Abstract
Circulating miRNA stability suggests potential utility of miRNA based biomarkers to monitor tumor burden and/or progression, particularly in cancer types where serial biopsy is impractical. Assessment of miRNA specificity and sensitivity is challenging within the clinical setting. To address this, circulating miRNAs were examined in mice bearing human SCLC tumor xenografts and SCLC patient derived circulating tumor cell explant models (CDX). We identified 49 miRNAs using human TaqMan Low Density Arrays readily detectable in 10 μl tail vein plasma from mice carrying H526 SCLC xenografts that were low or undetectable in non‐tumor bearing controls. Circulating miR‐95 measured serially in mice bearing CDX was detected with tumor volumes as low as 10 mm3 and faithfully reported subsequent tumor growth.
Having established assay sensitivity in mouse models, we identified 26 miRNAs that were elevated in a stage dependent manner in a pilot study of plasma from SCLC patients (n = 16) compared to healthy controls (n = 11) that were also elevated in the mouse models. We selected a smaller panel of 10 previously reported miRNAs (miRs 95, 141, 200a, 200b, 200c, 210, 335#, 375, 429) that were consistently elevated in SCLC, some of which are reported to be elevated in other cancer types. Using a multiplex qPCR assay, elevated levels of miRNAs across the panel were also observed in a further 66 patients with non‐small cell lung, colorectal or pancreatic cancers. The utility of this circulating miRNA panel as an early warning of tumor progression across several tumor types merits further evaluation in larger studies.
Keywords: miRNA, Cancer monitoring, Blood biomarker
Highlights
Simple miRNA assay developed suitable for plasma volumes as low as 10 μl.
Assay suitable for serial weekly monitoring of human xenograft growth in mice.
Can detect xenograft tumor volumes as low as 10 mm3 and correlates with tumor growth.
Selected miRNAs elevated in SCLC, NSCLC, colorectal and pancreatic cancers.
Abbreviations
- cfDNA
circulating cell-free DNA
- CTC
circulating tumor cell
- CDX
tumor xenografts derived from patient circulating tumor cells
- HNV
healthy normal volunteers
- SCLC
small cell lung cancer
- NSCLC
non-small cell lung cancer
- CRC
colorectal cancer
1. Introduction
Regular monitoring of tumor burden and evolution is an important component of personalized cancer treatment. Biomarkers that have a high sensitivity and specificity to monitor changes in disease status are required; but ideally these should be relatively non‐invasive and affordable allowing serial monitoring of dynamic changes following therapy. MicroRNAs are small (19–26 bp), non‐coding RNAs important in the epigenetic control of translation and transcription. miRNAs can act as both tumor suppressors and promoters (Ambros, 2004) and have been associated with carcinogenesis, metastasis, and drug resistance (Schwarzenbach et al., 2014). Analysis of miRNA expression in tumors can help to identify the primary site of origin in cancers with occult primaries (Ferracin et al., 2011), and in a number of disease settings miRNAs have been shown to have potential utility as diagnostic, prognostic, and predictive biomarkers (Schwarzenbach et al., 2014).
miRNAs are released from the cell within membrane bound particles (exosomes) or in conjunction with nucleotide binding proteins, such as Argonaute proteins, that protect them from degradation by circulating RNases (Arroyo et al., 2011). Therefore it has been anticipated that miRNAs would provide a stable circulating biomarker, which would provide a specific and sensitive readout of tumor behavior (Schwarzenbach et al., 2014). In support of this, miRNAs are easily quantified in the circulation and different profiles are seen between healthy controls and patients. Modeling based on placental miRNAs suggested that as little as 0.3 g of tumor tissue should be detectable in a 1 ml blood sample (Williams et al., 2013), whilst abnormalities in the circulating profile can be detected many years before the diagnosis of cancer (Boeri et al., 2011; Sozzi et al., 2014). Levels of putative tumor derived miRNAs have been found to correlate with the number of circulating tumor cells and levels of tumor derived proteins (Madhavan et al., 2012; Roth et al., 2011).
However, a number of issues have arisen that have delayed the implementation of these promising biomarkers into routine clinical practice. Choice of sample matrix (i.e. plasma or serum) and the assessment of the impact of variations in sample processing are vital prior to qualification of a biomarker (Greystoke et al., 2008; Wang et al., 2012). The presence in large amounts of miRNAs in normal components of the blood, including platelets and red blood cells, mean that the use of serum or hemolysis may unduly impact on subsequent analysis (Pritchard et al., 2012; Wang et al., 2012). Also as qPCR based analysis of miRNAs is a quasi‐quantitative assay and there is no recognized ‘house‐keeper’ circulating miRNA, there have been problems with the normalization of expression data and reproducibility between laboratories (Leidner et al., 2013).
The use of murine models, harboring human tumors, allows the identification of tumor derived miRNAs in the circulation distinct from the background mouse miRNA (Mitchell et al., 2008; Selth et al., 2012). For subcutaneous implants, the models could demonstrate if circulating miRNAs accurately reflect tumor growth (Waters et al., 2012). However, given the importance of miRNAs in epigenetic control, many miRNAs are highly conserved between species (Meunier et al., 2013), and the extent to which human miRNAs can be detected over the host background in the circulation of mice bearing xenografts has yet to be determined. Therefore we first set out to develop a robust protocol that allowed specific profiling of human tumor miRNAs in microliters of tail vein plasma. Having established base line sensitivities in the murine models, we then tested the methodology by applying it to clinical samples from 82 patients with tumors that are challenging to biopsy and where minimally invasive biomarkers are sought (SCLC, n = 16, colorectal cancer, n = 25, pancreatic cancer, n = 26, and NSCLC, n = 15).
2. Material and methods
2.1. Xenograft models
All procedures were carried out in accordance with Home Office Regulations (UK) and the UK Coordinating Committee on Cancer Research guidelines and by approved protocols (Home Office Project license no. 40‐3306). The CTC derived explants (CDX) were established using 10 ml of blood from patients with extensive SCLC. The sample was enriched for CTCs using the RosetteSep Circulating Epithelial Cell cocktail and the resulting cell suspension injected s.c. into one or both flanks of 8–16 week old female NOD.Cg‐Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (Jackson Laboratories) as previously described (Hodgkinson et al., 2014). Tumor growth was monitored along with three‐weekly tail bleeds (∼20 μl) from prior to implantation to death including in mice where no CDX developed.
2.2. Subject recruitment
The study was carried out according to the Declaration of Helsinki and International Conference for Harmonization of Good Clinical Practice guidelines. Following ethical approval, patients who gave informed consent were recruited who were receiving chemotherapy for SCLC, pancreatic cancer and colorectal cancer or receiving radiotherapy for NSCLC (Christie NHS Trust, Manchester and Clatterbridge Cancer Centre Liverpool, UK). Blood samples were taken immediately prior to therapy and in the patients with NSCLC and CRC approximately 6 weeks after the initiation of therapy. Healthy volunteers were recruited to donate blood samples according to an ethically approved protocol. The healthy individuals were age matched with the colorectal cancer patients, and were a mixture of male and female as for the patient cohort. Response was determined by a radiologist blinded to biomarker data using the Response Evaluation Criteria in Solid Tumours (RECIST).
2.3. Sample processing and microRNA extraction
Blood (6 ml) was collected in EDTA tubes (BD Vacutainer®) and processed within 4 h. Blood was centrifuged at 2000 × g for 10 min at 4 °C and the supernatant was centrifuged a second time (2000 × g for 10 min at 4 °C) to obtain the final plasma sample for storage (at −80 °C) and subsequent miRNA extraction.
MicroRNA was routinely extracted from 200 μl plasma, although up to 10 μl plasma was sufficient, using the miRNeasy kit (Qiagen) according to the manufacturer's instructions for purification of total RNA, including miRNA. Briefly, 5 volumes of QIAzol Lysis Reagent were added to the plasma samples (i.e. 1 ml QIAzol was added to 200 μl plasma), vortexed for 1 min and incubated at room temperature for 5 min. A synthetic plant (Arabidopsis thaliana, Life Technologies) aly‐miR‐159 ‘spike‐in’ (12.5 pM) was added as an exogenous control. Chloroform was then added an equal volume to the starting material, (i.e. for 200 μl plasma, 200 μl chloroform) and vortexed thoroughly for 15 s to obtain a homogenous mix. After incubation at room temperature for 3 min, samples were centrifuged at 12,000 × g for 15 min at 4 °C. The top, aqueous layer was transferred to a new collection tube and 1.5 volumes of 100% ethanol added with thorough mixing. The sample was applied to an RNeasy MinElute spin column and centrifuged ≥8000 × g for 30 s and the flow‐through discarded. This was followed by 3 washes, with Buffer RWT, Buffer RPE and 80% ethanol, using the above centrifugation parameters and discarding the flow‐through each time. The RNA (including the miRNA) was eluted in 14 μl of RNase‐free H2O and stored at −80 °C until further analysis.
2.4. Quantitative real‐time polymerase chain reaction (qRT‐PCR) for individual miRNAs
Reverse transcription was performed using the individual miRNA specific primers and the TaqMan miRNA Reverse Transcription kit (Life Technologies), as per the manufacturer's instructions. The relative levels of individual miRNAs were assessed using quantitative real‐time polymerase chain reaction on the specific cDNA generated, using miRNA‐specific TaqMan primer‐probes (Life Technologies), on an ABI Prism 7900HT Sequence detection system using 2× TaqMan Universal Master Mix II, no Uracil N‐glycosylase. The reactions were incubated at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Levels of the synthetic spike‐in, aly‐miR‐159, as well as the endogenous hsa/mmu‐miR‐16, were used to assess the efficacy of extraction. Levels of the 10‐plex were reported as the geometric mean of the 40‐Ct values for the 10 miRNAs (Geomean10).
2.5. TaqMan low density arrays (TLDAs)
miRNAs were profiled by the TaqMan Human MicroRNA array Card Set v3.0 and MegaPlex chemistries (Life Technologies) as per the manufacturer's instructions. Briefly, 3 μl of RNA was reverse transcribed, for each array, using the MegaPlex RT kit and the TaqMan miRNA Reverse Transcription kit and 2.5 μl of the resultant cDNA was used in the pre‐amplification step. The pre‐amplified product was diluted 1 in 4 in Tris–EDTA buffer, and the levels of aly‐miR‐159 and hsa/mmu‐miR‐16 were assessed, using the individual miRNA assays (as described above), to determine the accuracy of amplification. Of the diluted product, 9 μl of this was loaded onto the TaqMan Low Density Arrays (TLDAs) plates A and B, for relative quantitative PCR analysis. The qPCR was performed on the 7900HT Sequence detection system. Initial data output was in SDS software v2.4 (with a fixed threshold setting of 0.2).
2.6. Quality control and validation
Prior to a clinical study, it was important to set steps to ensure robust extraction and amplification and perform initial analysis on a data set, with technical replicates to validate the technique. With this in mind, two quality assessment points were implemented prior to proceeding to TLDA analysis, with Step 1 testing miRNA extraction efficiency and Step 2 testing MegaPlex RT and PreAmp efficiencies (Supplementary material Figure 1).
2.7. Normalization and statistical analysis
Normalization is a critical pre‐analysis step for gene expression data analysis, particularly for high‐throughput TLDA data. Initially, any miRNAs with no evidence of any significant expression in any subject's samples (40‐Ct < 4) were discarded as having limited clinical utility. Results were normalized to the geometric mean of all miRNAs and highly expressed miRNAs (40‐Ct > 10) to obtain the ΔCt value. Analysis of TLDA data including significance analysis of microarrays (SAM); hierarchal cluster analysis, and principle component analysis was performed in MultiExperiment Viewer (MeV, TM4.org). In order to reduce the possibility of false positives and highlight miRNAs that were most likely to be significantly elevated the SAM analysis was performed with thresholds set for an estimated false discovery rate of 0. Principle component analysis was performed in MEV using the recommended k‐Nearest Neighbour algorithm optimized for speed in median centering mode. Analysis of expression of miRNAs assessed by qRT‐PCR and receiver operating characteristic curves were performed in GraphPad Prism v6.0 (GraphPad Software, San Diego California USA, www.graphpad.com) with probability values of P < 0.05 considered significant throughout.
3. Results
3.1. Establishment of a microscale plasma miRNA assay and serial sampling of mouse tail vein blood
Initially, we determined the minimal volume required for reliable assessment of circulating miRNAs in mice in order to assess the feasibility of serial blood sampling over time during tumor growth. Tail vein blood collection can be used for weekly monitoring of mice harboring human tumors where the volume of plasma available is typically only 10–30 μl. We identified 2 endogenous miRNAs (hsa/mmu‐miR‐16, ‐21) with consistent expression in healthy volunteers and mice to assess the reliability of extraction and amplification outlined in Table 1. Both miR‐16 and miR‐21 are highly conserved with identical mature sequences in the mouse and human (Supplementary material Table 1).
Table Table 1.
Expression and reproducibility of miR‐16 and miR‐21 chosen as endogenous miRNAs to confirm efficient extraction and amplification prior to TaqMan Low Density Array analysis (expression given as 40‐Ct).
| Average expression | Inter‐subject variability | Intra‐subject reproducibility | |
|---|---|---|---|
| Healthy volunteers (n = 20) | |||
| hsa‐miR‐16 | 16.7 ± 0.6 | 16.8% | 13.2% |
| hsa‐miR‐21 | 10.4 ± 0.6 | 24.4% | 16.6% |
| Mice (n = 16) | |||
| mmu‐miR‐16 | 22.4 ± 0.2 | 4% | 1% |
| mmu‐miR‐21 | 17.0 ± 0.4 | 8.0% | 5.1% |
hsa/mmu‐Mir‐16 and hsa/mmu‐miR‐21 along with the exogenous aly‐miR‐159 were used to monitor extraction efficiencies and confirm the sensitivity of the assay developed (Supplementary material Figure 1 Step 1). Efficient miRNA extraction was seen down to 10 μl plasma input with an average 26 ± 9% change in ΔCt signal compared to 200 μl. This allowed us to proceed to the MegaPlex chemistry for TaqMan Low Density Arrays (TLDAs), with reverse transcription and pre‐amplification primer pools of ∼750 different miRNAs. A second step (Supplementary material Figure 1 Step 2) was performed to check the amplification efficiency; faithful amplification and good reproducibility was seen with all 3 miRNAs (hsa/mmu‐miR‐16, hsa/mmu‐miR‐21 and aly‐miR‐159) with an average 21 ± 4% change in the ΔCt signal. The final workflow is outlined in Supplementary material Figure 1. To ensure our developed assays were compatibility with routine analysis of small plasma volumes therefore all further mouse miRNAs analysis was carried out with a starting 10 μl plasma volume.
3.2. Identification of xenograft specific miRNAs
We assessed whether it was possible to detect circulating human miRNAs in the plasma of mice bearing s.c. SCLC xenografts. The profile of miRNAs in pooled plasma collected at the time of sacrifice from 10 SCID‐Beige mice bearing s.c. H526 xenografts measuring between 700 and 1200 mm3 was compared to plasma pooled from 5 non‐tumor bearing animals. TLDA analysis was performed on 10 μl of plasma miRNAs extracted from 3 separate aliquots to assess technical variability.
There was low variability in the plasma miRNA replicates analyzed both from tumor bearing and non‐tumor bearing animals. Three hundred and ten miRNAs were detectable in non‐tumor bearing animals, presumed to be murine miRNAs with sufficient homology to human miRNAs to be detected by the TLDA, with an average CV (defined as the standard deviation/mean for each individual miRNA) of 22 ± 1%. Four hundred and thirty one miRNAs were detectable in mice bearing tumors with an average CV of 27 ± 1%. Sixty five miRNAs were upregulated significantly (relative to non‐tumor bearing animals, P < 0.05); of these 49 were low or undetectable in non‐tumor bearing animals from two different strains of mouse (SCID‐Beige and NSG), suggesting human origin (Supplementary material Table 2).
3.3. Serial monitoring of miRNAs in SCLC CDX models
We recently developed SCLC patient derived CTC explant models that display similar histology to matched diagnostic biopsies and faithfully recapitulate patient responses to chemotherapy (Hodgkinson et al., 2014). We next asked whether the miRNAs elevated in blood of mice bearing traditional SCLC cell line xenograft tumors were also detectable in these CDX models. The circulating miRNA profiles from 4 CDX grown s.c in NSG mice were compared to miRNA profiles of 3 non‐tumor bearing animals and mice bearing xenografts from a further 2 established SCLC cell‐lines (H1048 and Cor‐106). Principle component analysis suggested a wide variability in miRNA profiles although profiles from non‐tumor bearing mice clustered together and separately from those mice bearing CDX or cell line xenografts tumor implants (Supplementary material Figure 2). Thirteen miRNAs were found to be significantly over‐expressed in the CDX models compared to non‐tumor bearing mice (Figure 1A).
Figure 1.
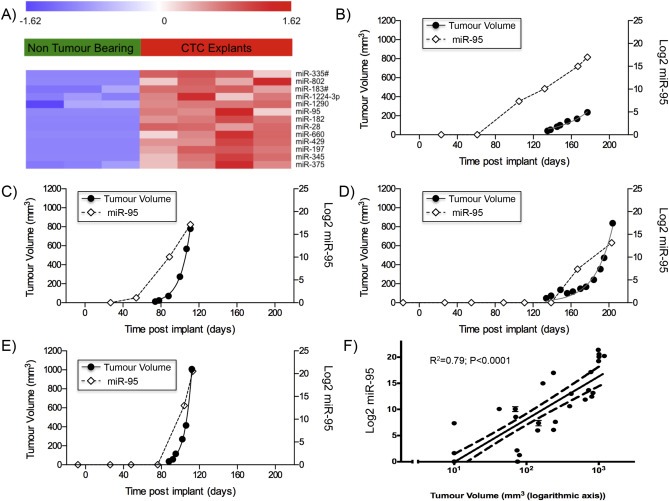
miRNA analysis of 10 μl plasma samples using TaqMan Low Density. A. Heat‐map of the circulating levels of the 13 miRNAs significantly upregulated in bearing NOD scid gamma (NSG) mice bearing explants derived from human SCLC circulating tumor cells (CDX explants; n = 4) compared to non‐tumor bearing animals (n = 3). B to E. Changes following implantation in the individual expression of circulating hsa‐miR‐95 in 10 μl of plasma from tail vein bleeds from 4 mice bearing explants derived from human SCLC circulating tumor cells compared to estimated tumor volume (measured with calipers). F. Correlation of circulating hsa‐miR‐95 in 10 μl of plasma compared to tumor volume across SCLC xenograft models (4 CTC explants and H526 cell line). miRNA levels were assessed using TaqMan Low Density Arrays (Human Pool A and B) and normalized to global expression of miRNA.
To determine the potential sensitivity of miRNAs for tumor monitoring, the relationship between tumor burden and circulating miRNAs was assessed in our preclinical models. For this purpose, levels of circulating hsa‐miR‐95 was chosen as this miRNA was undetectable in non‐tumor bearing mice, has no close murine homologue and was significantly elevated in all xenograft models (Figure 1A) and previously reported to be present in resected tumors from patients with SCLC (Miko et al., 2009).
In the fast growing H526 xenograft model used to initially screen for SCLC miRNAs (doubling time 4 days), circulating miRNA could be detected by day 4 post implant with an average tumor size of 60 mm3. Circulating hsa‐miR‐95 was also able to monitor tumor growth in tumor models with different growth kinetics. In the CDX models, where tumors took from 2.4 to 4.4 months to become palpable (doubling times from 5 to 21 days), circulating hsa‐miR‐95 was detectable shortly before or after measurable tumors were observed. In CDX1 and CDX2, hsa‐miR‐95 plasma levels increased prior to detection of palpable tumor (Figure 1B and 1C); in CDX3, hsa‐miR‐95 was detected as the tumor reached 150 mm3 (Figure 1D) and in CDX4 bearing the most rapidly growing tumor, the kinetics of increasing hsa‐miR‐95 levels mirrored those of tumor volume (Figure 1E). Linear regression of Log‐ hsa‐miR‐95 expression vs Log tumor volume in all mouse models evaluated, showed a significant correlation and suggested that the lower limit of detection of hsa‐miR‐95 was at approximately 10 mm3 of tumor (95% CI 6–15 mm3) (Figure 1F).
3.4. Identification of SCLC clinically relevant miRNAs
Having verified the ability of our protocol to identify miRNAs that accurately monitored tumor burden in mice bearing CDX, we analyzed blood samples donated by patients with SCLC prior to the initiation of chemotherapy. TLDA analysis was performed on 200 μl of plasma donated from 16 patients with SCLC (4 limited stage disease, LD; 12 extensive stage disease, ED). The circulating miRNA profile was compared to that observed in 200 μl of plasma from 11 healthy volunteers. Principle component analysis of the circulating miRNA profiles suggested extensive patient to patient variability amongst the 16 SCLC patients but with the 11 healthy volunteers clustering separately (Figure 2). The 4 LD patients also clustered together and tended to be closer to the profiles of healthy volunteers than ED patients. In order to determine which miRNAs distinguished between healthy volunteers, patients with LD and patients with ED; SAM analysis was performed with an estimated FDR of 0. This identified 29 miRNAs deregulated in a stage dependent manner (3 down regulated and 26 upregulated; Figure 3). The 26 upregulated miRNAs were also found to be significantly elevated in the 7 SCLC mouse models (xenografts and CDX) compared to non‐tumor bearing animals (Two‐way ANOVA; P < 0.0001).
Figure 2.
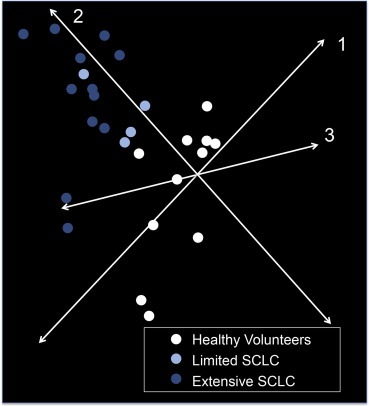
Principle component analysis of 754 miRNA levels in patient plasma. For each donor 200 μl of plasma was used for miRNA isolation. Samples shown are from patients with SCLC (4 with limited disease, 12 with extensive disease) and HNV controls (n = 11). miRNA levels were assessed using TaqMan Low Density Arrays (Human Pool A and B) and normalized to global expression of miRNA.
Figure 3.
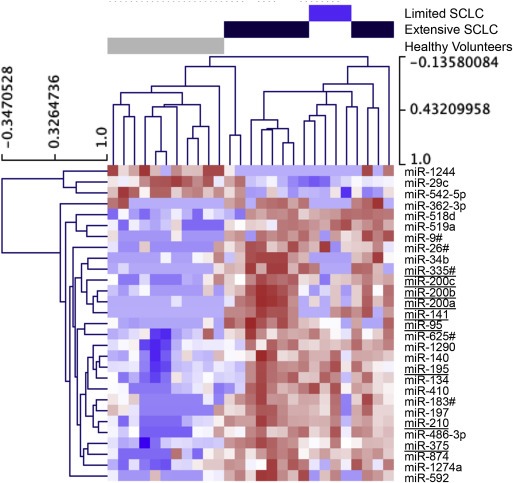
Heat‐map of the circulating levels of 29 miRNAs showing significant differences in expression between SCLC and HNV samples. miRNA levels assessed using TaqMan Low Density Arrays (Human Pool A and B) and normalized to global expression of miRNA. miRNAs subsequently selected for inclusion in the 10‐plex are underlined.
3.5. Establishment of a rapid sensitive clinical miRNA assay
TLDA analysis is not amenable for routine clinical use due to cost and slow singleplexed throughput. The qRT‐PCR protocol we established (described above), allows simultaneous assessment of 7 miRNAs across multiple clinical samples (2 μl of sample in a 15 μl RT reaction, containing enzyme, buffer, dNTPs and the 7 miRNA RT primers, followed by qRT‐PCR on 1 μl of the RT reaction, containing cDNA, in duplicate). The inclusion of both endogenous (hsa‐miR‐16) and exogenous controls (aly‐miR‐159) allowed assessment of 5 disease related miRNAs in each multiplexed assay. We decided on performing 2 multiplex assays for each clinical sample with each assay assessing 5 disease related miRNAs and the 2 reference miRNAs, allowing cheap and quick analysis on 10 miRNAs of interest.
We chose 10 miRNAs that we had either identified in the preclinical models or identified in patients' profiles and that had previously been reported as elevated either in SCLC tumors or cell‐lines (Supplementary material Figure 3 and Supplementary material Table 2). This included all 5 members of the hsa‐miR‐200 family (miR‐200a, miR‐200b, miR‐200c, miR‐141, and miR‐429) and also hsa‐miR‐95, hsa‐miR‐210, hsa‐miR‐335#, and hsa‐miR‐375.
Some of the miRNAs we had identified in this miRNA 10‐plex had been reported as elevated in the circulation of patients with other malignancies including CRC (Cazzoli et al., 2013, 2014, 2013, 2013, 2010, 2012, 2014). We hypothesized that these miRNAs might reflect disease burden in a range of patients with epithelial tumors. To explore this possibility, the miRNA 10‐plex was measured in plasma samples taken from 66 patients with pancreatic cancer, NSCLC and colorectal cancer, prior to therapy. This analysis confirmed similar elevation of these miRNAs across a number of diseases (Figure 4A and Supplementary material Table 3). In keeping with the potential to monitoring of disease burden, the 10‐plex was elevated in a stage specific manner in patients with colorectal cancer; patients with resected stage 3 cancer had significantly lower levels (geometric mean of 10‐plex expression Geomean10 6.0 ± 0.5) than patients with metastatic disease (Geomean10 9.1 ± 0.6) although still significantly higher than in healthy volunteers (Geomean10 0.9 ± 0.1) (P < 0.0003; Figure 4B).
Figure 4.
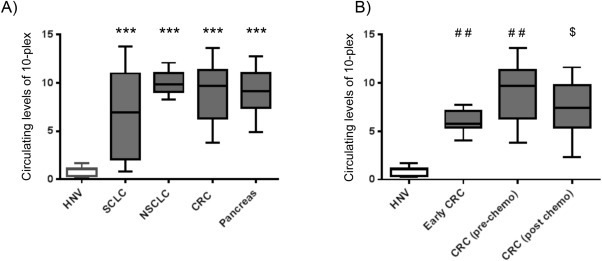
Combined expression of a 10 miRNA panel across a range of cancer types. A. Geometric mean of the selected miRNA panel in patients prior to therapy with SCLC (n = 12), NSCLC (n = 15), CRC (n = 24), pancreas cancer (n = 26) compared to HNV controls (n = 11). B. Geometric mean of the selected miRNA panel in Early CRC patients following resection for stage 3 cancer (n = 7) compared to patients with colorectal cancer prior to and following 6 weeks of cytotoxic chemotherapy (N = 24) and healthy volunteers (n = 11). For all samples described miRNA preparations were made from 200 μl plasma and a portion subjected to RT qPCR of for a panel of 10 miRNAs consisting of: miR‐200a, miR‐200b, miR‐200c, miR‐141 miR‐429, hsa‐miR‐95, hsa‐miR‐210, hsa‐miR‐335#, and hsa‐miR‐375 (see text for details). For each sample the geometric mean of all 10 miRNAs was calculated and *** indicates any clinical group significantly elevated compared to HNV controls (ANOVA; P < 0.0001).
The use of the 10‐plex improved the accuracy of the assay to distinguish between healthy volunteers and patients with low disease burden, and between these patients and those with stage 4 disease over the analysis of any single miRNA within the 10‐plex (Supplementary material Table 4). The 10‐plex has a sensitivity of 98% at a 100% specificity to distinguish patients with stage 4 cancer from healthy volunteers (AUC for the ROC 0.99 ± 0.1; P < 0.0001) and 82% sensitivity at a 100% specificity to detect patients with stage 3 disease from healthy volunteers (AUC for ROC 0.97 ± 0.03; P < 0.0001). The 10‐plex was able to distinguish between patients with stage 3 and stage 4 disease with a sensitivity of 73% at a specificity of 100% (AUC for ROC 0.88 ± 0.04; P < 0.0001).
Matched plasma samples were available from 6 weeks after the initiation of therapy in the patients with CRC and NSCLC. In the patients with CRC levels fell significantly 6 weeks following treatment with chemotherapy (mean change in Geomean10 – 1.7 ± 0.6; P < 0.01) towards levels seen in patients with stage 3 disease (Figure 4B). Larger decreases in miRNAs were seen in patients who derived clinical benefit from therapy (mean change in Geomean10 – 2.0 ± 0.6) whilst levels remained relatively static in patients with progressive disease (mean change in Geomean10 – +0.2 ± 0.6) at this 6 week time‐point; although in this small substudy this was not a significant change, it is provocative and warrants further investigation in a larger cohort. Less marked changes were seen in the patients with NSCLC treated with radiotherapy (mean change Geomean10 in patients with clinical benefit of −0.4 ± 0.4 compared to progressive disease +0.6 ± 0.3).
4. Discussion
We have developed and begun to evaluate a circulating miRNA panel intended as minimally invasive epithelial tumor monitoring tool; this was performed in a stepwise fashion building evidence from preclinical models and translating data to a pilot study in cancer patients. A number of studies have shown differences in circulating miRNA profiles between patients with cancer and healthy controls, but the clinical utility to monitor an individual patient course once diagnosed with cancer is not certain. In particular, the relative contributions to the miRNA profile from the tumor and from the host are unclear. Mice bearing human SCLC tumors supported our identification of tumor associated miRNAs in the circulation and demonstrated the close relationship between circulating miRNAs and tumor volume. The sensitivity of this approach is strongest evaluating miRNAs where the human sequence differs from the murine homolog.
Concern has been raised about analytical variability in measurement of miRNAs (Duttagupta et al., 2011; Pritchard et al., 2012; Wang et al., 2012). We identified quality controls that can be routinely used to assess robustness of extraction and amplification prior to expensive and time‐consuming TLDA analysis. When using appropriate quality controls, we showed that technical variability is small (similar to other studies such as Stratz et al., 2012) and able to pick out changes in an individual patient course.
In addition we have shown that this technology can be used robustly with as little sample volume as 10 μl plasma. This is lower than previous studies assessing miRNAs in mouse models (range 50–100 μl) (LaConti et al., 2011; Mitchell et al., 2008; Selth et al., 2012; Waters et al., 2012) and is more readily allows serial analysis in bloods taken from preclinical studies. Applications here include testing new therapeutics in preclinical models and identification of potential predictive or response miRNA biomarkers.
Previous models have identified tumor related miRNAs in xenograft models but have either not confirmed their utility in the patient population, or struggled to link tumor burden with circulating miRNA levels (Mitchell et al., 2008; Selth et al., 2012; Waters et al., 2012). We were able to measure circulating tumor miRNAs in mouse blood that were associated with tumor and detectable at the time when s.c. tumors became palpable. Our sensitivity for miRNA detection was higher than that reported for human circulating free DNA in a xenograft model (10 mm3 cf. to 60 mm3 – see Gorges et al., 2012).
miRNAs within the tumor are expressed in a tissue specific manner (Ferracin et al., 2011), but it is not certain whether miRNAs within the circulation differ according to tumor origin and histology. Unsurprisingly, different miRNA profiles are seen between the circulation and tumor (Waters et al., 2012); however several studies including the work presented here suggest that the circulating profile is at least in part tumor derived (LaConti et al., 2011). We have identified a small panel of circulating tumor miRNAs initially in SCLC using both clinical and preclinical models, which proved to also be elevated in patients with pancreatic, NSCLC, and colorectal cancers. These miRNAs have also been reported as elevated in the circulation in patients with a wider range of malignancies including NSCLC, ovarian, prostate, and breast cancer (Cazzoli et al., 2013; Chen et al., 2013; Cheng et al., 2013; Li et al., 2010; Madhavan et al., 2012; Toiyama et al., 2014). Of note, hsa‐miR‐200a, hsa‐miR‐200b, hsa‐miR‐200c, hsa‐miR‐210 and hsa‐miR‐375 correlated with CTC count (using the CellSearch platform) in breast cancer (Madhavan et al., 2012).
The miR‐200 family are important in the control of EMT, and are involved in a negative feedback loop with Zinc finger E‐box‐binding homeobox (ZEB) 1 and 2 (Hill et al., 2013). In spite of the fact that miR‐200 suppresses EMT (Zhu et al., 2012), elevated miR‐200 actually leads to increased levels of metastases in animal models (Korpal et al., 2011) suggesting that the miR‐200 family may be important in tumor EMT plasticity that is vital for metastasis (Hill et al., 2013). MiR‐210 is a hypoxia‐induced miRNA, a target of HIF‐1α, which is over‐expressed in the majority of solid tumors (Devlin et al., 2011). Circulating hsa‐miR‐210 has been reported to be elevated in patients with a number of malignancies including prostate cancer, CRC, and pancreatic cancer (Chen et al., 2014; Cheng et al., 2013; LaConti et al., 2011). In CRC, levels of hsa‐miR‐210 normalize following surgery or chemotherapy and then increase again with progression of the disease (Chen et al., 2014). Relatively little is known about hsa‐miR‐95 but it has been shown to be associated with proliferation and differentiation in a number of preclinical models (Huang et al., 2011; Li et al., 2012a) and elevated in SCLC tumors (Miko et al., 2009). The other miRNAs identified in the panel (hsa‐miR‐195, hsa‐miR‐335#, hsa‐miR‐375) were unexpected as they had previously been reported as a tumor‐suppressor miRNAs. Levels of hsa‐miR‐375 have been found to be decreased in a number of tumors particularly those related to smoking including NSCLC, head and neck, and esophageal cancers (Li et al., 2012b; Siow et al., 2013; Wu et al., 2013) and a recent report details lower levels of circulating hsa‐miR‐375 in patients with NSCLC (Yu et al., 2014). However, hsa‐miR‐375 has been also reported to be elevated in a number of tumor types including SCLC (Miko et al., 2009). In pancreatic cancer where levels were decreased in tumor compared to both premalignant lesions and normal pancreas, hsa‐miR‐375 levels were elevated in the circulation and fell following chemotherapy (LaConti et al., 2011). Similarly the tumor suppressor miR‐195 has been associated with the development of chemotherapy resistance (Dai et al., 2011; Ujifuku et al., 2010) and elevated circulating hsa‐miR‐195 has been reported in prostate cancer and breast cancer (Heneghan et al., 2010; Mahn et al., 2011).
The finding that miRNAs can be used to monitor tumor volume in murine models has immediate utility with the increasing development of patient derived orthotopic tumors, particularly where the primary tumor cells are difficult to label for intra‐vital imaging. A simple method of monitoring orthotopic tumor growth to determine the optimum time for drug dosing would be beneficial in this setting and could be performed by serial sampling of tail veins and miRNA analysis.
In the clinical setting circulating miRNAs may be particularly useful in low disease volume settings, if miRNA sensitivity is proven superior to detectable circulating tumor cells, proteins, and cfDNA. This could include identifying patients who have had potentially curative resections who would benefit from adjuvant therapy, or on the completion of palliative chemotherapy highlighting patients most likely to benefit from maintenance therapy. The potential of circulating miRNAs in this low tumor volume setting was recently shown by the added benefit of serum miRNAs over low dose CT in screening high‐risk patients for lung cancer (Sozzi et al., 2014). The potential of miRNA to monitor the tumor during therapy is less clear. The effects of chemotherapy on miRNA expression are complicated (Svoboda et al., 2008; Zhang et al., 2012), and even in non‐tumor bearing mice changes in possible biomarkers of disease burden can be seen (LaConti et al., 2011).
In summary, we have identified a panel of 10 circulating miRNAs that are elevated in patients with cancer. The identification of potential biomarkers with overlap in both preclinical and the clinical setting provides additional evidence for the potential uses of miRNAs. The pilot clinical substudy now warrants further evaluation of the panel of miRNAs as a measure of tumor progression in larger patient cohorts.
Authors' contributions
GB devised the study and co‐wrote the manuscript. CD co‐designed and co‐supervised the project. AG co‐wrote the manuscript contributed to planning, sample collection, and analysis. MA developed and refined most the miRNA methodology as well as contributing to sample process and analysis. DGR assisted in study design and method development. DM, DB, and NS contributed to sample collection, logging, and processing. CLH and CM provided mouse samples and devised tail vein blood collection. JV, LC, FB, and KA collected clinical samples and oversaw ethical permission and patient consent for blood samples. All authors read and approved the final manuscript.
Conflicts of interest
No conflicts of interest.
Supporting information
The following is the supplementary material related to this article:
Supplementary data
Acknowledgments
We would like to acknowledge the help of the CR‐UK Molecular Biology Core Facilities and members of the Clinical and Experimental Pharmacology Group for their support of this study. We also thank the patients and healthy volunteers who provided their blood samples for this study. This work was supported by core funding to CR‐UK Manchester Institute (C5759/A20971).
Supplementary material 1.
1.1.
Supplementary material related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2015.10.004.
Greystoke Alastair, Ayub Mahmood, Rothwell Dominic G., Morris Dan, Burt Deborah, Hodgkinson Cassandra L., Morrow Christopher J., Smith Nigel, Aung Kyaw, Valle Juan, Carter Louise, Blackhall Fiona, Dive Caroline, Brady Dive, (2016), Development of a circulating miRNA assay to monitor tumor burden: From mouse to man, Molecular Oncology, 10, doi: 10.1016/j.molonc.2015.10.004.
References
- Ambros, V. , 2004. The functions of animal microRNAs. Nature. 431, 350–355. [DOI] [PubMed] [Google Scholar]
- Arroyo, J.D. , Chevillet, J.R. , Kroh, E.M. , 2011. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. U.S.A. 108, (12) 5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeri, M. , Verri, C. , Conte, D. , Roz, L. , Modena, P. , Facchinetti, F. , Calabro, E. , Croce, C.M. , Pastorino, U. , Sozzi, G. , 2011. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc. Natl. Acad. Sci. U.S.A. 108, 3713–3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzoli, R. , Buttitta, F. , Di Nicola, M. , Malatesta, S. , Marchetti, A. , Rom, W.N. , Pass, H.I. , 2013. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J. Thorac. Oncol. 8, 1156–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J. , Wang, W. , Zhang, Y. , Chen, Y. , Hu, T. , 2014. Predicting distant metastasis and chemoresistance using plasma miRNAs. Med. Oncol. 31, 799–013-0799-x. Epub 2013 Dec 6 [DOI] [PubMed] [Google Scholar]
- Chen, Y. , Zhang, L. , Hao, Q. , 2013. Candidate microRNA biomarkers in human epithelial ovarian cancer: systematic review profiling studies and experimental validation. Cancer Cell. Int. 13, 86–2867-13-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, H.H. , Mitchell, P.S. , Kroh, E.M. , 2013. Circulating microRNA profiling identifies a subset of metastatic prostate cancer patients with evidence of cancer-associated hypoxia. PLoS One. 8, e69239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, Y. , Xie, C.H. , Neis, J.P. , Fan, C.Y. , Vural, E. , Spring, P.M. , 2011. MicroRNA expression profiles of head and neck squamous cell carcinoma with docetaxel-induced multidrug resistance. Head Neck. 33, 786–791. [DOI] [PubMed] [Google Scholar]
- Devlin, C. , Greco, S. , Martelli, F. , Ivan, M. , 2011. miR-210: more than a silent player in hypoxia. IUBMB Life. 63, 94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duttagupta, R. , Jiang, R. , Gollub, J. , Getts, R.C. , Jones, K.W. , 2011. Impact of cellular miRNAs on circulating miRNA biomarker signatures. PLoS One. 6, e20769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferracin, M. , Pedriali, M. , Veronese, A. , 2011. MicroRNA profiling for the identification of cancers with unknown primary tissue-of-origin. J. Pathol. 225, 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorges, T.M. , Schiller, J. , Schmitz, A. , Schuetzmann, D. , Schatz, C. , Zollner, T.M. , Krahn, T. , von Ahsen, O. , 2012. Cancer therapy monitoring in xenografts by quantitative analysis of circulating tumor DNA. Biomarkers. 17, 498–506. [DOI] [PubMed] [Google Scholar]
- Greystoke, A. , Cummings, J. , Ward, T. , 2008. Optimisation of circulating biomarkers of cell death for routine clinical use. Ann. Oncol. 19, (5) 990–995. [DOI] [PubMed] [Google Scholar]
- Heneghan, H.M. , Miller, N. , Kelly, R. , Newell, J. , Kerin, M.J. , 2010. Systemic miRNA-195 differentiates breast cancer from other malignancies and is a potential biomarker for detecting noninvasive and early stage disease. Oncologist. 15, 673–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, L. , Browne, G. , Tulchinsky, E. , 2013. ZEB/miR-200 feedback loop: at the crossroads of signal transduction in cancer. Int. J. Cancer. 132, 745–754. [DOI] [PubMed] [Google Scholar]
- Hodgkinson, C.L. , Morrow, C.J. , Li, Y. , 2014. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat. Med. 20, 897–903. [DOI] [PubMed] [Google Scholar]
- Huang, Z. , Huang, S. , Wang, Q. , Liang, L. , Ni, S. , Wang, L. , Sheng, W. , He, X. , Du, X. , 2011. MicroRNA-95 promotes cell proliferation and targets sorting Nexin 1 in human colorectal carcinoma. Cancer Res. 71, 2582–2589. [DOI] [PubMed] [Google Scholar]
- Korpal, M. , Ell, B.J. , Buffa, F.M. , 2011. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat. Med. 17, 1101–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaConti, J.J. , Shivapurkar, N. , Preet, A. , Deslattes Mays, A. , Peran, I. , Kim, S.E. , Marshall, J.L. , Riegel, A.T. , Wellstein, A. , 2011. Tissue and serum microRNAs in the Kras(G12D) transgenic animal model and in patients with pancreatic cancer. PLoS One. 6, e20687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidner, R.S. , Li, L. , Thompson, C.L. , 2013. Dampening enthusiasm for circulating microRNA in breast cancer. PLoS One. 8, e57841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, A. , Omura, N. , Hong, S.M. , Vincent, A. , Walter, K. , Griffith, M. , Borges, M. , Goggins, M. , 2010. Pancreatic cancers epigenetically silence SIP1 and hypomethylate and overexpress miR-200a/200b in association with elevated circulating miR-200a and miR-200b levels. Cancer Res. 70, 5226–5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W.G. , Yuan, Y.Z. , Qiao, M.M. , Zhang, Y.P. , 2012. High dose glargine alters the expression profiles of microRNAs in pancreatic cancer cells. World J. Gastroenterol. 18, 2630–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Jiang, Q. , Xia, N. , Yang, H. , Hu, C. , 2012. Decreased expression of microRNA-375 in nonsmall cell lung cancer and its clinical significance. J. Int. Med. Res. 40, 1662–1669. [DOI] [PubMed] [Google Scholar]
- Madhavan, D. , Zucknick, M. , Wallwiener, M. , 2012. Circulating miRNAs as surrogate markers for circulating tumor cells and prognostic markers in metastatic breast cancer. Clin. Cancer Res. 18, 5972–5982. [DOI] [PubMed] [Google Scholar]
- Mahn, R. , Heukamp, L.C. , Rogenhofer, S. , von Ruecker, A. , Muller, S.C. , Ellinger, J. , 2011. Circulating microRNAs (miRNA) in serum of patients with prostate cancer. Urology. 77, 1265.e9–1265.e16. [DOI] [PubMed] [Google Scholar]
- Meunier, J. , Lemoine, F. , Soumillon, M. , Liechti, A. , Weier, M. , Guschanski, K. , Hu, H. , Khaitovich, P. , Kaessmann, H. , 2013. Birth and expression evolution of mammalian microRNA genes. Genome Res. 23, 34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miko, E. , Czimmerer, Z. , Csanky, E. , Boros, G. , Buslig, J. , Dezso, B. , Scholtz, B. , 2009. Differentially expressed microRNAs in small cell lung cancer. Exp. Lung Res. 35, 646–664. [DOI] [PubMed] [Google Scholar]
- Mitchell, P.S. , Parkin, R.K. , Kroh, E.M. , 2008. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U.S.A. 105, 10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, C.C. , Kroh, E. , Wood, B. , Arroyo, J.D. , Dougherty, K.J. , Miyaji, M.M. , Tait, J.F. , Tewari, M. , 2012. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev. Res. (Phila). 5, 492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, C. , Kasimir-Bauer, S. , Pantel, K. , Schwarzenbach, H. , 2011. Screening for circulating nucleic acids and caspase activity in the peripheral blood as potential diagnostic tools in lung cancer. Mol. Oncol. 5, 281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzenbach, H. , Nishida, N. , Calin, G.A. , Pantel, K. , 2014. Clinical relevance of circulating cell-free microRNAs in cancer. Nat. Rev. Clin. Oncol. 11, (3) 145–156. [DOI] [PubMed] [Google Scholar]
- Selth, L.A. , Townley, S. , Gillis, J.L. , Ochnik, A.M. , Murti, K. , Macfarlane, R.J. , Chi, K.N. , Marshall, V.R. , Tilley, W.D. , Butler, L.M. , 2012. Discovery of circulating microRNAs associated with human prostate cancer using a mouse model of disease. Int. J. Cancer. 131, 652–661. [DOI] [PubMed] [Google Scholar]
- Siow, M. , Karen Ng, L. , Vincent Chong, V. , Jamaludin, M. , Abraham, M. , Abdul Rahman, Z. , Kallarakkal, T. , Yang, Y.H. , Cheong, S. , Zain, R. , 2013. Dysregulation of miR-31 and miR-375 expression is associated with clinical outcomes in oral carcinoma. Oral Dis. 20, (4) 345–351. [DOI] [PubMed] [Google Scholar]
- Sozzi, G. , Boeri, M. , Rossi, M. , 2014. Clinical utility of a plasma-based miRNA signature classifier within computed tomography lung cancer screening: a correlative mild trial study. J. Clin. Oncol. 32, (8) 768–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratz, C. , Nuhrenberg, T.G. , Binder, H. , Valina, C.M. , Trenk, D. , Hochholzer, W. , Neumann, F.J. , Fiebich, B.L. , 2012. Micro-array profiling exhibits remarkable intra-individual stability of human platelet micro-RNA. Thromb. Haemost. 107, 634–641. [DOI] [PubMed] [Google Scholar]
- Svoboda, M. , Izakovicova Holla, L. , Sefr, R. , Vrtkova, I. , Kocakova, I. , Tichy, B. , Dvorak, J. , 2008. Micro-RNAs miR125b and miR137 are frequently upregulated in response to capecitabine chemoradiotherapy of rectal cancer. Int. J. Oncol. 33, 541–547. [PubMed] [Google Scholar]
- Toiyama, Y. , Hur, K. , Tanaka, K. , Inoue, Y. , Kusunoki, M. , Boland, C.R. , Goel, A. , 2014. Serum miR-200c is a novel prognostic and metastasis-predictive biomarker in patients with colorectal cancer. Ann. Surg. 259, (4) 735–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujifuku, K. , Mitsutake, N. , Takakura, S. , 2010. miR-195, miR-455-3p and miR-10a( *) are implicated in acquired temozolomide resistance in glioblastoma multiforme cells. Cancer Lett. 296, 241–248. [DOI] [PubMed] [Google Scholar]
- Wang, K. , Yuan, Y. , Cho, J.H. , McClarty, S. , Baxter, D. , Galas, D.J. , 2012. Comparing the microRNA spectrum between serum and plasma. PLoS One. 7, e41561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters, P.S. , McDermott, A.M. , Wall, D. , Heneghan, H.M. , Miller, N. , Newell, J. , Kerin, M.J. , Dwyer, R.M. , 2012. Relationship between circulating and tissue microRNAs in a murine model of breast cancer. PLoS One. 7, e50459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, Z. , Ben-Dov, I.Z. , Elias, R. , Mihailovic, A. , Brown, M. , Rosenwaks, Z. , Tuschl, T. , 2013. Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proc. Natl. Acad. Sci. U.S.A. 110, (11) 4255–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X. , Ajani, J.A. , Gu, J. , Chang, D.W. , Tan, W. , Hildebrandt, M.A. , Huang, M. , Wang, K.K. , Hawk, E. , 2013. MicroRNA expression signatures during malignant progression from Barrett's esophagus to esophageal adenocarcinoma. Cancer Prev. Res. (Phila). 6, 196–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H. , Jiang, L. , Sun, C. , Li Guo, L. , Lin, M. , Huang, J. , Zhu, L. , 2014. Decreased circulating miR-375: a potential biomarker for patients with non-small-cell lung cancer. Gene. 534, 60–65. [PubMed] [Google Scholar]
- Zhang, H.L. , Ruan, L. , Zheng, L.M. , Whyte, D. , Tzeng, C.M. , Zhou, X.W. , 2012. Association between class III beta-tubulin expression and response to paclitaxel/vinorebine-based chemotherapy for non-small cell lung cancer: a meta-analysis. Lung Cancer. 77, 9–15. [DOI] [PubMed] [Google Scholar]
- Zhu, W. , Xu, H. , Zhu, D. , Zhi, H. , Wang, T. , Wang, J. , Jiang, B. , Shu, Y. , Liu, P. , 2012. miR-200bc/429 cluster modulates multidrug resistance of human cancer cell lines by targeting BCL2 and XIAP. Cancer Chemother. Pharmacol. 69, 723–731. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following is the supplementary material related to this article:
Supplementary data


