Abstract
Staphylococcus pseudintermedius is often associated with pyoderma, which can turn into a life-threatening disease. The dissemination of highly resistant isolates has occurred in the last 10 years and has challenged antimicrobial treatment of these infections considerably. We have compared the carriage of virulence genes and biofilm formation between methicillin-resistant and methicillin-susceptible S. pseudintermedius (MRSP and MSSP, respectively) isolates and their in vitro gene expression profiles by transcriptome sequencing (RNA-seq). Isolates were relatively unevenly distributed among the four agr groups, and agr type III predominated in MRSP. Five virulence genes were detected in all isolates. Only the spsO gene was significantly associated with MSSP isolates (P = 0.04). All isolates produced biofilm in brain heart infusion broth (BHIB)–4% NaCl. MSSP isolates produced more biofilm on BHIB and BHIB–1% glucose media than MRSP isolates (P = 0.03 and P = 0.02, respectively). Virulence genes encoding surface proteins and toxins (spsA, spsB, spsD, spsK, spsL, spsN, nucC, coa, and luk-I) and also prophage genes (encoding phage capsid protein, phage infection protein, two phage portal proteins, and a phage-like protein) were highly expressed in the MRSP isolate (compared with the MSSP isolate), suggesting they may play a role in the rapid and widespread dissemination of MRSP. This study indicates that MRSP may upregulate surface proteins, which may increase the adherence of MRSP isolates (especially sequence type 71 [ST71]) to corneocytes. MSSP isolates may have an increased ability to form biofilm under acidic circumstances, through upregulation of the entire arc operon. Complete understanding of S. pseudintermedius pathogenesis and host-pathogen signal interaction during infections is critical for the treatment and prevention of S. pseudintermedius infections.
INTRODUCTION
Methicillin-resistant Staphylococcus pseudintermedius (MRSP) isolates have emerged as one of the leading causes of infectious diseases (including pyoderma, otitis and urinary tract infections) in companion animals, accounting for 20% to 47% of all clinical S. pseudintermedius isolates from dogs and cats (1). Moreover, some MRSP isolates are resistant to the antimicrobials regularly used for treatment (β-lactams, fluoroquinolones, tetracyclines, lincosamides, and potentiated sulfonamides) in small animal practice (1, 2). The mecA gene, encoding resistance to β-lactams, has been acquired by several S. pseudintermedius clonal lineages on independent occasions; however, two clones, MRSP ST68-SCCmec V and MRSP ST71-SCCmec II-III, are the dominant ones and have spread globally (1, 3, 4). This dissemination was rapid, but the reasons for the fast emergence and success of these lineages are not fully understood (2). Genomic and proteomic studies conducted in the last few years are giving the first clues on the pathways by which MRSP isolates have become successful. A recent genomic report suggested that multidrug resistance evolved rapidly in MRSP due to the acquisition of a very limited number of mobile genetic elements and mutations (1). Thus, the use of different antimicrobial classes coselected for the spread and emergence of the multidrug-resistant MRSP isolates (1). The frequent carriage of prophages in the MRSP sequence type 71 (ST71) and ST68 genomes suggested they have a role in the fitness of MRSP and that the predominant transfer of genetic material in these isolates is through bacteriophage transduction, rather than plasmid conjugation, as happens in methicillin-resistant Staphylococcus aureus (MRSA) (1). MRSP isolates are able to produce biofilm, and MRSP ST71 isolates, in particular, are better biofilm producers than other MRSP clones (5, 6). The icaA gene can be significantly upregulated in biofilm samples, suggesting a role in the biofilm production by S. pseudintermedius (7). The ability to form biofilm may play an important role in the pathophysiology of bacterial infections and can be related to survival and persistence of S. pseudintermedius, namely, MRSP, in the environment (5, 6). The MRSP ST71 isolates also show greater adherence to corneocytes than MRSP non-ST71 and methicillin-susceptible S. pseudintermedius (MSSP) isolates, and thus it has been suggested that the enhanced adherence of ST71 might be a factor contributing to the epidemiological success of this MRSP lineage (2). Furthermore, an MRSP ST71 isolate of human origin adhered evenly well to canine and human corneocytes, implying that MRSP ST71 may also be capable of adapting to human skin (2). Two proteins, SpsD and SpsO, can mediate adherence to canine corneocytes (8); however, the genetic factors responsible for the enhanced in vitro adherence of MRSP ST71 are not yet known (2).
In order to understand the epidemiological success of MRSP isolates, our goal was to understand if the phenotypes (biofilm) and genotypes (virulence genes) related to virulence factors were different between MRSP and methicillin-susceptible S. pseudintermedius (MSSP) isolates. Furthermore, we compared the in vitro transcriptional profiles by transcriptome sequencing (RNA-seq) of one MRSP isolate and one MSSP isolate to test the hypothesis that MRSP could have altered expression of virulence genes, by comparison with MSSP, which could have contributed to its rapid spread.
MATERIALS AND METHODS
Genotypic characterization of the MRSP and MSSP isolates.
Twenty-one consecutive methicillin-resistant S. pseudintermedius (MRSP) isolates obtained over a 7-year period from 2007 to 2014 were included in the study. Twenty-one methicillin-susceptible S. pseudintermedius (MSSP) isolates matched in terms of isolation year, isolation site, and host were also included. These isolates were from 18 asymptomatic carriers (9 with MRSP and 9 with MSSP), 12 patients with pyoderma (6 with MRSP and 6 with MSSP), 6 patients with urinary tract infection (3 with MRSP and 3 with MSSP), 5 patients with otitis (2 with MRSP and 3 with MSSP), and 1 patient with a surgical site infection (MRSP). Five isolates were from cats, and 37 were from dogs. Isolates were characterized by multilocus sequence typing (MLST) (9). The eBURST algorithm identified groups of related sequence types (ST) (10).
Specific sequences for virulence genes involved in biofilm formation (bap, icaA, icaB, icaC, and icaD), enterotoxin production (se-int, seccanine, and seh), host adherence (ebpS, spsD, spsL, and spsO), and toxin production (lukS, lukF, siet, speta, expA, and expB) were detected by PCR on a Mastercycle thermocycler (Eppendorf, New York) with the primers, product sizes, and annealing temperatures shown in Table S1 in the supplemental material (11–16). The primers designed in this study were generated using the Primer-BLAST tool from NCBI. All PCR products were analyzed by electrophoresis through 1.2% agarose gels (NZYTech, Lisbon, Portugal). The primers agrD-F (5′-GGG GTA TTA TTA CAA TCA TTC -3′) and agrD-R (5′-CTG ATG CGA AAA TAA AGG ATT G -3′) (STABvida, Monte da Caparica, Portugal) were used as previously described to amplify a 300-bp agr fragment encompassing the 3′ end of agrB, all of agrD, and the 5′ end of agrC. Amplification was carried out on a Mastercycler thermocycler (Eppendorf) under the following conditions: an initial 5-min denaturation step at 94°C, followed by 30 cycles of 1 min of denaturation at 94°C, 1 min of annealing at 45°C, and 1 min of extension at 72°C, and a final extension step at 72°C for 10 min. The PCR products were purified by using NZYGelpure (NZYTech) and sequenced with the same primers used for the PCRs (STABvida). The 42 isolates were assigned to one of the four agr groups by comparing the predicted product of the agrD gene and the N-terminal half of the AgrC with those of four control isolates (GenBank accession no. EU157336, EU157366, EU157334, and EU157330).
Biofilm-producing ability on polystyrene.
The capacity of the isolates to form biofilm was investigated by a method described by Stepanovic and colleagues (17) and Pettit and colleagues (18) with minor modifications, and was determined by the ability of S. pseudintermedius isolates to adhere to 96-well polystyrene microtiter plates (Greiner bio-one, Frickenhausen, Germany). In brief, the study was carried out using brain heart infusion broth (BHIB [Biokar]), BHIB with 4% NaCl, and BHIB with 1% glucose as the growth media. The plates were incubated at 37°C for 24 h. Following incubation, the alamarBlue solution was added to each well. After 30 min at room temperature, the optical densities at 570 nm (OD570) were measured. Staphylococcus epidermidis strain RP62A (ATCC 35984) was used as a positive control. We defined the cutoff OD (ODC) for the microtiter plate test as 3 standard deviations (SD) above the mean OD of the negative control as described previously (17). All isolates were classified into the following categories based upon the ODs of bacterial films (17): nonadherent, OD ≤ ODC; weakly adherent, ODC < OD ≤ 2× ODC; moderately adherent, 2× ODC < OD ≤ 4× ODC; or strongly adherent, OD ≥ 4× ODC.
RNA isolation, sequencing and gene expression analyses.
To test the hypothesis that MRSP and MSSP isolates differ in their expression of virulence genes, we compared the in vitro transcriptional profiles of a clinical MRSP isolate and a clinical MSSP isolate using RNA-seq. We attempted to choose 2 representative isolates from the S. pseudintermedius collection. The isolates were obtained from skin swabs of dogs with pyoderma (the most frequent clinical specimen from which S. pseudintermedius was isolated), they were obtained during the same period of time, were agr type III (the most frequent agr type found in this study), at least one was ST71 (the most frequent ST found in this study), and they had similar virulence profiles (considering virulence genes tested by PCR). Bacterial cells were grown until the mid-log-phase of growth (OD600 of 0.5), since it has been shown that the majority of surface proteins are produced during this phase (19). RNA was isolated using the RNeasy kit from Qiagen (Hilden, Germany). Briefly, 2 × 108 cells were removed from growing cultures, 2 volumes of RNAprotect bacterial reagent (Qiagen) was added, and the mixture was incubated for 5 min at room temperature. Cells were then centrifuged and incubated with TE buffer (30 mM Tris-Cl, 1 mM EDTA, pH 8.0 [Sigma]) containing 0.5-mg/ml lysostaphin (Sigma) and 15-mg/ml lysozyme (Sigma) for 20 min at 37°C. Proteinase K (20 mg/ml [Sigma]) was added, and the mixture was incubated for 10 min. After this, the procedure was carried out according to the manufacturer's specifications. The purified RNA was quantified using a NanoDrop spectrometer (ThermoScientific). RNA quality was assessed by visualization on an agarose gel. The rRNA was removed using the MICROBExpress kit (Ambion). RNA quality was then evaluated on a ByoAnalyzer (Agilent). Bacterial mRNA was fragmented (yield fragments were in the size range of 200 to 250 bp), and the double-stranded cDNA was generated using the Ion Total RNA-seq kit v2 (Life Technologies, Thermo Fisher Scientific) according to the manufacturer's instructions. The samples were sequenced using the Ion PGM (Life Technologies, Thermo Fisher Scientific) sequencer at STABvida.
Mapping to the reference genomes and normalization of gene expression were performed by CLC Genomics Workbench v8.0.1. RNA-seq reads were aligned with the three available S. pseudintermedius reference genomes ED99 (ST25, agr type III, lacks spsF, spsO, and spsQ), HKU10-03 (ST308, agr type III, lacks nanB), and E140 (ST71, agr type III, lacks nanB, lukF, and lukS) (RefSeq accession no. CP002478, CP002439 and ANOI01000001, respectively). Gene expression was normalized by calculating reads per kilobase per million mapped reads (RKPM), given by dividing the total number of reads by the number of mapped reads (in millions) × the length in kilobases (20).
Differentially expressed genes were identified using Baggerly's test (binomial test), which compares the proportions of counts in a group of samples against those of another group of samples (21) with a false discovery rate (FDR) correction applied (22). Genes with an adjusted P value of ≤0.05 were identified as being differentially expressed. This study focused particularly in the expression of virulence genes, but expression of other relevant genes (e.g., antimicrobial resistance genes) was also evaluated.
Statistical analysis.
All data analysis was carried out using IBM SPSS Statistics version 20.0 (IBM, New York). Differences between the two groups MRSP and MSSP were calculated by Fisher's exact test for categorical comparisons and Student's t test for continuous outcome. A P value of ≤0.05 was considered statistically significant.
RESULTS
The results of MLST are shown in Table 1. The MSSP isolates were divided into 21 different STs, while 15 MRSP isolates were assigned to ST71, 3 to ST203, 1 to ST196, 1 to ST213, and 1 to ST195. Yet ST203 and ST195 belonged to clonal complex 71 (CC71), as detected by eBURST analysis. Equally, ST196 and ST213 differed by only one allele and belonged to CC196.
TABLE 1.
Epidemiological characteristics of the MRSP and MSSP isolates used in this study
| agr type | ST (no. of isolates) |
P valuea | |
|---|---|---|---|
| MSSP (21) | MRSP (21) | ||
| All | 0.025 | ||
| I | ST207 (1), ST215 (1) | No STs | >0.05 |
| II | ST201 (1), ST205 (1), ST206 (1), ST209 (1), ST217 (1) | ST196 (1), ST213 (1) | >0.05 |
| III | ST17 (1), ST197 (1), ST199 (1), ST200 (1), ST202 (1), ST204 (1), ST210 (1), ST211 (1), ST212 (1), ST214 (1), ST379 (1) | ST71 (15), ST195 (1), ST203 (3) | 0.014 |
| IV | ST198 (1), ST208 (1), ST216 (1) | No STs | >0.05 |
Shown are P values for differences between MRSP and MSSP.
All isolates were classified as part of one of the four agr groups, and the distribution was highly uneven, with 2 isolates belonging to agr group I, 7 belonging to group II, 30 belonging to group III, and 3 belonging to group IV (Table 1). There was a significant difference in the agr groups' distribution between MRSP and MSSP (P = 0.025), with allele III being significantly more associated with MRSP than with MSSP (P = 0.014).
The virulence genes detected in the MRSP and MSSP isolates are detailed in Table 2. Genes se-int, speta, siet, spsL, and ebpS were present in all 42 isolates. The genes lukF and lukS, encoding leukocidin Luk-I, were found in all isolates except for two MRSP isolates (ST196 and ST213). Gene expB was detected in only 3 isolates. Only two MSSP isolates carried the enterotoxin gene seccanine. No isolates harbored genes seh and expA. Eight isolates carried the spsO gene, and by statistical analysis, this gene was significantly more associated with MSSP than with MRSP (P = 0.04). No differences were found between clinical isolates and isolates from carriage.
TABLE 2.
Virulence traits of the MRSP and MSSP isolates
| Virulence genea | No. of isolates with virulence gene shown |
P value | ||
|---|---|---|---|---|
| Total (n = 42) | MRSP (n = 21) | MSSP (n = 21) | ||
| expA | 0 | 0 | 0 | >0.05 |
| expB | 3 | 0 | 3 | >0.05 |
| luk-I | 40 | 19 | 21 | >0.05 |
| seccanine | 2 | 0 | 2 | >0.05 |
| seh | 0 | 0 | 0 | >0.05 |
| spsD | 4 | 1 | 3 | >0.05 |
| spsO | 8 | 1 | 7 | 0.04 |
The genes ebpS, se-int, siet, speta, and spsL and the ica operon were positive in all isolates and were not included in this table.
Results related to the biofilm-forming ability on polystyrene are shown in Table 3. All isolates produced biofilm in the BHIB–4% NaCl medium. Two isolates did not produce biofilm on BHIB, and nine isolates did not produce biofilm on BHIB–1% glucose. Biofilm production in the BHIB and BHIB–1% glucose media was significantly higher in MSSP than in MRSP isolates (P = 0.03 and P = 0.02, respectively), but there were no differences between clinical isolates and isolates from carriage. The ica genes were detected in all 42 isolates.
TABLE 3.
Overall results of the microtiter plate test according to the pattern of methicillin resistance in S. pseudintermedius
| Medium | Mean OD570 ± SD | No. of isolates showing: |
|||
|---|---|---|---|---|---|
| No adherence | Weak adherence | Moderate adherence | Strong adherence | ||
| MRSP | |||||
| BHIB | 0.50 ± 0.052 | 1 | 5 | 15 | 0 |
| BHIB + 1% glucose | 0.28 ± 0.038 | 3 | 18 | 0 | 0 |
| BHIB + 4% NaCl | 0.44 ± 0.055 | 0 | 18 | 3 | 0 |
| MSSP | |||||
| BHIB | 0.57 ± 0.082 | 1 | 6 | 14 | 0 |
| BHIB + 1% glucose | 0.35 ± 0.081 | 6 | 15 | 0 | 0 |
| BHIB + 4% NaCl | 0.45 ± 0.098 | 0 | 13 | 8 | 0 |
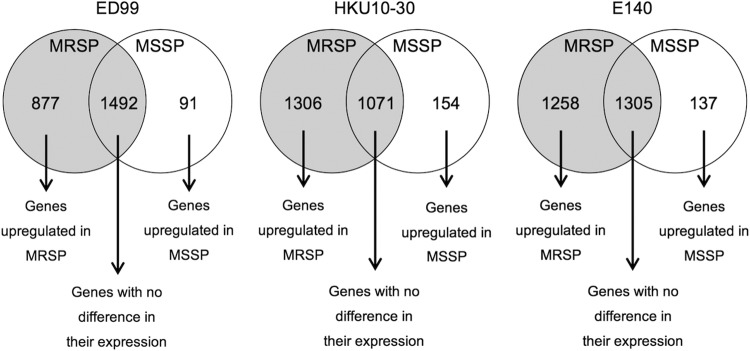
The numbers of mapped reads assigned by using each reference genome (ED99, HKU10-30, and E140) are shown in Table S2 in the supplemental material. Of these mapped reads, the number of S. pseudintermedius genes with altered expression also varied when using the three different reference genomes, as shown in Fig. 1. The MSSP isolate had higher expression in transcription of regulatory genes agrB and agrD. On the other hand, the MRSP isolate had higher transcription of regulatory genes sigB, srrA, sarA, rot, and the saeRS system. The signal transduction protein TRAP gene (traP) was also highly expressed. Considering genes encoding surface proteins, only one, spsC, encoding an autolysin, was highly expressed in the MSSP isolate, while 6, spsA, spsB, spsD, spsK, spsL, and spsN, were highly expressed in the MRSP isolate. The gamma-hemolysin component B gene (hlgB), both subunits of the luk-I gene (lukF-I and lukS-I), and the coagulase and thermonuclease genes (coa and nucC, respectively) were upregulated in the MRSP isolate. The arc genes (arcA, arcB, arcC, and arcD) were upregulated in the MSSP isolate. Several genes associated with antimicrobial resistance were highly expressed in the MRSP isolate: the norA, gyrA and gyrB genes associated with quinolone resistance, the aadE and the bifunctional aacA-aphD genes associated with aminoglycoside resistance, the mecA, mecR1, and blaI genes associated with β-lactam resistance, and the tet(M) gene associated with tetracycline resistance. The MRSP isolate upregulated several phage-associated genes (encoding phage capsid protein, phage infection protein, two phage portal proteins, and a phage-like protein), and an integrase gene located in the superantigen-encoding pathogenicity island SaPI (SPSINT_0063).
FIG 1.
Number of S. pseudintermedius genes with altered expression identified by Baggerly's test with the FDR correction applied, using the three different reference genomes available ED99 (ST25, agr III), HKU10-30 (ST308, agr III), and E140 (ST71, agr III).
DISCUSSION
In the last 10 years, MRSP isolates have become highly frequent in clinical samples from infected animals and as colonizers of healthy ones (1). However, it is still not clear why MRSP isolates, especially certain lineages like ST71, have spread so quickly. To understand the rapid evolution that led to the dissemination of MRSP isolates, we assessed the virulence determinants present in a collection of MSSP and MRSP isolates and compared their abilities to form biofilm in 3 different media. Finally, we performed in vitro gene expression analysis and compared the levels of expression of one MSSP isolate and one MRSP isolate.
Analysis of the virulence genotype of the MRSP and MSSP isolates revealed a strong conservation of genes: five genes (ebpS, se-int, siet, speta, and spsL) were carried by all S. pseudintermedius isolates, and five genes (expB, luk-I, seccanine, spsD, and spsO) were only present in some isolates. Two studies have reported the existence of some specific toxin genes (e.g., coa, lip, geh, htrA, nuc, clpX, hlb, se-int, speta, spsA, spsB, and spsC) present in several S. pseudintermedius isolates that might be important for the canine host tropism, in particular the skin (1, 23). However, variation was found in others (e.g., spsF, spsO, spsP, spsQ, luk-I, and nanB), suggesting that a difference in virulence factors in the core genome was probably lineage associated (1). For example, in one of these studies, the five ST71 isolates lacked the nanB and lukF and -S genes (1). Still, in our study and in a previous study conducted in Spain (24), all of the ST71 isolates carried the lukF and -S genes, suggesting that variation may also be related to the region of isolation. It would be interesting to collect a large collection of ST71 isolates from different countries to study these variations. In other lineages, however, this will be difficult to ascertain, since only a few isolates in each lineage have been reported so far.
The capacity of bacteria to form biofilms is an important virulence factor not only in the development of device-related infections but also in a range of chronic infections (23). This capacity might further complicate the treatment of already challenging infections due to the decrease in effectiveness of antimicrobials on biofilms (5). In one study, all S. pseudintermedius isolates produced biofilms, suggesting that biofilm production might be essential for the pathogenicity of S. pseudintermedius (6). Yet, the study failed to find differences in the biofilm formation between MRSP and MSSP isolates. The number of MSSP isolates that was studied was low, and the authors suggested that further experiments with a larger number of isolates were warranted (6). By using a larger set of isolates, we observed that biofilm production in the BHIB and BHIB–1% glucose media was significantly higher in MSSP than in MRSP isolates. This is a phenomenon that has been observed in S. aureus, when comparing methicillin-resistant and methicillin-susceptible isolates, and is due to different triggering mechanisms leading to biofilm formation, including ica-dependent and -independent mechanisms (25). In our study, all isolates produced biofilm, and all were positive for the ica genes, suggesting this operon has a crucial role in biofilm formation. However, the mechanisms triggering the higher biofilm production in the BHIB and BHIB–1% glucose media by MSSP strains remain unknown. One clue to this occurrence may be related to the upregulation of the entire arc operon in the MSSP isolate studied here. A similar operon has been found in other staphylococcal species, and in S. aureus, arcA (which belongs to the arc operon) encodes an arginine deaminase, which allows for enhanced survival in acidic environments (26). The upregulation of this operon may improve survival and promote biofilm formation of MSSP in acidic circumstances, such as in BHIB medium with glucose (which has a more acidic pH than BHIB medium alone or with NaCl).
During the early emergence of community-acquired MRSA, the USA300 (ST8) lineage disseminated rapidly and was considered hypervirulent, compared with lineages like MRSA USA400 (ST1) (26). However, USA300 does not contain many more virulence genes than USA400, but it does have an alteration in the expression of regulatory genes and increased expression of certain virulence genes (26). By microarray analysis, USA300 displayed an increased expression of genes encoding cell envelope proteins (including lipoproteins and superantigen-like proteins), genes residing in the prophage ϕSa3usa, several genes contained in pathogenicity islands vSAα and vSAß, genes encoding proteases, and the gene encoding the IgG binding protein Sbi (26). Interestingly our MRSP isolate also had increased expression of several genes, including spsK, which encodes the IgG binding protein Sbi, the toxin genes nucC and coa, prophage genes, and several virulence regulatory genes, including saeRS. The higher expression of the prophage genes might be one of the factors contributing to the rapid dissemination of MRSP, particularly ST71 isolates. The higher expression of the genes spsD and spsL (encoding fibronectin-binding proteins able to adhere to the extracellular matrix) found in this study may explain the higher adherence of MRSP ST71 isolates to corneocytes previously detected (2). We observed a very different expression of virulence regulatory genes between the two isolates, with agr highly expressed in MSSP and saeRS highly expressed in MRSP. This may explain the differences observed in the expression of the genes encoding surface proteins and toxins.
Among the most important bacterial defenses against uptake of foreign DNA are the restriction-modification (R-M) systems (27). These systems, comprising restriction endonucleases and methyltransferases, recognize and modify specific DNA sequences, protecting “own” DNA from restriction while eliminating potentially harmful foreign DNA (27). In S. pseudintermedius, type I R-M systems have been recognized, including one that was carried on all SCCmec II-III elements of MRSP ST71 (1). One study suggested that MRSP isolates were not more efficient or inefficient than MSSP isolates in acquiring mobile genetic elements due to the wide distribution of the type I and type II R-M systems in S. pseudintermedius isolates (1). In our study, however, we found that the type I restriction-modification system restriction subunit R (hsdR) was highly expressed in the MSSP isolate, suggesting it blocks DNA horizontal gene transfer into methicillin-susceptible isolates. Lower expression of subunit R in the MRSP isolate could also suggest a more efficient way of acquiring mobile genetic elements. In fact, it has been shown that MRSP genomes carry more prophages than MSSP isolates. Our results showed that the MRSP isolate also upregulates several phage-associated genes, which could be linked to the upregulation of the integrase located in the superantigen-encoding pathogenicity island, SaPI. The upregulation of prophage particles is also concordant with the suggestion that transfer in MRSP is predominantly made by transduction (1).
In summary, this is the first study to document the global transcription differences between the MSSP and MRSP isolates during in vitro growth. This study indicates that MRSP may upregulate surface proteins, which may increase the adherence of MRSP isolates (especially ST71) to corneocytes. Although MRSP and MSSP have the capacity to form biofilm, MSSP may have an increased ability to form biofilm under acidic circumstances, through upregulation of the entire arc operon. Complete understanding of S. pseudintermedius pathogenesis and host-pathogen signal interaction during infections is critical for the treatment and prevention of S. pseudintermedius infections.
Supplementary Material
ACKNOWLEDGMENTS
We thank Elena Gómez-Sanz, Myriam Zarazaga, and Carmen Torres for providing control isolates for some of the virulence genes and Rui Seixas from FMV-UL and Magdalena Lewicka from STABvida for excellent technical assistance.
Funding Statement
This work was funded by national funds through the FCT—Fundação para a Ciência e Tecnologia, Project PTDC/CVT-EPI/4345/2012 and Project UID/CVT/00276/2013, and a Ph.D. grant SFRH/BD/68864/2010 to Natacha Couto from the same institution. Manuela Oliveira is a researcher from the program “Ciência 2007” from FCT, Portugal.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01907-15.
REFERENCES
- 1.McCarthy AJ, Harrison EM, Stanczak-Mrozek K, Legget B, Waller A, Holmes MA, Lloyd DH, Lindsay JA, Loeffler A. 2015. Genomic insights into the rapid emergence and evolution of MDR in Staphylococcus pseudintermedius. J Antimicrob Chemother 70:997–1007. doi: 10.1093/jac/dku496. [DOI] [PubMed] [Google Scholar]
- 2.Latronico F, Moodley A, Nielsen SS, Guardabassi L. 2014. Enhanced adherence of methicillin-resistant Staphylococcus pseudintermedius sequence type 71 to canine and human corneocytes. Vet Res 45:70. doi: 10.1186/1297-9716-45-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perreten V, Kadlec K, Schwarz S, Grönlund Andersson U, Finn M, Greko C, Moodley A, Kania SA, Frank LA, Bemis DA, Franco A, Iurescia M, Battisti A, Duim B, Wagenaar JA, van Duijkeren E, Weese JS, Fitzgerald JR, Rossano A, Guardabassi L. 2010. Clonal spread of methicillin-resistant Staphylococcus pseudintermedius in Europe and North America: an international multicentre study. J Antimicrob Chemother 65:1145–1154. doi: 10.1093/jac/dkq078. [DOI] [PubMed] [Google Scholar]
- 4.Kadlec K, Schwarz S, Perreten V, Andersson UG, Finn M, Greko C, Moodley A, Kania SA, Frank LA, Bemis DA, Franco A, Iurescia M, Battisti A, Duim B, Wagenaar JA, van Duijkeren E, Weese JS, Fitzgerald JR, Rossano A, Guardabassi L. 2010. Molecular analysis of methicillin-resistant Staphylococcus pseudintermedius of feline origin from different European countries and North America. J Antimicrob Chemother 65:1826–1828. doi: 10.1093/jac/dkq203. [DOI] [PubMed] [Google Scholar]
- 5.Osland AM, Vestby LK, Fanuelsen H, Slettemeås JS, Sunde M. 2012. Clonal diversity and biofilm-forming ability of methicillin-resistant Staphylococcus pseudintermedius. J Antimicrob Chemother 67:841–848. doi: 10.1093/jac/dkr576. [DOI] [PubMed] [Google Scholar]
- 6.Singh A, Walker M, Rosseau J, Weese SJ. 2013. Characterization of the biofilm forming ability of Staphylococcus pseudintermedius from dog. BMC Vet Res 9:93. doi: 10.1186/1746-6148-9-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crawford EC, Singh A, Metcalf D, Gibson TWG, Weese SJ. 2014. Identification of appropriate reference genes for qPCR studies in Staphylococcus pseudintermedius and preliminary assessment of icaA gene expression in biofilm-embedded bacteria. BMC Res Notes 7:451. doi: 10.1186/1756-0500-7-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bannoehr J, Brown JK, Shaw DJ, Fitzgerald RJ, van den Broek AHM, Thoday KL. 2012. Staphylococccus pseudintermedius surface proteins SpsD and SpsO mediate adherence to ex vivo canine corneocytes. Vet Dermatol 23:119–124. doi: 10.1111/j.1365-3164.2011.01021.x. [DOI] [PubMed] [Google Scholar]
- 9.Solyman SM, Black CC, Duim B, Perreten V, van Duijkeren E, Wagenaar JA, Eberlein LC, Sadeghi LN, Videla R, Bemis DA, Kania SA. 2013. Multilocus sequence typing for characterization of Staphylococcus pseudintermedius. J Clin Microbiol 51:306–354. doi: 10.1128/JCM.02421-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spratt BG, Hanage WP, Li B, Aanensen DM, Feil EJ. 2004. Displaying the relatedness among isolates of bacterial species—the eBURST approach. FEMS Microbiol Lett 241:129–134. doi: 10.1016/j.femsle.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 11.Becker K, Roth R, Peters G. 1998. Rapid and specific detection of toxigenic Staphylococcus aureus: use of two multiplex PCR enzyme immunoassays for amplification and hybridization of staphylococcal enterotoxin genes, exfoliative toxin genes, and toxic shock syndrome toxin 1 gene. J Clin Microbiol 36:2548–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monday SR, Bohach GA. 1999. Use of multiplex PCR to detect classical and newly described pyrogenic toxin genes in staphylococcal isolates. J Clin Microbiol 37:3411–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Futagawa-Saito K, Sugiyama T, Karube S, Sakurai N, Ba-Thein W, Fukuyasu T. 2004. Prevalence and characterization of leukotoxin-producing Staphylococcus intermedius in isolates from dogs and pigeons. J Clin Microbiol 42:5324–5326. doi: 10.1128/JCM.42.11.5324-5326.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cucarella C, Solano C, Valle J, Amorena B, Lasa I, Penadés JR. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J Bacteriol 183:2888–2896. doi: 10.1128/JB.183.9.2888-2896.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lautz S, Kanbar T, Alber J, Lämmler C, Weiss R, Prenger-Berninghoff E, Zschöck M. 2006. Dissemination of the gene encoding exfoliative toxin of Staphylococcus intermedius among isolates isolated from dogs during routine microbiological diagnostics. J Vet Med B Infect Dis Vet Public Health 53:434–438. doi: 10.1111/j.1439-0450.2006.00999.x. [DOI] [PubMed] [Google Scholar]
- 16.Futagawa-Saito K, Makino S, Sunaga F, Kato Y, Sakurai-Komada N, Ba-Thein W, Fukuyasu T. 2009. Identification of first exfoliative toxin in Staphylococcus pseudintermedius. FEMS Microbiol Lett 301:176–180. doi: 10.1111/j.1574-6968.2009.01823.x. [DOI] [PubMed] [Google Scholar]
- 17.Stepanovic S, Vukovic D, Dakic I, Savic B, Svabic-Vlahovic M. 2000. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods 40:175–179. doi: 10.1016/S0167-7012(00)00122-6. [DOI] [PubMed] [Google Scholar]
- 18.Pettit RK, Weber CA, Kean MJ, Hoffmann H, Pettit GR, Tan R, Franks KS, Horton ML. 2005. Microplate Alamar Blue assay for Staphylococcus epidermidis biofilm susceptibility testing. Antimicrob Agents Chemother 49:2612–2617. doi: 10.1128/AAC.49.7.2612-2617.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bannoehr J, Ben Zakour NL, Reglinski M, Inglis NF, Prabhakaran S, Fossum E, Smith DG, Wilson GJ, Cartwright RA, Haas J, Hook M, van den Broek AH, Thoday KL, Fitzgerald JR. 2011. Genomic and surface proteomic analysis of the canine pathogen Staphylococcus pseudintermedius reveals proteins that mediate adherence to the extracellular matrix. Infect Immun 79:3074–3086. doi: 10.1128/IAI.00137-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods 5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 21.Baggerly K, Deng L, Morris J, Aldaz C. 2003. Differential expression in SAGE: accounting for normal between-library variation. Bioinformatics 19:1477–1483. doi: 10.1093/bioinformatics/btg173. [DOI] [PubMed] [Google Scholar]
- 22.Pawitan Y, Michiels S, Koscielny S, Gusnanto A, Ploner A. 2005. False discovery rate, sensitivity and sample size for microarray studies. Bioinformatics 21:3017–3024. [DOI] [PubMed] [Google Scholar]
- 23.Ben Zakour NL, Beatson SA, van den Broek AHM, Thoday KL, Fitzgerald JR. 2012. Comparative genomics of the Staphylococcus intermedius group of animal pathogens. Front Cell Infect Microbiol 2:44. doi: 10.3389/fcimb.2012.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gómez-Sanz E, Torres C, Lozano C, Zarazaga M. 2013. High diversity of Staphylococcus aureus and Staphylococcus pseudintermedius lineages and toxigenic traits in healthy pet-owning household members. Underestimating normal household contact? Comp Immunol Microbiol Infect Dis 36:83–94. doi: 10.1016/j.cimid.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 25.O'Neill E, Humphreys H, O'Gara JP. 2009. Carriage of both the fnbA and fnbB genes and growth at 37°C promote FnBP-mediated biofilm development in methicillin-resistant Staphylococcus aureus clinical isolates. J Med Microbiol 58:399–402. doi: 10.1099/jmm.0.005504-0. [DOI] [PubMed] [Google Scholar]
- 26.Jones MB, Montgomery CP, Boyle-Vavra S, Shatzkes K, Maybank R, Frank BC, Peterson SN, Daum RS. 2014. Genomic and transcriptomic differences in community acquired methicillin resistant Staphylococcus aureus USA300 and USA400 isolates. BMC Genomics 15:1145. doi: 10.1186/1471-2164-15-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murray NE. 2000. Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle). Microbiol Mol Biol Rev 64:412–434. doi: 10.1128/MMBR.64.2.412-434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.