Abstract
We confirmed strong association of rs78378222:A>C (per allele odds ratio [OR] = 3.14; P = 6.48 × 10−11), a germline rare single-nucleotide polymorphism (SNP) in TP53, via imputation of a genome-wide association study of glioma (1,856 cases and 4,955 controls). We subsequently performed integrative analyses on the Cancer Genome Atlas (TCGA) data for GBM (glioblastoma multiforme) and LUAD (lung adenocarcinoma). Based on SNP data, we imputed genotypes for rs78378222 and selected individuals carrying rare risk allele (C). Using RNA sequencing data, we observed aberrant transcripts with ~3 kb longer than normal for those individuals. Using exome sequencing data, we further showed that loss of haplotype carrying common protective allele (A) occurred somatically in GBM but not in LUAD. Our bioinformatic analysis suggests rare risk allele (C) disrupts mRNA termination, and an allelic loss of a genomic region harboring common protective allele (A) occurs during tumor initiation or progression for glioma.
Keywords: glioma, TP53, rare SNP, TCGA
Approximately 80% of primary adult malignant brain tumors are gliomas, which carry a poor prognosis (5-year relative survival rate 33.8%). The most common histological subtype, glioblastoma multiforme (GBM) has a median survival rate of less than 15 months [Dolecek et al., 2012]. Genetic studies suggest an inherited component of risk for glioma, both in the general population and in rare familial syndromes, as observed in TP53 (MIM #191170) gene mutations in Li–Fraumeni syndrome [Melean et al., 2004; Hemminki et al., 2009] as well as POT1 (MIM #606478) gene mutations described in glioma family [Bainbridge et al., 2015]. Genome-wide association studies (GWAS) have conclusively identified eight distinct genomic regions with plausible candidate genes including 3q26.2 (TERC), 5p15.33 (TERT), 8q24.21 (CCDC26), 9p21.3 (CDKN2A-CDKN2B), 11q23.3 (PHLDB1), 20q13.33 (RTEL1), and two independent association signals at 7p11.2 (EGFR) [Shete et al., 2009; Wrensch et al., 2009; Rajaraman et al., 2012; Walsh et al., 2014]. In addition, a moderately penetrant risk locus marked by the rare single-nucleotide polymorphism (SNP) rs78378222 in the 3′untranslated region (3′-UTR) of TP53 was recently reported [Stacey et al., 2011; Egan et al., 2012; Enciso-Mora et al., 2013; Walsh et al., 2013]. It was further suggested that this susceptibility allele could be associated with a poor prognosis [Egan et al., 2012], but subsequent study has not confirmed this observation (Enciso-Mora et al., 2013].
To investigate the previous findings of the association of the rs78378222 with glioma risk, we performed an imputation analysis using our previously reported GWAS of 1,856 cases and 4,955 controls [Rajaraman et al., 2012]. After additional quality control, metrics were applied to the previously reported genotype data; specifically for the region of chromosome 17 bounded by 70717528071752 (hg19), we performed an imputation analysis based on a hybrid reference set including both the 1000 Genomes Project data release v3 and the Division of Cancer Epidemiology and Genetics (DCEG) Reference Set version 1 [Wang et al., 2012].
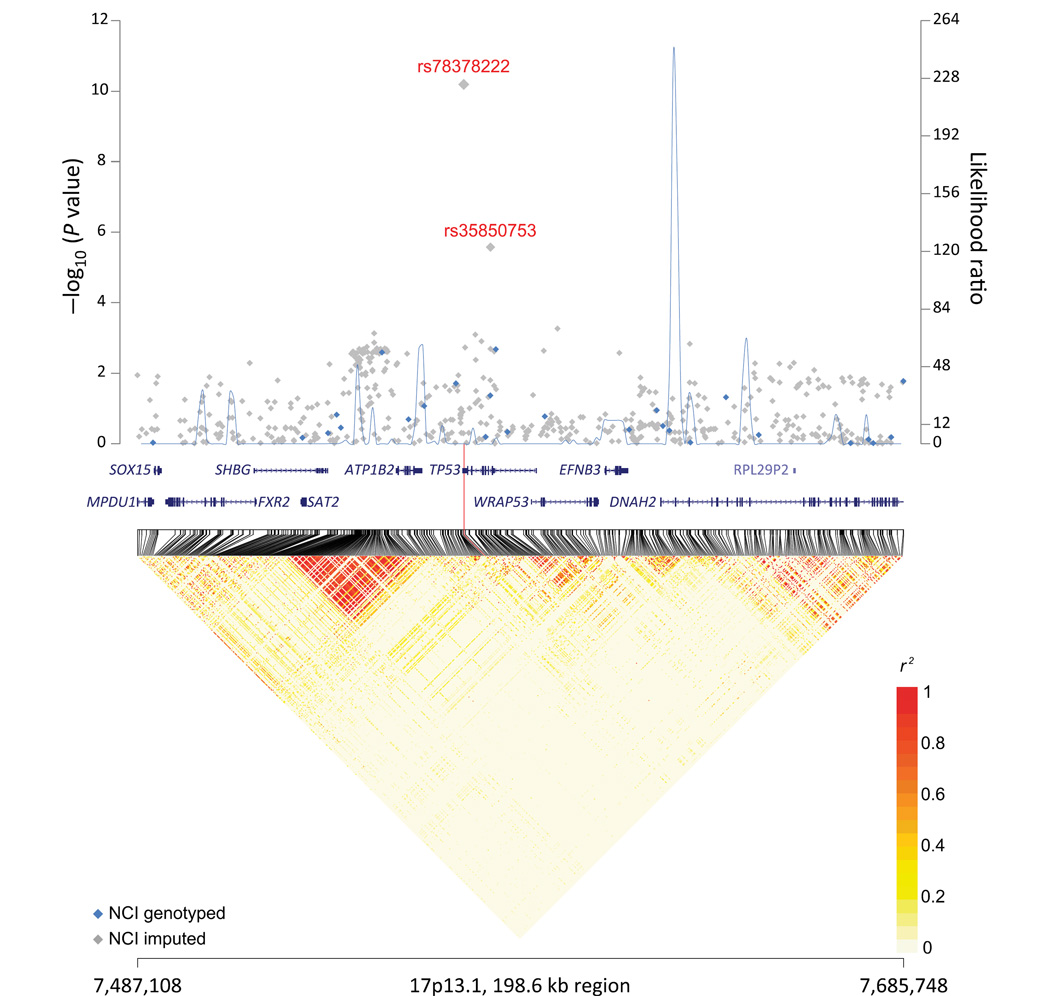
Association analyses were subsequently performed on both genotyped and imputed SNPs using logistic regression on the allelic trend effect with adjustments for age, gender, study group, and significant eigenvectors for the following groups: (1) overall; (2) cohort; (3) case-control studies; (4) GBM cases; (5) non-GBM cases; (6) early-onset cases; (7) late-onset cases; (8) cases with brain tumor family history; and (9) cases without brain tumor family history. Included in the analysis was a set of 2,943 SNPs with imputation INFO score >0.3 and minor allele frequency (MAF) >0.01. The SNP marker, rs78378222, was shown to be the strongest associated SNP within this region with a P value of 6.48 × 10−11 and per allele OR of 3.14 (95% confidence interval [CI]: 2.23–4.43); rs35850753 was ranked the second top SNP with a P value of 2.68 × 10−6 and per allele OR of 1.88 (95% CI: 1.45–2.45) (Table 1; and Fig. 1). Both MAFs and ORs are comparable to published results [Stacey et al., 2011; Enciso-Mora et al., 2013; Walsh et al., 2013]. Both SNPs were imputed with high accuracies. Moreover, TaqMan validation for rs78378222 performed with 236 samples (including 86 heterozygotes) showed high correlation between the imputed and TaqMan genotypes (r2 = 0.96), thus confirming the accuracy of imputation. It is noteworthy that the imputation INFO score for rs78378222 and rs35850753 were 0.92 and 0.95, respectively, compared with 0.66 and 0.61 previously reported [Enciso-Mora et al., 2013]. The substantial improvement of the imputation accuracy could be attributed to a more recent 1000 Genomes Project data release used as the reference as well as the benefit of the DCEG Reference Set, which has previously been observed to substantially boost the performance of imputation, especially for the SNPs with MAF ranging from 1% to 10% [Wang et al., 2012]. There was no appreciable difference in the ORs between the case-control studies (1,300 cases and 2,014 controls) and cohort studies (543 cases and 2,939 controls) (Phet = 0.92; 0.63). Similarly, the ORs for GBM cases and non-GBM cases models are comparable (Phet = 0.24; 0.17). In addition, we performed the stratified analyses by age at diagnosis or brain tumor family history (Table 1). We observed no statistically significant difference between the early onset (age at diagnosis <45 years) and the late onset (age at diagnosis ≥45 years) (Phet = 0.99; 0.85), or between cases with brain cancer family history and cases without brain cancer family history (Phet = 0.39; 0.47).
Table 1.
Association Between Glioma Risk and Selected SNPS (rs78378222 and rs35850753) for Glioma Overall, and Stratified by Study Design, Tumor Type, Age of Onset, and Family History of Brain Tumor
| Locus (reference allele, effect allele) |
Chr | Pos (hg19) | Model | Effect allele frequency in cases |
Effect allele frequency in controls |
Cases | Controls | P value | OR | (95% CI) | Heterogeneity P valuea |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs78378222 (A,C) | 17 | 7,571,752 | Overall | 0.0292 | 0.0125 | 1,856 | 4,955 | 6.48 × 10−11 | 3.14 | (2.23–4.43) | |
| Case-control | 0.0302 | 0.0122 | 1,300 | 2,014 | 3.06 × 10−8 | 3.31 | (2.17–5.06) | 0.92 | |||
| Cohort | 0.0274 | 0.0128 | 543 | 2,939 | 1.59 × 10−4 | 3.19 | (1.75–5.83) | ||||
| GBM | 0.0314 | 0.0125 | 970 | 4,955 | 6.75 × 10−10 | 4.17 | (2.65–6.56) | 0.24 | |||
| Non-GBM | 0.0268 | 0.0125 | 885 | 4,955 | 9.06 × 10−7 | 3.56 | (2.15–5.92) | ||||
| Early onset | 0.0299 | 0.0125 | 502 | 4,955 | 1.46 × 10−6 | 4.45 | (2.42–8.17) | 0.99 | |||
| Late onset | 0.029 | 0.0125 | 1,353 | 4,955 | 3.72 × 10−10 | 3.23 | (2.24–4.67) | ||||
| FH positive | 0.0364 | 0.0086 | 105 | 59 | 1.13 × 10−1 | 3.31 | (0.75–14.53) | 0.39 | |||
| FH negative | 0.0273 | 0.0115 | 1,384 | 2,220 | 1.19 × 10−7 | 2.82 | (1.92–4.14) | ||||
| rs35850753 (G,A) | 17 | 7,578,671 | Overall | 0.0403 | 0.0243 | 1,856 | 4,955 | 2.68 × 10−6 | 1.88 | (1.45–2.45) | |
| Case-control | 0.0406 | 0.0232 | 1,299 | 2,015 | 3.52 × 10−5 | 2.02 | (1.45–2.82) | 0.63 | |||
| Cohort | 0.0404 | 0.0251 | 543 | 2,939 | 1.43 × 10−2 | 1.76 | (1.12–2.75) | ||||
| GBM | 0.0441 | 0.0243 | 970 | 4,954 | 2.85 × 10−6 | 2.24 | (1.60–3.13) | 0.17 | |||
| Non-GBM | 0.0361 | 0.0243 | 885 | 4,954 | 1.15 × 10−2 | 1.61 | (1.11–2.32) | ||||
| Early onset | 0.0396 | 0.0243 | 502 | 4,954 | 1.28 × 10−3 | 2.05 | (1.32–3.18) | 0.85 | |||
| Late onset | 0.0405 | 0.0243 | 1,352 | 4,954 | 6.16 × 10−7 | 2.03 | (1.54–2.68) | ||||
| FH positive | 0.0472 | 0.009 | 104 | 59 | 5.64 × 10−2 | 3.53 | (0.97–12.87) | 0.47 | |||
| FH negative | 0.0378 | 0.0217 | 1,385 | 2,220 | 3.23 × 10−5 | 1.88 | (1.39–2.53) |
Heterogeneity P value is trend effect test based on case–case comparison between each pair of groups.
Figure 1.
17p13.1 regional plot. Results of trend test for association for genotyped (blue diamond) and imputed (gray diamond) SNPs were plotted on a negative log scale (left y-axis) against genomic coordinates (x-axis, 17p13.1:7487108–7685748, hg19). Blue line graph depicts recombination hotspots inferred from the 1000 Genomes Project phase 1 European data (100 random sampled) in likelihood ratio statistics (right y-axis). Lower panel depicts linkage disequilibrium heat map based on r2 using the 1000 Genomes Project phase 1 European data (n = 379).
We conducted an integrative analysis of the region including rs78378222 based on the rich set of Cancer Genome Atlas (TCGA) data to investigate the contribution of this germline variation in somatic settings. In particular, we examined both GBM and LUAD data from TCGA for a comparison because the rare allele (C) of rs78378222 does not seem to be associated with risk for LUAD. We imputed the genotypes for rs78378222 (see Supp. Methods) based on the Affymetrix 6.0 SNP array data for blood DNA samples from both GBM and LUAD sets, respectively, and then selected individuals with either heterozyogous (AC) or homozygous (AA) genotype for rs7837822. All together, we were able to identify a total of four GBM individuals and three LUAD individuals with AC genotype based on imputation results, and then we randomly chose two GBM individuals and three LUAD individuals with AA genotype for a total of six samples drawn from each set. We first analyzed the RNASeq data of a pair of GBM individuals, TCGA-12-0618 (AC) and TCGA-02-2483 (AA) as well as a pair of LUAD samples, TCGA-55-8505 (AC), and TCGA-69-7973 (AA). We observed a significant abundance of “run-on” transcripts with extended 3′UTR in TCGA-12-0618 but not in TCGA-02-2483 (Supp. Fig. S1 upper two panels). The aberrant transcripts have reads that map to ~3 kb region further downstream of normal 3′UTR of TP53. Although the existence of such aberrant transcripts has previously been demonstrated with RT-PCR with “run-on” primer designed at ~300 bp away from the end of normal 3′-UTR, and the expression ratio between normal transcript and aberrant transcript was shown to be 70% and 30% [Stacey et al., 2011], that functional study was only based on mRNA obtained from blood or adipose instead of tumor tissue, and was also limited by applying RT-PCR in characterizing the aberrant transcript. Since the aberrant transcripts almost exclusively appear in GBM with AC genotype for rs78378222, it is likely that the aberrant transcripts would also carry the risk allele C, which was known to disrupt the proper termination of transcription as well as polyadenylation of TP53 mRNA. For the LUAD set, although there are some noticeable aberrant transcripts covering the same 3-kb region in TCGA-55-8505 (AC) but not in TCGA-69-7973 (AA) (Supp. Fig. S1 lower two panels), the abundance is much lower as compared with GBM across the extended 3′UTR region. The same pattern was also seen for other GBM or LUAD samples with or without the rare risk allele C (Supp. Fig. S2 and 3).
We analyzed allele-specific expression (ASE) by categorizing the number of reads covering either A or C allele (Supp. Table S1). We observed that for the total mRNA, among all four GBM individuals with heterozygous genotype for rs78378222, there is striking ASE between the rare risk allele C and the common protective allele A. The number of reads carrying the risk allele C at rs78378222 is predominant, approximately 9.5 times on average more than those with the A allele. Interestingly, in three tumor samples from LUAD, the fold change (0.68) goes in the opposite direction (Supp. Table S1).
The predominant aberrant transcript in the total TP53 mRNA in GBM suggests that the chromosome fragment carrying the common allele (A) might have been deleted in glioma. To verify this, we checked the raw reads and variant calls based on whole-exome sequencing data from both tumor and matched blood sample for several neighboring SNPs with each of their rare alleles sharing the same haplotype with the rare allele (C) of rs78378222 (pairwise d′ = 1); and for three out of four GBM samples with heterozygous genotype in blood, there appears to be a deletion of the common (reference) allele in matched tumor (Supp. Table S2).
Our bioinformatic analysis suggests a functional mechanism for rs78378222, a germline rare variant in TP53 (Supp. Fig. S4). In glioma, this germline rare risk allele disrupts the proper mRNA termination and 3′-end processing, and simultaneously, a copy loss of the protective common allele for the same genomic region also occurs during tumor initiation or progression. The observed deletion in the second allele, as seen in the TCGA somatic analyses, is suggestive of a “2-hit” hypothesis, proposed by Knudson (1971), for which both alleles are altered, one at the germline and the second by somatic alteration.
In this study, we present further independent confirmation that a rare susceptibility allele, marked by rs78378222 in TP53, confers glioma risk of approximately the same effect size. We observed no evidence that the association differed by study type, major tumor subtype, age at onset, or family history of brain tumors (although the lack of finding for family story could be due to the lack of statistical power for that analysis). Nonsynonymous mutations in TP53 are one of the most frequent somatic events in glioma. In addition to glioma, rs78378222 has also been shown to be associated with multiple other cancers including basal cell carcinoma, prostate cancer, colorectal adenoma, esophageal squamous cell carcinoma in Huaian Han Chinese [Zhou et al., 2012], squamous cell carcinoma of head and neck [Guan et al., 2013], and pediatric neuroblastoma [Diskin et al., 2014].
We have shown that imputation can be useful not only in fine mapping but also in the discovery of new susceptibility alleles, especially those with lower MAFs, not necessarily well covered by commercial SNP arrays. Since the strongest signal for a genotyped SNP in this region from our GWAS was rs8075459 (MAF = 0.104 in all controls of our study) with a P value of only 1.7 × 10−3, well below the genome-wide significance threshold, so it is unlikely common genotyped markers alone would identify this signal. As noted by Enciso-Mora et al. (2013), the allele frequency of rs78378222 is only approximately 1% in the European population, but it accounts for 6% of the familial risk of glioma. Therefore, imputation analysis based on the 1000 Genomes Project data can be a cost-effective approach to search for variants with MAF between 0.005 and 0.05 to conduct a more comprehensive investigation of the underlying architecture of genetic susceptibility. rs78378222 has an estimated MAF of 1% in the European population but it is monomorphic in African, Asian, or American populations based on the 1000 Genomes Project data. Interestingly, the same rare allele C was also seen in a Han Chinese population [Zhou et al., 2012]. Further deep resequencing will be required to map the background haplotype to trace its population origin.
It is notable that the rare SNP, rs78378222 (A>C), could have a functional effect because it changes the fifth nucleotide of the TP53 polyadenylation signal (AATAAA>AATACA), resulting in an impaired 3′-end processing of TP53 mRNA, the downregulation of both p53 mRNA and protein level as well as reduction of cellular apoptosis with potential implication in prognosis [Li et al., 2013]. We performed an integrative analysis using multidimensional TCGA data and provide preliminary evidence for the functional underpinning of rs78378222 as a glioma susceptibility allele. Using the SNP array data of the blood DNA samples from the GBM or LUAD TCGA set, we applied the same approach to impute this rare SNP and then select individual heterozygous for rs78378222. Based on the RNASeq data, we observed aberrant transcripts with 3 kb longer than usual, and the aberrant transcripts were predominantly expressed in GBM individuals with heterozygote genotype due to the loss of the common protective allele in glioma. Our study also demonstrates in general the utility of analyses of TCGA for functional follow-up for GWAS. Moreover, the lack of somatic LOH in LUAD could possibly explain the observed effect of the rare variant on glioma risk but not LUAD risk. Further work is needed to investigate the possible mechanisms underlying somatic alterations in glioma in this specific region as well as functional work to illustrate how the germline variant informs our understanding of the somatic landscape of glioma.
Supplementary Material
Acknowledgments
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the National Institute for Occupational Safety and Health. Cancer incidence data were provided by the Maryland Cancer Registry, Center for Cancer Surveillance and Control, Department of Health and Mental Hygiene, 201 W. Preston Street, Room 400, Baltimore, MD 21201, http://phpa.dhmh.maryland.gov/cancer, 410-767-4055. We acknowledge the State of Maryland, the Maryland Cigarette Restitution Fund, and the National Program of Cancer Registries of the Centers for Disease Control and Prevention for the funds that support the collection and availability of the cancer registry data.
Contract grant sponsors: NCI; NIH; Department of Health and Human Services; NCI; NIH (N01-C0-12400).
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- Bainbridge MN, Armstrong GN, Gramatges MM, Bertuch AA, Jhangiani SN, Doddapaneni H, Lewis L, Tombrello J, Tsavachidis S, Liu Y, Jalali A, Plon SE, et al. Germline mutations in shelterin complex genes are associated with familial glioma. J Natl Cancer Inst. 2015;107(1):dju384. doi: 10.1093/jnci/dju384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diskin SJ, Capasso M, Diamond M, Oldridge DA, Conkrite K, Bosse KR, Russell MR, Iolascon A, Hakonarson H, Devoto M, Maris JM. Rare variants in TP53 and susceptibility to neuroblastoma. J Natl Cancer Inst. 2014;106(4):dju047. doi: 10.1093/jnci/dju047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolecek TA, Propp JM, Stroup NE, Kruchko C. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro Oncol. 2012;14(Suppl 5):v1–v49. doi: 10.1093/neuonc/nos218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan KM, Nabors LB, Olson JJ, Monteiro AN, Browning JE, Madden MH, Thompson RC. Rare TP53 genetic variant associated with glioma risk and outcome. J Med Genet. 2012;49:420–421. doi: 10.1136/jmedgenet-2012-100941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enciso-Mora V, Hosking FJ, Di Stefano AL, Zelenika D, Shete S, Broderick P, Idbaih A, Delattre JY, Hoang-Xuan K, Marie Y, Labussiere M, Alentorn A, et al. Low penetrance susceptibility to glioma is caused by the TP53 variant rs78378222. Br J Cancer. 2013;108:2178–2185. doi: 10.1038/bjc.2013.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X, Wang LE, Liu Z, Sturgis EM, Wei Q. Association between a rare novel TP53 variant (rs78378222) and melanoma, squamous cell carcinoma of head and neck and lung cancer susceptibility in non-Hispanic Whites. J Cell Mol Med. 2013;17:873–878. doi: 10.1111/jcmm.12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemminki K, Tretli S, Sundquist J, Johannesen TB, Granstrom C. Familial risks in nervous-system tumours: a histology-specific analysis from Sweden and Norway. Lancet Oncol. 2009;10:481–488. doi: 10.1016/S1470-2045(09)70076-2. [DOI] [PubMed] [Google Scholar]
- Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Gordon MW, Xu-Monette ZY, Visco C, Tzankov A, Zou D, Qiu L, Montes-Moreno S, Dybkaer K, Orazi A, Zu Y, Bhagat G, et al. Single nucleotide variation in the TP53 3’ untranslated region in diffuse large B-cell lymphoma treated with rituximab-CHOP: a report from the International DLBCL Rituximab-CHOP Consortium Program. Blood. 2013;121:4529–4540. doi: 10.1182/blood-2012-12-471722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melean G, Sestini R, Ammannati F, Papi L. Genetic insights into familial tumors of the nervous system. Am J Med Genet C Semin Med Genet. 2004;129C:74–84. doi: 10.1002/ajmg.c.30022. [DOI] [PubMed] [Google Scholar]
- Rajaraman P, Melin BS, Wang Z, McKean-Cowdin R, Michaud DS, Wang SS, Bondy M, Houlston R, Jenkins RB, Wrensch M, Yeager M, Ahlbom A, et al. Genome-wide association study of glioma and meta-analysis. Hum Genet. 2012;131:1877–1888. doi: 10.1007/s00439-012-1212-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, Simon M, Marie Y, Boisselier B, Delattre JY, Hoang-Xuan K, El Hallani S, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41:899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey SN, Sulem P, Jonasdottir A, Masson G, Gudmundsson J, Gudbjartsson DF, Magnusson OT, Gudjonsson SA, Sigurgeirsson B, Thorisdottir K, Ragnarsson R, Benediktsdottir KR, et al. A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nat Genet. 2011;43:1098–1103. doi: 10.1038/ng.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh KM, Anderson E, Hansen HM, Decker PA, Kosel ML, Kollmeyer T, Rice T, Zheng S, Xiao Y, Chang JS, McCoy LS, Bracci PM, et al. Analysis of 60 reported glioma risk SNPs replicates published GWAS findings but fails to replicate associations from published candidate-gene studies. Genet Epidemiol. 2013;37:222–228. doi: 10.1002/gepi.21707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh KM, Codd V, Smirnov IV, Rice T, Decker PA, Hansen HM, Kollmeyer T, Kosel ML, Molinaro AM, McCoy LS, Bracci PM, Cabriga BS, et al. Variants near TERT and TERC influencing telomere length are associated with high-grade glioma risk. Nat Genet. 2014;46:731–735. doi: 10.1038/ng.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Jacobs KB, Yeager M, Hutchinson A, Sampson J, Chatterjee N, Albanes D, Berndt SI, Chung CC, Diver WR, Gapstur SM, Teras LR, et al. Improved imputation of common and uncommon SNPs with a new reference set. Nat Genet. 2012;44:6–7. doi: 10.1038/ng.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrensch M, Jenkins RB, Chang JS, Yeh RF, Xiao Y, Decker PA, Ballman KV, Berger M, Buckner JC, Chang S, Giannini C, Halder C, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41:905–908. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Yuan Q, Yang M. A functional germline variant in the P53 polyadenylation signal and risk of esophageal squamous cell carcinoma. Gene. 2012;506:295–297. doi: 10.1016/j.gene.2012.07.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.