Abstract
Mucopolysaccharidosis VI (MPS VI) is an autosomal recessive inborn error of metabolism caused by mutations in the arylsulfatase B gene (ARSB) and consequent deficient activity of ARSB, a lysosomal enzyme. We present here the results of a study undertaken to identify the mutations in ARSB in MPS VI patients in India. Around 160 ARSB mutations, of which just 4 are from India, have been reported in the literature. Our study covered nine MPS VI patients from eight families. Both familial mutations were found in seven families, and only one mutation was found in one family. Seven mutations were found — four novel (p.G38_G40del3, p.C91R, p.L98R and p.R315P), two previously reported from India (p.D53N and p.W450C), and one reported from outside India (p.R160Q). One mutation, p.W450C, was present in two families, and the other six mutations were present in one family each. Analysis of the molecular structure of the enzyme revealed that most of these mutations either cause loss of an active site residue or destabilize the structure of the enzyme. The only previous study on mutations in ARSB in Indian MPS VI patients, by Kantaputra et al. 2014 [1], reported four novel mutations of which two (p.D53N and p.W450C) were found in our study as well. Till date, nine mutations have been reported from India, through our study and the Kantaputra study. Eight out of these nine mutations have been found only in India. This suggests that the population studied by us might have its own typical set of mutations, with other populations equally likely to have their own set of mutations.
Abbreviations: ARSB, arylsulfatase B; ERT, enzyme replacement therapy; GALNS, N-acetyl galactosamine 6-sulfatase; GAG, glycosaminoglycan; HSCT, hematopoietic stem cell transplantation; HGMD, Human Gene Mutation Database; LSD, lysosomal storage disorder; MPS, mucopolysaccharidosis; PCT, pharmacological chaperone therapy; VUS, variants of unknown significance
Keywords: Mucopolysaccharidosis VI (MPS VI), Maroteaux–Lamy syndrome, Arylsulfatase B (ARSB), Inborn error of metabolism (IEM), Lysosomal storage disorder (LSD), Lysosomal enzyme, Mutations, India, Active site
1. Introduction
Mucopolysaccharidosis VI (MPS VI, OMIM: 253200), first identified by Maroteaux and Lamy in 1963, is a rare, autosomal recessive lysosomal storage disorder (LSD) [2]. Estimate of incidence has been determined in only a few countries and it ranges from 0 in Northern Ireland [3] and 0.04:1,00,000 live births in Poland [4], to 8:1,00,000 live births in the Eastern Province in Saudi Arabia [5]. Prevalence, where estimated, ranges from 0 in Northern Ireland [3] to 1:5000 in Monte Santo county in Northeast Brazil [6]. Incidence and prevalence in India are yet to be estimated.
MPS VI is caused by mutations in the Arylsulfatase B gene (ARSB). The gene codes for the ARSB enzyme, also called N-acetylgalactosamine-4-sulfatase (EC: 3.1.6.12), a member of the human sulfatase family. The enzyme catalyzes the desulfation of C4-linked sulfate esters, a step in the intralysosomal degradation of N-acetyl galactosamine residues in the glycosaminoglycans (GAGs) dermatan sulfate and chondroitin sulfate. Deficient activity of the enzyme leads to accumulation of the GAGs in lysosomes and causes lysosomal dysfunction, and the disease. Dermatan sulfate is present in high amounts in connective tissue, and its accumulation there may explain the bone and joint deformities found in MPS VI patients [7].
The characteristic clinical symptoms of MPS VI include short stature, coarse facial features, dysostosis multiplex, joint stiffness, respiratory and cardiac abnormalities, enlarged liver and spleen, and clouding of cornea, varying from patient to patient. Patients usually have normal intelligence. Specific treatment options available for this disorder are allogeneic Hematopoietic Stem Cell Transplantation (HSCT) [8] and Enzyme Replacement Therapy (ERT) [9].
The ARSB gene is located on chromosome 5 (q11–q13) [10]. It has eight exons. The precursor of the ARSB enzyme has 533 amino acid residues, 36 of which constitute the signal peptide. Study of the crystal structure (PDB code: 1FSU) of the folded mature form of ARSB (MW 66 kDa) [11] reveals that the enzyme is a glycoprotein with two domains. The N-terminal domain (Domain 1) belongs to the α/β class and houses the active site. The C-terminal domain (Domain 2), whose function is not known, consists of a β sheet made of four antiparallel β strands and an α helix orthogonal to the sheet [11]. The active site pocket, located at the base of a cleft in Domain 1, has 10 evolutionarily conserved amino acid residues. The cysteine 91 in the active site undergoes conversion to an aldehyde, 3-oxoalanine (2-amino-3-oxopropanoic acid), also called formylglycine. This post-translational modification is vital for the activity of the enzyme [12]. A Ca2 + ion, present in the active site, acts as the cofactor and coordinates with seven atoms from the side chain groups of as many active site residues, and two oxygen atoms from the sulfate derivative of formylglycine [11].
Missense mutations form the largest group among the more than 160 mutations in the ARSB gene reported worldwide. Small insertions, splice site mutations, small and gross deletions and frameshift mutations comprise the remainder, according to the Human Gene Mutation Database (HGMD) [13]. The allelic heterogeneity observed in ARSB is high, which might, in part, explain the variable expressivity of MPS VI with respect to age of onset, rate of progression and clinical phenotypes [14]. Most mutations are “private”. Some mutations are common, and a few of them have been attributed to founder effect [1], [15], [16], [17], [18], [19], [20], [21], [22], [23] ([20] cited from Abstract). Study of influence of missense mutations on the overall structure and folding of ARSB helps to predict disease severity [24], [25]. It might also help in guiding the choice of personalized therapies for individual patients, when more therapies become available.
The present study was initiated to identify mutations in ARSB in MPS VI patients in India. Prior to this study, around 160 mutations in ARSB have been reported, of which just four were from Indian patients [1]. Our study has led to the identification of seven mutations, including four novel ones. We also carried out a computational study of the impact of the mutations on the structure of the protein. The mutations seem to affect the structure and function of the enzyme through various mechanisms.
2. Methods
2.1. Approval from bioethics committees
Approval for conduct of this study was obtained from the Bioethics Committees of CHG, Bangalore, India and FCRF, Chennai, India. Informed consent was obtained from participants or from their parents or legal guardians.
2.2. Patients and control subjects
The study covered nine patients (P1–P3 and P5–P10) with MPS VI from eight families (F1–F3 and F5–F9), together with their parents and control subjects. Samples from patients and their parents were collected through two clinical centers in South India, in Chennai and Bangalore, over a period of 2 years and 2 months. Patients were identified as having MPS VI, through assay of activity of ARSB, either during this period or earlier. Clinical phenotypes of patients were recorded. Control subjects, not born of consanguineous parents, were chosen from among individuals with ethnic backgrounds similar to those of patients with the respective novel mutations. Demographic data, family medical history, and blood samples were collected from participants.
2.3. Mutation analysis
The term “mutation” is used in this study to describe a disease-causing genetic variation. A “novel” mutation or Variant of Unknown Significance (VUS) refers to a mutation not reported previously. The literature, and databases like HGMD (basic), ClinVar, ENSEMBL, UNIPROT and dbSNP were checked for previously reported mutations or VUS to determine whether a mutation is novel.
Genomic DNA was isolated from blood samples of study participants. The eight exons and exon–intron boundaries of ARSB were amplified from DNA samples from patients (except in the case of P9), with primers designed manually or by using Primer3Plus [26]. Mutation in one patient (P9, deceased), from whom sample was not available, was inferred from mutation analysis of DNA samples from his parents. PCR products were subjected to Sanger sequencing. Mutations were confirmed by bi-directional sequencing of products of independent PCR reactions. Samples from parents were analyzed for the presence of the mutations found in patients. To ascertain the pathogenicity of putative novel mutations, we analyzed samples from at least 50 control subjects (i.e., 100 chromosomes).
Differences between nucleotide sequences as observed in our results and the reference sequence of human ARSB (GenBank Accession number: NG_007089.1) were documented. Variants in the sequence of the gene and predicted sequence of the protein were named according to the guidelines of the Human Genome Variation Society, using human ARSB mRNA and protein sequences as references (GenBank Accession numbers: NM_000046.3 and NP_000037.2, respectively).
2.4. Bioinformatics studies
Structural and bioinformatics studies were undertaken to ascertain the effects of mutations found through this study. The bioinformatics tools employed were: MutPred [27], Polymorphism Phenotyping v2 (PolyPhen-2) [28], Sorting Tolerant from Intolerant (SIFT) [29], and Protein Variation Effect Analyzer (PROVEAN) [30].
Multiple sequence alignment of ARSB in a few vertebrate species was carried out using ClustalW server and plotted using ESPript (Easy Sequencing of Postscript) 3.0 (http://espript.ibcp.fr) [31]. The species and GenBank accession numbers of the ARSB sequences used are: Homo sapiens — NP_000037.2, Canis lupus — NP_001041598.1, Mus musculus — P50429.3, Anolis carolinensis — XP_008101121.1 (predicted), Gallus gallus — XP_003642960.1 (predicted), Xenopus tropicalis — XP_002940244.2 (predicted), and Danio rerio — XP_003200848.2 (predicted). Information on the secondary structure of ARSB was inferred from the PDB coordinates (code 1FSU). Hydropathy bar was calculated using Kyte and Doolittle method with a 3-residue window averaging [32].
Molecular models of ARSB (based on the crystal structure of wild type ARSB) incorporating the mutations identified were constructed using molecular graphics software suite COOT [33] ensuring that the side chain rotamers did not create any steric hindrance. Superposition of the molecular structures of the wild type and the mutants, and the intramolecular interactions at the sites of mutations were analyzed using the program PyMol (The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrodinger, LLC).
3. Results
3.1. Patients
The study covered nine MPS VI patients from eight families. One patient (P2) comes from Assam and the rest are from Andhra Pradesh, Karnataka and Tamil Nadu in South India. Five patients are female, and four male. Patients in 6/8 families (75%) were born to consanguineous parents. The present age of the eight patients alive ranges from 2 to 19 years.
3.2. Clinical phenotypes of patients
Patients had received supportive care and general treatment (such as surgery), and not ERT or HSCT. The presence of extensive Mongolian patches was the only symptom noticed in P7. All other patients had developed the standard clinical symptoms of MPS VI (Table S1) and had short stature, coarse facies, dolichocephaly, hirsutism, corneal clouding, pectus carinatum and brachydactyly. Age of onset of clinical symptoms ranged from 5 months to 5 years. Age at diagnosis ranged from 1 year to 14 years. Time from onset to diagnosis (excluding the preclinical patient P7) ranged from 7 months to 11 years. Disease severity was categorized qualitatively as Attenuated, Moderate or Severe. The following parameters were taken into account to rate disease severity: cardiac and respiratory involvement, ambulation or need for cervical surgery, number of surgeries, disease in joints, and the overall quality of life. Apart from Patient P7, who had not yet developed clinical symptoms, disease severity in patients ranged from Attenuated to Severe (Table S1 and Table 1). One patient, P9, had the severe form of the disease. Cardiovascular symptoms were not noticed in two patients, P8 and P10.
Table 1.
Mutations in ARSB found in patients, through this study. Mutations, demographic particulars and parental consanguinity of patients are listed. Patients P6 & P7 belong to the same family (F6). Mutations shown in bold font are novel ones. Mutation in the second allele was not found in P1.
| Family ID | Patient ID | State (language) |
Parental consanguinity (degree) |
Present Age (years) |
Mutation |
Disease severity in patient | |||
|---|---|---|---|---|---|---|---|---|---|
| Nucleotide change | Predicted amino acid change | Homozygous/heterozygous | Original reference | ||||||
| F1 | P1 | Tamil Nadu (Tamil/Telugu) |
No | 19 | c.1348G > C/? | p.W450C/? | Heterozygous | [1] | Moderate |
| F2 | P2 | Assam (Hindi) |
No | 7 | c.293 T > G | p.L98R | Homozygous | This study | Attenuated |
| F3 | P3 | Tamil Nadu (Tamil) |
Yes (4°) | 19 | c.1348G > C | p.W450C | Homozygous | [1] | Attenuated |
| F5 | P5 | Andhra Pradesh (Telugu) | Yes (2°) | 6 | c.157G > A | p.D53N | Homozygous | [1] | Moderate |
| F6 | P6 | Karnataka (Kannada) |
Yes (5°) | 4 | c.944G > C | p.R315P | Homozygous | This study | Moderate |
| F6 | P7 | Karnataka (Kannada) |
Yes (5°) | 2 | c.944G > C | p.R315P | Homozygous | This study | NA |
| F7 | P8 | Karnataka (Kannada) |
Yes (3°) | 16 | c.479G > A | p.R160Q | Homozygous | [34] | Moderate |
| F8 | P9 | Andhra Pradesh (Telugu) |
Yes (3°) | Deceased | c.271 T > Ca | p.C91Ra | Homozygousa | This study | Severe |
| F9 | P10 | Tamil Nadu (Tamil) |
Yes (3°) | 7 | c.113_121del9 | p.G38_G40del3 | Homozygous | This study | Attenuated |
NA = Not applicable. Patient P7, who is homozygous for her familial ARSB mutation, has not yet developed clinical symptoms of the disease. Patient P4 (F4) was not a subject of this study. Refer to Section 3.2 for the parameters used to rate disease severity.
Mutation inferred from genotypes of parents.
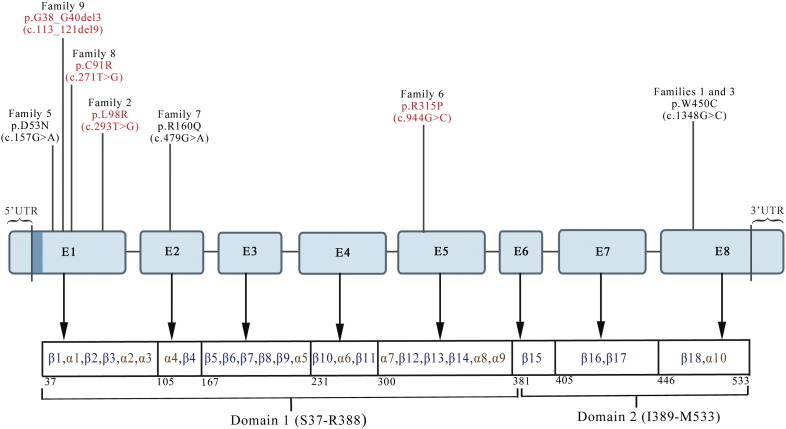
A total of seven mutations were found: p.G38_G40del3, p.D53N, p.C91R, p.L98R, p.R160Q p.R315P and p.W450C. Four mutations were located in Exon 1, and one mutation each in Exons 2, 5 and 8 of ARSB (Fig. 1). Six mutations were missense, and one was a small deletion. Four of the mutations are novel: p.G38_G40del3, p.C91R, p.L98R and p.R315P. These were absent in control individuals studied (100 individuals for p.G38_G40del3, p.C91R and p.L98R; and 55 individuals for p.R315P) and in the results of the 1000 Genomes Project. Among the other mutations found by us, two (p.D53N and p.W450C) were previously reported in patients from India [1] and one (p.R160Q) from outside India [34]. As expected, the p.D53N mutation was absent in the 100 control individuals whose samples were analyzed for the coding part of Exon 1.
Fig. 1.
Schematic representation of mutations in ARSB identified through this study. Rectangles labeled E1–E8 represent exons, with mutations identified marked above them, novel ones in red, known ones in black. Exon-wise locations of Domains 1 and 2 and the secondary structures in the protein are also shown. The region encoding the signal peptide (aa 1–37), located in Exon 1, is shown as a dark blue stripe to the right of 5′ UTR. Figure not to scale.
The p.W450C mutation was found in 2/8 families, and all other mutations in one family each. Patients born to consanguineous parents were homozygous for mutations in ARSB, as expected. One patient (P2) had a homozygous mutation, even though her parents were not known to be consanguineous. Thus, patients in 7/8 families (88%) had homozygous mutations.
Several VUS and two non-synonymous polymorphisms (p.V358M and p.V376M) [35], [36] were found in patients and controls (Table S3). The p.V358M polymorphism has an adverse effect on the activity of wild type ARSB and mutant ARSB, whereas p.V376M does not [37]. Information on which individual patient has which of the two polymorphisms is given in Table S2. In controls, p.V358M was present in 25/110 alleles analyzed (23%) and p.V376M in 16/110 alleles analyzed (15%).
It may be noted that in parents and controls, unlike in patients, only certain parts of the coding region were sequenced. In parents, only those regions were sequenced in which mutations were found in their children. In controls, only Exon 5 (in 55 individuals) and the coding part of Exon 1 (in 100 individuals) were sequenced, as these are the regions where the novel mutations found in patients are present.
The novel missense mutations are likely to be pathogenic, going by the predictions made by bioinformatics tools used, viz., MutPred, PolyPhen-2, SIFT and PROVEAN. The predicted effects of the mutations on the structure and function of the protein are shown in Table 2. Previously reported mutations, included as positive controls, were also predicted to be pathogenic. Non-synonymous polymorphisms p.V358M and p.V376M, included as negative controls, were predicted to be non-pathogenic by all tools except PolyPhen-2.
Table 2.
Effect of the missense mutations and non-synonymous polymorphisms on the structure and function of ARSB, as predicted by use of bioinformatics tools. Novel mutations are in bold font. Non-synonymous polymorphisms, used as negative controls, are italicized. Scores in bold indicate non-deleterious. All the mutations are likely to be pathogenic, based on predictions by all the tools used. The non-synonymous polymorphisms are not likely to be so, based on predictions by all tools except PolyPhen-2. In MutPred, probability > 0.75 is considered a “very confident hypothesis” for a mutation to be causative of disease. In PolyPhen-2, the scores for neutral to damaging range from 0 to 1, with 1 being “probably damaging”. According to SIFT, a mutation is “damaging” if the score is ≤ 0.05 and tolerated if the score is > 0.05. In PROVEAN, an amino acid substitution with a score < − 2.5 is deleterious.
| Mutation/ polymorphism |
Scores obtained through use of bioinformatics tools |
|||
|---|---|---|---|---|
| MutPred | PolyPhen-2 | SIFT | PROVEAN | |
| p.D53N | 0.914 | 1.000 | 0.00 | − 4.413 |
| p.C91R | 0.927 | 1.000 | 0.00 | − 10.563 |
| p.L98R | 0.900 | 1.000 | 0.00 | − 5.193 |
| p.R160Q | 0.883 | 1.000 | 0.05 | − 3.981 |
| p.R315P | 0.790 | 1.000 | 0.00 | − 6.633 |
| p.W450C | 0.798 | 1.000 | 0.15 | − 10.605 |
| p.V358M | 0.148 | 0.997 | 0.06 | − 2.178 |
| p.V376M | 0.213 | 0.831 | 0.37 | − 1.159 |
3.3. Identification of mutations
Samples from patients other than P9 and from parents of the deceased patient P9 (from whom sample was not available), were analyzed to identify mutations. A genetic variant is taken to be a mutation if most or all of the following criteria are satisfied: (1) it has been previously reported as a mutation; (2) it is present in the homozygous state, or in the compound heterozygous state with another disease-causing mutation in a patient; (3) it is absent in the homozygous state in parents; (4) it is likely to adversely affect the structure or function of the protein; (5) if the site of the amino acid substitution is highly conserved across vertebrate orthologues; and (6) it is absent in at least 50 control subjects of matched ethnicity (i.e., 100 chromosomes).
Mutations in both ARSB alleles were identified in each of seven families, whereas only one heterozygous mutation was identified in Patient 1 (Family 1). The mutations found are represented schematically in Fig. 1. Detailed information on mutations found, and data on parental consanguinity and ethnicity of patients is provided in Table 1, and chromatograms in Fig. S1. Parents of all patients except P1 were heterozygous for their respective familial mutations in ARSB (Fig. S2). Mother of P1 was heterozygous for the mutation identified in P1.
A total of seven mutations were found: p.G38_G40del3, p.D53N, p.C91R, p.L98R, p.R160Q p.R315P and p.W450C. Four mutations were located in Exon 1, and one mutation each in Exons 2, 5 and 8 of ARSB (Fig. 1). Six mutations were missense, and one was a small deletion. Four of the mutations are novel: p.G38_G40del3, p.C91R, p.L98R and p.R315P. These were absent in control individuals studied (100 individuals for p.G38_G40del3, p.C91R and p.L98R; and 55 individuals for p.R315P) and in the results of the 1000 Genomes Project. Among the other mutations found by us, two (p.D53N and p.W450C) were previously reported in patients from India [1] and one (p.R160Q) from outside India [34]. As expected, the p.D53N mutation was absent in the 100 control individuals whose samples were analyzed for the coding part of Exon 1.
The p.W450C mutation was found in 2/8 families, and all other mutations in one family each. Patients born to consanguineous parents were homozygous for mutations in ARSB, as expected. One patient (P2) had a homozygous mutation, even though her parents were not known to be consanguineous. Thus, patients in 7/8 families (88%) had homozygous mutations.
Several VUS and two non-synonymous polymorphisms (p.V358M and p.V376M) [35], [36] were found in patients and controls (Table S3). The p.V358M polymorphism has an adverse effect on the activity of wild type ARSB and mutant ARSB, whereas p.V376M does not [37]. Information on which individual patient has which of the two polymorphisms is given in Table S2. In controls, p.V358M was present in 25/110 alleles analyzed (23%) and p.V376M in 16/110 alleles analyzed (15%).
It may be noted that in parents and controls, unlike in patients, only certain parts of the coding region were sequenced. In parents, only those regions were sequenced in which mutations were found in their children. In controls, only Exon 5 (in 55 individuals) and the coding part of Exon 1 (in 100 individuals) were sequenced, as these are the regions where the novel mutations found in patients are present.
The novel missense mutations are likely to be pathogenic, going by the predictions made by bioinformatics tools used, viz., MutPred, PolyPhen-2, SIFT and PROVEAN. The predicted effects of the mutations on the structure and function of the protein are shown in Table 2. Previously reported mutations, included as positive controls, were also predicted to be pathogenic. Non-synonymous polymorphisms p.V358M and p.V376M, included as negative controls, were predicted to be non-pathogenic by all tools except PolyPhen-2.
3.4. Evolutionary conservation of sites of mutations
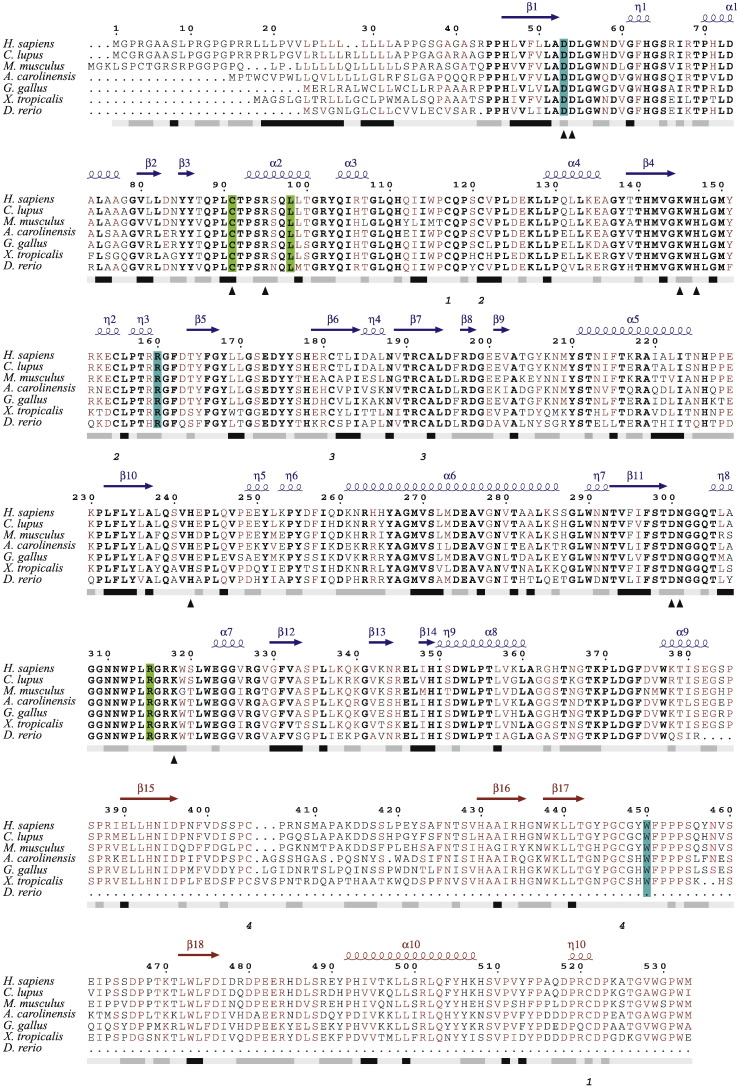
Multiple sequence alignment of ARSB in the vertebrate species selected for study shows strict conservation of active site residues (Fig. 2). It is evident from the figure that differences among the sequences are prominent in the signal peptide region and are present to a lower extent between β15 and β16 in Domain 2. It could be seen that with the exception of W450, amino acid residues at which missense mutations were found in this cohort of patients are conserved across all organisms queried. W450 was present in all these organisms except D. rerio, which lacks Domain 2. One novel mutation, p.L98R, is in an α-helical region (α2). None of the other mutations occurs at the secondary structure regions.
Fig. 2.
Multiple sequence alignment of ARSB. Multiple sequence alignment of ARSB of a few vertebrate species including the human. Bold font represents strictly conserved amino acid residues, sites with sequence identities of 70% or more are in maroon. The bar below the sequence block represents the hydropathy plot. Hydrophobic regions are marked in black, hydrophilic ones in gray & the intermediate regions in faint gray. Cysteine residues forming disulfide bridges are shown below the hydropathy bar. Mutations identified through this study are highlighted in green (novel mutations) or cyan (previously reported). The secondary structure depicted on top of the alignment blocks are inferred from the crystal structure of human ARSB (PDB code: 1FSU). Blue and maroon in the secondary structure cartoon denote the domains 1 and 2, respectively.
3.5. Structural effects of the mutations
Molecular models of individual mutants were superposed on to the crystal structure of native ARSB [11] (PDB code: 1FSU) using the software PyMol. The intramolecular interactions of the native residues were analyzed to decipher the impact of the mutations on the structure of the protein (Fig. 3). The mutations found by us were not confined to any particular region in the molecular structure of ARSB. In this respect, the set of mutations found by us appear to be fairly representative of mutations in ARSB found across the global population. Six mutations found through this study are in Domain 1 (Figs. 1, 3). Two of these mutations (p.D53N and p.C91R) are substitutions in active site residues, and interestingly, both have been found only from India so far. One mutation is in Domain 2. Predicted effects of the mutations are discussed below.
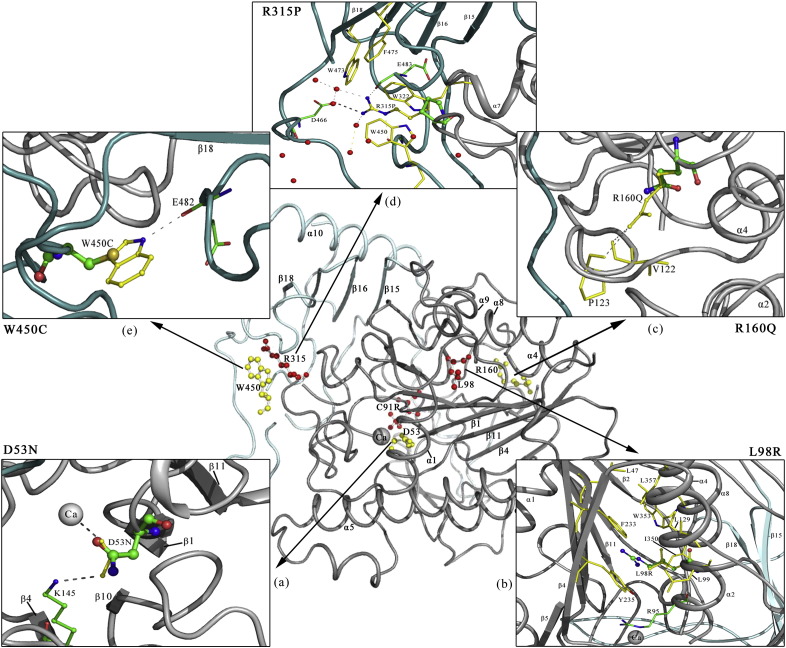
Fig. 3.
Three-dimensional structure of ARSB showing the localization of missense mutations identified in this study (represented with ball and stick model). Calcium ion bound at the active site of ARSB is shown as a gray sphere marked 'Ca'. Novel mutations are shown in red and previously reported ones in yellow ball-and-stick representations. Zoomed in images show the localized interactions of a) p.D53N, b) p.L98R, c) p.R160Q, d) p.R315P, and e) p.W450C. The orientations of the zoomed in structures may not be the same as in the total molecule, and a few residues might have been omitted in them for clarity.
3.5.1. p.G38_G40del3
This novel in-frame deletion spanning four codons is amidst runs of the nucleotides G and C. The deletion would cause loss of three amino acid residues, 38–40, which are next to the first residue of the mature enzyme (Fig. S3). This is not expected to have a strong structural impact. Also, post-translational modifications are not known to occur in this region. In some precursor proteins, the region beyond the signal peptide, a part of the mature protein, is important for the cleavage of the signal peptide [38], [39]. If this is true of ARSB, the p.G38_G40del3 mutation would adversely influence the maturation of the enzyme. Alternatively, the mutation may exert its effect at the level of mRNA. Disease severity was “attenuated” in patient P10, who is homozygous for this mutation.
3.5.2. p.D53N
This active site mutation has been reported previously in an Indian patient [1]. Structural analysis reveals (Fig. 3a) that D53 forms a salt bridge with K145, while the carboxyl oxygen atom interacts with the Ca2 + ion present at the active site [11]. Mutations at the active site residues are expected to result in loss of activity of the enzyme. Due to the presence of a carbonyl group and an amide group in the asparagine residue, the p.D53N mutation would not affect Ca2 + ion coordination at the active site. The hydrogen bonding interaction with K145 would also be retained, though it would not be as strong as the salt bridge in the native structure. The overall geometry of the active site may not change drastically. The only noticeable impact would be the loss of a charged residue at the active site. Disease severity was “moderate” in Patient P5, who is homozygous for this mutation.
3.5.3. p.C91R
This novel mutation is at the cysteine residue of ARSB that is highly conserved in the active sites of sulfatases from bacteria to humans [40]. Sulfatases can become active as a catalyst only if this cysteine residue is converted to formylglycine, by formylglycine-generating enzyme [41], [42]. This post-translational modification cannot occur in ARSB if C91 is mutated into an arginine residue. In addition, this mutation might cause other impairments, such as defective intracellular trafficking of ARSB caused by p.C91T, an artificially induced mutation at the same residue [43]. Two artificially induced substitutions at C91 – p.C91S and p.C91T – resulted in loss of enzyme activity [44]. A different mutation, p.C91Y, has been reported at C91 in an MPS VI patient, in the compound heterozygous state [4].
Patient P9 had the severe form of the disease, not surprising considering the drastic biochemical effect of this mutation. This patient expired at 10 years of age. His younger brother, who also had MPS VI but was not a subject of this study, expired at two years of age. Prenatal diagnosis and genetic counseling, before the birth of the second child, would have been helpful in this case.
3.5.4. p.L98R
In this novel mutation, a neutral, relatively small nonpolar amino acid is replaced by a basic residue with a bulkier side chain group. This substitution would cause steric hindrance at the hydrophobic core of the protein. L98 is located at the helical segment (α2) of the N-terminal domain (Fig. 3b) and it has hydrophobic interactions with the adjacent β sheet through β1 and β14. Its mutation into a hydrophilic residue would destabilize the fold of the protein. Also, L98 lies adjacent to the active site residue R95. The presence of two basic residues could also disrupt the conformation of the active site. Two other mutations have been reported at L98: p.L98Q and p.L98P. Patients with these substitutions had severe clinical phenotype and rapidly progressing disease, respectively [15], [45] (cited from [46]). The vulnerability of ARSB to substitutions in L98, as in p.L98Q and p.L98P, also supports the pathogenic nature of the p.L98R mutation found through this study. Disease severity was “attenuated” in the patient P2, who is homozygous for the p.L98R mutation.
3.5.5. p.R160Q
This mutation was found in the homozygous state in our study, in patient P8. This mutation was previously reported from four unrelated patients, two from Belarus and one each from Russia and Spain, in all of whom it was in the compound heterozygous state [4], [18], [23], [34]. The R160Q substitution results from a transition in a CpG dinucleotide [46]. The mechanism of loss of function of the enzyme due to this mutation is not readily evident from the structural perspective, as described previously [46]. No significant impact is expected, apart from the loss of a charged residue on the surface of the enzyme (Fig. 3c). Disease severity was “moderate” in P8, who is homozygous for the p.R160Q mutation.
3.5.6. p.R315P
The fourth novel mutation was found in R315. As inferred from the crystal structure, R315 forms a salt bridge with D466 and has hydrogen bonding interactions with the main chain carbonyl groups of G448 and E483, stabilizing the inter-domain conformation of the protein. Substitution of R315 by a proline residue would result in a loss of the above interactions, and hence affect inter-domain stability. The guanidium group of R315 is exposed to the solvent surface (Fig. 3d) while the rest of the groups in the side chain are shielded by three tryptophan residues (W322, W450 and W472) and a phenylalanine (F475) residue. The p.R315P mutation would increase the hydrophobicity of the surface exposed to the solvent. It could thus have an adverse impact on the folding of the protein. Also, proline isomerization (cis-trans interconversion) influences the rate of protein folding and affects its conformational rigidity [47]. The p.R315Q mutation, another missense mutation at R315, resulted in early onset and rapid progression of disease [15], [48]. In this light, our finding of the novel mutation p.R315P suggests that ARSB is vulnerable to substitutions at R315. Patient P6 has the “moderate” form of the disease. Her younger sister, P7, has not yet developed clinical symptoms. Early start of ERT or early HSCT would be ideal in her case.
3.5.7. p.W450C
In this mutation, a hydrophilic amino acid residue is incorporated in place of a hydrophobic one. W450 is located at the solvent accessible surface shielding the hydrophobic core the protein. Tryptophan residues have a high propensity to be at the juncture of polar and nonpolar regions. Loss of a tryptophan residue at such places, especially when mutated into hydrophilic residues, can cause misfolding of the protein. The side chain of W450 forms a hydrogen bond with E482, (Fig. 3e) which would not be formed upon substitution by a cysteine residue. Also, introduction of a cysteine residue might lead to the formation of nonspecific disulfide cross-linking during protein folding. Thus p.W450C is expected to have an adverse effect on the folding and structure of ARSB. Disease severity was “attenuated” in Patient P3, who is homozygous for this mutation.
4. Discussion
Findings from this study lend support to the hypothesis that parental consanguinity contributes to occurrence of MPS VI in South India. Of the eight families with MPS VI patients who participated in this study, seven are from South India. The rate of parental consanguinity in these seven families is 86% (6/7 families). This is higher than the background rate of consanguineous marriages (30–38%) in the three South Indian states from which these seven families come [49], [50]. In one family (F2), the patient (P2) had homozygous mutations though not born of consanguineous parents. This is surprising, considering that most mutations in ARSB are rare and family specific. The reason for this is not clear. This might be because parents may be distantly related, or because certain mutations may be more frequent in some communities or geographic locations. Patients in this study are from varied geographic locations.
In a couple of patients (P8 and P9), MPS VI was diagnosed several years after symptoms first appeared (11 years for P8 and 6 years for P9). Clearly, greater awareness among physicians and patients about MPS VI is essential for early diagnosis and better management of the disease.
In one patient, P1, we could find only one of the two mutations expected to be present. The unlocated mutation might be in regulatory or intronic regions not analyzed during this study, and it may affect synthesis of functional mRNA or regulation of its levels. It could also be of a type of mutation (such as a type of deletion, duplication or translocation) that cannot be detected by the PCR assays used in this study. Levels of mRNA could not be measured as RNA samples from patients were not available.
Genotype–phenotype correlations could not be done because the number of patients was small and most mutations were found in single unrelated families. Further, the disease is still in its early stage in many patients, and the full complement of symptoms may not yet have developed to enable a definitive correlation. Patients would have to be followed up longer to allow disease progression to full term to obtain a true picture of disease severity.
A few different mutations were observed earlier at some of the same sites in ARSB in which mutations were found in the present study. For example, all three substitutions at L98 known to date – p.L98Q, p.L98P, and the novel one p.L98R – resulted from mutations in c.293T (Table S4). Similarly, both known substitutions at R315 – the common mutation p.R315Q and the novel one p.R315P – resulted from mutations in c.944G (Table S4), which is in a CpG dinucleotide [46]. Together, these facts suggest that c.293T and c.944G of ARSB are hot spots for mutations.
Prior to this study, six mutations were known to be present in active site residues of ARSB (Table S5). Identification of p.C91R through this study takes the number to seven. Patients with p.C91R had the severe form of the disease. A mutation similar to p.C91R of ARSB is known to occur at the conserved cysteine residue in the active site of GALNS [51], a paralogue of ARSB, whose deficient activity is the cause of MPS IVA. The p.C79R mutation in GALNS also results from a T > C change in the TGC codon, as does p.C91R of ARSB. This substitution in GALNS has also been reported only from India.
We analyzed data available on all mutations known from India to date. Nine mutations have been found so far from MPS VI patients in India. Five of these mutations were found in this study alone, two in the Kantaputra study alone [1], and two in both studies. Eight of these mutations have been found from this country alone (Table 3). Thus, the mutations in ARSB in MPS VI patients in India may be typical of this population. A larger study will provide a clear picture of the mutation spectrum of MPS VI patients in India.
Table 3.
ARSB mutations found from India. The table summarizes the exonic distribution of the mutations found from India, and the studies through which they were found. “+” indicates that mutation was found through a particular study, and “−” indicates mutation was not found through a particular study. The p.R160Q mutation was first reported from outside India. Families F1–F3 (Patients P1–P3) and F5–F9 (Patients P5–P10) were participants in this study. Families FA–FD refer to families from India in the earlier study [1].
| Serial no. | Mutation | Exon | No. of families in India | Families in India in which the mutations were found | This study | Kantaputra Study [1] | Studies from outside India |
|---|---|---|---|---|---|---|---|
| 1 | p.G38_G40del3 | 1 | 1 | F9 (P10) | + | − | − |
| 2 | p.D53N | 1 | 2 | F5 (P5), FD (PD) | + | + | − |
| 3 | p.C91R | 1 | 1 | F8 (P9) | + | − | − |
| 4 | p.L98R | 1 | 1 | F2 (P2) | + | − | − |
| 5 | c.1208delC (p.Ser403fs) |
1 | 1 | FA (PA) | − | + | − |
| 6 | p.R160Q | 2 | 1 | F7 (P8) | + | − | + |
| 7 | p.R315P | 5 | 1 | F6 (P6, P7) | + | − | − |
| 8 | p.P445L | 7 | 1 | FB (PB) | − | + | − |
| 9 | p.W450C | 8 | 4 | F3 (P3), F1 (P1), FB (PB), FC (PC) | + | + | − |
Twelve families with MPS VI patients have been studied from India: eight through this study and four through the Kantaputra study. The p.W450C mutation, present in 4/12 families (33%), is the most common mutation in ARSB (found in 2/8 families from this study, and 2/4 families from the Kantaputra study) (Table 3). This mutation, reported so far only from India, may be a common one in this country. Mutations are clustered in either Exon 8 or in the coding region of Exon 1 in 9/12 families (75%) studied from India (6/8 families from this study, and 3/4 families from the Kantaputra study) (Table 3). Identification of regions of the gene which harbor mutations in a large number of families would aid in carrier-testing and DNA-based diagnosis.
New treatment options are being developed by the science community for treatment of LSDs to complement existing ones (like ERT and HSCT for MPS VI). Pharmacological chaperone therapy (PCT) is one such option. PCT has shown promise in some LSDs other than MPS VI [52], [53]. In PCT, a small molecule that acts as a specific chaperone enables the proper folding of a mutant enzyme, which would otherwise undergo misfolding. Thus, PCT, used alone or in combination with ERT, would be useful for patients with mutations that only cause misfolding or instability of the enzyme, but not for patients with other types of mutations. Among patients in this study, those with p.R315P or p.W450C may benefit from PCT, in addition to ERT or HSCT. Patients with the active site mutation p.C91R would not benefit from PCT.
Determining the incidence and prevalence of MPS VI, study of natural history of MPS VI, identification of mutations in ARSB and study of genotype-correlations in a larger number of patients from this subcontinent would contribute to a better understanding of the MPS VI disorder as a whole. A comprehensive study would help to achieve this.
The following are the supplementary data related to this article.
Acknowledgments
We thank the patients, parents and control individuals for participating in this study. We are grateful to Dr. Sashikala Sathyamurthy, Dr. Madhuri Maganthi and Dr. Sanjeeva GN for assistance in collection of blood samples from patients and analysis of phenotype data, and Ms. Gaganashree S for technical assistance. Thanks are due to Dr. S. J. Patil of Narayana Hrudayalaya Hospital, Bangalore, who referred Patient P2 to the clinic in Chennai. We thank the Sequencing Facility of CHG.
This study was funded by the Department of Biotechnology, Government of India through a grant (BT/PR4224/MED/97/60/2011) to SS and SMJ. The bioinformatics part of this work was supported by the institutional grant to IBAB from Department of Electronics and Information Technology, Government of India, and the Department Information Technology, Biotechnology and Science & Technology, Government of Karnataka.
Conflict of interest: The authors declare that they have no conflict of interest.
References
- 1.Kantaputra P.N., Kayserili H., Guven Y., Kantaputra W., Balci M.C., Tanpaiboon P., Tananuvat N., Uttarilli A., Dalal A. Clinical manifestations of 17 patients affected with mucopolysaccharidosis type VI and eight novel ARSB mutations. Am. J. Med. Genet. A. 2014;164A:1443–1453. doi: 10.1002/ajmg.a.36489. [DOI] [PubMed] [Google Scholar]
- 2.E. Neufeld, J. Muenzer, The online metabolic and molecular basis of inherited disease, The Mucopolysaccharidoses; Chapter 136, McGraw-Hill Medical, 2013, pp. 3421–3452.
- 3.Nelson J. Incidence of the mucopolysaccharidoses in Northern Ireland. Hum. Genet. 1997;101:355–358. doi: 10.1007/s004390050641. [DOI] [PubMed] [Google Scholar]
- 4.Jurecka A., Zakharova E., Cimbalistiene L., Gusina N., Malinova V., Rozdzynska-Swiatkowska A., Golda A., Kulpanovich A., Kaldenovna Abdilova G., Voskoboeva E., Tylki-Szymanska A. Mucopolysaccharidosis type VI in Russia, Kazakhstan, and Central and Eastern Europe. Pediatrics International: Official Journal of the Japan Pediatric Society. 2014;56:520–525. doi: 10.1111/ped.12281. [DOI] [PubMed] [Google Scholar]
- 5.Moammar H., Cheriyan G., Mathew R., Al-Sannaa N. Incidence and patterns of inborn errors of metabolism in the Eastern Province of Saudi Arabia, 1983–2008. Ann. Saudi Med. 2010;30:271–277. doi: 10.4103/0256-4947.65254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costa-Motta F.M., Acosta A.X., Abe-Sandes K., Bender F., Schwartz I.V., Giugliani R., Leistner-Segal S. Genetic studies in a cluster of mucopolysaccharidosis type VI patients in northeast Brazil. Mol. Genet. Metab. 2011;104:603–607. doi: 10.1016/j.ymgme.2011.09.017. [DOI] [PubMed] [Google Scholar]
- 7.H.H. Freeze, Genetic disorders of glycan degradation. In: Varki A, Cummings RD, Esko JD, et al., editors. Essentials of Glycobiology. 2nd edition. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2009. Chapter 41 Available from: http://www.ncbi.nlm.nih.gov/books/NBK1934/. Accessed April 8, 2015.
- 8.Turbeville S., Nicely H., Rizzo J.D., Pedersen T.L., Orchard P.J., Horwitz M.E., Horwitz E.M., Veys P., Bonfim C., Al-Seraihy A. Clinical outcomes following hematopoietic stem cell transplantation for the treatment of mucopolysaccharidosis VI. Mol. Genet. Metab. 2011;102:111–115. doi: 10.1016/j.ymgme.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harmatz P., Whitley C.B., Waber L., Pais R., Steiner R., Plecko B., Kaplan P., Simon J., Butensky E., Hopwood J.J. Enzyme replacement therapy in mucopolysaccharidosis VI (Maroteaux–Lamy syndrome) J. Pediatr. 2004;144:574–580. doi: 10.1016/j.jpeds.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 10.Litjens T., Baker E.G., Beckmann K.R., Morris C.P., Hopwood J.J., Callen D.F. Chromosomal localization of ARSB, the gene for human N-acetylgalactosamine-4-sulphatase. Hum. Genet. 1989;82:67–68. doi: 10.1007/BF00288275. [DOI] [PubMed] [Google Scholar]
- 11.Bond C.S., Clements P.R., Ashby S.J., Collyer C.A., Harrop S.J., Hopwood J.J., Guss J.M. Structure of a human lysosomal sulfatase. Structure. 1997;5:277–289. doi: 10.1016/s0969-2126(97)00185-8. [DOI] [PubMed] [Google Scholar]
- 12.Schmidt B., Selmer T., Ingendoh A., von Figura K. A novel amino acid modification in sulfatases that is defective in multiple sulfatase deficiency. Cell. 1995;82:271–278. doi: 10.1016/0092-8674(95)90314-3. [DOI] [PubMed] [Google Scholar]
- 13.Stenson P.D., Mort M., Ball E.V., Shaw K., Phillips A., Cooper D.N. The human gene mutation database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum. Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valayannopoulos V., Nicely H., Harmatz P., Turbeville S. Mucopolysaccharidosis VI. Orphanet J. Rare Dis. 2010;5:5. doi: 10.1186/1750-1172-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karageorgos L., Brooks D.A., Pollard A., Melville E.L., Hein L.K., Clements P.R., Ketteridge D., Swiedler S.J., Beck M., Giugliani R., Harmatz P., Wraith J.E., Guffon N., Leao Teles E., Sa Miranda M.C., Hopwood J.J. Mutational analysis of 105 mucopolysaccharidosis type VI patients. Hum. Mutat. 2007;28:897–903. doi: 10.1002/humu.20534. [DOI] [PubMed] [Google Scholar]
- 16.Petry M.F., Dieter T., Burin M., Giugliani R., Leistner S. Identification of a novel mutation in the ARSB gene that is frequent among Brazilian MPSVI patients. Genet. Test. 2003;7:347–349. doi: 10.1089/109065703322783743. [DOI] [PubMed] [Google Scholar]
- 17.Petry M.F., Nonemacher K., Sebben J.C., Schwartz I.V., Azevedo A.C., Burin M.G., de Rezende A.R., Kim C.A., Giugliani R., Leistner-Segal S. Mucopolysaccharidosis type VI: identification of novel mutations on the arylsulphatase B gene in south American patients. J. Inherit. Metab. Dis. 2005;28:1027–1034. doi: 10.1007/s10545-005-0020-2. [DOI] [PubMed] [Google Scholar]
- 18.Garrido E., Chabas A., Coll M.J., Blanco M., Dominguez C., Grinberg D., Vilageliu L., Cormand B. Identification of the molecular defects in Spanish and Argentinian mucopolysaccharidosis VI (Maroteaux–Lamy syndrome) patients, including 9 novel mutations. Mol. Genet. Metab. 2007;92:122–130. doi: 10.1016/j.ymgme.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 19.Lin W.D., Lin S.P., Wang C.H., Hwu W.L., Chuang C.K., Lin S.J., Tsai Y., Chen C.P., Tsai F.J. Genetic analysis of mucopolysaccharidosis type VI in Taiwanese patients. Clin. Chim. Acta. 2008;394:89–93. doi: 10.1016/j.cca.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 20.Zheng J., Huang Y., Zhao X., Sheng H., Cheng J., Zhou Z., Li X., Mao X., Liu L. Analysis of clinical features and arylsulfatase B gene mutation in thirteen Chinese children with mucopolysaccharidosis type VI. Zhonghua Er Ke Za Zhi. 2014;52:403–408. [PubMed] [Google Scholar]
- 21.Zanetti A., Onenli-Mungan N., Elcioglu N., Ozbek M.N., Kor D., Lenzini E., Scarpa M., Tomanin R. Molecular analysis of Turkish Maroteaux–Lamy patients and identification of one novel mutation in the arylsulfatase B (ARSB) gene. JIMD Rep. 2014;14:1–9. doi: 10.1007/8904_2013_276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costa-Motta F.M., Bender F., Acosta A., Abe-Sandes K., Machado T., Bomfim T., Boa Sorte T., da Silva D., Bittles A., Giugliani R., Leistner-Segal S. A community-based study of mucopolysaccharidosis type VI in Brazil: the influence of founder effect, endogamy and consanguinity. Hum. Hered. 2014;77:189–196. doi: 10.1159/000358404. [DOI] [PubMed] [Google Scholar]
- 23.Jurecka A., Piotrowska E., Cimbalistiene L., Gusina N., Sobczynska A., Czartoryska B., Czerska K., Ounap K., Wegrzyn G., Tylki-Szymanska A. Molecular analysis of mucopolysaccharidosis type VI in Poland, Belarus, Lithuania and Estonia. Mol. Genet. Metab. 2012;105:237–243. doi: 10.1016/j.ymgme.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 24.Saito S., Ohno K., Sugawara K., Sakuraba H. Structural and clinical implications of amino acid substitutions in N-acetylgalactosamine-4-sulfatase: insight into mucopolysaccharidosis type VI. Mol. Genet. Metab. 2008;93:419–425. doi: 10.1016/j.ymgme.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 25.Saito S., Ohno K., Sekijima M., Suzuki T., Sakuraba H. Database of the clinical phenotypes, genotypes and mutant arylsulfatase B structures in mucopolysaccharidosis type VI. J. Hum. Genet. 2012;57:280–282. doi: 10.1038/jhg.2012.6. [DOI] [PubMed] [Google Scholar]
- 26.Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., Leunissen J.A. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–W74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li B., Krishnan V.G., Mort M.E., Xin F., Kamati K.K., Cooper D.N., Mooney S.D., Radivojac P. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics. 2009;25:2744–2750. doi: 10.1093/bioinformatics/btp528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng P.C., Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi Y., Sims G.E., Murphy S., Miller J.R., Chan A.P. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 33.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Voskoboeva E., Isbrandt D., von Figura K., Krasnopolskaya X., Peters C. Four novel mutant alleles of the arylsulfatase B gene in two patients with intermediate form of mucopolysaccharidosis VI (Maroteaux–Lamy syndrome) Hum. Genet. 1994;93:259–264. doi: 10.1007/BF00212019. [DOI] [PubMed] [Google Scholar]
- 35.Jin W.D., Desnick R.J., Schuchman E.H. A common polymorphism in the human arylsulfatase B (ARSB) gene at 5q13–q14. Nucleic Acids Res. 1991;19:4305. doi: 10.1093/nar/19.15.4305-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wicker G., Prill V., Brooks D., Gibson G., Hopwood J., von Figura K., Peters C. Mucopolysaccharidosis VI (Maroteaux–Lamy syndrome). An intermediate clinical phenotype caused by substitution of valine for glycine at position 137 of arylsulfatase B. J. Biol. Chem. 1991;266:21386–21391. [PubMed] [Google Scholar]
- 37.Garrido E., Cormand B., Hopwood J.J., Chabas A., Grinberg D., Vilageliu L. Maroteaux–Lamy syndrome: functional characterization of pathogenic mutations and polymorphisms in the arylsulfatase B gene. Mol. Genet. Metab. 2008;94:305–312. doi: 10.1016/j.ymgme.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 38.Andrews D.W., Perara E., Lesser C., Lingappa V.R. Sequences beyond the cleavage site influence signal peptide function. J. Biol. Chem. 1988;263:15791–15798. [PubMed] [Google Scholar]
- 39.Choo K.H., Ranganathan S. Flanking signal and mature peptide residues influence signal peptide cleavage. BMC Bioinformatics. 2008;9(Suppl 12):S15. doi: 10.1186/1471-2105-9-S12-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diez-Roux G., Ballabio A. Sulfatases and human disease. Annu. Rev. Genomics Hum. Genet. 2005;6:355–379. doi: 10.1146/annurev.genom.6.080604.162334. [DOI] [PubMed] [Google Scholar]
- 41.Dierks T., Schmidt B., Borissenko L.V., Peng J., Preusser A., Mariappan M., von Figura K. Multiple sulfatase deficiency is caused by mutations in the gene encoding the human C(alpha)-formylglycine generating enzyme. Cell. 2003;113:435–444. doi: 10.1016/s0092-8674(03)00347-7. [DOI] [PubMed] [Google Scholar]
- 42.Cosma M.P., Pepe S., Annunziata I., Newbold R.F., Grompe M., Parenti G., Ballabio A. The multiple sulfatase deficiency gene encodes an essential and limiting factor for the activity of sulfatases. Cell. 2003;113:445–456. doi: 10.1016/s0092-8674(03)00348-9. [DOI] [PubMed] [Google Scholar]
- 43.Bradford T.M., Gething M.J., Davey R., Hopwood J.J., Brooks D.A. Processing of normal lysosomal and mutant N-acetylgalactosamine 4-sulphatase: BiP (immunoglobulin heavy-chain binding protein) may interact with critical protein contact sites. Biochem. J. 1999;341(Pt 1):193–201. [PMC free article] [PubMed] [Google Scholar]
- 44.Brooks D.A., Robertson D.A., Bindloss C., Litjens T., Anson D.S., Peters C., Morris C.P., Hopwood J.J. Two site-directed mutations abrogate enzyme activity but have different effects on the conformation and cellular content of the N-acetylgalactosamine 4-sulphatase protein. Biochem. J. 1995;307(Pt 2):457–463. doi: 10.1042/bj3070457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Voskoboeva E., Krasnopol'skaia K.D., Peters K., von Figura K. Identification of mutations in the arylsulfatase B gene in Russian mucopolysaccharidosis type VI patients. Genetika. 2000;36:837–843. [PubMed] [Google Scholar]
- 46.Litjens T., Hopwood J.J. Mucopolysaccharidosis type VI: structural and clinical implications of mutations in N-acetylgalactosamine-4-sulfatase. Hum. Mutat. 2001;18:282–295. doi: 10.1002/humu.1190. [DOI] [PubMed] [Google Scholar]
- 47.Reimer U., Scherer G., Drewello M., Kruber S., Schutkowski M., Fischer G. Side-chain effects on peptidyl-prolyl cis/trans isomerisation. J. Mol. Biol. 1998;279:449–460. doi: 10.1006/jmbi.1998.1770. [DOI] [PubMed] [Google Scholar]
- 48.Villani G.R., Balzano N., Vitale D., Saviano M., Pavone V., Di Natale P. Maroteaux–Lamy syndrome: five novel mutations and their structural localization. Biochim. Biophys. Acta. 1999;1453:185–192. doi: 10.1016/s0925-4439(98)00099-4. [DOI] [PubMed] [Google Scholar]
- 49.Bittles A.H. Endogamy, consanguinity and community genetics. J. Genet. 2002;81:91–98. doi: 10.1007/BF02715905. [DOI] [PubMed] [Google Scholar]
- 50.Bittles A.H. Population stratification and genetic association studies in south Asia. Journal of Molecular and Genetic Medicine: An International Journal of Biomedical Research. 2005;1:43–48. doi: 10.4172/1747-0862.1000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bidchol A.M., Dalal A., Shah H., Nampoothiri S., Kabra M., Gupta N., Danda S., Gowrishankar K., Phadke S.R., Kapoor S., Kamate M., Verma I.C., Puri R.D., Sankar V.H., Devi A.R., Patil S.J., Ranganath P., Jain S.J., Agarwal M., Singh A., Mishra P., Tamhankar P.M., Gopinath P.M., Nagarajaram H.A., Satyamoorthy K., Girisha K.M. GALNS mutations in Indian patients with mucopolysaccharidosis IVA. Am. J. Med. Genet. A. 2014;164A:2793–2801. doi: 10.1002/ajmg.a.36735. [DOI] [PubMed] [Google Scholar]
- 52.Parenti G. Treating lysosomal storage diseases with pharmacological chaperones: from concept to clinics. EMBO Mol. Med. 2009;1:268–279. doi: 10.1002/emmm.200900036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parenti G., Andria G., Ballabio A. Lysosomal storage diseases: from pathophysiology to therapy. Annu. Rev. Med. 2015;66:471–486. doi: 10.1146/annurev-med-122313-085916. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.