Abstract
Histiotrophic nutrition pathways (HNPs) are processes by which the organogenesis-stage conceptus obtains nutrients, amino acids, vitamins, and cofactors required for protein biosynthesis and metabolic activities. Nutrients are captured from the maternal milieu as whole proteins and cargoes via receptor-mediated endocytosis in the visceral yolk sac (VYS), degraded by lysosomal proteolysis, and delivered to the developing embryo (EMB). Several nutrients obtained by HNPs are required substrates for one-carbon (C1) metabolism and supply methyl groups required for epigenetic processes, including DNA and histone methylation. Increased availability of methyl donors has been associated with reduced risk for neural tube defects (NTDs). Here we show that mono-2-ethylhexyl phthalate (MEHP) treatment (100 or 250 µM) alters HNPs, C1 metabolism, and epigenetic programming in the organogenesis-stage conceptus. Specifically, 3 h MEHP treatment of mouse embryos in whole culture resulted in dose dependent reduction of HNP activity in the conceptus. To observe nutrient consequences of decreased HNP function, C1 components and substrates and epigenetic outcomes were quantified at 24 h. Treatment with 100 µM MEHP resulted in decreased dietary methyl donor concentrations, while treatment with 100 or 250 µM MEHP resulted in dose-dependent elevated C1 products and substrates. In MEHP-treated EMBs with NTDs, H3K4 methylation was significantly increased, while no effects were seen in treated VYS. DNA methylation was reduced in MEHP-treated EMB with and without NTDs. This research suggests that environmental toxicants such as MEHP decrease embryonic nutrition in a time-dependent manner, and that epigenetic consequences of HNP disruption may be exacerbated in EMB with NTDs.
Keywords: histiotrophic nutrition, one-carbon metabolism, DNA methylation, histone methylation
1. Introduction
The processes by which mammalian embryos obtain nutrients and substrates for growth and development during embryogenesis and early organogenesis are known as histiotrophic nutrition pathways (HNPs). Because the placenta has not yet become fully functional at this stage, the active maternal-embryonic exchange of micronutrient substrates and metabolic cofactors from maternal circulation is not yet possible. HNPs facilitate the receptor-mediated endocytosis (RME) of bulk and carrier proteins along with their bound nutrient and cofactor cargoes through the visceral yolk sac (VYS) brush border. Subsequent proteolysis of these proteins in secondary vesicles (lysosomes) releases nutrient cargoes required for various cellular processes and supplies nearly 100% of all amino acids required for protein biosynthesis and enzymatic function in the VYS and in the embryo (EMB) proper [1, 2].
HNPs are responsible for providing and transporting many substrates to the EMB, including methyl donors for one-carbon (C1) metabolism such as methionine, folate, choline, and betaine [3–11]. C1 metabolism is the process by which dietary methyl donors are utilized to synthesize S-adenosylmethionine (SAM), the primary molecular methyl donor for cells. Through C1 pathway activity, SAM is made available for numerous processes, including post-translational modifications and epigenetic regulation. Thus, the nutritional state of the conceptus likely influences epigenetic reprogramming and regulation of genetic processes [12].
During the first trimester of pregnancy, cells are rapidly dividing to facilitate overall growth and anatomical form in order to complete embryogenesis. During this time, DNA undergoes substantial epigenetic programming, from near-complete loss of methylation marks after fertilization to reprogramming of methylation marks during embryogenesis and early organogenesis [13, 14]. Histone methylation, on the other hand, is variable; methylation increases over developmental time at the histone 3 lysine 4 (H3K4) locus and decreases at the histone 3 lysine 27 (H3K27) locus [15]. Because these processes occur during the phase of HNP-supplied nutrition and provide the necessary methyl donors for epigenetic marks, it is possible that perturbation of HNPs may result in abnormal epigenetic programming. Previous work has demonstrated that inhibition of HNP lysosomal function with leupeptin resulted in decreased DNA methylation in the EMB and VYS during early organogenesis [16].
Common dietary methyl donors and cofactors for C1 metabolism, including folate, betaine, choline, and vitamin B12, are required for normal growth and development. Maternal deficiencies in folate and choline during pregnancy have been associated with increased risk for neural tube defects (NTDs) [17–19]. Likewise, folate supplementation of maternal diet, via the fortification of foods or prenatal vitamins, has been associated with nearly a 30% decreased risk for NTDs [20–23]. However, the mechanism by which folate is beneficial is unknown [24, 25].
Mono-2-ethylhexyl phthalate (MEHP) is the primary metabolite of the ubiquitous plasticizing agent di-2-ethylhexyl phthalate (DEHP), a compound that has been found almost ubiquitously in consumer, industrial, and medical products including PVC products, medical tubing, and children’s toys. Numerous studies investigating exposures to DEHP have found detectable levels of its metabolites in a great majority of individuals sampled, including pregnant women [26–28]. Previous studies have demonstrated that MEHP exposure during early organogenesis increased the number of embryos with NTDs [29, 30]. Thus, the purpose of this exploratory work is to identify whether this increased number of NTDs due to MEHP exposure is concordant with decreased nutrient bioavailability to the EMB and whether this may have epigenetic consequences. It is hypothesized that MEHP exposure will reduce the activity of HNPs, decreasing the availability of methyl donors and C1 metabolism, and that these changes will result in decreased DNA and histone methylation.
2. Materials & Methods
2.1 Chemicals and Reagents
Bicinchoninic acid, dimethyl sulfoxide (HPLC grade), Tyrode’s balanced salt solution (TBSS), penicillin/streptomycin (10,000 units/ml penicillin, 10,000 µg/ml streptomycin sulfate), choline chloride, betaine, folic acid, D,L-homocysteine, D,L-methionine, L-cysteine hydrochloride, N,N,N-Trimethyl-d9-Glycine HCl (Betaine-d9) and Homocysteine-d8 were obtained from Sigma/Aldrich (St. Louis, MO). L-Methionine (methyl-d3) and D,L-cysteine (3,3-d2) were purchased from Cambridge Isotope Laboratories (Andover, MA, USA), and S-Adenosyl-L-methionine-d3 (S-methyl-d3) tetra (p-toluenesulfonate) salt was from C/D/N Isotopes (Pointe-Claire, Quebec, Canada). Hanks balanced salt solution (HBSS) was purchased from GIBCO/Life Technologies (Grand Island, NY). Mono-2-ethylhexyl phthalate was obtained from AccuStandard (New Haven, CT).
2.2 Mouse Whole Embryo Culture
Mouse embryo culture (mWEC) was performed following the conditions outlined in [31], and described in [30]. Briefly, female CD-1 mice were time-mated and obtained from Charles River (Portage, MI). The morning of a positive vaginal smear was designated as gestational day (GD) 0. Animals were maintained on a 12 h light-12h dark cycle and were supplied food and water ad libitum. Pregnant mice were euthanized with CO2, supplied at 10–30%, on GD 8 and the uterus was removed. Conceptuses were explanted from the uterus, randomized, and placed into 10 ml culture bottles. Each culture bottle contained 5 ml of 75% heat-inactivated rat serum/25% Tyrode’s balanced salt solution (TBSS), and 21.5 µl penicillin/streptomycin (Invitrogen, Carlsbad, CA 10,000 U/ml penicillin; 10,000 µg/streptomycin sulfate). Bottles were placed on a continuous-gassing carousel and initially supplied with 5% O2/5% CO2/90% N2. The gas mixture was changed after 6 hours to 20% O2/5% CO2/75% N2 to optimize conceptal growth. All experiments were performed in strict concordance with protocols approved by the University Committee on Use and Care of Animals.
2.3 Exposures and Sample Collection
MEHP was suspended in DMSO to increase solubility and stored at a concentration of 1 mg/ml. This MEHP mixture was diluted in DMSO and added to each treatment culture bottle to produce a final concentration of 100 or 250 µg/ml. These concentrations were optimized from previous experiments [30], based on viability and morphology, as well as reproducibility from other phthalate WEC experiments [29, 32]. Control bottles received an equivalent volume of DMSO only and the total volume added was no greater than 1 µl/ml culture medium. Conceptuses remained in culture for 24 h.
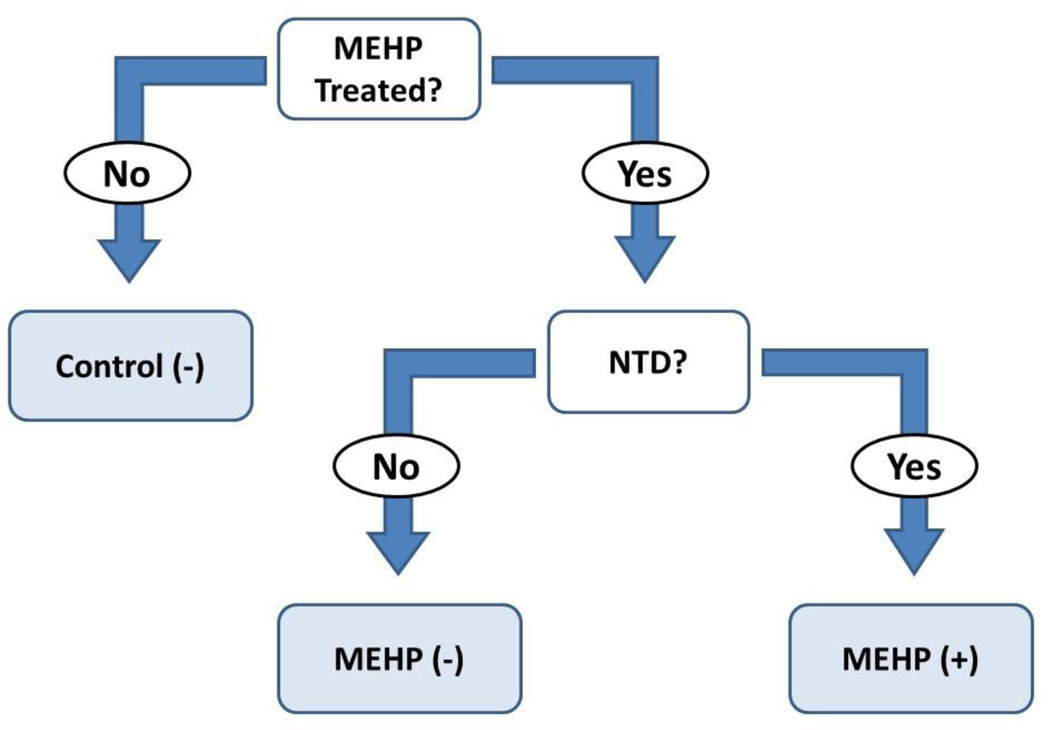
Following the culture period, conceptuses for DNA and histone analysis were thoroughly rinsed in HBSS and briefly inspected, at which time anatomical malformations and delayed closure of the embryonic neural tube were noted. EMBs with closed neural tubes were denoted as NTD (−) while those with open neural tubes were denoted as NTD (+) (Fig. 1). At the time of collection, no control samples were found to have abnormal neural tube closure. Conceptuses providing tissues for DNA isolation were dissected into EMB and VYS, snap frozen, and stored at −76°C until use. Conceptuses for histone isolation were also dissected into EMB and VYS, but histone proteins were immediately extracted using the EpiQuik™ Total Histone Extraction Kit (Epigentek). Following extraction, proteins were stored at −20°C overnight until completion of histone methylation experiments the following day.
Figure 1.
Schematic representing the sampling strategy utilized in this study. The first layer of stratification separated untreated conceptuses from those treated with MEHP. Visceral yolk sacs were removed from each intact concept and evaluated separately at each stratification level. Control embryos exhibiting open neural tubes (NTD) occur normally at a very low incidence. Only control conceptuses without NTD [Control (−)] were used in this stratification model. MEHP-treated conceptuses were further separated according to their NTD status. Those with embryos exhibiting an open neural tube [MEHP (+)] were separated from exposed conceptuses where no open neural tube was observed [MEHP (−)].
Conceptuses used for HNP analysis were randomized and explanted into culture on GD 8. After 24 h in culture (GD 9), fluorescence tagged FITC-albumin was added to the culture media at a nontoxic total concentration of 100 µg/ml [33, 34]. At this time, 100 or 250 µg/ml MEHP was added. Treated conceptuses were returned to the culture apparatus and grown in complete darkness for 3 h. At the conclusion, conceptuses were removed from culture and thoroughly rinsed in HBSS. Ethanol (6 mg/ml) was added to concurrent cultures and HNP assays were performed as a positive control because it is a known inhibitor of endocytosis in the HNP [35, 36]. Following culture, conceptuses were pooled in duplicate and transferred into a 150 µl drop containing 50 mM sodium phosphate buffer (pH=6.0) for dissection. The VYS was first removed and collected into 0.1% Triton X-100, followed by the EMB. The remaining extra-embryonic fluids (EEF) were then released into the sodium phosphate buffer and also collected.
Whole conceptuses for analysis for C1 component quantification or quantification of methyl donors were treated with 100 or 250 µg/ml MEHP on GD 8 and grown for 24 h in culture. At the conclusion of the culture period, whole conceptuses were carefully rinsed and stored intact in polypropylene tubes at – 80 C for later analysis. Samples for analysis of C1 components and for choline and betaine were quantitated as described below using two separate by LC-MS/MS techniques. Conceptuses for immunoassay quantification of folate and vitamin B12 were collected into 500 µl 6% trichloroacetic acid (TCA).
2.4 Histiotrophic Nutrition Assay
Histiotrophic nutrition assays were conducted using a modified version of the protocol outlined in [33]. Even though its contribution to total fluorescence is usually minimal, fluorescence in the EMB fraction of the conceptus was also measured. After collection of tissues and fluids, samples were sonicated to mix. A 20 µl aliquot of each sample and of culture media were immediately collected and set aside for measurement of total fluorescence, following addition of 230 µl of Triton X-100 and 750 ml of 6% TCA with 1% SDS. 750 µl of 6% TCA was added to the remainder of each sample, after which they were vortexed and allowed to precipitate at 4°C overnight.
The next morning, samples were centrifuged at 9,500 × g for 10 min. The supernatant of each sample was transferred to a new tube for quantification of soluble fraction fluorescence. One ml of 500 mM and 150 µl of 1N sodium hydroxide were added to each tube before setting aside for fluorescence quantification. The insoluble fractions were precipitated in 150 µl of 1N NaOH at room temperature for 1 h, prior to addition of Tris buffer, sodium hydroxide, and 6% TCA in equivalent amounts to the soluble fractions.
Soluble, insoluble, and total fluorescence were quantified by fitting sample fluorescence to a standard curve, ranging from 1–250 µg/ml FITC-albumin. Samples were plated onto opaque, black 96-well plates in triplicate replicates of 200 µl for each sample. Plates were read by spectrofluorometer, with excitation and emission wavelengths set at 495 and 520 nm, respectively. Normalization of results to sample protein concentrations was performed following bicinchoninic acid (BCA) assay. Clearance, defined as the volume of culture media cleared of FITC-albumin per min per mg of conceptal protein, was calculated for each fraction and tissue, and the cumulative total clearance was also determined by adding the clearance values from all measured compartments.
2.5 C1 Metabolism Component Quantification
Quantification of C1 components was performed as previously described in [16]. Liquid chromatography-tandem mass spectrometry was performed using a TSQ Quantum Ultra AM triple quadrupole mass spectrometer (Thermo Scientific, Rockford, IL) coupled to a Waters 2695 Separations Module (Waters, Milford, MA). These were fitted with a Luna C18(2) 5-µM 250 × 4.6-mm column and 4 mm C18 guard column (Phenomenex, Torrence, CA). All data was recorded using Xcalibur software (Thermo Scientific).
Briefly, isotopically labeled internal standards (ISTD) for SAM, methionine, cysteine, and homocysteine were added to each sample in known quantities following the incubation in reducing agent. To precipitate protein in the samples, 0.1% formic acid and 0.05% trifluoroacetic acid in acetonitrile was added to the samples before centrifugation and transfer of the supernatant to high-performance liquid chromatographic (HPLC) vials and 10 uL was injected in the positive ion electrospray LC-MS/MS analysis. The pellet was resolubilized in 0.25M sodium hydroxide and total protein was quantified using BCA assay. The gradient method was programmed to flow 1 mL/min of water 0.1% formic acid changing linearly to 10 % acetonitrile 0.1% formic at 10 minutes, then back to initial conditions and held until 30 minutes. All ion source and precursor/fragment ion transitions were optimized by direct infusion experiments. Concentrations of each analyte were calculated by ratio of the measured analyte to the known quantity of the isotopic standard in each sample. All data was normalized to the total protein concentration of the conceptus to account for the differences in size and morphology due to MEHP treatment [16, 30].
2.6 Quantification of Choline and Betaine
Quantification of choline and betaine was performed using a modified version of the protocol outlined in [37]. Briefly, 100 ul water was added to tubes containing each conceptus, and the contents were tip sonicated for suspension. The conceptus volume varies from about 30–50 ul, and volumes are typically smaller for those exposed to DEHP and MEHP. To compensate for volume differences, 50 ul of the suspension was removed for protein quantitation by the BCA assay, and all values were normalized to total protein concentrations. To 50 ul of the remaining suspension, 150 ul of acetonitrile was added and vortexed to precipitate proteins. After 10 min at room temperature, the sample was centrifuged at 14000 rpm. Fifty ul of the supernatant was removed and added to 100 ul of 4 uM of internal standards (d9-choline and d9-betaine) in 75:25 acetonitrile:water and 850 uL of 75:25 acetonitrile:water. The final concentration of ISTD was 0.4 uM and 2 ul was injected onto the LC-MS/MS system. The concentration of choline and betaine was calculated as the ratio of the analyte to the known quantity of isotopic standard. To generate measured “response factors” a solvent standard containing choline and betaine and their ISTD’s each at 0.4 uM in 75:25 acetonitrile:water was injected. These measured response factors were applied to calculate the concentrations of choline and betaine in conceptus samples.
The same instrumentation used to measure C1 components was utilized (Waters 2695 HPLC, Thermo Scientific TSQ Quantum Ultra AM), but outfitted with an Atlantis HILIC Silica 3-µm 4.6 × 30-mm column (Waters) and 4-mm guard column. A linear gradient method flowing at 2.0 ml/min was programmed for mobile phases. Mobile phase A was comprised of water, 10 mM ammonium acetate, and 0.15% formic acid. Mobile phase B was 100% acetonitrile. The gradient began at 20% A:80% B for 1 min, ramped to 60% A from 1 to 2 min, and then held until 2.3 min until returning to initial conditions at 2.4 min, and total run time was 5.5 minutes (Suppl. Fig. 1). The needle wash used was 9:1 methanol:water, and carryover was very minimal. Flow was split 4:1, sending 500 ul/min to the mass spectrometer set to ESI positive ion mode. The following mass spectrometer settings were applied: 3500 spray voltage, 55 units sheath gas, 30 units auxiliary gas, capillary 300 C and 35 V, and 1.5 mtorr argon collision pressure. The m/z precursor to product ion transitions were optimized for detection of analytes and isotopic standards as follows: choline 104>60+, choline-d9 113>69+ (collision energy 16V, tubelens 62 V), betaine 118>59+ and betaine-d9 127>68 (collision energy 21V, tubelens 75 V). Dwell times were 100 msec. To account for potential differences in the amount of analytes supplied in the culture media, paired culture medium was also sampled from each culture bottle before and after the culture period. No differences in the amounts of choline and betaine were discovered between culture bottles.
2.7 Quantification of Folate and Vitamin B12
Samples collected in 6% TCA were briefly sonicated then centrifuged at 9,500 × g for 5 minutes. The supernatant was transferred to a new tube, snap frozen, and shipped to the Gastrointestinal Laboratory at the Texas A&M University Department of Veterinary Small Animal Clinical Sciences. Immunoassays for folate and vitamin B12 were conducted using the IMMULITE 2000 system (Siemens Healthcare). The pellet was re-suspended in 0.25M sodium hydroxide and total protein was determined using BCA assay. Folate and vitamin B12 measures concentrations were reported after normalization to total conceptal protein. No differences were seen in the amounts of folate and vitamin B12 that were detected in the culture medium between culture bottles sampled before or after the culture period.
2.8 Global Histone Methylation
Total histone protein was quantified using the BCA assay and equivalent amounts of histone protein were used for methylation analysis. Histone methylation at the H3K4 and H3K27 loci was probed using EpiQuik™ Global Pan-Methyl Histone Quantification Kits (Epigentek). Fluorescence associated with methyl markers at the loci was detected and used to quantify percent methylation. H3K4 histone methylation is typically associated with activation of genes, making chromatin more accessible, and H3K27 methylation is typically considered a repressive marker, encouraging the tight packaging of chromatin [15]. These pan-methyl kits were selected to probe the effects of altered nutrition on histone methylation instead of specifically investigating the effects on tri-methylation at these loci.
2.9 DNA Isolation
Genomic DNA from EMB and VYS samples exposed for 24 h to MEHP (100 or 250 µg/ml) or controls (DMSO) was isolated using phenol-chloroform extraction. Briefly, thawed samples were sonicated in Buffer ATL (Qiagen) and digested overnight at 50µC after addition of Proteinase K (Qiagen). The following morning, RNase A was added before repeated phenol-chloroform isoamyl alcohol extractions in phase-lock gel tubes (5 PRIME). A choloroform-only wash was performed, followed by repeated ethanol precipitations. The DNA pellet was then dried before storage in Tris buffer (pH 8.0) until use.
2.10 LUmiometric Methylation Assay (LUMA)
LUMA was utilized as explained in [38, 39] and previously performed in [16]. This is an extension assay which relies on differential cleavage of the DNA by enzymes that are sensitive or insensitive to methylation. Briefly, genomic DNA isolated from EMB and VYS samples was subjected to restriction enzyme digestion for 4 hours at 37µC. Samples were separately digested with either methylation-sensitive HpaII or methylation–insensitive MspI restriction enzymes (Invitrogen), which both cleave genomic DNA at CCGG sites. This provides differential numbers of cleavage sites for extension based upon methylation, and the ratio can be used to quantify the relative percent global methylation. EcoRI (Invitrogen) was also added to each digest to function as an internal control. Following digestion, Annealing buffer (Qiagen) was added to the products and these were analyzed using the PyroMark™ Q96 MD system (Qiagen). Samples were run in triplicate to account for variation. Ratios for MspI and HpaII relative to their internal EcoRI controls were used to quantify overall percent methylation, using the following equation: 1-[(HpaII/EcoRI)/(MspI/EcoRI)]x100.
2.11 Statistical Analysis
Results are presented as mean ± SEM. Unpaired t-tests and ANOVA with Tukey’s post-hoc test were used to probe statistical significance. Due to the exploratory nature of this study, many of these experiments were performed on a small or unbalanced sample size. An F-test was performed to assess the variance of each sample group prior to hypothesis testing. When appropriate, a Welch’s t-test was utilized to assess differences between groups with unequal variance. A confidence level of 95% was used throughout the study, and all tests were performed using SPSS. Statistical outliers were removed if data points fell into a range that was at least 1.5 times the interquartile range and outside of the first and third quartile values.
3. Results
3.1 Histiotrophic Nutrition Assay
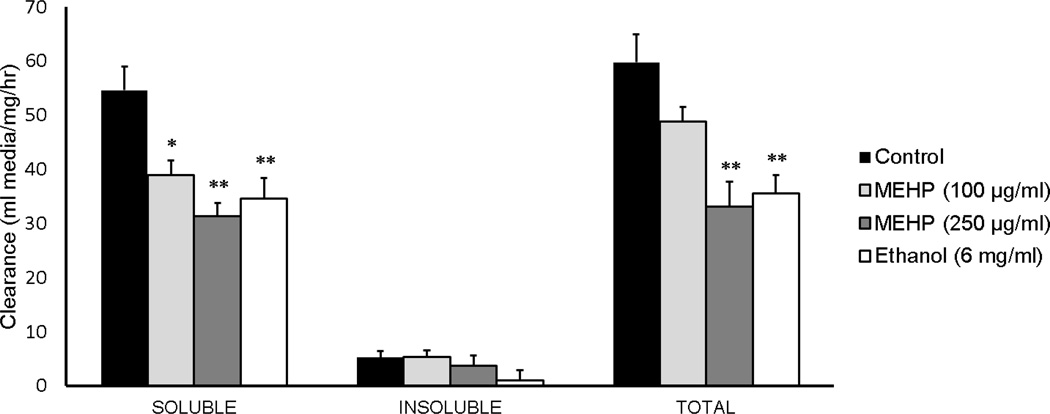
Total clearance, defined as the sum of VYS, EEF, and EMB values, was quantified to determine the extent to which MEHP reduced the histiotrophic uptake of proteins in the whole conceptus (Fig. 2). Ethanol was selected as a positive control because it is a known selective inhibitor of RME [35, 36]. Ethanol-exposed conceptuses had significant reductions (p<0.01) in soluble (34.5 ± 3.9; n=9) and total (35.5 ± 3.3; n=9) clearance fractions when compared to their respective soluble (54.6 ± 4.3 ml media/mg protein/h; n=9) and total (soluble + insoluble) (59.8 ± 5.2; n=9) FITC clearance values. No significant changes were observed due to any MEHP or ethanol treatment for the insoluble fractions (p>0.05). Exposure to MEHP at the 100 µg/ml dose significantly decreased clearance as measured by the soluble fraction (38.9 ± 2.7; n=8; p=0.02), though there was no statistically significant decrease of FITC-albumin clearance of the total fraction (48.8 ± 2.7; n=8; p=0.28). Exposure to MEHP at the 250 µg/ml dose significantly reduced both soluble (31.3 ± 2.5; n=9) and total (33.1 ± 4.7; n=9) fraction clearance (p<0.01). These decreases in clearance for the high MEHP exposure group were even greater than those of the ethanol positive control. The acid soluble fraction of total cleared FITC was small (10%) relative to the corresponding acid insoluble fraction. This indicates that lysosomal proteolytic activity was not inhibited by MEHP, and suggests that MEHP inhibition of RME is likely to be solely responsible for observed decreases in nutrient uptake. Overall, MEHP significantly reduced histiotrophic nutrient uptake of the conceptus.
Figure 2.
Histiotrophic nutrient uptake is hindered by MEHP treatment. Clearance is measured as µl media cleared into conceptus per mg protein per hour, and thus is the rate of uptake. Ethanol was used as a positive control, as it has been previously demonstrated to decreased endocytotic processes. *p<0.05, **p<0.01, as compared to controls
3.2 Quantification of Methyl Donors and C1 Metabolism Components
C1 metabolism components and dietary methyl group donors were quantified to determine whether the observed decreases in HNP result in a loss of methyl donor availability in the form of SAM (Table 1). MEHP concentration had no significant effect on SAM concentrations, though there was an increasing trend overall (ANOVA p=0.11). These same increasing trends were evident for cysteine and methionine (ANOVA p=0.05 and 0.04, respectively). Methionine concentrations were significantly elevated in the 250 µg/ml group (1427.0 ± 235.6; n=5) compared to controls (635.1 ± 44.0 ng/mg protein; n=4; p=0.04). Homocysteine was statistically elevated in the 250 µg/ml group (97.4 ± 8.5 ng/mg protein; n=4) compared to controls (70.1 ± 5.1; n=5; p=0.02) and the 100 µg/ml (63.8 ± 3.4; n=4; p<0.01).
Table 1.
Measurements of dietary methyl donors and C1 metabolism components in whole conceptuses following 24 h in WEC. All measurements were normalized, and are reported as the concentration of each nutrient per mg total protein.
| Control | MEHP (100 µg/ml) | MEHP (250 µg/ml) | |
|---|---|---|---|
| Folate (ng/mg protein) | 0.66±0.04 (n=4) |
0.51±0.03 (n=4) |
0.74±0.08 (n=5) |
| Vitamin B12 (pg/mg protein) | 17.00±0.09 (n=3) |
12.25±0.41*** (n=3) |
15.23±0.47*$$ (n=5) |
| Choline (ng/mg protein) | 2.15±0.41 (n=5) |
1.35±0.54 (n=5) |
3.04±0.87$$ (n=4) |
| Betaine (ng/mg protein) | 4.23±0.39 (n=4) |
2.13±0.95** (n=5) |
2.95±0.42 (n=4) |
| SAM (ng/mg protein) | 820.93±87.27 (n=4) |
1487.44±358.57 (n=5) |
2380.10±659.50 (n=5) |
| Cysteine (ng/mg protein) | 1467.39±192.00 (n=5) |
2338.37±377.96 (n=5) |
2506.89±265.18 (n=5) |
| Homocysteine (ng/mg protein) | 70.10±5.07 (n=5) |
63.75±3.36 (n=4) |
97.37±8.50*$$ (n=4) |
| Methionine (ng/mg protein) | 635.06±43.95 (n=4) |
956.51±171.79 (n=5) |
1426.98±235.62* (n=5) |
| Total protein (mg/sample) | 2.68±0.09 (n=5) |
2.84±0.09 (n=5) |
2.40±0.11$ (n=5) |
Comparisons between treatments and controls:
p<0.05
p<0.01
p<0.001.
Comparisons between low- and high-dose groups:
p<0.05
p<0.01.
Folate concentrations fluctuated at low and high MEHP concentrations, but were not significantly changed. Choline concentrations were unchanged by 100 µg/ml treatment (1.35 ± 0.54; n=5) compared to controls, but were significantly elevated in the 250 ug/ml compared to the 100 µg/ml group (3.04 ± 0.87; n=4; p<0.01). Betaine concentrations (4.23 ± 0.39 ng/mg protein; n=4) are significantly decreased in conceptuses exposed to 100 µg/ml MEHP (2.13 ± 0.95; n=5; p<0.01). Though betaine in the 250 µg/ml conceptuses is still slightly reduced compared to controls, this relationship is not statistically significant (p=0.06).
Vitamin B12 control concentrations (17.00 ± 0.09 pg/mg protein; n=3) are significantly decreased at the 100 µg/ml MEHP dose (12.25 ± 0.41; n=3; p<0.001). Vitamin B12 in conceptuses treated with 250 µg/ml MEHP (15.23 ± 0.47; n=5) is significantly elevated compared to the 100 µg/ml dose (p<0.01), and remains decreased compared to the control measure (p=0.05).
3.3 Global Histone Methylation
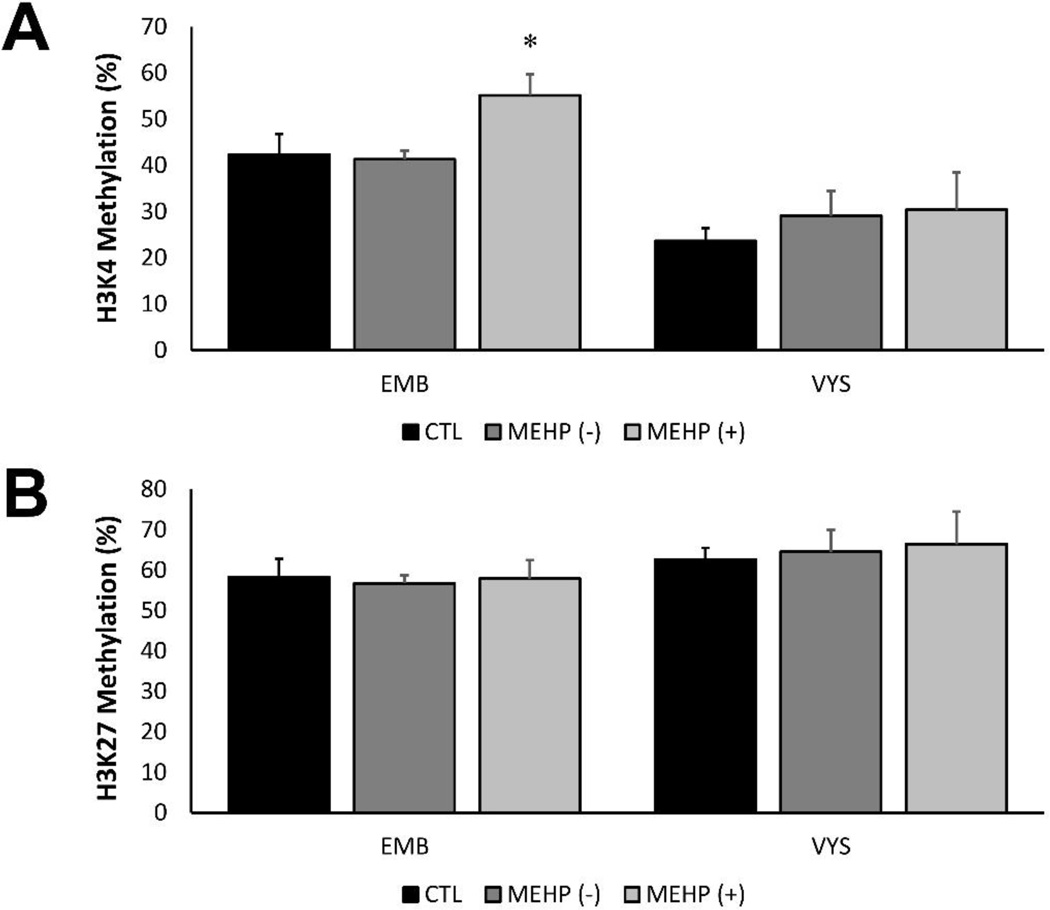
Global histone pan-methylation was measured in both control and MEHP-exposed (100 µg/ml) EMB and VYS, also stratified by NTD status (Fig. 3). Pan methylation is defined as the cumulative mono-, di-, and tri-methylation at a locus. Pan-methylation was examined at the H3K4 and H3K27 loci after 24 h in culture to determine whether the accompanying nutritional modifications may result in methylation changes. H3K4 methylation was significantly increased in MEHP-treated, NTD+ EMB (55.1 ± 4.5; n=4) compared to MEHP-treated NTD- EMB (41.3 ± 1.9; n=6; p=0.01), though not significantly elevated compared to the controls (Fig. 3A). No significant H3K4 methylation changes were observed in the VYS. H3K27 pan methylation was also measured in the EMB and VYS (Fig. 3B). No statistically significant changes in H3K27 methylation were observed in the EMB or VYS.
Figure 3.
Global histone H3K4 and H3K27 methylation percentages in the EMB and VYS. MEHP-treated (100 µg/ml) EMB with NTDs have elevated H3K4 methylation compared to controls and MEHP-treated NTD negative EMB (A). No significant changes in H3K4 methylation were observed in the VYS. No significant changes in H3K27 methylation were observed in EMB or VYS (B). *p<0.05, as compared to controls
3.4 LUminometric Methylation Assay (LUMA)
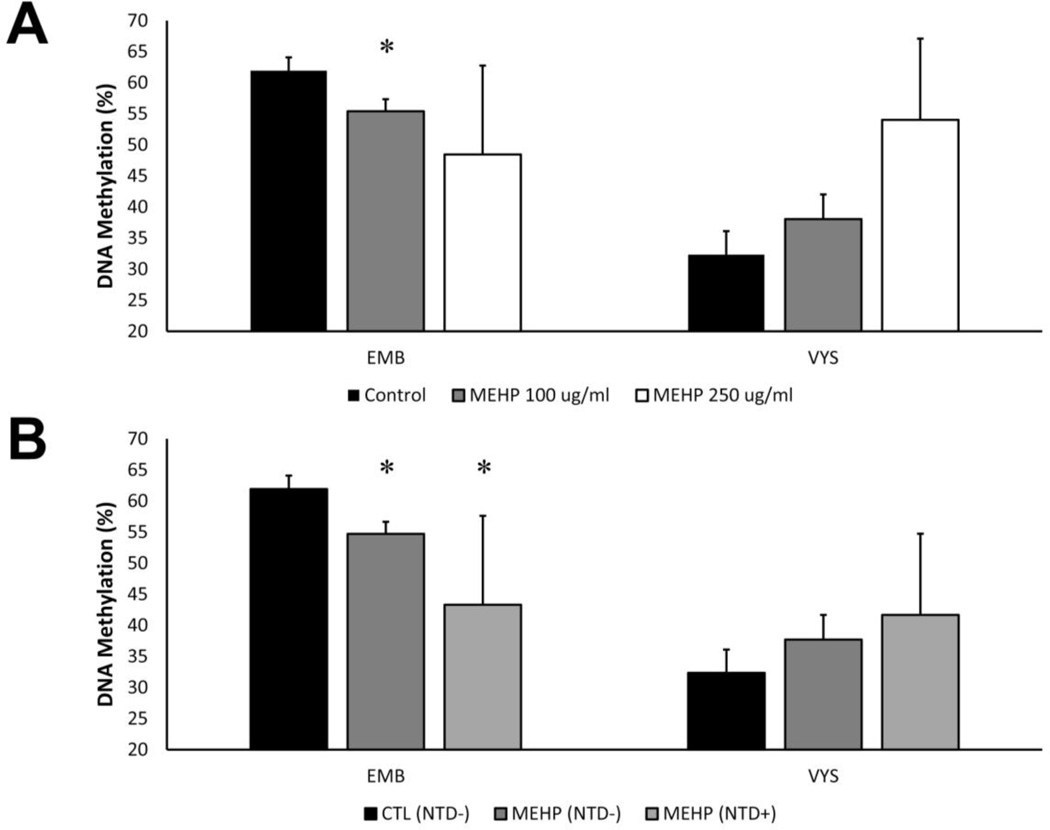
The effect of MEHP treatment was examined by treating conceptuses with 100 or 250 µg/ml MEHP in addition to the controls (Fig. 4A). MEHP 100 µg/ml treatment significantly reduced global DNA methylation in the EMB (55.4 ± 1.9; n=13; p=0.01) compared to controls (61.9 ± 2.2; n=5). High variability of global DNA methylation was observed in EMB treated with 250 µg/ml MEHP, thus, no statistically significant further decrease was observed (p=0.4). No statistically significant changes were observed in the VYS, though there was an increasing trend. These changes are consistent with methylation changes previously observed due to ethanol, which is also an inhibitor of RME (unpublished results).
Figure 4.
Global DNA methylation is modified by MEHP treatment and in conceptuses with NTDs. DNA methylation decreases in the EMB and increases in the VYS with increasing MEHP dose (A). EMB exposed to MEHP at 100 µg/ml concentrations in culture had significantly lower global DNA methylation percentages than controls, regardless of NTD status (B). EMB that were MEHP treated and NTD+ had further reduced DNA methylation. In the VYS, MEHP treatment and NTD status did not significantly alter DNA methylation, though there was an increasing trend. *p<0.05, as compared to controls
Global DNA methylation was also measured in both control and MEHP-exposed (100 µg/ml) EMB and VYS following stratification by NTD status (Fig. 4B). MEHP exposure in the absence of NTD significantly reduced (p=0.03) DNA methylation in the EMB (54.7 ± 1.8; n=10) after 24 h when compared to controls (61.9 ± 2.2; n=5). DNA methylation was further reduced in MEHP-treated EMB with NTD (43.3 ± 7.7; n=3) compared to both controls (p=0.03) and MEHP-treated NTD negative EMB (p=0.05). Though VYS methylation was much lower than EMB DNA methylation overall, there were no statistically significant changes in methylation in the VYS.
4. Discussion
The relationship between prenatal nutrition and growth and development has been well studied, although the long-term consequences of nutritional deficiencies are highly diverse. Previous work from our laboratory has demonstrated that exposure to MEHP during early organogenesis may result in stunted growth and delayed or halted neurulation [30]. In this study, nutritional deficiencies were observed as changes in nutrient and cofactor uptake and availability after exposure to MEHP, suggesting that embryonic exposures to toxicants may reduce essential nutrient supplies and produce a canonical stunting of growth.
Many studies have utilized toxicological and pharmacological agents to modify RME [40], however, relatively few studies have demonstrated a link between toxicological or pharmacological agents and disruption of HNPs during development [35, 41–48]. These previous studies encompass a wide range of chemical agents, with diverse structures and functions, and yet many of these exposures have the same outcomes. In this study, both ethanol and MEHP have been shown to disrupt the RME functions of HNPs, but it is unknown whether they are acting via the same mechanisms and transporters. Different classes of persistent compounds may directly or indirectly affect HNPs, but the lack of comparative structure/function data and complete ontogeny profiles for HNP-mediated nutrient clearance preclude the development of predictive models for nutrient availability in the presence of environmental chemicals. In this study, total HNP clearance was measured, representing the total amount of media cleared of FITC-albumin by the VYS, EEF, and EMB. However, very little fluorescence is found in the EMB following a 3-h uptake period and long-term exposure data using this model is not possible due to inviability in culture beyond GD 10.
Megalin and cubilin, known as the multiligand endocytotic receptor complex (MERC), are major receptors involved in HNPs, which are localized to polarized epithelia such as the VYS [10, 49, 50]. Although the rodent VYS is inverted, both rodent and human VYS and neuroepitehelia highly express megalin and cubilin, and have been directly implicated in neurulation and neurodevelopment [51–55]. The MERC is responsible for the uptake of a vast array of nutrients and is also a major transporter of the developmental ligands crucial for neurulation such as Sonic Hedgehog (Shh) and retinoic acid [10, 53–59]. Vitamin B12 and folate are also co-transported across epithelia via the MERC by carrier proteins, suggesting a link between methyl donor metabolism and the MERC [9, 10]. Solute carrier (SLC) families of transporters are also crucial for the transport of nutrients across membranes, including choline, betaine, folate, and amino acids required for C1 metabolism. In a companion study, we have shown that the expression of many SLC genes is altered by 6 h MEHP treatment, but the specific effects of MEHP treatment on nutrient transporters and whether responses are time- or stage-dependent is unknown [30]. Work with transgenic Lrp2-deficient or Cubn-deficient models could provide insight into this mechanism, as well as an ontogeny of MERC and SLC expression following MEHP exposure.
Decreases in HNP function following treatment with the protease inhibitor leupeptin result in C1 deficiencies 6 h after initial exposure [16]. Here, however, C1 metabolic components follow an increasing trend with increasing MEHP concentrations in culture after 24 h. These results were unexpected because HNP activity is reduced in a dose-dependent fashion after 3 h, and it was believed that methyl donors and C1 components should decrease initially. It is possible that exposed conceptuses are able to adapt and compensate by the end of the 24 h exposure period. The primary focus of this work was to examine nutrition during embryogenesis and early organogenesis, and does not attempt to draw any conclusions about the long-term fetal and neonatal consequences of this deficiency. A growing body of epidemiological studies have suggested that in utero deficiencies may result in the programming of a “thrifty” phenotype, resulting in predispositions to metabolic syndromes later in life [60]. Though this work suggests that growth is immediately stunted, it is possible that adaptation to these deficiencies may result in a thrifty phenotype.
The epigenetic literature has canonically correlated increased methyl donor supplementation and availability with increased global methylation measures [12]. Pharmacological inhibition of HNPs was associated with decreased EMB and VYS SAM concentrations after 6 h and decreased global DNA methylation at both 6- and 24-h post initial treatment [16]. Here, 24 h treatment with increasing concentrations of MEHP increased C1 component concentrations but had variable effects on methyl donor concentrations. As a result, VYS DNA methylation was increased as would be expected, but EMB DNA methylation was decreased. However, the concentrations of methyl donors and C1 metabolic components were measured in whole conceptuses inclusive of the VYS, EMB and all EEF.
During embryogenesis, histone methylation is reprogrammed. H3K4 methylation is absent in the zygote and increases through to organogenesis, while H3K27 methylation becomes progressively demethylated between the zygote and organogenesis stages [15]. In the VYS, both treatment with MEHP and presence of NTD are associated with increased H3K27 methylation. It is possible that treatment with MEHP or presence of NTD prevent the demethylation processes required for normal methylation programming at the H3K27 locus. H3K4 global histone methylation was increased by both MEHP treatment and presence of NTD in both EMB and VYS, most significantly in NTD+ EMBs. Since H3K4 methylation is an activating epigenetic mark, it is possible that this is a mechanism of gene dysregulation that leads to teratogenesis.
Pan methylation of H3K4 and H3K27 histone loci was assayed, rather than tri-methylation, in order to establish a nutritional framework. It was expected that decreased HNPs would result in decreased SAM biosynthesis and decreased methylation of histones and DNA. However, this was not the case at the time point examined. It is likely that the tight control of gene regulation during embryonic development is prioritized and more conserved. Though there were slight modifications to histone methylation, it is unknown whether these changes will lead to altered epigenetic regulation.
Like histone methylation, DNA methylation also undergoes substantial reprogramming during the first trimester of development. After fertilization, DNA is substantially demethylated and then remethylated later during embryogenesis and early organogenesis [13–15]. In this study, treatment with MEHP was initiated and continued during this de novo remethylation phase of development. It was expected that MEHP would decrease HNP activity and deplete methyl donor availability, and that this would lead to decreased SAM generation and resultant decreased methylation. This is concordant with findings of a previous study [16], but was not consistently supported here. MEHP did in fact reduce EMB DNA methylation; however, DNA methylation was increased in the MEHP-treated VYS, though this change was not statistically significant. These findings are consistent with previous studies conducted in EMB and VYS treated with ethanol, another inhibitor of endocytosis. It is unknown why DNA methylation would even modestly increase in the VYS if HNP function is reduced, although increased DNA methylation due to treatment in the terminally-differentiated VYS could be a mechanism to conserve other cellular resources for the developing EMB.
It is also possible that this outcome is due to lineage-specific effects of phthalate exposure, since growth-related genes in the VYS are both maternally- and paternally-imprinted [61, 62]. It has been previously demonstrated that impaired activity of the de novo DNA methyltransferase (Dnmt) enzyme, Dnmt3a, results in EMB with open neural tubes, loss of maternal imprints throughout the genome, and yet most of the paternal imprints and methylation at repetitive elements were maintained [63]. Therefore, the increase in NTD, loss of methylation in the EMB, and increases in the degree of methylation in the VYS that follow MEHP treatment may be related to the function of Dnmt3a. In previous studies, leupeptin, an inhibitor of the proteolysis stage of HNPs was unable to modify Dnmt3a expression in the EMB or VYS and we confirmed that there was no significant change in Dnmt3b expression following 6-h MEHP exposure in WEC [16, 30]. However, HNP disruptors such as MEHP may alter Dnmt “activity” rather than gene expression, and thus characterization of the specific activities of Dnmts after disruption of HNPs during development would be necessary to uncover this relationship.
Histiotrophic nutrition activity was decreased linearly with increasing MEHP concentrations but this trend did not correlate with conceptal micronutrient concentrations. Cysteine, methionine, and SAM were actually increased with increasing MEHP, which was contrary to our initial hypothesis. Because the compounds measured in this study are known metabolic products of C1 metabolism, it is possible that their individual metabolism is altered to compensate for decreased nutrient uptake. Furthermore, other nutrients followed a nonmonotonic concentration curve based upon MEHP exposure. It is likely that the timing and compartmental responses of these nutrients are complex, and an ontogeny of their uptake and metabolism is required to elucidate a specific mechanism. Separate quantification of methyl donors and C1 metabolic components should be done in order to determine whether the nutritional consequences observed in the whole conceptus are consistent with each compartment of the conceptus. It is possible that nutrient concentrations are maintained in the VYS and potentially even the EEF, but that these are not properly taken up by the EMB.
The use of WEC in this study provided unique tools for the investigation of nutrient uptake. Though WEC conditions mimic many of the in utero conditions, the translation of this work to in vivo studies would allow further examination of the link between nutrition and epigenetic regulation with a more complex environment. The uterine glands secrete nutrients, which are transported through the trophoblast to the coelomic fluid and ultimately undergo the HNPs via the VYS [64, 65]. In vivo studies of these relationships would also provide insight into the effects of MEHP treatment on the function of the uterine glands and intermediate transport steps before entering the conceptal HNPs. Because conceptuses must share these nutrients with litter mates in the same uterus, in vivo studies would also provide information about the potential for resource competition due to altered nutrition pathways.
In summary, this exploratory study has demonstrated the ability of MEHP to modify the conceptal nutrition environment and ultimately the epigenetic landscape of the developing embryo. Histiotrophic clearance of nutrients was decreased with increasing MEHP treatment concentrations after 3 h, although measured conceptal methyl donor concentrations were variable and relatively unchanged after 24 h. C1 metabolism components were increased with increasing MEHP concentration in culture after 24 h, including increased SAM. DNA methylation of the EMB was decreased at 24 h, although there was not significant change in the VYS. Histone methylation at the H3K4 locus was increased in EMB that were treated and positive for NTDs, though MEHP treatment alone had little effect. HNP sensitivity to toxicants during this window of development may be a mechanism by which developmental susceptibility is induced. Toxicants could disrupt HNPs in either the endocytosis or proteolysis stage, and these modulations of nutrient environment may manifest as different epigenetic patterns. Further characterization of the mechanisms and timing by which toxicants alter HNPs is necessary and may provide predictive models of toxicant-nutrient disruption for future compounds.
Supplementary Material
Acknowledgments
We acknowledge Tamara R. Jones and Muna S. Nahar for their efforts in methylation assay design and optimization. Laboratory support was provided in part by Erin Scarlett, Lindsey Jacobs, and Grace Kuan.
Funding Information: This work was supported by the University of Michigan National Institute of Environmental Health Sciences (NIEHS) Core Center “Lifestage Exposures and Adult Disease” (P30 ES017885) with generous support from the UM School of Public Health (SPH) and the SPH Department of Environmental Health Sciences as well as NIEHS grants R01 ES017524 (D.C.D.) and T32 ES007062 (K.E.S.). Additional funding was provided by the Bill and Melinda Gates Foundation, Grand Challenges Explorations-Round 7.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Beckman DA, et al. Sources of amino acids for protein synthesis during early organogenesis in the rat. 1. Relative contributions of free amino acids and of proteins. Placenta. 1990;11(2):109–121. doi: 10.1016/s0143-4004(05)80173-0. [DOI] [PubMed] [Google Scholar]
- 2.Lloyd JB, Brent RL, Beckman DA. Sources of amino acids for protein synthesis during earlyorganogenesis in the rat. 3. Methionine incorporation. Placenta. 1996;17(8):629–634. doi: 10.1016/s0143-4004(96)80081-6. [DOI] [PubMed] [Google Scholar]
- 3.Jauniaux E, et al. Transfer of folic acid inside the first-trimester gestational sac and the effect of maternal smoking. American Journal of Obstetrics & Gynecology. 197(1):58.e1–58.e6. doi: 10.1016/j.ajog.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 4.Maddox DM, et al. Reduced-folate carrier (RFC) is expressed in placenta and yolk sac, as well as in cells of the developing forebrain, hindbrain, neural tube, craniofacial region, eye, limb buds and heart. BMC Dev Biol. 2003;3:6. doi: 10.1186/1471-213X-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambers BJ, et al. Methionine overcomes neural tube defects in rat embryos cultured on sera from laminin-immunized monkeys. J Nutr. 1995;125(6):1587–1599. doi: 10.1093/jn/125.6.1587. [DOI] [PubMed] [Google Scholar]
- 6.Polliotti BM, Panigel M, Miller RK. Free vitamin B12 and transcobalamin II-vitamin B12 complex uptake by the visceral yolk sac of the Sprague-Dawley rat: effect of inhibitors. Reprod Toxicol. 1997;11(4):617–626. doi: 10.1016/s0890-6238(97)89180-0. [DOI] [PubMed] [Google Scholar]
- 7.Padykula HA, Deren JJ, Wilson TH. Development of structure and function in the mammalian yolk sac. I. Developmental morphology and vitamin B12 uptake of the rat yolk sac. Dev Biol. 1966;13(3):311–348. doi: 10.1016/0012-1606(66)90053-4. [DOI] [PubMed] [Google Scholar]
- 8.Kozyraki R, et al. The intrinsic factor-vitamin B12 receptor, cubilin, is a high-affinity apolipoprotein A-I receptor facilitating endocytosis of high-density lipoprotein. Nat Med. 1999;5(6):656–661. doi: 10.1038/9504. [DOI] [PubMed] [Google Scholar]
- 9.Gelineau-van Waes J, et al. Microarray analysis of E9.5 reduced folate carrier (RFC1; Slc19a1) knockout embryos reveals altered expression of genes in the cubilin-megalin multiligand endocytic receptor complex. BMC Genomics. 2008;9(1):156. doi: 10.1186/1471-2164-9-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moestrup SK, Verroust PJ. Megalin- and Cubilin-Mediated Endocytosis of Protein-Bound Vitamins, Lipids, and Hormones in Polarized Epithelia. Annual Review of Nutrition. 2001;21(1):407–428. doi: 10.1146/annurev.nutr.21.1.407. [DOI] [PubMed] [Google Scholar]
- 11.Birn H, et al. Characterization of an Epithelial ~460-kDa Protein That Facilitates Endocytosis of Intrinsic Factor-Vitamin B12 and Binds Receptor-associated Protein. Journal of biological chemistry. 1997;272(42):26497–26504. doi: 10.1074/jbc.272.42.26497. [DOI] [PubMed] [Google Scholar]
- 12.Anderson OS, Sant KE, Dolinoy DC. Nutrition and epigenetics: an interplay of dietary methyl donors, one-carbon metabolism and DNA methylation. The Journal of Nutritional Biochemistry. 2012;23(8):853–859. doi: 10.1016/j.jnutbio.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reik W, Dean W, Walter J. Epigenetic Reprogramming in Mammalian Development. Science. 2001;293(5532):1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 14.Faulk C, Dolinoy DC. Timing is everything: The when and how of environmentally induced changes in the epigenome of animals. Epigenetics. 2011;6(7):791–797. doi: 10.4161/epi.6.7.16209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447(7143):425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 16.Sant KE, et al. Inhibition of proteolysis in histiotrophic nutrition pathways alters DNA methylation and one-carbon metabolism in the organogenesis-stage rat conceptus. The Journal of Nutritional Biochemistry. 2013;24(8):1479–1487. doi: 10.1016/j.jnutbio.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeisel SH. Importance of methyl donors during reproduction. Am J Clin Nutr. 2009;89(2):673S–677S. doi: 10.3945/ajcn.2008.26811D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imbard A, Benoist JF, Blom HJ. Neural tube defects, folic acid and methylation. Int J Environ Res Public Health. 2013;10(9):4352–4389. doi: 10.3390/ijerph10094352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osterhues A, Ali NS, Michels KB. The role of folic acid fortification in neural tube defects: a review. Crit Rev Food Sci Nutr. 2013;53(11):1180–1190. doi: 10.1080/10408398.2011.575966. [DOI] [PubMed] [Google Scholar]
- 20.Pitkin RM. Folate and neural tube defects. The American journal of clinical nutrition. 2007;85(1):285S–288S. doi: 10.1093/ajcn/85.1.285S. [DOI] [PubMed] [Google Scholar]
- 21.Centers for Disease Control and Prevention. Folic Acid Helps Prevent Neural Tube Defects. National Center on Birth Defects and Developmental Disabilities, Editor. 2014 [Google Scholar]
- 22.Carmichael SL, Yang W, Shaw GM. Periconceptional nutrient intakes and risks of neural tube defects in California. Birth Defects Research Part A: Clinical and Molecular Teratology. 2010;88(8):670–678. doi: 10.1002/bdra.20675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang B-Y, et al. Correlation Between Birth Defects and Dietary Nutrition Status in a High Incidence Area of China. Biomedical and Environmental Sciences. 2008;21(1):37–44. doi: 10.1016/S0895-3988(08)60005-7. [DOI] [PubMed] [Google Scholar]
- 24.Pitkin RM. Folate and neural tube defects. Am J Clin Nutr. 2007;85(1):285S–288S. doi: 10.1093/ajcn/85.1.285S. [DOI] [PubMed] [Google Scholar]
- 25.Denny KJ, et al. Neural tube defects, folate, and immune modulation. Birth Defects Res A Clin Mol Teratol. 2013;97(9):602–609. doi: 10.1002/bdra.23177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whyatt RM, et al. Prenatal di(2-ethylhexyl)phthalate exposure and length of gestation among an inner-city cohort. Pediatrics. 2009;124(6):e1213–e1220. doi: 10.1542/peds.2009-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silva MJ, et al. Measurement of eight urinary metabolites of di(2-ethylhexyl) phthalate as biomarkers for human exposure assessment. Biomarkers. 2006;11(1):1–13. doi: 10.1080/13547500500382868. [DOI] [PubMed] [Google Scholar]
- 28.Kato K, et al. Mono(2-ethyl-5-hydroxyhexyl) phthalate and mono-(2-ethyl-5-oxohexyl) phthalate as biomarkers for human exposure assessment to di-(2-ethylhexyl) phthalate. Environ Health Perspect. 2004;112(3):327–330. doi: 10.1289/ehp.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson JF, et al. Dose-response analysis of phthalate effects on gene expression in rat whole embryo culture. Toxicology and Applied Pharmacology. 2012;264(1):32–41. doi: 10.1016/j.taap.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 30.Sant KE, et al. Mono-2-ethylhexyl phthalate disrupts neurulation and modifies the embryonic redox environment and gene expression [Submitted] doi: 10.1016/j.reprotox.2016.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris C. Rodent Whole Embryo Culture. In: Harris C, Hansen JM, editors. Developmental Toxicology. Humana Press; 2012. pp. 215–237. [Google Scholar]
- 32.Janer G, et al. Use of the rat postimplantation embryo culture to assess the embryotoxic potency within a chemical category and to identify toxic metabolites. Toxicology in Vitro. 2008;22(7):1797–1805. doi: 10.1016/j.tiv.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 33.Ambroso J, Harris C. Assessment of Histiotrophic Nutrition Using Fluorescent Probes. In: Harris C, Hansen JM, editors. Developmental Toxicology. Humana Press; 2012. pp. 407–423. [DOI] [PubMed] [Google Scholar]
- 34.Ambroso JL, et al. Fluorometric analysis of endocytosis and lysosomal proteolysis in the rat visceral yolk sac during whole embryo culture. Teratology. 1997;56(3):201–209. doi: 10.1002/(SICI)1096-9926(199709)56:3<201::AID-TERA3>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 35.Steventon GB, Williams KE. Ethanol-induced inhibition of pinocytosis and proteolysis in rat yolk sac in vitro. Development. 1987;99(2):247–253. doi: 10.1242/dev.99.2.247. [DOI] [PubMed] [Google Scholar]
- 36.Jilek JL, et al. Ethanol Attenuates Histiotrophic Nutrition Pathways and Alters the Intracellular Redox Environment and Thiol Proteome during Rat Organogenesis [Submitted] doi: 10.1093/toxsci/kfv145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holm PI, et al. Determination of choline, betaine, and dimethylglycine in plasma by a high-throughput method based on normal-phase chromatography-tandem mass spectrometry. Clin Chem. 2003;49(2):286–294. doi: 10.1373/49.2.286. [DOI] [PubMed] [Google Scholar]
- 38.Karimi M, Johansson S, Ekström T. Using LUMA: a Luminometric-Based Assay for Global DNA-Methylation. Epigenetics. 2006;1(1):45–48. doi: 10.4161/epi.1.1.2587. [DOI] [PubMed] [Google Scholar]
- 39.Karimi M, et al. LUMA (LUminometric Methylation Assay)—A high throughput method to the analysis of genomic DNA methylation. Experimental Cell Research. 2006;312(11):1989–1995. doi: 10.1016/j.yexcr.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 40.Ivanov AI. Pharmacological inhibition of endocytic pathways: is it specific enough to be useful? Methods Mol Biol. 2008;440:15–33. doi: 10.1007/978-1-59745-178-9_2. [DOI] [PubMed] [Google Scholar]
- 41.Claussen U, et al. The embryotoxicity of cyclophosphamide in rabbits during the histiotrophic phase of nutrition. Teratog Carcinog Mutagen. 1985;5(2):89–100. doi: 10.1002/tcm.1770050203. [DOI] [PubMed] [Google Scholar]
- 42.Beck F. Induced cell injury and cell death as a cause of congenital malformations in rats. Histochem J. 1981;13(4):667–679. doi: 10.1007/BF01002718. [DOI] [PubMed] [Google Scholar]
- 43.Holson JF, et al. Mode of action: yolk sac poisoning and impeded histiotrophic nutrition--HBOC-related congenital malformations. Crit Rev Toxicol. 2005;35(8–9):739–745. doi: 10.1080/10408440591007412. [DOI] [PubMed] [Google Scholar]
- 44.Ambroso J, Harris C. In vitro embryotoxicity of the cysteine proteinase inhibitors benzyloxycarbonyl-phenylalanine-alanine-diazomethane (Z-Phe-Ala-CHN2) and benzyloxycarbonyl-phenylalanine-phenylalanine-diazomethane (Z-Phe-Phe-CHN2) Teratology. 1994;50(3):214–228. doi: 10.1002/tera.1420500307. [DOI] [PubMed] [Google Scholar]
- 45.Ambroso JL, Harris C. Chloroquine accumulation and alterations of proteolysis and pinocytosis in the rat conceptus in vitro. Biochemical Pharmacology. 1994;47(4):679–688. doi: 10.1016/0006-2952(94)90131-7. [DOI] [PubMed] [Google Scholar]
- 46.Hunter ES, 3rd, et al. Altered visceral yolk sac function produced by a low-molecular-weight somatomedin inhibitor. Teratology. 1991;43(4):331–340. doi: 10.1002/tera.1420430408. [DOI] [PubMed] [Google Scholar]
- 47.Brent RL, et al. Experimental yolk sac dysfunction as a model for studying nutritional disturbances in the embryo during early organogenesis. Teratology. 1990;41(4):405–413. doi: 10.1002/tera.1420410406. [DOI] [PubMed] [Google Scholar]
- 48.Balkan W, et al. Role of the mouse visceral yolk sac in nutrition: inhibition by a somatomedin inhibitor. J Exp Zool. 1989;249(1):36–40. doi: 10.1002/jez.1402490108. [DOI] [PubMed] [Google Scholar]
- 49.Christensen E, Verroust P. Megalin and cubilin, role in proximal tubule function and during development. Pediatric Nephrology. 2002;17(12):993–999. doi: 10.1007/s00467-002-0956-5. [DOI] [PubMed] [Google Scholar]
- 50.Muller D, Nykjaer A, Willnow TE. From holoprosencephaly to osteopathology: role of multifunctional endocytic receptors in absorptive epithelia. Ann Med. 2003;35(5):290–299. doi: 10.1080/07853890310006488. [DOI] [PubMed] [Google Scholar]
- 51.Burke KA, et al. Expression and immunolocalisation of the endocytic receptors megalin and cubilin in the human yolk sac and placenta across gestation. Placenta. 2013;34(11):1105–1109. doi: 10.1016/j.placenta.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Willnow TE, et al. Defective forebrain development in mice lacking gp330/megalin. Proc Natl Acad Sci U S A. 1996;93(16):8460–8464. doi: 10.1073/pnas.93.16.8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fisher CE, Howie SEM. The role of megalin (LRP-2/Gp330) during development. Developmental Biology. 2006;296(2):279–297. doi: 10.1016/j.ydbio.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 54.Zohn IE, Sarkar AA. The visceral yolk sac endoderm provides for absorption of nutrients to the embryo during neurulation. Birth Defects Research Part A: Clinical and Molecular Teratology. 2010;88(8):593–600. doi: 10.1002/bdra.20705. [DOI] [PubMed] [Google Scholar]
- 55.Kozyraki R, Gofflot F. Multiligand endocytosis and congenital defects: roles of cubilin, megalin and amnionless. Curr Pharm Des. 2007;13(29):3038–3046. doi: 10.2174/138161207782110507. [DOI] [PubMed] [Google Scholar]
- 56.Duester G. Retinoic acid synthesis and signaling during early organogenesis. Cell. 2008;134(6):921–931. doi: 10.1016/j.cell.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maden M. Retinoids and spinal cord development. J Neurobiol. 2006;66(7):726–738. doi: 10.1002/neu.20248. [DOI] [PubMed] [Google Scholar]
- 58.Copp AJ, Greene ND. Neural tube defects--disorders of neurulation and related embryonic processes. Wiley Interdiscip Rev Dev Biol. 2013;2(2):213–227. doi: 10.1002/wdev.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shimamura K, et al. Longitudinal organization of the anterior neural plate and neural tube. Development. 1995;121(12):3923–3933. doi: 10.1242/dev.121.12.3923. [DOI] [PubMed] [Google Scholar]
- 60.Hales CN, Barker DJ. The thrifty phenotype hypothesis. Br Med Bull. 2001;60:5–20. doi: 10.1093/bmb/60.1.5. [DOI] [PubMed] [Google Scholar]
- 61.Rastan S, Cattanach BM. Interaction between the Xce locus and imprinting of the paternal X chromosome in mouse yolk-sac endoderm. Nature. 1983;303(5918):635–637. doi: 10.1038/303635a0. [DOI] [PubMed] [Google Scholar]
- 62.Davis TL, Tremblay KD, Bartolomei MS. Imprinted expression and methylation of the mouse H19 gene are conserved in extraembryonic lineages. Dev Genet. 1998;23(2):111–118. doi: 10.1002/(SICI)1520-6408(1998)23:2<111::AID-DVG3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 63.Kaneda M, et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429(6994):900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 64.Burton GJ, Hempstock J, Jauniaux. E. Nutrition of the human fetus during the first trimester--a review. Placenta. 2001;22(Suppl A):S70–S77. doi: 10.1053/plac.2001.0639. [DOI] [PubMed] [Google Scholar]
- 65.Burton GJ, et al. Uterine glands provide histiotrophic nutrition for the human fetus during the first trimester of pregnancy. J Clin Endocrinol Metab. 2002;87(6):2954–2959. doi: 10.1210/jcem.87.6.8563. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.