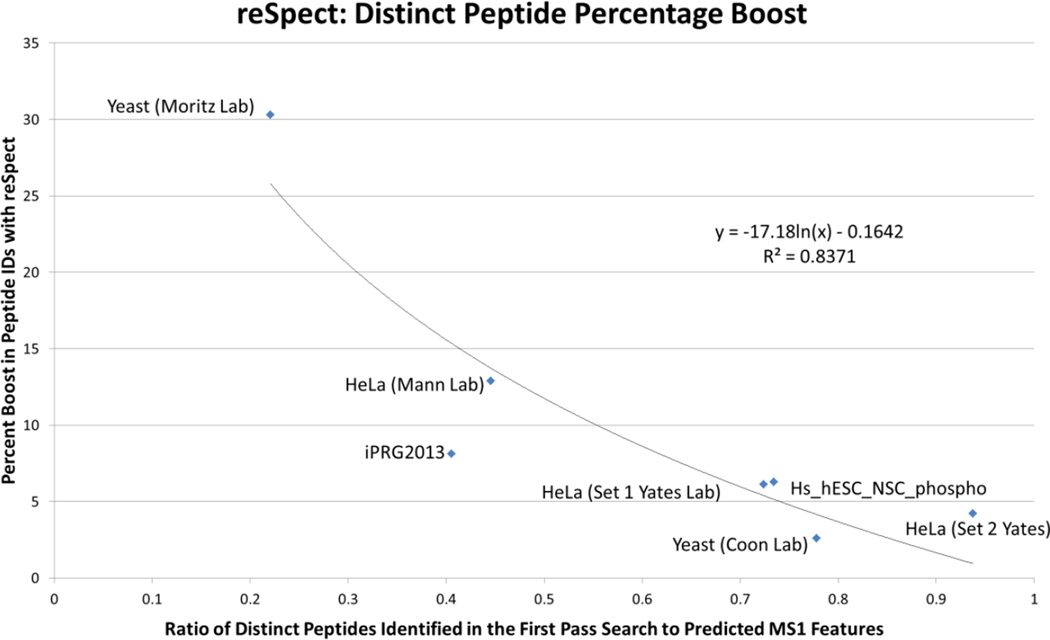
Although there is a wide variation among datasets in the achievable benefit from the use of the reSpect algorithm, the benefit is significant in all of the datasets we tested, even in highly fractionated datasets. We further explored the data using the analysis workflow shown in Figure 5 to estimate the number of peptide features that are seen in the MS signal of a dataset. Briefly, the Hardklör [34] algorithm was used to pick the peaks in each precursor spectrum (MS1), followed by Krönik [34] to count persistent features (i.e. a series of peaks over time at nearly the same m/z value) in each MS run, followed by a script called kronikCount.pl that we wrote to count persistent features across all files of a dataset. This method was applied to establish the maximum number of peptides that we should expect to identify by MS2 spectra. We ran this on all datasets and compared the results to the number of distinct peptides identified by MS2 spectra and the percentage boost yielded by the application of reSpect. As can be seen from Figure 6, there is a strong negative correlation between the number of distinct peptides seen in MS2 as a fraction of MS1 features that are estimated from the dataset, and the reSpect percentage boost. In other words, as the ratio of MS2 identifications to MS1 features in the data rises, the percentage of new peptides that can be seen by applying reSpect decreases. Interestingly, Datasets 1 and 6 of yeast tryptic digests show the greatest polarity in terms of the fraction of MS1 features estimated and the percent boost to MS2 identifications after using reSpect, despite the similarity of the samples analyzed. Inspection of the data acquisition methods provides insight into these differences. The datasets were acquired using different instruments and the acquisition parameters also show several differences. Most notable among them are the dynamic exclusion duration and the isolation window width. Dataset 6 uses a much longer dynamic exclusion duration (45 s vs. 10 s), minimizing the chance that a peptide ion will be reselected after expiration from the exclusion list. Dataset 1 used a wider isolation window (3.0 m/z vs. 0.7 m/z), increasing the likelihood that multiple precursor ions are fragmented at the same time. The scan speed of the two instruments used perform at different data rates and the Orbitrap Fusion instrument provides a deeper dataset for the yeast digest analyzed from the Coon lab (i.e., Q Exactive ~12Hz; Fusion Tribrid instrument ~20 Hz). These method parameter and instrument differences influence both the coverage of the entire sample and the potential to observe chimeric MS2 spectra. However, there is no golden rule for data acquisition; instrumentation, sample complexity, and LC gradient duration must be considered when optimizing sample coverage. Application of reSpect allows increasing the sample coverage in all situations, particularly when the optimal acquisition parameters cannot be met. Table 1 lists each of the test datasets along with the most important attributes of the datasets, the analyses, and the results. Importantly, because the results of reSpect are additive, it is able to boost the counts of proteins that can be identified in a given sample. It can do this by identifying new peptides that can distinguish previously indistinguishable proteins, and it can identify new peptides for proteins that have not been seen before. The identification of confident peptides by applying reSpect with additional error-rate control using PeptideProphet, iProphet and ProteinProphet increases the number of proteins that can be confidently identified. For example, on the Moritz lab yeast dataset the number of proteins went from ~650 at 1% decoy-estimated error-rate to ~710 at 1% decoy-estimated error-rate (Supplementary Figures 1A and 1B). Also, at the same probability cutoff of 90% (corresponding to an error-rate of 1.1% for the original analysis and 0.4% for the reSpect analysis) the number of proteins goes up from 608 without reSpect to 616 with reSpect while the number of single hit proteins goes down from 95 without reSpect to 29 with reSpect. reSpect is able to increase both the depth and the breadth of sample coverage.
Figure 5. Overview of a workflow for estimating the number of potential MS1 features present in a given dataset.
First the MS data is processed using Hardklör to model the features in each MS1 spectrum. Then, Krönik is used to determine the number of ion features that are persistent over several spectra in each MS run. Finally, a script called kronikCount.pl is used to count the union of MS1 features across all runs of a given dataset.
Figure 6. Percentage increase in the number of distinct peptide sequences identified after using reSpect.
The results of newly identified peptides in MS2 attenuated spectra are plotted against the fraction of counted MS1 features over the count of identified distinct peptide sequences in the initial search. An approximate trend line that would be exponentially large at X~0 and that goes approximately to 0 near X=1 is overlaid. There appears to be a high correlation between these two metrics, implying that the likely effectiveness of reSpect in recovering additional peptide sequences may be estimated based on these metrics, which can be calculated after the first search.
Additional analysis compared features within the reSpect algorithm, and performance of the reSpect algorithm compared to a similar tools. To illustrate the differences between attenuation and deletion of PSM matched fragment ion peaks, reSpect was operated in DELETE mode for Dataset 1, and the results are presented in Supplementary Figure 2. In DELETE mode reSpect removes the matched peaks rather than attenuate them. In general, the two methods are very similar. The attenuation approach performs slightly better, although this may not be significant unless the minimum probability of peptides that are subjected to reSpect is lowered to 0. Additionally, we compared the performance of reSpect to DeMix, using the DeMix example dataset (Supplementary Figure 3). Out of a combined total of 469 distinct peptides seen by DeMix and TPP iProphet with reSpect pipeline, 82 were only seen by DeMix, while TPP iProphet with reSpect identified 117 that were missed by DeMix. Thus, the two approaches are comparable in performance and complementary.
An important feature of reSpect is its ability to identify low intensity peptides. These peptides don’t necessarily have an isotopic pattern in the MS1 signal and are therefore unlikely to be picked up by tools such as Hardklör and Krönik. These peptides are also less likely to be targeted by the mass-spectrometer due to their low intensities. However, the fragments for them exist in the MS/MS spectra of other peptides that were targeted. The fragments of peptides that are not targeted can be orders of magnitude smaller than the target peptide fragments, and identification of these relies on the ability of reSpect to significantly attenuate the signal of the dominant peptide in the fragment spectrum, not necessarily on the existence of an MS1 peptide feature as one may not exist.
Integrating reSpect into existing analysis pipelines will serve to improve the coverage and depth of proteomics datasets. Computationally, its execution is linear in complexity to the number of peaks being processed, typically taking just a few minutes per MS run and proportional in time to the number of spectra in the input pepXML file. Subsequent sequence database searches add to the computational burden. However, the implementation of reSpect as a standalone tool makes it possible to integrate it into existing complex analysis workflows. The availability of cheap computational cycles on the cloud make the additional computational cost more manageable, especially, at the benefit of identifying more peptides from the same data.
The application of reSpect methodology provides confident identification of otherwise unmatched peptides that co-elute and co-fragment with identified peptides that are more abundant, and the fragments for which are easier to observe. Interestingly, using reSpect allows the identification of PTM containing peptides that are not seen by a single pass search. One such example is presented in Supplementary Figures 4A and 4B. The first pass peptide is identified by the spectrum in Supplementary Figure 4A with a high probability of over 99%. The use of peak attenuation with reSpect followed by a second search of the processed spectra and TPP validation using PeptideProphet and iProphet yields a second confident PTM containing peptide shown in Supplementary Figure 4B having a probability of over 98%.
Chimeric spectra are also an important consideration for quantitation. For isobaric labeling techniques, the effect of co-fragmenting multiple peptide ions causes a compression in the range of the reporter ions [35, 36]. This effect can be somewhat mitigated by not using spectra for which multiple peptides are identified. For isotopic labeling or label-free ion intensity techniques extra care must be taken that elution profiles are extracted from the MS1 scans using very narrow tolerances to avoid being contaminated by signal from the co-eluting peptide ions with very similar precursor m/z values in their respective isotopic envelopes. The reSpect workflow presents an improvement for spectrum counting techniques, since additional instances of peptide ions can be recovered, increasing the overall numbers of counts beyond the one-peptide-per-spectrum paradigm.
The analysis results for all datasets can be downloaded from PeptideAtlas at the following link: http://www.peptideatlas.org/PASS/PASS00704. The spectral matches for the new peptides found in the iPRG2013 data are provided for viewing in the Supplementary Material.
Conclusion
We have presented a new post sequence searching tool, called reSpect, to attenuate peaks from dominant peptide ions identified to be present in mass spectra via a common sequence search engine with the aim of enabling the identification of additional peptide ions that are also represented at lower levels in chimeric spectra from isobaric or near-isobaric precursor ions. It is compatible with all other search engines supported by TPP, including sequence search engines and spectral library search engines. Although previously-presented tools have demonstrated their effectiveness on datasets where the improvement is very large, we find that the degree to which processing might benefit from properly handling chimeric spectra varies enormously from dataset to dataset, as one would expect. With some datasets, the increase in the number of identified distinct peptides is quite large (over 30% more in one of our examples), but the increase is more modest yet significant in other datasets. We find a significant correlation between the increase in distinct peptide identifications and the ratio of total MS1 features over identifications in the initial search. This estimator can be used to determine if there would be significant benefit in using reSpect for additional iterative processing.
The reSpect tool is integrated into TPP, and therefore is easy to use in conjunction with many different search engines and interoperable with the many other TPP tools, including iProphet and ProteinProphet. This makes reSpect ideal for use as part of an organized workflow such as the TPP, although this is not required and can be run as a standalone tool. Such workflow systems are becoming more prevalent, and TPP has been adapted [37] to the Taverna [38] workflow platform, as well as others. Since reSpect is a component of TPP, it is available for all platforms. Additional information, documentation, and downloads are available at the main TPP website http://tools.proteomecenter.org/TPP.
Supplementary Material
Acknowledgments
We would like to thank the contributors of the samples analyzed for this manuscript. We would also like to thank Mr. Joseph Slagel, Dr. Kristian Swearingen, current, and former members of the Moritz Lab for their meaningful discussions.
This work was funded in part by National Institutes of Health from the National Institute of General Medical Sciences under Grant Nos. R01GM087221, S10RR027584 and the 2P50 GM076547/Center for Systems Biology and the Dept. of Defense CDMRP grant W81XWH-11-1-0487.
REFERENCES
- 1.Yates JR, Ruse CI, Nakorchevsky A. Proteomics by mass spectrometry: approaches, advances, and applications. Annu Rev Biomed Eng. 2009;11:49–79. doi: 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- 2.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 3.Nilsson T, Mann M, Aebersold R, Yates JR, 3rd, Bairoch A, Bergeron JJ. Mass spectrometry in high-throughput proteomics: ready for the big time. Nat Methods. 2010;7:681–685. doi: 10.1038/nmeth0910-681. [DOI] [PubMed] [Google Scholar]
- 4.Hauser S, Wulfken LM, Holdenrieder S, Moritz R, Ohlmann CH, Jung V, et al. Analysis of serum microRNAs (miR-26a-2*, miR-191, miR-337-3p and miR-378) as potential biomarkers in renal cell carcinoma. Cancer epidemiology. 2012;36:391–394. doi: 10.1016/j.canep.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Deutsch EW, Lam H, Aebersold R. Data analysis and bioinformatics tools for tandem mass spectrometry in proteomics. Physiol Genomics. 2008;33:18–25. doi: 10.1152/physiolgenomics.00298.2007. [DOI] [PubMed] [Google Scholar]
- 6.Eng J, McCormack AL, Yates JR. An Approach to Correlate Tandem Mass Spectral Data of Peptides with Amino Acid Sequences in a Protein Database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 7.Eng JK, Jahan TA, Hoopmann MR. Comet: an open-source MS/MS sequence database search tool. Proteomics. 2013;13:22–24. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 8.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 9.Lam H, Deutsch EW, Eddes JS, Eng JK, Stein SE, Aebersold R. Building consensus spectral libraries for peptide identification in proteomics. Nat Methods. 2008;5:873–875. doi: 10.1038/nmeth.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eng JK, Searle BC, Clauser KR, Tabb DL. A face in the crowd: recognizing peptides through database search. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.R111.009522. R111 009522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shteynberg D, Nesvizhskii AI, Moritz RL, Deutsch EW. Combining results of multiple search engines in proteomics. Mol Cell Proteomics. 2013;12:2383–2393. doi: 10.1074/mcp.R113.027797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keller A, Eng J, Zhang N, Li XJ, Aebersold R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol Syst Biol. 2005;1 doi: 10.1038/msb4100024. 2005 0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deutsch EW, Mendoza L, Shteynberg D, Farrah T, Lam H, Tasman N, et al. A guided tour of the Trans-Proteomic Pipeline. Proteomics. 2010;10:1150–1159. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nuhn P, May M, Fritsche HM, Buchner A, Brookman-May S, Bolenz C, et al. External validation of disease-free survival at 2 or 3 years as a surrogate and new primary endpoint for patients undergoing radical cystectomy for urothelial carcinoma of the bladder. European journal of surgical oncology : the journal of the European Society of Surgical Oncology and the British Association of Surgical Oncology. 2012;38:637–642. doi: 10.1016/j.ejso.2012.02.187. [DOI] [PubMed] [Google Scholar]
- 15.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 16.Shteynberg D, Deutsch EW, Lam H, Eng JK, Sun Z, Tasman N, et al. iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.007690. M111 007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 18.Zhang N, Li XJ, Ye M, Pan S, Schwikowski B, Aebersold R. ProbIDtree: an automated software program capable of identifying multiple peptides from a single collision-induced dissociation spectrum collected by a tandem mass spectrometer. Proteomics. 2005;5:4096–4106. doi: 10.1002/pmic.200401260. [DOI] [PubMed] [Google Scholar]
- 19.Wang J, Perez-Santiago J, Katz JE, Mallick P, Bandeira N. Peptide identification from mixture tandem mass spectra. Mol Cell Proteomics. 2010;9:1476–1485. doi: 10.1074/mcp.M000136-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Bourne PE, Bandeira N. Peptide identification by database search of mixture tandem mass spectra. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.010017. M111 010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang B, Pirmoradian M, Chernobrovkin A, Zubarev RA. DeMix workflow for efficient identification of cofragmented peptides in high resolution data-dependent tandem mass spectrometry. Mol Cell Proteomics. 2014;13:3211–3223. doi: 10.1074/mcp.O114.038877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gillet LC, Navarro P, Tate S, Rost H, Selevsek N, Reiter L, et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.O111.016717. O111 016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li GZ, Vissers JP, Silva JC, Golick D, Gorenstein MV, Geromanos SJ. Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics. 2009;9:1696–1719. doi: 10.1002/pmic.200800564. [DOI] [PubMed] [Google Scholar]
- 24.Keller A, Bader SL, Shteynberg D, Hood L, Moritz RL. Automated Validation of Results and Removal of Fragment Ion Interferences in Targeted Analysis of Data-independent Acquisition Mass Spectrometry (MS) using SWATHProphet. Mol Cell Proteomics. 2015;14:1411–1418. doi: 10.1074/mcp.O114.044917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rost HL, Rosenberger G, Navarro P, Gillet L, Miladinovic SM, Schubert OT, et al. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat Biotechnol. 2014;32:219–223. doi: 10.1038/nbt.2841. [DOI] [PubMed] [Google Scholar]
- 26.Lam H, Deutsch EW, Eddes JS, Eng JK, King N, Stein SE, et al. Development and validation of a spectral library searching method for peptide identification from MS/MS. Proteomics. 2007;7:655–667. doi: 10.1002/pmic.200600625. [DOI] [PubMed] [Google Scholar]
- 27.Desiere F, Deutsch EW, Nesvizhskii AI, Mallick P, King NL, Eng JK, et al. Integration with the human genome of peptide sequences obtained by high-throughput mass spectrometry. Genome Biol. 2004;6:R9. doi: 10.1186/gb-2004-6-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farrah T, Deutsch EW, Omenn GS, Sun Z, Watts JD, Yamamoto T, et al. State of the human proteome in 2013 as viewed through PeptideAtlas: comparing the kidney, urine, and plasma proteomes for the biology- and disease-driven Human Proteome Project. J Proteome Res. 2014;13:60–75. doi: 10.1021/pr4010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huth-Schwarz A, Settele J, Moritz RF, Kraus FB. Factors influencing Nosema bombi infections in natural populations of Bombus terrestris (Hymenoptera: Apidae) Journal of invertebrate pathology. 2012;110:48–53. doi: 10.1016/j.jip.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 30.Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, Kelso J, et al. Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol. 2011;7:548. doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen R, Mias GI, Li-Pook-Than J, Jiang L, Lam HY, Chen R, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148:1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boutet E, Lieberherr D, Tognolli M, Schneider M, Bairoch A. UniProtKB/Swiss-Prot. Methods Mol Biol. 2007;406:89–112. doi: 10.1007/978-1-59745-535-0_4. [DOI] [PubMed] [Google Scholar]
- 34.Hoopmann MR, MacCoss MJ, Moritz RL. Identification of peptide features in precursor spectra using Hardklor and Kronik. Current protocols in bioinformatics / editoral board, Andreas D. Baxevanis … [et al.] 2012;13(Unit13 18) doi: 10.1002/0471250953.bi1318s37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karp NA, Huber W, Sadowski PG, Charles PD, Hester SV, Lilley KS. Addressing accuracy and precision issues in iTRAQ quantitation. Mol Cell Proteomics. 2010;9:1885–1897. doi: 10.1074/mcp.M900628-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Savitski MM, Mathieson T, Zinn N, Sweetman G, Doce C, Becher I, et al. Measuring and managing ratio compression for accurate iTRAQ/TMT quantification. J Proteome Res. 2013;12:3586–3598. doi: 10.1021/pr400098r. [DOI] [PubMed] [Google Scholar]
- 37.Huang Q, Kryger P, Le Conte Y, Moritz RF. Survival and immune response of drones of a Nosemosis tolerant honey bee strain towards N. ceranae infections. Journal of invertebrate pathology. 2012;109:297–302. doi: 10.1016/j.jip.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 38.Wolstencroft K, Haines R, Fellows D, Williams A, Withers D, Owen S, et al. The Taverna workflow suite: designing and executing workflows of Web Services on the desktop, web or in the cloud. Nucleic Acids Res. 2013;41:W557–W561. doi: 10.1093/nar/gkt328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.