Abstract
A novel antifungal drug candidate, the 1-tetrazole-based agent VT-1161 [(R)-2-(2,4-difluorophenyl)-1,1-difluoro-3-(1H-tetrazol-1-yl)-1-{5-[4-(2,2,2-trifluoroethoxy)phenyl]pyridin-2-yl}propan-2-ol], which is currently in two phase 2b antifungal clinical trials, was found to be a tight-binding ligand (apparent dissociation constant [Kd], 24 nM) and a potent inhibitor of cytochrome P450 sterol 14α-demethylase (CYP51) from the protozoan pathogen Trypanosoma cruzi. Moreover, VT-1161 revealed a high level of antiparasitic activity against amastigotes of the Tulahuen strain of T. cruzi in cellular experiments (50% effective concentration, 2.5 nM) and was active in vivo, causing >99.8% suppression of peak parasitemia in a mouse model of infection with the naturally drug-resistant Y strain of the parasite. The data strongly support the potential utility of VT-1161 in the treatment of Chagas disease. The structural characterization of T. cruzi CYP51 in complex with VT-1161 provides insights into the molecular basis for the compound's inhibitory potency and paves the way for the further rational development of this novel, tetrazole-based inhibitory chemotype both for antiprotozoan chemotherapy and for antifungal chemotherapy.
INTRODUCTION
Chagas disease is a vector-borne zoonosis caused by a genetically diverse population of the protozoan parasite Trypanosoma cruzi (1, 2). The infection is transmitted to more than 150 mammalian species by triatomine insects, often called “kissing bugs.” The other most frequent ways of transmission to humans involve blood transfusion, organ transplantation, oral ingestion via contaminated food or drinks, and transmission from mother to child. The disease is endemic in Central and South America. With human migration and HIV coinfections, Chagas disease is now also found in all other parts of the globe. According to the World Health Organization, worldwide, an estimated 6 million to 8 million people are infected with Trypanosoma cruzi, with 24,000 fatalities occurring each year (3). The situation is becoming particularly alarming in North America, due to the broadening of the area of vector habitat (4) and, accordingly, the wide geographic spread of the naturally infected wild animals that form the disease reservoir (5). Some estimates indicate that there are up to 1 million cases of Chagas disease in the United States, with most of them remaining undetected (6–9). Kissing bug bites have been reported in 43 states of the United States.
In the meantime, Chagas disease remains essentially incurable, with two nitroheterocyclic compounds, benznidazole and nifurtimox, being the only drugs available on the market, and these are available only in Latin America. The mechanism of their action is not completely clear, though it is believed to involve oxidative stress via the formation of free radicals and electrophilic metabolites that are generated when the nitroheterocyclic group of the compounds undergoes nitroreductase-mediated activation (10–12). Due to their quite serious toxicity and insufficient efficiency (13), benznidazole and nifurtimox are not approved for use by the U.S. Food and Drug Administration (FDA), and therefore, they are not prescribed in the United States. Although it has been reported that these drugs can potentially be obtained by special request from the Centers for Disease Control and Prevention (8), http://www.cdc.gov/parasites/chagas/health_professionals/tx.html), to our knowledge, most physicians are unaware of such an opportunity.
There has been hope that repurposing of two antifungal drugs (posaconazole and ravuconazole) may resolve the problem (4, 14), though thus far the results of clinical trials of their use for the treatment of Chagas disease have been quite controversial (they have a treatment failure rate of ∼80%, and patients experience numerous side effects [15, 16]). Both posaconazole and ravuconazole are 1-(1,2,4-triazole)-based inhibitors of fungal sterol 14α-demethylase (CYP51), the cytochrome P450 enzyme essential for the production of sterols, which are required for the formation of viable fungal membranes (17). Like fungi, T. cruzi is completely dependent on endogenously synthesized sterols (18). However, because T. cruzi is an intracellular parasite and has a complex life cycle involving so-called quiescent (dormant) forms with reduced metabolic activity (15, 19), it is reasonable to presume that alternative CYP51 inhibitors with optimized pharmacological properties (e.g., higher bioavailability and cellular permeability, broader tissue distribution, lower toxicity) and, in particular, lower production costs, which would easily allow treatment for longer periods of time, should be seriously considered as potential antichagasic agents.
In the study described here, we characterized as a T. cruzi CYP51 ligand and inhibitor the novel 1-tetrazole-based antifungal agent VT-1161 (20), which is currently undergoing phase 2b antifungal clinical trials. As we reported previously, this low-affinity metal-binding group (MBG)-bearing agent displays high target selectivity (it has only a weak inhibitory effect on human drug-metabolizing cytochromes P450) (21, 22) and excellent activity when it is administered orally (23). Orally administered therapy with this new drug candidate targets recurrent vulvovaginal candidiasis and onychomycosis. The initial antifungal program focused on Candida yeasts and dermatophytes, ultimately resulting in the selection of VT-1161 for clinical phase 1 pharmacokinetic studies and phase 2a efficacy studies. The safety and pharmacokinetics in humans have mirrored the data from preclinical studies with animals, insofar as VT-1161 appeared to raise no safety concerns and achieved an excellent pharmacokinetic profile when administered orally, exhibiting an extended half-life (23–25). All data obtained to date support the potential for VT-1161 to be a best-in-class CYP51 antifungal, overcoming the side effect profiles of the currently marketed fungal CYP51 inhibitors (26), which offer limited dosing options. The results of this study indicate the noteworthy anti-T. cruzi potency of VT-1161.
MATERIALS AND METHODS
Proteins.
All proteins were engineered with a His tag at the C terminus, and the genes were subcloned into the pCW expression vector and expressed in Escherichia coli strain HMS174(DE3) (Novagen). For enzymatic assays, including ligand binding and reconstitution of sterol 14α-demethylase activity in vitro, we used the full-length CYP51 of T. cruzi Tulahuen (GenBank accession number AY856083) (27). The enzyme was purified by affinity chromatography on Ni2+-nitrilotriacetic acid (NTA) agarose (Qiagen), followed by cation-exchange chromatography on a carboxymethyl (CM) Sepharose Fast Flow cation exchanger (GE Healthcare) (27). The samples of CYP51 enzymes from Aspergillus fumigatus and Candida albicans were expressed and purified as previously described (28, 29). The CYP51 electron donor partner NADPH-cytochrome P450 reductase (CPR) was from T. brucei and rat for protozoan and fungal orthologs, respectively (29–31). For crystallographic experiments, the T. cruzi CYP51 was truncated to replace the 30-amino-acid membrane anchor sequence at the N terminus (up to residue P31) with the more polar 5-amino-acid sequence fragment MAKKT (32) and purified in three steps, including anion-exchange chromatography on DEAE-Sepharose (GE Healthcare), affinity chromatography on Ni2+-NTA agarose, and cation-exchange chromatography on the CM Sepharose Fast Flow cation exchanger. Complexes with VT-1161 were obtained by saturating the protein with the inhibitor during the last step of purification (33) by adding a 20 mM stock solution of VT-1161 in dimethyl sulfoxide (DMSO) to the washing and elution buffers at a final concentration of 10 μM.
Spectroscopic measurements and ligand binding assay.
UV-visible absorption spectra were recorded using a dual-beam Shimadzu UV-2401PC spectrophotometer in 50 mM potassium phosphate buffer (pH 7.2) containing 10% (vol/vol) glycerol. P450 concentrations were estimated from the Soret band intensity using an absolute molar extinction coefficient ε417 of 117 mM−1 cm−1 (27) or a difference molar extinction coefficient Δε(450–490) of 91 mM−1 cm−1 for the reduced carbon monoxide difference spectra (34). Titrations with VT-1161 were carried out at a 0.3 μM P450 concentration in 5-cm-optical-path-length cuvettes, with ligand binding being monitored as a type II spectral response that reflects the coordination of a basic heterocyclic nitrogen to the P450 heme iron (a red shift in the Soret band maximum from 417 nm to 421-427 nm [35], depending on the basicity of the coordinating nitrogen [33]). Difference spectra were generated by recording the absorbance of P450 in a sample cuvette versus the absorbance in a reference cuvette, both of which contained the same amount of the protein. Aliquots of VT-1161 dissolved in DMSO were added to the sample cuvette over a concentration range of 0.1 to 1.0 μM, with each titration step being a concentration of 0.1 μM. At each step, the corresponding volume of DMSO was added to the reference cuvette. The apparent dissociation constants (Kds) of the CYP51–VT-1161 complex were calculated in GraphPad Prism (version 6) software (GraphPad, La Jolla, CA) by fitting the data for the ligand-induced changes in the absorbance of the difference spectra [Δ(Amax − Amin), where Amax is the maximum absorbance and Amin is the minimum absorbance] versus the ligand concentration to quadratic equation 1 (tight-binding ligands [33])
| (1) |
where ΔAmax is the change in the maximum absorbance and L and E are the total concentrations of ligand and enzyme used for the titration, respectively.
Reconstitution of catalytic activity and CYP51 inhibition assay.
The enzymatic activity of T. cruzi CYP51 was reconstituted in vitro as described previously using eburicol (24-methylenedihydrolanosterol) as the substrate (27). Briefly, the reaction mixture contained 1 μM CYP51, 2 μM CPR, 100 μM dilauroyl-α-phosphatidylcholine, 0.4 mg/ml isocitrate dehydrogenase, and 25 mM sodium isocitrate in 20 mM MOPS (morpholinepropanesulfonic acid; pH 7.4), 50 mM KCl, 5 mM MgCl2, and 10% glycerol. After addition of the 3H-radiolabeled sterol substrate (∼2,000 cpm/nmol; final concentration, 50 μM), the mixture was preincubated for 5 min at 37°C; the reaction was initiated by addition of 100 μM NADPH and stopped by extraction of the sterols with ethyl acetate. The enzymatic activities of the CYP51 orthologs from C. albicans (30) and A. fumigatus (29) were reconstituted with lanosterol and eburicol, respectively. The extracted sterols were dried, dissolved in methanol, and analyzed by a reverse-phase high-pressure liquid chromatography system (Waters) equipped with a β-RAM detector (INUS Systems, Inc.) using a Nova-Pak C18 column and a linear gradient from H2O-CH3CN-CH3OH (1.0:4.5:4.5, vol/vol/vol) (solvent A) to CH3OH (solvent B) increasing from 0 to 100% solvent B for 30 min at a flow rate of 1.0 ml/min. The inhibitory potencies of VT-1161 on CYP51 activity were compared on the basis of the decreases in substrate conversion in 60-min reactions (31–33) at a substrate/enzyme/inhibitor molar ratio of 50:1:2 (33, 36).
T. cruzi cellular growth inhibition assay.
A cellular T. cruzi infection assay was performed using the highly invasive 20A clone of the Tulahuen strain of the parasite (37). T. cruzi trypomastigotes expressing green fluorescent protein (GFP) were generated as described previously (32). Trypomastigotes were used to infect cardiomyocyte monolayers in 48-well tissue culture plates and in 8-well Labtech tissue culture chambers in triplicate at a ratio of 10 parasites per cell as described previously (38, 39). Cultures were incubated in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) as described previously (38). Unbound trypomastigotes were removed by washing the cellular monolayers with DMEM, and infected monolayers were exposed to several concentrations of VT-1161 (from 1 to 500 nM), dissolved in DMSO-DMEM free of phenol red in triplicate at 24 h of infection, and cocultured in DMEM with 10% FBS for 48 h to observe parasite multiplication. At 72 h after infection, the cardiomyocyte monolayers were washed with phosphate-buffered saline, and the infection was fluorimetrically quantified as the number of relative fluorescence units (RFU) using a Synergy HT fluorometer (BioTek Instruments) (39). For fluorescence microscopy observation, the infection assays were performed in 8-well Labtech tissue culture chambers in triplicate. At 72 h after infection, the cardiomyocyte monolayers were fixed with 2.5% paraformaldehyde and stained with 4′,6-diamidino-2-phenylindole to visualize the DNA and with Alexa Fluor 546 phalloidin (Invitrogen) to visualize cardiomyocyte actin myofibrils (38).
In vivo assay for efficacy.
Female Swiss mice (weight, 18 to 20 g) were obtained from the animal facilities of the Oswaldo Cruz Foundation (CECAL, Rio de Janeiro, Brazil). Mice were housed at six per cage and kept in a conventional room at 20 to 24°C under a 12-h light and 12-h dark cycle. The animals were provided with sterilized water and food ad libitum. All procedures and experimental protocols were conducted in accordance with the guidelines issued by the FIOCRUZ Committee of Ethics for the Use of Animals (CEUA LW16/14). Animals were inoculated with 104 trypomastigotes of the Y strain of T. cruzi by intraperitoneal (i.p.) injection. Three experimental groups were established: (i) 6 mice treated with VT-1161 at 50 mg/kg of body weight once a day, (ii) 6 mice treated with 0.5% carboxymethyl cellulose (vehicle), and (iii) 6 mice maintained as infected and untreated controls. VT-1161 was suspended in 0.5% (wt/vol) carboxymethyl cellulose, and each treated animal received 0.2 ml of drug suspension by gavage. The treatment was started at 5 days postinfection (p.i.), which corresponds to the onset of parasitemia in this experimental mouse model, and lasted up to 9 days p.i. (i.e., five consecutive daily doses were given) to cover the peak of parasitemia for infection with the Y strain (day 8 p.i.). Parasitemia was evaluated by microscopic examination of fresh blood samples (direct microscopic counting of parasites in 5 μl of tail blood) and performed on day 4, to select only the animals that revealed observable parasitemia, and then on days 7 and 8.
X-ray crystallography.
The initial screening of the crystallization conditions was carried out using Hampton Research crystallization kits. The crystals were obtained by the hanging-drop vapor diffusion method at 18 ° C. Equal volumes of 300 μ M T. cruzi CYP51–VT-1161 complex in 20 mM K-phosphate buffer, pH 7.2, containing 100 mM NaCl, 0.1 mM EDTA, 10% glycerol, 5.8 mM Tris (carboxyethyl)phosphine (TCEP), and 0.048 mM n -tridecyl-beta-d -maltoside were mixed with the mother liquor (0.2 M lithium sulfate, 0.1 M HEPES [pH 7.4], 25% [wt/vol] polyethylene glycol 3350) and equilibrated against the reservoir solution. Crystals appeared after several days and were cryoprotected by plunging them into a drop of reservoir solution supplemented with 40% (vol/vol) glycerol, flash-cooled in liquid nitrogen, and then prescreened on Bruker Microstar microfocus rotating-anode X-ray generator/Proteum PT135 charge-coupled-device (CCD) area detectors. Crystals that diffracted to an ∼ 3.0-Å resolution were subsequently used for data collection. The data were collected on the 21-ID-F beamline of the Life Sciences Collaborative Access Team (LS-CAT) at the Advanced Photon Source (APS), Argonne National Laboratory (Argonne, IL), at 100 K and a wavelength of 0.9786 Å using a MAR225 CCD detector. The diffraction images were integrated using the Mosflm program and scaled with the Aimless program (CCP4 program suite, version 6.3.0 [40]) in the trigonal P3(1)21 space group to a maximum resolution of 2.75 Å. The solvent content was estimated with a Matthews probability calculator (40). The crystal structure was determined by molecular replacement in the PhaserMR program using the atomic coordinates of the posaconazole-bound T. cruzi CYP51 structure (PDB accession number 3K1O) as the search model. An iterative model of the protein-inhibitor complex was then built with the Coot program (41) and refined with Refmac5 in the CCP4 program suite (40). Data collection and refinement statistics are shown in Table 1. The accession numbers of the other T. cruzi CYP51 structures discussed in this work are 3K1O (CYP51 in complex with the antifungal drug triazole posaconazole [32]), 4CK8 {CYP51 in complex with the experimental imidazole derivative (R)-1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl 4-[4-(3,4-dichlorophenyl)piperazin-1-yl]phenylcarbamate (LFD) (36)}, and 3ZG2 (CYP51 in complex with the fenarimol derivative pyridine (S)-2-(4-chlorophenyl)-2-(pyridin-3-yl)-1-{4-[4-(trifluoromethyl)phenyl]piperazin-1-yl}ethanone (UDO) [33]). Structure superimpositions were done in the LSQkab program of the CCP4 program suite. Molecular volumes and surface areas were calculated in the Accelrys Discovery Studio Visualizer (version 2.5) program (probe radius, 1.4 Å). Figures were prepared with the PyMOL and Chimera programs.
TABLE 1.
Data collection and refinement statistics
| Parametera | Value(s) for T. cruzi CYP51–VT-1161b |
|---|---|
| Data collection statistics | |
| Wavelength (Å) | 0.9786 |
| Unit cell dimensions | |
| a, b, c (Å) | 63.06, 63.06, 221.75 |
| α, β, γ (°) | 90.00, 90.00, 120.00 |
| No. of molecules per asymmetric unit | 1 |
| Solvent content (%) | 47.8 |
| Resolution (outer shell) (Å) | 54.6–2.75 (2.82–2.75)c |
| No. of reflections (outer shell) | 14,063 (1,526)c |
| Rmerge (outer shell) | 0.065 (0.695)c |
| I/σ (outer shell) | 23.1 (2.7)c |
| Completeness (outer shell) (%) | 99.8 (100)c |
| Redundancy (outer shell) | 8.0 (4.6)c |
| Refinement statistics | |
| Rwork | 0.259 |
| Rfree | 0.270 |
| RMSD from ideal geometry | |
| Bond length (Å) | 0.002 |
| Bond angle (°) | 0.89 |
| Ramachandran plot (%) | |
| Residues in favorable/allowed regions | 96.4/99.8 |
| Outliers | 0.2 |
| No. of atoms (mean B factor) (Å) | 3,672 (118.5)d |
| No. of residues | |
| Protein (mean B factor, Å) | 450 (120.7)d |
| Heme (mean B factor, Å) | 1 (76.4)d |
| Ligand (mean B factor, Å) | VT1 1 (99.6)d |
| PDB accession no. | 5AJR |
I, intensity of a reflection; RMSD, root mean square deviation.
The PDB code for the VT-1161 compound is VT1.
Values in parentheses are for the highest-resolution shell.
Values in parentheses are the mean B factor (in angstroms).
Protein structure accession number.
The atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession number 5AJR.
RESULTS
Characterization of VT-1161 as a T. cruzi CYP51 heme-coordinating ligand.
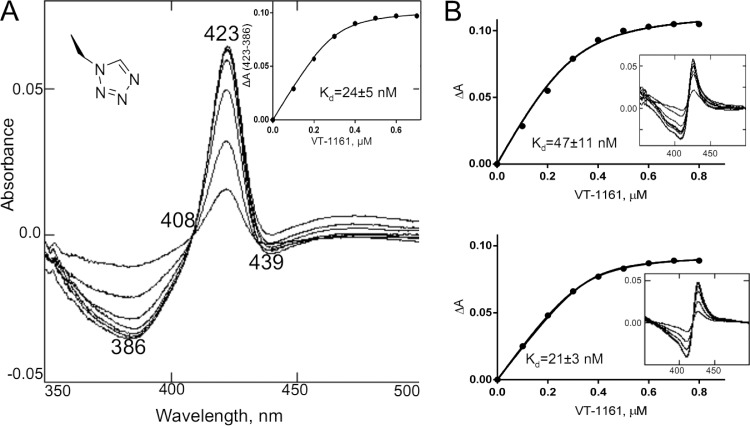
Titration of T. cruzi CYP51 with VT-1161 caused a red shift in the Soret band absorbance, which in the difference spectra was expressed as the peak, the trough, and the isosbestic point at 423, 386, and 408 nm, respectively (Fig. 1A), indicating expulsion of a water molecule from the cytochrome P450 active site and coordination of a basic nitrogen atom of the ligand to the heme iron. Although the shape of the type II spectral response (35) suggested that the length of the N-Fe coordination bond in the enzyme-ligand complex is likely to be relatively longer than that which is usually observed in CYP51 complexes with imidazole-based ligands (2.0 to 2.04 Å, peak at 428 nm) or triazole-based ligands (2.07 to 2.15 Å, peak at 426 nm) (33), the apparent binding affinity of VT-1161 was rather high, with the calculated spectral dissociation constant (Kd) being in the lower nanomolar range (24 nM), a value comparable to that obtained upon spectral titration of C. albicans CYP51 (21 nM) and about two times lower than the Kd calculated for CYP51 from A. fumigatus (47 nM) (Fig. 1B).
FIG 1.
(A) Spectral response of T. cruzi CYP51 to the addition of the heme-coordinating ligand 1-tetrazole VT-1161, shown as the difference in the type 2 binding spectra. The P450 concentration was 0.3 μM, and the optical path length was 5 cm. (Inset) The titration curve obtained using equation 1. ΔA(423-386), the difference in absorption between A423 and A386. (B) The corresponding titration curves and type 2 spectral responses (insets) of the A. fumigatus (top) and C. albicans (bottom) CYP51 orthologs.
VT-1161 as an inhibitor of enzymatic activity of sterol 14α-demethylase.
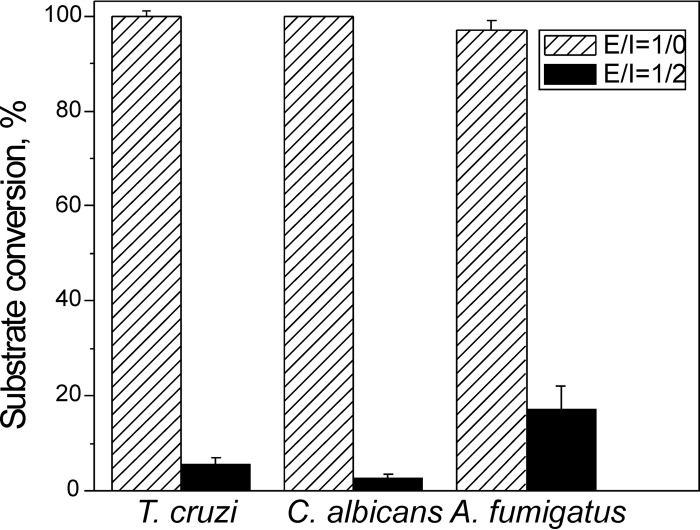
At a 2-fold molar excess over the concentration of the enzyme, VT-1161 inhibited 94% of substrate conversion by T. cruzi CYP51 (Fig. 2), thus showing an inhibitory effect comparable to its effect on the activity of CYP51 from C. albicans (affording 2.5% substrate conversion) and an effect somewhat stronger than the effect on the activity of CYP51B from A. fumigatus (17% substrate conversion). The results correlate with the higher activity of VT-1161 against yeast than against filamentous fungi and imply that the compound can potentially serve as a promising antichagasic agent.
FIG 2.
Inhibitory effects of VT-1161 on the enzymatic activities of sterol 14α-demethylases from the protozoan parasite T. cruzi and the opportunistic fungal pathogens C. albicans and A. fumigatus. The incubation time was 60 min. E/I, molar ratio enzyme/inhibitor. P450 concentration was 0.5 µM; the concentration of the substrate (see Materials and Methods) was 25 µM. The experiments were performed in triplicate, and the results are presented as means ± SEs.
Antiparasitic effect of VT-1161 in T. cruzi cells.
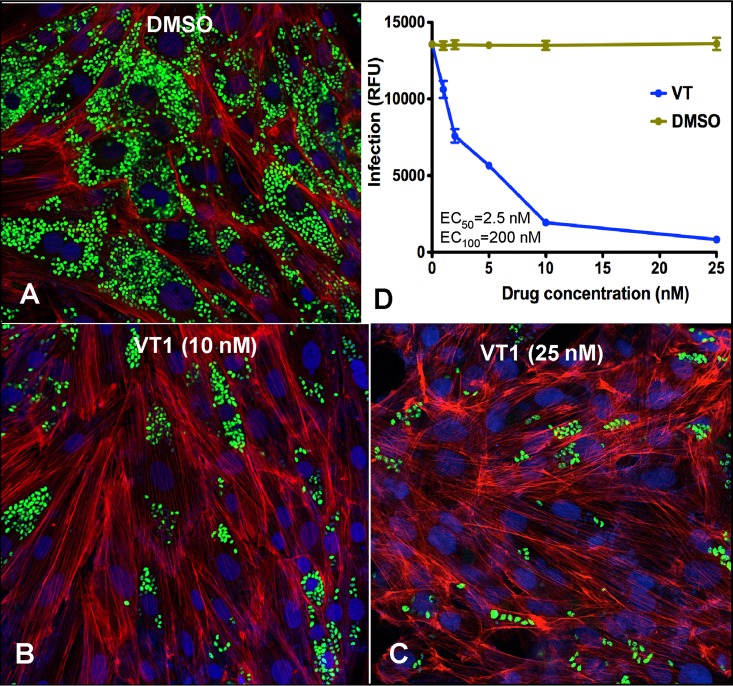
Cellular experiments were performed using the highly invasive 20A clone of the Tulahuen strain of T. cruzi because it infects >98% of exposed cardiomyocytes (37) (Fig. 3A). The antiparasitic activity of VT-1161 against the clinically most relevant form of the parasite, intracellular amastigotes, was analyzed by quantifying the rate of their replication within infected cardiomyocytes (examples are shown in Fig. 3B and C). The dose-response curve obtained (Fig. 3D) indicates that the effect of the drug was already seen at a 1 nM concentration. The VT-1161 concentration capable of reducing the infection by 50% compared with the level of infection in the nontreated infected controls (the 50% effective concentration [EC50]) was ∼2.5 nM, which is comparable to the anti-Tulahuen T. cruzi activity reported for one of the most potent sterol biosynthetic inhibitors, posaconazole (EC50 = 1 nM [15]), and is about 3 orders of magnitude lower than the EC50 determined for benznidazole (2.4 μM) (12). At a concentration of 200 nM, VT-1161 killed all parasites within cardiomyocytes. The same effect was seen at 500 nM (data not shown). Thus, VT-1161 demonstrated significant efficacy against T. cruzi and was therefore further tested in in vivo experiments.
FIG 3.
Cellular effects of VT-1161 in T. cruzi Tulahuen-infected cardiomyocytes. Cardiomyocyte monolayers were exposed to green fluorescent protein-expressing trypomastigotes (10 parasites per cell) for 24 h and then treated with VT-1161 or with the corresponding volumes of DMSO. (A to C) Fluorescence microscopic observations of T. cruzi inside cardiomyocytes treated with DMSO (control) (A) or with 10 nM (B) and 25 nM (C) VT-1161 at 72 h after infection. Green, T. cruzi amastigotes; blue, cardiomyocyte nuclei; red, cardiomyocyte actin myofibrils. (D) Dose-dependent clearance of the parasite. The infection was quantified by determination of the fluorescence, indicated as relative fluorescence units (RFU), at 72 h. The experiments were performed in triplicate, and the results are presented as means ± SEs.
Activity of VT-1161 in a murine model of T. cruzi infection.
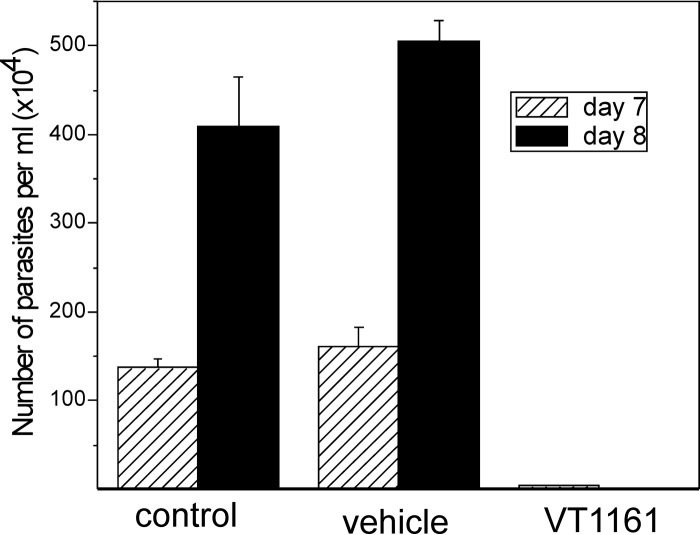
In these short-term in vivo experiments, we evaluated the ability of VT-1161 to suppress parasitemia during the acute phase of infection of mice with the Y strain of T. cruzi, because the Y strain is known to be naturally moderately resistant to nitro derivatives, such as benznidazole and nifurtimox (1), and has decreased susceptibility to the CYP51 inhibitors posaconazole (42, 43) and VNI (44). Besides, infection of Swiss mice using 104 bloodstream forms of the Y strain of T. cruzi reaches peak parasitemia on day 8 (44), which allows the relatively fast selection of potentially promising compounds. Although treatment with VT-1161 was started on day 5 (corresponding to the onset of parasitemia), after 2 days of drug administration the infection was restrained to 2.3% relative to the level of parasitemia in the control group of mice, while a 3-day treatment with VT-1161 caused >99.8% suppression of parasitemia (Fig. 4). After 5 days of treatment, all treated mice survived (not shown).
FIG 4.
VT-1161 suppresses parasitemia in mice infected with T. cruzi. Groups of Swiss female mice (n = 6) were infected i.p. with 104 blood trypomastigotes of the Y strain of T. cruzi and not treated (control) or treated with 0.5% carboxymethyl cellulose (vehicle) or 50 mg/kg of VT-1161. The treatment was started on day 5 after infection and performed by oral gavage. The bars represent means ± SEs. Three days of treatment with VT-1161 caused >99.8% suppression of parasitemia.
X-ray costructure of T. cruzi CYP51–VT-1161 complex.
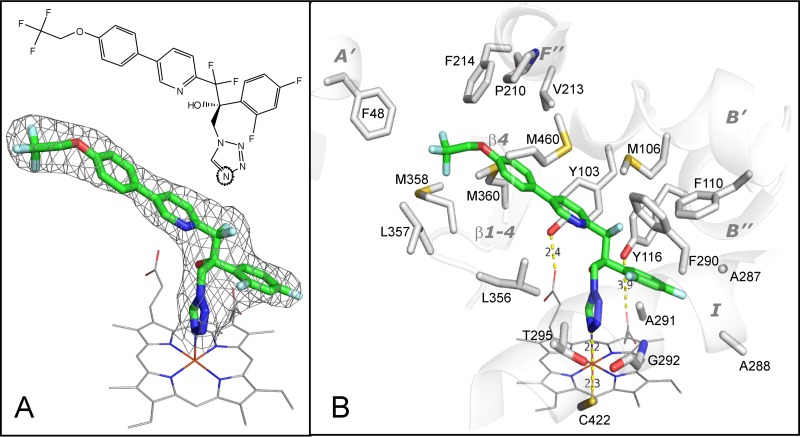
In order to better understand the potency and selectivity of VT-1161, we determined the X-ray costructure of VT-1161 in complex with T. cruzi CYP51. The complex has one monomer in the asymmetric unit. The protein chain is seen from Lys-29 (KKTP in the N-terminal MAKKTP sequence) to Lys-478 (the fifth residue from the C terminus), and one residue (Pro-222, in the G″G loop) is missing. The electron density for VT-1161 is well defined, showing a single orientation of the inhibitor molecule within the enzyme substrate binding cavity and full occupancy. The 2Fo-Fc electron density map weighted at 1.3σ is shown in Fig. 5A, and the structural formula of VT-1161 in the same orientation is given at the top of Fig. 5A.
FIG 5.
(A) Structural formula (top) and 2Fo-Fc electron density map (bottom) of VT-1161 coordinated to the T. cruzi CYP51 heme iron. Here and in Fig. 6, VT-1161 is presented as a stick model, and carbon atoms are green. The map is shown as a gray mesh and contoured at 1.3σ. The heme is depicted as a wire model, and carbon atoms are gray. (B) The 19 amino acid residues that surround VT-1161 in the T. cruzi CYP51 active site. The corresponding secondary structural elements of the enzyme are presented as semitransparent ribbons and marked. (The corresponding residues in the aligned sequences of CYP51 from C. albicans and A. fumigatus are listed in Table 2).
In the T. cruzi CYP51 active site, the VT-1161 molecule (ligand PDB ID VT1; molecular weight, 527; partition coefficient [log P], 5.07) occupies a volume of 572 Å3, with the surface area being 439 Å2. The tetrazole ring binds to the CYP51 heme iron via the N-4 nitrogen, which forms the sixth axial (distal) coordination bond, and the sulfur atom of C-422 serves as the canonical P450 fifth axial (proximal) ligand (Fig. 5B). The length of the N-Fe coordination bond is 2.2 Å, which is in good agreement with the shape of the type II spectral response and reflects a moderate basicity of the N-4–tetrazole nitrogen (between that of N-4 in triazoles [corresponding bond length, 2.07 to 2.15 Å] and N-1 in pyridines [corresponding bond length, 2.30 to 2.35 Å] [33, 45]), thus providing the molecular background for the high selectivity of this compound for CYP51 and its weak inhibitory effect on human drug-metabolizing cytochromes P450 (21).
At a distance of 5 Å, VT-1161 was found to be contacted by 19 amino acid residues of T. cruzi CYP51 (Fig. 5B). Of these residues, five (F48, P210, V213, F214, and M460) form the entry into the CYP51 substrate access channel, possibly playing a role in the ligand recognition function (46). All these residues are conserved across protozoan CYP51s and often phylum specific (46). Thus, F48 (A′ helix) is invariant across most biological kingdoms but is a Y residue in all fungal CYP51 sequences. P210 (F″ helix) is conserved in protozoan, fungal, and plant CYP51s but corresponds to H in vertebrates. V213 (F″ helix) is conserved in protozoa and plants but aligns with F in fungi and W in vertebrates. F214 (F″ helix) is M or V in fungi, always L in vertebrates, and L or I in plants, while M460 (β4 hairpin) is replaced by L only in some filamentous fungi. These five residues contact the distal portion of the long arm of the VT-1161 molecule.
Fourteen other VT-1161-contacting residues line the inner surface of the CYP51 substrate binding cavity. Four residues are from cytochrome P450 substrate recognition site 1 (SRS1) (47). Y103 and F110 (B′ helix) are invariant in the whole family (>300 sequenced proteins), and Y116 (B″ helix) is conserved in vertebrates, fungi, and protozoa yet is replaced by F in all plants. In the ligand-free and sterol-bound protozoan CYP51 structures (PDB accession numbers 3G1Q [48] and 3P99 [49]), Y103 and Y116 form hydrogen bonds with the heme ring A and D propionates, respectively. The binding of some heme-coordinating ligands, however, was found to disrupt these H bonds (32, 33, 36, 48, 50), which is likely to enhance the inhibitory potency of the compounds by weakening the P450 heme support from the protein moiety (51). As shown in Fig. 5B, in the complex with T. cruzi CYP51, VT-1161 also disrupts the interaction between the heme and Y116 by intercalating its β-phenyl ring between the bulky tyrosine side chain and the heme ring D propionate. F110 contacts the β-phenyl ring of VT-1161 from the top. M106 (which aligns with T in fungal CYP51s) lies above the pyridine ring and two fluorine atoms of VT-1161. Four I helix residues (SRS4), A287, A288, F290, and A291, encircle the opposite side of the VT-1161 β-phenyl ring, while two I helix residues, G292 and T295, interact with the tetrazole ring. The remaining four VT-1161-contacting residues (L356, L357, M358, and M360) are from SRS5 (K/β1-4 loop and β1-4 strand). They surround the two-ring area of the inhibitor.
The overall view of VT-1161 bound to T. cruzi CYP51 is shown in Fig. 6. The phenoxy arm of the inhibitor protrudes toward the distal surface of the protein and can be seen through the entry into the substrate access channel (helices A′ and F″ and the β4 hairpin) in the surface representation model (Fig. 6A and B). Such an orientation is quite typical for CYP51 inhibitors (Fig. 6C), indicating that further elongation of the VT-1161 two-ring arm is possible and may well result in compounds with even higher antiprotozoan activity. Moreover, crystallographic analysis and the results in Table 2 suggest that an elongation of this arm might also be favorable for inhibition of A. fumigatus, as both CYP51 orthologs in this fungal pathogen have a flexible methionine (Table 2, boldface) instead of bulky phenylalanine (F214) in this key position around the channel entry. The highest potency of VT-1161 for inhibition of the activity of C. albicans CYP51 (shown in Fig. 2) may be connected to the C. albicans F380 residue (Table 2, boldface) in the β1-4 strand (M360 in T. cruzi, M/L in A. fumigatus), as in the T. cruzi CYP51 costructure this residue closely approaches the aromatic ring of VT-1161. Taking into account the high degree of structural similarity of CYP51 enzymes across phylogeny (29, 46), phenylalanine in this position is quite likely to form π-π stacking interactions with the inhibitor, thus significantly strengthening the VT-1161–C. albicans CYP51 complex. On the other side of the CYP51 binding cavity, the para-Cl atom in the β-phenyl ring of VT-1161 is directed toward A288. The possibility that a longer side chain of isoleucine (I304 in C. albicans CYP51) (Table 2, boldface) is more favorable here is not excluded because it can form a larger number of van der Waals contacts with the inhibitor, while a leucine residue (L290 and L304 in A. fumigatus CYP51A and -B, respectively) might already be too bulky, creating some steric hindrances and therefore altering the VT-1161 orientation.
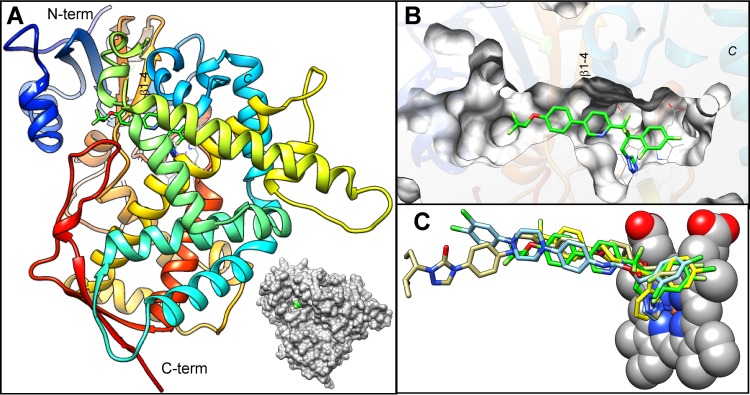
FIG 6.
VT-1161 (shown as a stick model with carbon atoms in green) bound to T. cruzi CYP51. (A) Overall view. The protein backbone is depicted as a rainbow ribbon color succession of the secondary structure from blue (N terminus) to red (C terminus). A surface representation is shown alongside to outline the view of VT-1161 through the entrance into the enzyme substrate access channel. (B) Enlarged view of panel A as a slice through the protein surface showing the location of VT-1161 within the CYP51 active-site cavity. The orientation is about the same as that in panel A. (C) Superimposition of VT-1161 with imidazole LFD (blue), the triazole posaconazole (khaki), and pyridine UDO (yellow) in the T. cruzi CYP51 active site. The PDB accession numbers of the corresponding structures are 4CK8, 3K1O, and 3ZG2, respectively.
TABLE 2.
VT-1161-contacting residues in T. cruzi CYP51 and the corresponding residues in the aligned CYP51 orthologs from two opportunistic fungal pathogens, C. albicans (yeast) and A. fumigatus
| Secondary structural element | Residuea |
|||
|---|---|---|---|---|
| T. cruzi | C. albicans |
A. fumigatusb |
||
| A | B | |||
| A′ helix | F48 | Y64 | Y53 | Y68 |
| B′ helix | Y103 | Y118 | Y107 | Y122 |
| M106 | T122 | T111 | T126 | |
| F110 | F126 | F115 | F130 | |
| B′C loop | Y116 | Y132 | Y121 | Y136 |
| F″ helix | P210 | P230 | P216 | P231 |
| V213 | F233 | F219 | F234 | |
| F214 | V234 | M220 | M235 | |
| I helix | A287 | G303 | T289 | A303 |
| A288 | I304 | L290 | L304 | |
| F290 | M306 | M292 | M306 | |
| A291 | G307 | A293 | A307 | |
| G292 | G308 | G294 | G308 | |
| T295 | T311 | S297 | S311 | |
| K/β1-4 loop | L356 | L376 | I364 | I373 |
| L357 | H377 | H365 | H374 | |
| M358 | S378 | S366 | S375 | |
| β1-4 strand | M360 | F380 | M368 | I377 |
| β4 hairpin | M460 | M508 | L494 | L503 |
Boldfacing is explained in the last paragraph of Results.
The filamentous fungus A. fumigatus has two CYP51 genes, A and B (24).
DISCUSSION
A recent report demonstrated that a prospective, multicenter, randomized study called BENEFIT involving 2,854 patients with Chagas cardiomyopathy who received benznidazole or placebo (for up to 80 days) and who were followed for a mean of 5.4 years showed that although this drug largely decreased the rate of detection of circulating parasites (by quantitative PCR analysis of blood), it was not able to reduce clinical progression to cardiomyopathy (52), strengthening the need for alternative therapies for the millions of chagasic patients who are at the later stage of disease (the chronic phase).
Sterols are essential components of eukaryotic cells. They contribute to the stability, permeability, and fluidity of the membrane and participate in multiple regulatory processes which are crucial for cell division, growth, and multiplication. Therefore, the sterol biosynthetic pathway is highly conserved across biological kingdoms. Among all the enzymes involved in this pathway, inhibitors of the sterol 14α-demethylase (CYP51) are the most efficient antifungal agents in clinical medicine and agriculture (18). Because the CYP51 orthologs appear to preserve their conserved biological function by maintaining a high degree of structural similarity of their substrate binding cavity at the secondary and tertiary levels (46), inhibitors of fungal CYP51 (such as the triazoles posaconazole and ravuconazole) are also often potent inhibitors of the orthologous enzyme in protozoa, which makes drug repurposing possible and sometimes quite effective. In this study, we explored for the first time the antifungal clinical candidate VT-1161, a 1-tetrazole-based agent, as an inhibitor of CYP51 from the protozoan pathogen T. cruzi and found that the drug has a strong potential to be an antichagasic agent.
Historically, there has been relatively little variation in CYP51 inhibitor metal-binding groups (MBGs). First-generation antifungal drugs, such as miconazole and ketoconazole, utilized the 1-imidazole, a high-affinity ligand for heme iron (53). These drugs also inhibited off-target human hepatic cytochrome P450 enzymes, leading to severe and sometimes fatal liver problems (54). Second-generation azole antifungal drugs (e.g., itraconazole, voriconazole) utilized a 1,2,4-triazole. Compared to 1-imidazole, the 1,2,4-triazole was a lower-affinity ligand for heme iron, and this drug with an alternative MBG had improved tolerability, but liver toxicity and drug-drug interactions remained problematic (26). VT-1161 was discovered using a strategy to investigate new, more selective agents that focused on alternative, low-affinity MBGs. In addition to its advantageous drug metabolism and pharmacokinetic properties and excellent safety profile, VT-1161 is readily prepared in seven synthetic transformations from commercially available reagents (21). Structural characterization of VT-1161 in complex with the target enzyme opens new opportunities for the rational, structure-directed design and optimization of new tetrazole-based CYP51 inhibitors.
In summary, our present findings demonstrate the potency of VT-1161 against the T. cruzi CYP51 enzyme as well as its phenotypic efficacy against T. cruzi infection in in vitro and in vivo biological assays, thus revealing a novel class of protozoan CYP51 inhibitors. An antichagasic agent with robust pharmacokinetic characteristics has the potential to exhibit an improved profile within the CYP51 inhibitor class. The marked affinity of the example 1-tetrazole disclosed in detail in this paper, coupled with its excellent in vivo exposure upon oral administration, could provide guidance toward the discovery and development of a novel and more efficient drug candidate(s) for the aforementioned neglected illness.
ACKNOWLEDGMENTS
This work was supported by Viamet Pharmaceuticals, Inc. (Durham, NC), and in part by National Institutes of Health grant R01 GM067871 (to G.I.L.). Vanderbilt University is a member institution of the Life Science Collaborative Team (LS-CAT) at Sector 21 of the Advanced Photon Source (APS), Argonne, IL. Use of the APS at Argonne National Laboratory was supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. DE-AC02-06CH11357. The use of LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (grant 085P1000817). The use of the Confocal Microscopy Facility at Meharry Medical College was supported by MD007593 (to F.V.). M.D.N.C.S. is research fellow of CNPq and CNE (FAPERJ).
REFERENCES
- 1.Filardi LS, Brener Z. 1987. Susceptibility and natural resistance of Trypanosoma cruzi strains to drugs used clinically in Chagas disease. Trans R Soc Trop Med Hyg 81:755–759. doi: 10.1016/0035-9203(87)90020-4. [DOI] [PubMed] [Google Scholar]
- 2.Lepesheva GI. 2013. Design or screening of drugs for the treatment of Chagas disease: what shows the most promise? Expert Opin Drug Discov 8:1479–1489. doi: 10.1517/17460441.2013.845554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. 2015. Third WHO report on neglected tropical diseases 2015: investing to overcome the global impact of neglected tropical diseases. Report WHO/HTM/NTD 2015.1. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 4.Leslie M. 2011. Drug developers finally take aim at a neglected disease. Science 333:933–935. doi: 10.1126/science.333.6045.933. [DOI] [PubMed] [Google Scholar]
- 5.Esch KJ, Petersen CA. 2013. Transmission and epidemiology of zoonotic protozoal diseases of companion animals. Clin Microbiol Rev 26:58–85. doi: 10.1128/CMR.00067-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hotez PJ. 2008. Neglected infections of poverty in the United States of America. PLoS Negl Trop Dis 2:e256. doi: 10.1371/journal.pntd.0000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hotez P, Dumonteil E, Betancourt Cravioto M, Bottazzi M, Tapia-Conyer R. 2013. An unfolding tragedy of Chagas disease in North America. PLoS Negl Trop Dis 7:e2300. doi: 10.1371/journal.pntd.0002300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bern C, Kjos S, Yabsley MJ, Montgomery SP. 2011. Trypanosoma cruzi and Chagas' disease in the United States. Clin Microbiol Rev 24:655–681. doi: 10.1128/CMR.00005-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barry MA, Bezek S, Serpa JA, Hotez PJ, Woc-Colburn L. 2012. Neglected infections of poverty in Texas and the rest of the United States: management and treatment options. Clin Pharmacol Ther 92:170–181. doi: 10.1038/clpt.2012.85. [DOI] [PubMed] [Google Scholar]
- 10.Wilkinson SR, Taylor MC, Horn D, Kelly JM, Cheeseman I. 2008. A mechanism for cross-resistance to nifurtimox and benznidazole in trypanosomes. Proc Natl Acad Sci U S A 105:5022–5027. doi: 10.1073/pnas.0711014105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maya JD, Cassels BK, Iturriaga-Vásquez P, Ferreira J, Faúndez M, Galanti N, Ferreira A, Morello A. 2007. Mode of action of natural and synthetic drugs against Trypanosoma cruzi and their interaction with the mammalian host. Comp Biochem Physiol A Mol Integr Physiol 146:601–620. doi: 10.1016/j.cbpa.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 12.Papadopoulou MV, Bloomer WD, Lepesheva GI, Rosenzweig HS, Kaiser M, Aguilera-Venegas B, Wilkinson SR, Chatelain E, Ioset J-R. 2015. Novel 3-nitrotriazole-based amides and carbinols as bifunctional antichagasic agents. J Med Chem 58:1307–1319. doi: 10.1021/jm5015742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Apt W. 2010. Current and developing therapeutic agents in the treatment of Chagas disease. Drug Des Dev Ther 4:243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clayton J. 2010. Chagas disease: pushing through the pipeline. Nature 465:S12–S15. doi: 10.1038/nature09224. [DOI] [PubMed] [Google Scholar]
- 15.Molina I, Salvador F, Sánchez-Montalvá A. 2015. The use of posaconazole against Chagas disease. Curr Opin Infect Dis 28:397–407. doi: 10.1097/QCO.0000000000000192. [DOI] [PubMed] [Google Scholar]
- 16.Molina I, Gómez i Prat J, Salvador F, Treviño B, Sulleiro E, Serre N, Pou D, Roure S, Cabezos J, Valerio L, Blanco-Grau A, Sánchez-Montalvá A, Vidal X, Pahissa A. 2014. Randomized trial of posaconazole and benznidazole for chronic Chagas' disease. N Engl J Med 370:1899–1908. doi: 10.1056/NEJMoa1313122. [DOI] [PubMed] [Google Scholar]
- 17.Lepesheva GI, Waterman MR. 2007. Sterol 14alpha-demethylase cytochrome P450 (CYP51), a P450 in all biological kingdoms. Biochim Biophys Acta 1770:467–477. doi: 10.1016/j.bbagen.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lepesheva GI, Villalta F, Waterman MR. 2011. Targeting Trypanosoma cruzi sterol 14α-demethylase (CYP51). Adv Parasitol 75:65–87. doi: 10.1016/B978-0-12-385863-4.00004-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chatelain E, Konar N. 2015. Translational challenges of animal models in Chagas disease drug development: a review. Drug Des Dev Ther 9:4807–4823. doi: 10.2147/DDDT.S90208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denning DW, Bromley MJ. 2015. How to bolster the antifungal pipeline. Science 347:1414–1416. doi: 10.1126/science.aaa6097. [DOI] [PubMed] [Google Scholar]
- 21.Hoekstra WJ, Garvey EP, Moore WR, Rafferty SW, Yates CM, Schotzinger RJ. 2014. Design and optimization of highly-selective fungal CYP51 inhibitors. Bioorg Med Chem Lett 24:3455–3458. doi: 10.1016/j.bmcl.2014.05.068. [DOI] [PubMed] [Google Scholar]
- 22.Warrilow AGS, Hull CM, Parker JE, Garvey EP, Hoekstra WJ, Moore WR, Schotzinger RJ, Kelly DE, Kelly SL. 2014. The clinical candidate VT-1161 is a highly potent inhibitor of Candida albicans CYP51 but fails to bind the human enzyme. Antimicrob Agents Chemother 58:7121–7127. doi: 10.1128/AAC.03707-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garvey EP, Hoekstra WJ, Moore WR, Schotzinger RJ, Long L, Ghannoum MA. 2015. VT-1161 dosed once daily or once weekly exhibits potent efficacy in treatment of dermatophytosis in a guinea pig model. Antimicrob Agents Chemother 59:1992–1997. doi: 10.1128/AAC.04902-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garvey EP, Hoekstra WJ, Schotzinger RJ, Sobel JD, Lilly EA, Fidel PL. 2015. Efficacy of the clinical agent VT-1161 against fluconazole-sensitive and -resistant Candida albicans in a murine model of vaginal candidiasis. Antimicrob Agents Chemother 59:5567–5573. doi: 10.1128/AAC.00185-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shubitz LF, Trinh HT, Galgiani JN, Lewis ML, Fothergill AW, Wiederhold NP, Barker BM, Lewis ERG, Doyle AL, Hoekstra WJ, Schotzinger RJ, Garvey EP. 2015. Evaluation of VT-1161 for treatment of coccidioidomycosis in murine infection models. Antimicrob Agents Chemother 59:7249–7254. doi: 10.1128/AAC.00593-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pitman SK, Drew RH, Perfect JR. 2011. Addressing current medical needs in invasive fungal infection prevention and treatment with new antifungal agents, strategies and formulations. Expert Opin Emerg Drugs 16:559–586. doi: 10.1517/14728214.2011.607811. [DOI] [PubMed] [Google Scholar]
- 27.Lepesheva GI, Zaitseva NG, Nes WD, Zhou W, Arase M, Liu J, Hill GC, Waterman MR. 2006. CYP51 from Trypanosoma cruzi: a phyla-specific residue in the B′ helix defines substrate preferences of sterol 14alpha-demethylase. J Biol Chem 281:3577–3585. doi: 10.1074/jbc.M510317200. [DOI] [PubMed] [Google Scholar]
- 28.Lepesheva GI, Podust LM, Bellamine A, Waterman MR. 2001. Folding requirements are different between sterol 14α-demethylase (CYP51) from Mycobacterium tuberculosis and human or fungal orthologs. J Biol Chem 276:28413–28420. doi: 10.1074/jbc.M102767200. [DOI] [PubMed] [Google Scholar]
- 29.Hargrove TY, Wawrzak Z, Lamb DC, Guengerich FP, Lepesheva GI. 2015. Structure-functional characterization of cytochrome P450 sterol 14α-demethylase (CYP51B) from Aspergillus fumigatus and molecular basis for the development of antifungal drugs. J Biol Chem 290:23916–23934. doi: 10.1074/jbc.M115.677310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lepesheva GI, Nes WD, Zhou W, Hill GC, Waterman MR. 2004. CYP51 from Trypanosoma brucei is obtusifoliol-specific. Biochemistry 43:10789–10799. doi: 10.1021/bi048967t. [DOI] [PubMed] [Google Scholar]
- 31.Lepesheva GI, Ott RD, Hargrove TY, Kleshchenko YY, Schuster I, Nes WD, Hill GC, Villalta F, Waterman MR. 2007. Sterol 14 alpha-demethylase as a potential target for antitrypanosomal therapy: enzyme inhibition and parasite cell growth. Chem Biol 14:1283–1293. doi: 10.1016/j.chembiol.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lepesheva GI, Hargrove TY, Anderson S, Kleshchenko Y, Furtak V, Wawrzak Z, Villalta F, Waterman MR. 2010. Structural insights into inhibition of sterol 14 alpha-demethylase in the human pathogen Trypanosoma cruzi. J Biol Chem 285:25582–25590. doi: 10.1074/jbc.M110.133215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hargrove TY, Wawrzak Z, Alexander PW, Chaplin JH, Keenan M, Charman SA, Waterman MR, Chatelain E, Lepesheva GI. 2013. Complexes of Trypanosoma cruzi sterol 14α-demethylase (CYP51) with two pyridine-based drug candidates for Chagas disease: structural basis for pathogen selectivity. J Biol Chem 288:31602–31615. doi: 10.1074/jbc.M113.497990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Omura T, Sato R. 1964. The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem 239:2370–2378. [PubMed] [Google Scholar]
- 35.Schenkman JB, Remmer H, Estabrook RW. 1967. Spectral studies of drug interaction with hepatic microsomal cytochrome. Mol Pharmacol 3:113–123. [PubMed] [Google Scholar]
- 36.Friggeri L, Hargrove TY, Rachakonda G, Williams AD, Wawrzak Z, Di Santo R, De Vita D, Waterman MR, Tortorella S, Villalta F, Lepesheva GI. 2014. Structural basis for rational design of inhibitors targeting Trypanosoma cruzi sterol 14α-demethylase: two regions of the enzyme molecule potentiate its inhibition. J Med Chem 57:6704–6717. doi: 10.1021/jm500739f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lima MF, Villalta F. 1989. Trypanosoma cruzi trypomastigote clones differentially express a parasite cell adhesion molecule. Mol Biochem Parasitol 33:159–170. doi: 10.1016/0166-6851(89)90030-3. [DOI] [PubMed] [Google Scholar]
- 38.Villalta F, Dobish MC, Nde PN, Kleshchenko YY, Hargrove TY, Johnson CA, Waterman MR, Johnston JN, Lepesheva GI. 2013. VNI cures acute and chronic experimental Chagas disease. J Infect Dis 208:504–511. doi: 10.1093/infdis/jit042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson CA, Rachakonda G, Kleshchenko YY, Nde PN, Madison MN, Pratap S, Cardenas TC, Taylor C, Lima MF, Villalta F. 2013. Cellular response to Trypanosoma cruzi infection induces secretion of defensin α-1, which damages the flagellum, neutralizes trypanosome motility, and inhibits infection. Infect Immun 81:4139–4148. doi: 10.1128/IAI.01459-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 41.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cencig S, Coltel N, Truyens C, Carlier Y. 2012. Evaluation of benznidazole treatment combined with nifurtimox, posaconazole or AmBisome® in mice infected with Trypanosoma cruzi strains. Int J Antimicrob Agents 40:527–532. doi: 10.1016/j.ijantimicag.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 43.Buckner F, Bahia MT, Suryadevara PK, White KL, Shackleford DM, Chennamaneni NK, Hulverson MA, Laydbak JU, Chatelain E, Scandale I, Verlinde CL, Charman SA, Lepesheva GI, Gelb MH. 2012. Pharmacological characterization, structural studies, and in vivo activity of anti-Chagas disease lead compounds derived from tipifarnib. Antimicrob Agents Chemother 56:4914–4921. doi: 10.1128/AAC.06244-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soeiro MDNC, de Souza EM, da Silva CF, Batista DDGJ, Batista MM, Pavão BP, Araújo JS, Lionel J, Britto C, Kim K, Sulikowski G, Hargrove TY, Waterman MR, Lepesheva GI. 2013. In vitro and in vivo studies of the antiparasitic activity of sterol 14α-demethylase (CYP51) inhibitor VNI against drug-resistant strains of Trypanosoma cruzi. Antimicrob Agents Chemother 57:4151–4163. doi: 10.1128/AAC.00070-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cherkesova TS, Hargrove TY, Vanrell MC, Ges I, Usanov SA, Romano PS, Lepesheva GI. 2014. Sequence variation in CYP51A from the Y strain of Trypanosoma cruzi alters its sensitivity to inhibition. FEBS Lett 588:3878–3885. doi: 10.1016/j.febslet.2014.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lepesheva GI, Waterman MR. 2011. Structural basis for conservation in the CYP51 family. Biochim Biophys Acta 1814:88–93. doi: 10.1016/j.bbapap.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gotoh O. 1992. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem 267:83–90. [PubMed] [Google Scholar]
- 48.Lepesheva GI, Park HW, Hargrove TY, Vanhollebeke B, Wawrzak Z, Harp JM, Sundaramoorthy M, Nes WD, Pays E, Chaudhuri M, Villalta F, Waterman MR. 2010. Crystal structures of Trypanosoma brucei sterol 14 alpha-demethylase and implications for selective treatment of human infections. J Biol Chem 285:1773–1780. doi: 10.1074/jbc.M109.067470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hargrove TY, Wawrzak Z, Liu J, Waterman MR, Nes WD, Lepesheva GI. 2012. Structural complex of sterol 14α-demethylase (CYP51) with 14α-methylenecyclopropyl-Δ7-24,25-dihydrolanosterol. J Lipid Res 53:311–320. doi: 10.1194/jlr.M021865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hargrove TY, Wawrzak Z, Liu J, Nes WD, Waterman MR, Lepesheva GI. 2011. Substrate preferences and catalytic parameters determined by structural characteristics of sterol 14α-demethylase (CYP51) from Leishmania infantum. J Biol Chem 286:26838–26848. doi: 10.1074/jbc.M111.237099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lepesheva GI, Waterman MR. 2011. Sterol 14alpha-demethylase (CYP51) as a therapeutic target for human trypanosomiasis and leishmaniasis. Curr Top Med Chem 11:2060–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morillo CA, Marin-Neto JA, Avezum A, Sosa-Estani S, Rassi A, Rosas F, Villena E, Quiroz R, Bonilla R, Britto C, Guhl F, Velazquez E, Bonilla L, Meeks B, Rao-Melacini P, Pogue J, Mattos A, Lazdins J, Connolly SJ, Yusuf S. 2015. Randomized trial of benznidazole for chronic Chagas' cardiomyopathy. N Engl J Med 373:1295–1306. doi: 10.1056/NEJMoa1507574. [DOI] [PubMed] [Google Scholar]
- 53.Heeres J, Meerpoel L, Lewi P. 2010. Conazoles. Molecules 15:4129–4188. doi: 10.3390/molecules15064129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lewis RE. 2011. Current concepts in antifungal pharmacology. Mayo Clin Proc 86:805–817. doi: 10.4065/mcp.2011.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]