Abstract
Despite the existing treatment options for onychomycosis, there remains a strong demand for potent topical medications. ME1111 is a novel antifungal agent that is active against dermatophytes, has an excellent ability to penetrate human nails, and is being developed as a topical agent for onychomycosis. In the present study, we investigated its mechanism of action. Trichophyton mentagrophytes mutants with reduced susceptibility to ME1111 were selected in our laboratory, and genome sequences were determined for 3 resistant mutants. The inhibitory effect on a candidate target was evaluated by a spectrophotometric enzyme assay using mitochondrial fractions. Point mutations were introduced into candidate genes by a reverse genetics approach. Whole-genome analysis of the 3 selected mutants revealed point mutations in the structural regions of genes encoding subunits of succinate dehydrogenase (complex II). All of the laboratory-generated resistant mutants tested harbored a mutation in one of the subunits of succinate dehydrogenase (SdhB, SdhC, or SdhD). Most of the mutants showed cross-resistance to carboxin and boscalid, which are succinate dehydrogenase inhibitors. ME1111 strongly inhibited the succinate-2,6-dichlorophenolindophenol reductase reaction in Trichophyton rubrum and T. mentagrophytes (50% inhibitory concentrations [IC50s] of 0.029 and 0.025 μg/ml, respectively) but demonstrated only moderate inhibition of the same reaction in human cell lines. Furthermore, the target protein of ME1111 was confirmed by the introduction of point mutations causing the amino acid substitutions in SdhB, SdhC, and SdhD found in the laboratory-generated resistant mutants, which resulted in reduced susceptibility to ME1111. Thus, ME1111 is a novel inhibitor of the succinate dehydrogenase of Trichophyton species, and its mechanism of action indicates its selective profile.
INTRODUCTION
Onychomycosis (also known as tinea unguium) is a progressive fungal infection of nails and nail beds that leads to the destruction and deformity of nails, causing pain and discomfort. The disease affects around 10% of the adult population in the United States and other countries (1–4), and the prevalence of the disease increases in the elderly population. One-third of people over 60 years of age have been reported to have onychomycosis (1). Although the standard treatment for onychomycosis is the use of oral agents (terbinafine and itraconazole), the potential side effects, such as drug-drug interactions and liver toxicity, make topical agents, such as ciclopirox and amorolfine, an alternative option for patients with mild to moderate onychomycosis. Recently, new topical agents (efinaconazole and tavaborole) were launched in the United States (5, 6). The mechanisms of action of current drugs for onychomycosis can be classified into inhibition of ergosterol biosynthesis (terbinafine, amorolfine, itraconazole, and efinaconazole), chelation of polyvalent cations (ciclopirox), inhibition of leucyl-tRNA synthetase (tavaborole), and interaction with microtubules (griseofulvin) (7, 8). Despite the existing treatment options for onychomycosis, there remains an urgent need for a new chemical class of topical agents with greater efficacy and fewer systemic side effects.
Aerobic respiration is the most efficient path for production of ATP in eukaryotes. The electron transport chain in mitochondria is the final stage of aerobic respiration. First, electrons are delivered from either NADH:ubiquinone oxidoreductase (complex I) or succinate dehydrogenase (complex II) to the cytochrome bc1 complex (complex III), and then they are transferred to cytochrome c oxidase (complex IV). ATP synthase utilizes the generated proton potential to phosphorylate ADP to ATP. Succinate dehydrogenase (complex II), which is localized to the inner membranes of mitochondria, also catalyzes a reaction in the citric acid cycle (succinate + ubiquinone → fumarate + ubiquinol). The enzyme consists of 4 subunits: SdhA, SdhB, SdhC, and SdhD. Agricultural chemicals, such as carboxin and boscalid, exert their antifungal activity by binding to the ubiquinone-binding site of succinate dehydrogenase, which is a cleft formed by amino acid residues from SdhB, SdhC, and SdhD (9–11). Moreover, several inhibitors that block mitochondrial electron transport have been used in humans as anti-infectives. Atovaquone is a structural analog of ubiquinone which inhibits the cytochrome bc1 complex and demonstrates broad-spectrum antiprotozoal activity (12). Bedaquiline is a specific inhibitor of mycobacterial ATP synthase and was recently approved for the treatment of multidrug-resistant tuberculosis (13).
ME1111 [2-(3,5-dimethyl-1H-pyrazol-1-yl)-5-methylphenol] (Fig. 1) is a new agent discovered by Meiji Seika Pharma Co., Ltd. (Tokyo, Japan), that possesses potent in vitro antifungal activity against dermatophytes, such as Trichophyton rubrum and Trichophyton mentagrophytes, which are common causative organisms of onychomycosis (14, 15). A small molecular weight is reported to be one of the important factors of a compound for nail penetration ability (16). The small molecular weight of ME1111 (202.25) enables it to penetrate human nails very efficiently. Indeed, in our previous in vitro studies, ME1111 demonstrated greater nail penetration than that of ciclopirox (Penlac nail lacquer) (17, 18). General toxicity studies, including repeated-dose toxicity studies, safety pharmacology studies, and genotoxicity studies, were conducted in compliance with good laboratory practice (GLP), and no concerns about ME1111's safety were observed (19). Since ME1111 is a new chemical class of antifungal agent with an unknown mechanism of action, we aimed to identify the target protein of ME1111 by using genetic analyses, enzyme assays, and a reverse genetics approach involving introduction of mutations into the genes encoding the succinate dehydrogenase subunits.
FIG 1.
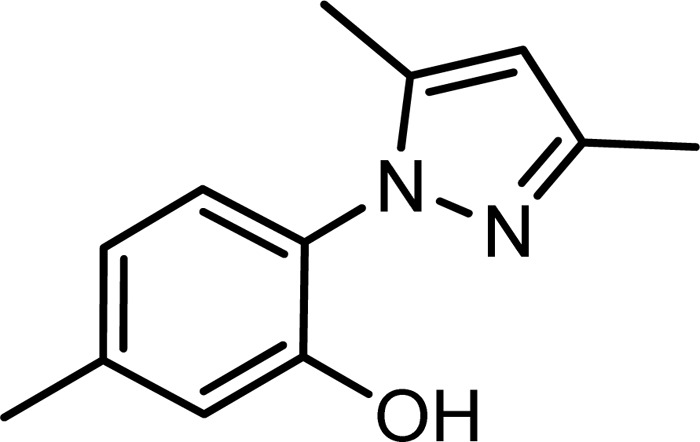
Chemical structure of ME1111.
(This study was presented in part at the 54th Interscience Conference on Antimicrobial Agents and Chemotherapy, 5 to 9 September 2014, Washington, DC [20], and the 55th Interscience Conference on Antimicrobial Agents and Chemotherapy, 17 to 21 September 2015, San Diego, CA [21].)
MATERIALS AND METHODS
Chemicals, fungal strains, and MIC testing.
ME1111 was synthesized at Meiji Seika Pharma Co., Ltd. Carboxin and boscalid were purchased from Wako Pure Chemical Industries, Ltd. (Tokyo, Japan). Ciclopirox olamine and amorolfine hydrochloride were obtained from Sigma-Aldrich Co. (St. Louis, MO) and LKT Laboratories, Inc. (St. Paul, MN), respectively. T. mentagrophytes ATCC 18748, T. mentagrophytes ATCC MYA-4439, and T. rubrum ATCC MYA-4438 were obtained from the American Type Culture Collection (Manassas, VA). T. mentagrophytes (anamorph of Arthroderma vanbreuseghemii) TmL28, a DNA ligase 4 (Lig 4)-defective mutant of T. mentagrophytes TIMM2789 (22), was used as a host strain for transformation. K562 (a human erythromyeloblastoid leukemia cell line) and HepG2 (a human hepatocellular carcinoma cell line) cells were obtained from DS Pharma Biomedical Co., Ltd. (Suita, Japan). MICs were determined by the broth microdilution method of the Clinical and Laboratory Standards Institute (23).
Selection of spontaneous ME1111-resistant T. mentagrophytes mutants.
A conidial suspension of T. mentagrophytes ATCC 18748 was applied to Sabouraud dextrose agar (SDA) plates containing 1 μg/ml of ME1111 and then cultured at 28°C for 2 weeks. Colonies grown on SDA with ME1111 were then subcultured separately onto fresh ME1111-containing SDA plates, and the resulting 17 colonies were identified as laboratory-generated ME1111-resistant mutants (M1 to M17).
Comparative analysis of the genomes of the parent strain and laboratory-selected ME1111-resistant mutants.
Genomic DNAs of T. mentagrophytes ATCC 18748 and 3 ME1111-resistant mutants selected in vitro (M1, M8, and M15) were purified using DNeasy Plant Maxi kits (Qiagen, Venlo, The Netherlands). Approximately 2 g (wet weight) of each mycelium, collected from 200 ml of Sabouraud dextrose broth (SDB) cultures, was first digested with 150 U/ml of Zymolyase 20T (Seikagaku Corporation, Tokyo, Japan) in 1 M sorbitol–0.1 M EDTA (pH 8)–14 mM 2-mercaptoethanol. After cell wall digestion, the genomic DNA was extracted by using the kit according to the manufacturer's protocol. Genome sequencing was performed at TaKaRa Bio Inc. (Otsu, Japan). A draft genome sequence of the parent strain was first constructed from pyrosequencing fragments and paired-end libraries of its genomic DNA by using an FLX sequencing system (Roche Diagnostics, Basel, Switzerland). Shotgun sequencing was then performed by use of a HiSeq 2000 system (Illumina Inc., San Diego, CA) to confirm and correct the sequences. Gene prediction was based on the genomic sequence of Coccidioides immitis. Genome sequencing of the 3 resistant mutants was performed with the HiSeq 2000 system, and their sequence data were mapped to the final genome sequence of the parent strain in order to identify mutated bases.
Sequencing of the sdhB, sdhC, and sdhD genes of the laboratory-generated ME1111-resistant mutants.
Genomic DNAs were extracted from T. mentagrophytes ATCC 18748 and the 17 laboratory-generated ME1111-resistant mutants by use of RNeasy Plant mini kits (Qiagen). Briefly, cells grown in 100 ml of SDB were collected by filtration, and about 0.1 g (wet weight) of mycelium from each strain was suspended in 1 ml of 0.6 M ammonium sulfate containing 2% Yatalase (TaKaRa Bio Inc.). The solutions were incubated at 30°C for 2 h and then centrifuged (5,000 × g, 10 min) to collect the cells. The cells were then resuspended in 0.45 ml of buffer RLT, contained in the kit, and genomic DNA was extracted according to the protocol provided with the kit. Primers used for amplification of the genes encoding three of the four subunits constituting succinate dehydrogenase (sdhB, sdhC, and sdhD) are shown in Table 1. DNA fragments of the sdhB, sdhC, or sdhD gene were amplified under the following PCR conditions: 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min. Using one of the amplification primers or sequencing primers shown in Table 1, sequencing was carried out with a BigDye Terminator v1.1 cycle sequencing kit (Applied Biosystems, Waltham, MA) and a model 3730 DNA analyzer (Applied Biosystems).
TABLE 1.
PCR primers used for genetic analysis of laboratory-generated ME1111-resistant mutants
| Purpose | Gene target | Primer sequences (5′–3′) |
|---|---|---|
| Amplification | sdhB | GGTTCAAGCGCTCTCATGC and GTGGTAGCCAAGGGTATTTGC |
| sdhC | CAATTCGATCAGACGCGAAG and GTCCATACTTCGATAAGGAAC | |
| sdhD | GCTTCCCGAGGATGTTTTTCG and GGGTACAAAAAAGACAGAAATACAGG | |
| Sequencing | sdhB | GAACATTGACGGAGTAAACAC, GCTGTACGAATGTATTCTCTG, and GTGTTTACTCCGTCAATGTTC |
| sdhC | TATACCGACCGCAGATAACC and CTGGCGAAGATATAGAGACC | |
| sdhD | AACCACTAACGATGCTGCTCC and GCAACGACTCTCTCAAAGG |
Preparation of mitochondrial fractions of T. mentagrophytes and T. rubrum.
Mitochondrial fractions of T. rubrum ATCC MYA-4438, T. mentagrophytes ATCC 18748, and 6 laboratory-generated ME1111-resistant mutants (M1, M11, M13, M15, M16, and M17) were prepared from mycelia cultured in SDB at 28°C for 3 to 7 days. The collected cells were incubated at 30°C for about 3 h in 0.6 M ammonium sulfate containing 1% Yatalase and then resuspended in 10 ml of 0.25 M sucrose-20 mM HEPES buffer-10 mM potassium chloride-0.1 mM EDTA-0.15% bovine serum albumin (BSA). The cells were disrupted by an ultrasonic cell disruptor (Bioruptor; Cosmo Bio Co., Ltd., Tokyo, Japan) and then centrifuged (3,700 × g, 5 min). The supernatants obtained were further centrifuged (5,000 × g, 15 min) to precipitate the mitochondria. The resultant pellets were resuspended in 1 ml of 0.25 M sucrose-20 mM HEPES buffer, and the suspensions were centrifuged (2,000 × g, 5 min) to collect the supernatants. These supernatants were further centrifuged (7,700 × g, 20 min) to precipitate the mitochondria, which were then resuspended in 1 ml of 0.25 M sucrose-20 mM HEPES buffer. The protein concentrations of these mitochondrial fractions were determined by the Bradford method, using a commercial protein assay (Bio-Rad Laboratories, Inc., Hercules, CA).
Preparation of mitochondrial fractions from human cell lines.
K562 and HepG2 cells were cultured at 37°C and 5% CO2 in RPMI 1640 and minimal essential medium (MEM), respectively. The adherent HepG2 cells were treated with 0.25% trypsin-EDTA at 37°C for about 5 min. Collected cells were resuspended in about 10 ml of ice-cold 0.25 M sucrose–3 mM Tris-HCl (pH 7.6)–0.1 mM EDTA, and mitochondrial fractions were obtained in the same way as that described for T. mentagrophytes and T. rubrum.
Measurement of succinate dehydrogenase (complex II) activity.
Succinate dehydrogenase activity was measured by observing the reduction of 2,6-dichlorophenolindophenol (DCIP), an artificial electron acceptor, by using a previously described method (24), with slight modifications. Upon receiving electrons, DCIP is reduced and its color changes from blue to colorless. Reaction solutions, which were prepared to reach final concentrations of 50 mM phosphate buffer solution (pH 7.4), 1 mM EDTA, 0.1 mM DCIP, and 30 mM disodium succinate, were applied to wells of a 96-well plate in aliquots of 186 μl. ME1111 and reference compounds were dissolved in ethanol, and each solution was mixed with the same amount of distilled water to obtain a 50% ethanol solution. The compounds diluted with 50% ethanol were added to wells in aliquots of 4 μl, and the solutions were mixed. Four microliters of 50% ethanol was added to drug-free wells. Ten microliters of the mitochondrial fraction, prepared to reach a final protein concentration of 20 to 25 μg/ml, was added to initiate the reaction. The absorbance at a wavelength of 595 nm (optical density at 595 nm [OD595]) was measured using a plate reader (EnVision 2102 multilabel reader; PerkinElmer, Inc., Waltham, MA) immediately after addition of the mitochondrial fraction and again after incubation at 28°C for 20 to 60 min. The succinate-DCIP reductase activity was calculated as a percentage by dividing the decrease in the OD595 of the drug-treated wells by the average decrease in the OD595 of the drug-free wells. The concentration at which 50% inhibition was observed compared to that of the drug-free wells was considered the 50% inhibitory concentration (IC50).
Construction of gene replacement cassettes.
For homologous recombination of the sdhB, sdhC, and sdhD loci in T. mentagrophytes TmL28, two DNA fragments were first amplified by PCR. One fragment contained the 5′-untranslated region and the coding region of the target gene, and the other fragment contained the 3′-untranslated region of the target gene (each fragment was approximately 2 to 3 kb). PCR primers (Table 2) were designed based on the whole-genome sequence of T. mentagrophytes TIMM2789 (unpublished data). Total DNA was extracted from mycelia of TmL28 according to the method of Girardin and Latge (25) and used as a template for PCR.
TABLE 2.
PCR primers used to construct gene replacement cassettes
| Purpose | Locus | Primer | Sequence (5′–3′) |
|---|---|---|---|
| Amplification of 5′-untranslated region and coding region | SdhB | SdhB-F1/SpeI | AAACTAGTACCTCCAGATTCAAACGGAGGC |
| SdhB | SdhB-R4/ApaI | CCGGGCCCAGTGGTAGCCAAGGGTATTTGC | |
| SdhC | SdhC-F3/SpeI | CCACTAGTCCGCTGACAAGATGCGCAAGC | |
| SdhC | SdhC-R6/ApaI | AGCCGGGCCCTCCATACTTCGATAAGGAAC | |
| SdhD | SdhD-F1/SpeI | AAACTAGTTCCAGTACCCGAGCAGAGACAC | |
| SdhD | SdhD-R4/ApaI | TAGGGCCCATATACATTTACGCCTTCCAGA | |
| Amplification of 3′-untranslated region | SdhB | SdhB-F2/BamHI | CCGGATCCTCTCATAGCTTTCCACGTGTCT |
| SdhB | SdhB-R2/SacI | GAGAGCTCGAACGCAGCAGCAGGTGGTAGT | |
| SdhC | SdhC-F8/BamHI | GTGGATCCTCTTGTTTGAAACCGATCATCC | |
| SdhC | SdhC-R2/SacI | AGTACGAGCTCACAACCTGATATGCGTACA | |
| SdhD | SdhD-F2/BamHI | CGTGGATCCGTCTTCGGTCCCATGGATACT | |
| SdhD | SdhD-R2/SacI | ACGGAGCTCTTTCCACGGTGCCAATGATCT | |
| Introduction of a point mutation | SdhB(His234Leu) | SdhB-F3(H234L) | GTGTACAGATGCCTCACCATTC |
| SdhB(His234Leu) | SdhB-R3(H234L) | GAATGGTGAGGCATCTGTACAC | |
| SdhB(Asn238Lys) | SdhB(N238K)F | ACCATTCTTAAATGCTCGAGAACT | |
| SdhB(Asn238Lys) | SdhB(N238K)R | AGTTCTCGAGCATTTAAGAATGGT | |
| SdhC(Thr83Asn) | SdhC-F5(T83N) | CCGCAGATAAACTGGTACCTGTCCAGT | |
| SdhC(Thr83Asn) | SdhC-R3(T83N) | ACAGGTACCAGTTTATCTGCGGTCGGTA | |
| SdhC(Ser88Gly) | SdhC-F6(S88G) | GTACCTGTCCGGTCTTAACCGTATTACTG | |
| SdhC(Ser88Gly) | SdhC-R4(S88G) | CAGTAATACGGTTAAGACCGGACAGGTA | |
| SdhC(Asn90Lys) | SdhC-F7(N90K) | CCTGTCCAGTCTTAAGCGTATTACTGGTGC | |
| SdhC(Asn90Lys) | SdhC-R5(N90K) | CCAGTAATACGCTTAAGACTGGACAGGTA | |
| SdhD(Asp161His) | SdhD-F3(D161H) | CATGATTGCCCATTACTTCCGT | |
| SdhD(Asp161His) | SdhD-R3(D161H) | ACGGAAGTAATGGGCAATCATGGC | |
| SdhB(Gly250Arg) | Sdh(G250R)R | AAAGGGCCCAGTGGTAGCCAAGGGTATTTGCATTTTAGTTGAAGGACATCATCTTCTTGATTTCAGCAATGGCCTTTCTGGGGTTCAGGCCCTT |
A point mutation leading to an amino acid substitution in one of the subunits was introduced into the coding region of each construct by the overlap extension PCR method. For this purpose, two DNA fragments were amplified separately by PCR, using a forward primer (either SdhB-F1/SpeI, SdhC-F3/SpeI, or SdhD-F1/SpeI) and a reverse primer for the mutation or a forward primer for the mutation and a reverse primer (either SdhB-R4/ApaI, SdhC-R6/ApaI, or SdhD-R4/ApaI) (Table 2). The two DNA fragments thus obtained were mixed and used as the template for the next PCR, which was performed using the forward and reverse primer pairs shown for the amplification of the 5′-untranslated region and the coding region. In the case of the Gly250Arg substitution in SdhB, PCR was performed using primers SdhB-F1/SpeI and Sdh(G250R)R to introduce a point mutation. The resulting DNA fragments were sequenced to verify introduction of the correct point mutation at the target site.
The two DNA fragments obtained for each gene were subcloned into the binary vector pAg1-hph (26) after digestion with restriction enzymes: the upstream fragment carrying the point mutation was subcloned into the SpeI-ApaI sites upstream of the hph cassette, and the downstream fragment was inserted into the BamHI-SacI sites downstream of the hph cassette (Fig. 2). Groups of constructs carrying point mutations at different positions in the sdhB, sdhC, and sdhD loci were designated the pAg1h-SdhB/T, pAg1h-SdhC/T, and pAg1h-SdhD/T series, respectively. The constructs were transfected into Escherichia coli DH5α cells, and plasmid DNA was extracted from E. coli by the miniprep method and then sequenced.
FIG 2.
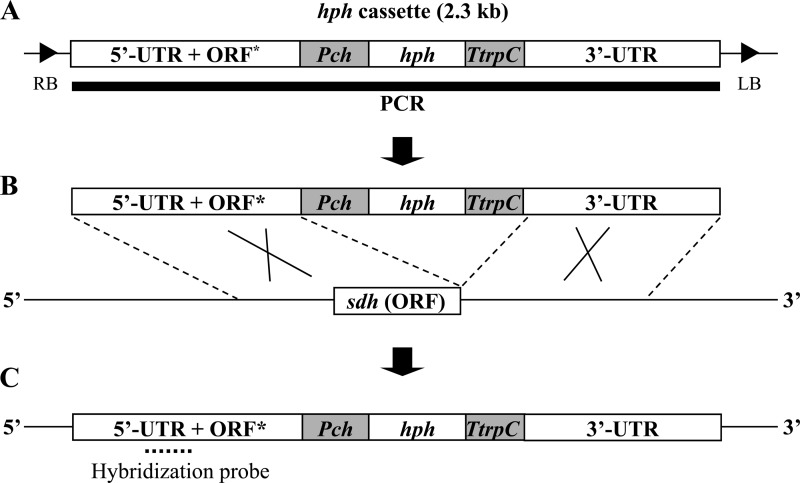
Introduction of a point mutation into the sdhB, sdhC, or sdhD locus by transformation of the respective DNA cassette. (A) Schematic presentation of part of the pAg1-hph vector carrying a gene replacement cassette. The 5′-untranslated region (UTR) plus an open reading frame containing a point mutation of the sdhB, sdhC, or sdhD locus (ORF*) and the 3′-UTR of the respective locus were inserted upstream and downstream of the hph cassette, respectively. RB, right border sequence; LB, left border sequence; Pch, Cochliobolus heterostrophus promoter 1; hph, E. coli hygromycin B phosphotransferase gene; TtrpC, terminator sequence of Aspergillus nidulans tryptophan C gene. (B) Schematic presentation of the gene cassette, shown above the wild-type locus and the target open reading frame of the sdhB, sdhC, or sdhD gene [sdh (ORF)]. (C) Schematic presentation of the locus harboring a point mutation after homologous recombination.
Transformation and molecular analysis.
Plasmid DNAs of the pAg1h-SdhB/T, pAg1h-SdhC/T, and pAg1h-SdhD/T series were used as templates for PCR to amplify the sequences indicated in Fig. 2 (bold line). Primer pairs used for PCR were as follows: for the sdhB locus, SdhB-F1/SpeI and SdhB-R2/SacI; for the sdhC locus, SdhC-F3/SpeI and SdhC-R2/SacI; and for the sdhD locus, SdhD-F1/SpeI and SdhD-R2/SacI. The resultant DNA fragments were collected and purified by ethanol precipitation and then were introduced into TmL28 cells by the protoplast-polyethylene glycol (PEG) method (27). After the PEG treatment, protoplasts were inoculated onto SDA supplemented with 1.2 M d-sorbitol and 0.5% (wt/vol) yeast extract containing 250 μg/ml hygromycin B, and colonies grown on the selective agar were isolated for further investigation. Total DNAs were extracted from the respective cultured mycelia and digested with one of the following restriction enzymes: EcoRI (for sdhB), PstI (for sdhC), or XhoI (for sdhD). The digested DNA fragments were fractionated in 0.8% (wt/vol) agarose gels, transferred to Hybond-N+ membranes (GE Healthcare UK Ltd., Buckinghamshire, United Kingdom), and analyzed by Southern hybridization using the ECL direct nucleic acid labeling and detection system (GE Healthcare UK Ltd.). Hybridization probes were prepared by PCR amplification, using total DNA of strain TmL28 and the following specific primer pairs for the sdhB, sdhC, and sdhD loci: for the sdhB locus, 5′-GACCTTGTTCCAGATATGAC-3′ and SdhB-R4/ApaI; for the sdhC locus, 5′-AGGCACCTTGCTCGCGTGAC-3′ and 5′-TTCATCTCAGACCGGCTTGT-3′; and for the sdhD locus, 5′-CTGTAGCTAACGATGCTGCTCCA-3′ and SdhD-R4/ApaI.
Sequence data.
The DNA sequences of the sdhB, sdhC, and sdhD genes for T. mentagrophytes ATCC 18748 have been deposited under DDBJ/ENA/GenBank accession no. LC068589, LC068590, and LC068591, respectively. The DNA sequences of the following laboratory-generated ME1111-resistant T. mentagrophytes mutants are also available in the DDBJ database (accession numbers are indicated in parentheses): sdhB gene mutants, M5 (LC068592), M8 and M9 (LC068593), M10 (LC068594), M11 (LC068595), M13 (LC068596), and M14 (LC068597); sdhC gene mutants, M1 and M2 (LC068598), M3 and M17 (LC068599), M7 and M16 (LC068600), and M15 (LC068601); and sdhD gene mutants, M4 and M6 (LC068602) and M12 (LC068603).
RESULTS
Genetic analysis of laboratory-generated ME1111-resistant mutants.
To identify the molecular target of ME1111, spontaneous ME1111-resistant mutants were generated in vitro from T. mentagrophytes ATCC 18748. Seventeen mutants (M1 to M17) that grew on SDA containing 1 μg/ml of ME1111 were selected for further investigation. At this concentration, the frequency of resistance to ME1111 was 7.6 × 10−8. Susceptibility to ME1111 was first investigated with the 17 mutants to confirm their resistance. As shown in Table 3, the MICs of ME1111 against the selected mutants increased 8- to 32-fold compared to that for the parent strain. The complete genome sequences of the 3 representative laboratory-generated resistant mutants (M1, M8, and M15) were compared to that of the parent strain. Whole-genome analysis revealed that only a single coding region differed from that of the parent strain by a genetic mutation, resulting in an amino acid substitution in one of the subunits of succinate dehydrogenase. The genetic mutations led to an amino acid substitution in SdhC (Thr83Asn) in M1, one in SdhB (His234Leu) in M8, and one in SdhC (Asn90Lys) in M15.
TABLE 3.
Amino acid substitutions in succinate dehydrogenase and susceptibilities of spontaneous ME1111-resistant T. mentagrophytes mutants selected in vitro
| Strain(s) | Amino acid substitutiona |
MIC (μg/ml) |
||||
|---|---|---|---|---|---|---|
| SdhB | SdhC | SdhD | ME1111 | Carboxin | Boscalid | |
| ATCC 18748 (parent strain) | 0.5 | 32 | 1 | |||
| M13 | Pro187Ser | — | — | 4 | 128 | 2 |
| M8, M9 | His234Leu | — | — | 8 | 256 | >64 |
| M11 | His234Tyr | — | — | 4 | 128 | >64 |
| M5, M14 | Asn238Lys | — | — | 4 | 32 | 1–2 |
| M10 | Gly250Arg | — | — | 4 | 32 | 1 |
| M1, M2 | — | Thr83Asn | — | 8 | 128 | 8 |
| M3, M17 | — | Thr83Ile | — | 4 | 64 | 8 |
| M7, M16 | — | Ser88Gly | — | 4 | 16 | 1 |
| M15 | — | Asn90Lys | — | 4 | 256 | >64 |
| M4, M6 | — | — | Asp161His | 4 | 128 | 4 |
| M12 | — | — | Tyr162Cys | 16 | 32 | 4 |
—, amino acid sequence identical to that of the parent strain.
Since this suggested that the target enzyme of ME1111 is succinate dehydrogenase, the entire coding sequences of the genes encoding the SdhB, SdhC, and SdhD subunits of succinate dehydrogenase in all of the spontaneous resistant mutants were determined. The results are shown in Table 3. Sequencing analysis showed that M5, M8, M9, M10, M11, M13, and M14 each contained a genetic mutation causing a single amino acid substitution in SdhB, M1, M2, M3, M7, M15, M16, and M17 each harbored a genetic mutation causing a single amino acid substitution in SdhC, and M4, M6, and M12 each possessed a genetic mutation causing a single amino acid substitution in SdhD. This confirmed that each of the mutants carried a genetic mutation causing an amino acid substitution in either SdhB, SdhC, or SdhD. When the susceptibilities of the laboratory-generated ME1111-resistant mutants to carboxin and boscalid were tested by the broth microdilution method, more than half of the mutants (M1, M2, M3, M4, M6, M8, M9, M11, M15, and M17) showed cross-resistance to these agents (Table 3).
Inhibitory effect of ME1111 on succinate dehydrogenase (complex II) activity.
Enzyme assays were performed to evaluate the inhibitory effect of ME1111 on succinate dehydrogenase. The inhibitory activities of ME1111 and the reference compounds (carboxin and boscalid) on the succinate-DCIP reductase reaction in T. rubrum, T. mentagrophytes, and two human cell lines (K562 and HepG2) are shown in Table 4. ME1111 exerted a strong inhibitory effect on succinate-DCIP reductase activity in T. rubrum (IC50 = 0.029 μg/ml) and T. mentagrophytes (IC50 = 0.025 μg/ml), indicating that complex II is the primary target of ME1111. On the other hand, ME1111 demonstrated a relatively weak inhibitory effect on the corresponding enzyme activity in human K562 (IC50 = 1.4 μg/ml) and HepG2 (IC50 = 0.94 μg/ml) cells, revealing approximately 30- to 60-fold greater selectivity in its inhibition of succinate-DCIP reductase in Trichophyton species than in human cells.
TABLE 4.
Inhibitory effects of ME1111 and reference compounds on succinate-DCIP reductase
| Organism | IC50 (μg/ml) (mean ± SE) |
||
|---|---|---|---|
| ME1111 | Carboxin | Boscalid | |
| T. rubrum | 0.0287 ± 0.0087 | 4.09 ± 0.44 | 0.0322 ± 0.0030 |
| T. mentagrophytes | 0.0247 ± 0.0034 | 2.45 ± 0.58 | 0.0189 ± 0.0027 |
| Human (K562 cells) | 1.40 ± 0.15 | 1.27 ± 0.18 | 0.965 ± 0.185 |
| Human (HepG2 cells) | 0.941 ± 0.082 | 0.733 ± 0.080 | 0.704 ± 0.137 |
The inhibitory activities of ME1111 on succinate-DCIP reductase activity in ME1111-resistant T. mentagrophytes mutants selected in vitro and in their parent strain are shown in Table 5. The inhibitory effects of ME1111 on the enzyme activity of 6 ME1111-resistant mutants (M1, M11, M13, M15, M16, and M17) were 8.5- to 26-fold weaker than that on the parent strain. The laboratory-generated ME1111-resistant T. mentagrophytes mutants harboring single amino acid substitutions in subunits of succinate dehydrogenase were less susceptible to the inhibitory effect of ME1111 on succinate-DCIP reductase activity.
TABLE 5.
Inhibitory effects of ME1111 on succinate-DCIP reductases from laboratory-generated ME1111-resistant T. mentagrophytes mutants
| Strain | Amino acid substitution | IC50 (μg/ml) (mean ± SE) | Fold change relative to parent strain |
|---|---|---|---|
| ATCC 18748 | None | 0.0274 ± 0.0039 | 1.0 |
| M13 | SdhB(Pro187Ser) | 0.565 ± 0.071 | 21 |
| M11 | SdhB(His234Tyr) | 0.232 ± 0.049 | 8.5 |
| M1 | SdhC(Thr83Asn) | 0.460 ± 0.057 | 17 |
| M17 | SdhC(Thr83Ile) | 0.708 ± 0.054 | 26 |
| M16 | SdhC(Ser88Gly) | 0.247 ± 0.008 | 9.0 |
| M15 | SdhC(Asn90Lys) | 0.499 ± 0.048 | 18 |
Introduction of point mutations in the coding regions of the sdhB, sdhC, and sdhD genes of T. mentagrophytes.
In order to investigate the effects of single amino acid substitutions in succinate dehydrogenase on ME1111 resistance, DNA cassettes containing point mutations leading to single amino acid substitutions in SdhB (i.e., His234Leu, Asn238Lys, and Gly250Arg), SdhC (i.e., Thr83Asn, Ser88Gly, and Asn90Lys), and SdhD (Asp161His) were introduced into T. mentagrophytes TmL28. Several attempts of transformation using the protoplast-PEG method led to successful production of three types, two types, and one type of clone carrying point mutations in the sdhB, sdhC, and sdhD gene, respectively. Two clones each for the His234Leu, Asn238Lys, and Gly250Arg substitutions in SdhB and the Thr83Asn and Ser88Gly substitutions in SdhC were obtained and further investigated. Clones harboring the Asn90Lys substitution in SdhC were not obtained despite several transformations. Only one transformant (JD8-#12-10) was obtained as a clone harboring the Asp161His substitution in the SdhD subunit.
Susceptibilities of clones harboring amino acid substitutions in SdhB, SdhC, or SdhD to ME1111.
The MICs of ME1111 and reference compounds against clones carrying point mutations in the sdhB, sdhC, or sdhD gene were measured using the CLSI broth microdilution method (Table 6). Transformants of the respective hph cassettes lacking point mutations in the coding sequence were used as control strains. The susceptibilities of the clones carrying amino acid substitutions in SdhB, SdhC, or SdhD were compared to those of the respective control strains (B6-6-#5M, C4-5-#8, or JD8-#16, respectively). The His234Leu, Asn238Lys, and Gly250Arg amino acid substitutions in SdhB increased the MIC of ME1111 64- to 128-, 32-, and 8-fold, respectively. The Thr83Asn and Ser88Gly substitutions in SdhC resulted in increases in the MIC of ME1111 of 16- to 32- and 8- to 32-fold, respectively. The Asp161His substitution in SdhD increased the MIC of ME1111 16-fold. All clones were at least 8-fold less susceptible to ME1111 than the relevant control strain. Most of the clones demonstrated significant reductions in susceptibility to the succinate dehydrogenase inhibitor carboxin. No apparent difference was observed in susceptibility to the reference compounds, ciclopirox and amorolfine, between clones harboring amino acid substitutions and the control strains.
TABLE 6.
Susceptibilities of clones harboring amino acid substitutions in SdhB, SdhC, or SdhD to ME1111 and reference compounds
| Strain | Target protein in transformation | Amino acid substitution | MIC (μg/ml) |
|||
|---|---|---|---|---|---|---|
| ME1111 | Carboxin | Ciclopirox | Amorolfine | |||
| ATCC MYA-4439 | None | None | 0.5 | 64 | 0.25 | 0.5 |
| TmL28 | None | None | 0.5 | 32 | 0.25 | 0.5 |
| B6-6-#5M | SdhB | None | 0.06 | 1 | 0.25 | 0.25 |
| JB2-#1M | SdhB | His234Leu | 4 | 256 | 0.12 | 0.25 |
| JB2-#5M | SdhB | His234Leu | 8 | 256 | 0.25 | 0.5 |
| B6-6-#3M | SdhB | Asn238Lys | 2 | 32 | 0.12 | 0.25 |
| B6-6-#8 | SdhB | Asn238Lys | 2 | 32 | 0.12 | 0.25 |
| B7-#11 | SdhB | Gly250Arg | 0.5 | 4 | 0.06 | 0.25 |
| B7-#14 | SdhB | Gly250Arg | 0.5 | 4 | 0.06 | 0.25 |
| C4-5-#8 | SdhC | None | 0.06 | 2 | 0.25 | 0.12 |
| J1-2-#3 | SdhC | Thr83Asn | 2 | 64 | 0.25 | 0.12 |
| J1-2-#11 | SdhC | Thr83Asn | 1 | 64 | 0.25 | 0.12 |
| J2-8-#8 | SdhC | Ser88Gly | 2 | 8 | 0.25 | 0.25 |
| J2-8-#13 | SdhC | Ser88Gly | 0.5 | 1 | 0.25 | 0.25 |
| JD8-#16 | SdhD | None | 0.03 | 0.5 | 0.25 | 0.12 |
| JD8-#12-10 | SdhD | Asp161His | 0.5 | 32 | 0.12 | 0.12 |
DISCUSSION
The molecular target of the novel antifungal agent ME1111 was shown here to be succinate dehydrogenase (complex II) of the mitochondrial electron transport system. This enzyme is also responsible for a reaction in the citric acid cycle and consists of 4 subunits (SdhA, SdhB, SdhC, and SdhD). In previous reports, amino acid substitutions in SdhB, SdhC, or SdhD were detected in strains resistant to succinate dehydrogenase inhibitors, such as carboxin and boscalid (28, 29). In this study, in vitro-generated ME1111-resistant T. mentagrophytes mutants possessed missense mutations in the genes encoding SdhB, SdhC, and SdhD. As shown in Table 3, most of the ME1111-resistant mutants demonstrated cross-resistance to carboxin and boscalid, which have been reported to bind to the ubiquinone-binding site surrounded by SdhB, SdhC, and SdhD. These results suggest that the binding site of ME1111 partially overlaps that of those agrichemicals, and thus it plausibly is either the ubiquinone-binding site or located adjacent to this site. The amino acid substitutions observed in the laboratory-generated ME1111-resistant mutants correspond to the ubiquinone-binding site of the protein as revealed by structural studies with E. coli and porcine models (10, 30). We speculate that the amino acid substitutions found in the spontaneous resistant mutants hamper binding of ME1111 to this pocket, while affinity for the natural substrate, ubiquinone, is retained. Although succinate dehydrogenase is a mitochondrial inner membrane protein and analysis of its crystal structure is challenging, crystallographic analysis of the succinate dehydrogenase-ME1111 complex is warranted.
The inhibitory effect of ME1111 on succinate dehydrogenase was measured by monitoring succinate-DCIP reductase activity in mitochondrial fractions. As shown in Table 4, ME1111 strongly inhibited the enzymatic activity of T. rubrum and T. mentagrophytes, both of which are major etiologies of onychomycosis (2, 31). In comparison, ME1111 showed relatively weak inhibitory effects in two human cell lines, and the IC50s against the human cell lines were 30- to 60-fold higher than those for Trichophyton species. The growth-inhibitory effects of ME1111 (IC50) in human K562 cells and HepG2 cells have been reported to be 47 and 37 μg/ml, respectively (18). These values are about 150-fold higher than its MIC90 against T. rubrum and T. mentagrophytes (0.25 μg/ml), and the inhibitory effect of ME1111 on the target enzyme correlates well with its growth-inhibitory activity. Therefore, the differences in the inhibitory effects of ME1111 on succinate dehydrogenases derived from dermatophytes and human cells can explain its selective antidermatophyte activity. Although residues that constitute the ubiquinone-binding pocket in succinate dehydrogenase are highly conserved between dermatophytes and humans, 3 amino acid residues in SdhC (Leu-75, Trp-84, and Ser-88 in T. mentagrophytes) are different in humans. As in the case of the laboratory-generated ME1111-resistant mutants harboring the Ser88Gly substitution in SdhC, the reduced susceptibility of human succinate dehydrogenase to ME1111 may be due to conformational changes of the protein because of Ile-88 and other residues (Ile-75 and Met-84) in human SdhC.
The molecular target of ME1111 was confirmed by introducing point mutations into the genes encoding SdhB, SdhC, and SdhD and evaluating the susceptibility of clones carrying amino acid substitutions in SdhB, SdhC, or SdhD to ME1111. Introduction of a single amino acid substitution into a subunit of succinate dehydrogenase decreased the susceptibility to ME1111 by 1/8 or less. The results strongly support the evidence that the primary target protein of ME1111 is indeed succinate dehydrogenase. Transformants harboring one of the wild-type sequence gene cassettes (B6-6-#5M, C4-5-#8, and JD8-#16) were more susceptible to ME1111 than the recipient strain (TmL28) or a reference strain (ATCC MYA-4439). This may be because the introduction of a gene cassette reduced the expression of the gene encoding one of the subunits of succinate dehydrogenase, and therefore the strain became susceptible to ME1111, although this requires further investigation.
In conclusion, ME1111 is a novel inhibitor of succinate dehydrogenase (complex II). The fungicidal activity of ME1111 (14) seems to be due to its inhibition of this enzyme, leading to the blockade of ATP production.
ACKNOWLEDGMENTS
We thank Kanako Sawada, Kazue Nagano, and Akane Soramoto for their technical support.
Funding Statement
Sho Takahata, Natsuki Kubota, Naomi Takei-Masuda, Yuji Tabata, and Kazunori Maebashi are full-time employees of Meiji Seika Pharma Co., Ltd.
The work performed by Tsuyoshi Yamada, Mari Maeda, Mohamed Mahdi Alshahni, and Shigeru Abe was financially supported by Meiji Seika Pharma Co., Ltd.
REFERENCES
- 1.Elewski BE, Charif MA. 1997. Prevalence of onychomycosis in patients attending a dermatology clinic in northeastern Ohio for other conditions. Arch Dermatol 133:1172–1173. [PubMed] [Google Scholar]
- 2.Ghannoum MA, Hajjeh RA, Scher R, Konnikov N, Gupta AK, Summerbell R, Sullivan S, Daniel R, Krusinski P, Fleckman P, Rich P, Odom R, Aly R, Pariser D, Zaiac M, Rebell G, Lesher J, Gerlach B, Ponce-De-Leon GF, Ghannoum A, Warner J, Isham N, Elewski B. 2000. A large-scale North American study of fungal isolates from nails: the frequency of onychomycosis, fungal distribution, and antifungal susceptibility patterns. J Am Acad Dermatol 43:641–648. doi: 10.1067/mjd.2000.107754. [DOI] [PubMed] [Google Scholar]
- 3.Gupta AK, Jain HC, Lynde CW, Macdonald P, Cooper EA, Summerbell RC. 2000. Prevalence and epidemiology of onychomycosis in patients visiting physicians' offices: a multicenter Canadian survey of 15,000 patients. J Am Acad Dermatol 43:244–248. doi: 10.1067/mjd.2000.104794. [DOI] [PubMed] [Google Scholar]
- 4.Heikkilä H, Stubb S. 1995. The prevalence of onychomycosis in Finland. Br J Dermatol 133:699–703. [DOI] [PubMed] [Google Scholar]
- 5.Elewski BE, Rich P, Pollak R, Pariser DM, Watanabe S, Senda H, Ieda C, Smith K, Pillai R, Ramakrishna T, Olin JT. 2013. Efinaconazole 10% solution in the treatment of toenail onychomycosis: two phase III multicenter, randomized, double-blind studies. J Am Acad Dermatol 68:600–608. doi: 10.1016/j.jaad.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 6.Toledo-Bahena ME, Bucko A, Ocampo-Candiani J, Herz-Ruelas ME, Jones TM, Jarratt MT, Pollak RA, Zane LT. 2014. The efficacy and safety of tavaborole, a novel, boron-based pharmaceutical agent: phase 2 studies conducted for the topical treatment of toenail onychomycosis. J Drugs Dermatol 13:1124–1132. [PubMed] [Google Scholar]
- 7.Rock FL, Mao W, Yaremchuk A, Tukalo M, Crépin T, Zhou H, Zhang YK, Hernandez V, Akama T, Baker SJ, Plattner JJ, Shapiro L, Martinis SA, Benkovic SJ, Cusack S, Alley MR. 2007. An antifungal agent inhibits an aminoacyl-tRNA synthetase by trapping tRNA in the editing site. Science 316:1759–1761. doi: 10.1126/science.1142189. [DOI] [PubMed] [Google Scholar]
- 8.Subissi A, Monti D, Togni G, Mailland F. 2010. Ciclopirox: recent nonclinical and clinical data relevant to its use as a topical antimycotic agent. Drugs 70:2133–2152. doi: 10.2165/11538110-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 9.Horsefield R, Yankovskaya V, Sexton G, Whittingham W, Shiomi K, Omura S, Byrne B, Cecchini G, Iwata S. 2006. Structural and computational analysis of the quinone-binding site of complex II (succinate-ubiquinone oxidoreductase). J Biol Chem 281:7309–7316. doi: 10.1074/jbc.M508173200. [DOI] [PubMed] [Google Scholar]
- 10.Ruprecht J, Yankovskaya V, Maklashina E, Iwata S, Cecchini G. 2009. Structure of Escherichia coli succinate:quinone oxidoreductase with an occupied and empty quinone-binding site. J Biol Chem 284:29836–29846. doi: 10.1074/jbc.M109.010058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scalliet G, Bowler J, Luksch T, Kirchhofer-Allan L, Steinhauer D, Ward K, Niklaus M, Verras A, Csukai M, Daina A, Fonné-Pfister R. 2012. Mutagenesis and functional studies with succinate dehydrogenase inhibitors in the wheat pathogen Mycosphaerella graminicola. PLoS One 7:e35429. doi: 10.1371/journal.pone.0035429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baggish AL, Hill DR. 2002. Antiparasitic agent atovaquone. Antimicrob Agents Chemother 46:1163–1173. doi: 10.1128/AAC.46.5.1163-1173.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koul A, Dendouga N, Vergauwen K, Molenberghs B, Vranckx L, Willebrords R, Ristic Z, Lill H, Dorange I, Guillemont J, Bald D, Andries K. 2007. Diarylquinolines target subunit c of mycobacterial ATP synthase. Nat Chem Biol 3:323–324. doi: 10.1038/nchembio884. [DOI] [PubMed] [Google Scholar]
- 14.Ghannoum M, Isham N, Long L. 2015. In vitro antifungal activity of ME1111, a new topical agent for onychomycosis, against clinical isolates of dermatophytes. Antimicrob Agents Chemother 59:5154–5158. doi: 10.1128/AAC.00992-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takei-Masuda N, Takahata S, Tabata Y, Maebashi K, Isham N, Long L, Ghannoum MA. 2014. In vitro antifungal activity of and assessment of resistance to ME1111, a new antifungal agent for topical treatment of onychomycosis, abstr F-1587 Abstr 54th Intersci Conf Antimicrob Agents Chemother American Society for Microbiology, Washington, DC. [Google Scholar]
- 16.Murdan S. 2002. Drug delivery to the nail following topical application. Int J Pharm 236:1–26. doi: 10.1016/S0378-5173(01)00989-9. [DOI] [PubMed] [Google Scholar]
- 17.Hui X, Maibach H. 2014. In vitro human nail plate penetration of a novel antifungal agent, ME1111, abstr F-1588 Abstr 54th Intersci Conf Antimicrob Agents Chemother American Society for Microbiology, Washington, DC. [Google Scholar]
- 18.Tabata Y, Nomoto M, Chikada T, Kubota N, Takei-Masuda N, Takahata S, Maebashi K. 2014. In vitro and in vivo nail penetration of ME1111, a novel antifungal agent for topical treatment of onychomycosis, abstr F-1589 Abstr 54th Intersci Conf Antimicrob Agents Chemother American Society for Microbiology, Washington, DC. [Google Scholar]
- 19.Ago K, Nomoto M, Chikada T, Uchida M, Tsuchiya T, Shibasaki S, Maebashi K, Hiratsuka K. 2014. Nonclinical safety assessment of ME1111, a novel antifungal agent for the topical treatment of onychomycosis, abstr F-1593 Abstr 54th Intersci Conf Antimicrob Agents Chemother American Society for Microbiology, Washington, DC. [Google Scholar]
- 20.Takahata S, Tabata Y, Takei-Masuda N, Kubota N, Maebashi K. 2014. Mechanism of action of ME1111, a new antifungal agent for topical treatment of onychomycosis, abstr F-1585 Abstr 54th Intersci Conf Antimicrob Agents Chemother American Society for Microbiology, Washington, DC. [Google Scholar]
- 21.Takahata S, Yamada T, Alshahni MM, Abe S, Kubota N, Takei-Masuda N, Tabata Y, Maebashi K. 2015. Identifying the target protein of ME1111 in Trichophyton mentagrophytes using a reverse genetics approach, abstr F-741 Abstr 55th Intersci Conf Antimicrob Agents Chemother American Society for Microbiology, Washington, DC. [Google Scholar]
- 22.Alshahni MM, Yamada T, Takatori K, Sawada T, Makimura K. 2011. Insights into a nonhomologous integration pathway in the dermatophyte Trichophyton mentagrophytes: efficient targeted gene disruption by use of mutants lacking ligase IV. Microbiol Immunol 55:34–43. doi: 10.1111/j.1348-0421.2010.00283.x. [DOI] [PubMed] [Google Scholar]
- 23.CLSI. 2008. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi; approved standard, 2nd ed. CLSI document M38-A2 CLSI, Wayne, PA. [Google Scholar]
- 24.Ulrich JT, Mathre DE. 1972. Mode of action of oxathiin systemic fungicides. J Bacteriol 110:628–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Girardin H, Latge JP. 1994. DNA extraction and quantification, p 5–9. In Maresca B, Kobayashi GS (ed), Molecular biology of pathogenic fungi, 2nd ed Telos Press, New York, NY. [Google Scholar]
- 26.Yamada T, Makimura K, Satoh K, Umeda Y, Ishihara Y, Abe S. 2009. Agrobacterium tumefaciens-mediated transformation of the dermatophyte, Trichophyton mentagrophytes: an efficient tool for gene transfer. Med Mycol 47:485–494. doi: 10.1080/13693780802322240. [DOI] [PubMed] [Google Scholar]
- 27.Yamada T, Makimura K, Uchida K, Yamaguchi H. 2005. Reproducible genetic transformation system for two dermatophytes, Microsporum canis and Trichophyton mentagrophytes. Med Mycol 43:533–544. doi: 10.1080/13693780500057619. [DOI] [PubMed] [Google Scholar]
- 28.Avenot HF, Sellam A, Karaoglanidis G, Michailides TJ. 2008. Characterization of mutations in the iron-sulphur subunit of succinate dehydrogenase correlating with boscalid resistance in Alternaria alternata from California pistachio. Phytopathology 98:736–742. doi: 10.1094/PHYTO-98-6-0736. [DOI] [PubMed] [Google Scholar]
- 29.Shima Y, Ito Y, Kaneko S, Hatabayashi H, Watanabe Y, Adachi Y, Yabe K. 2009. Identification of three mutant loci conferring carboxin-resistance and development of a novel transformation system in Aspergillus oryzae. Fungal Genet Biol 46:67–76. doi: 10.1016/j.fgb.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 30.Sun F, Huo X, Zhai Y, Wang A, Xu J, Su D, Bartlam M, Rao Z. 2005. Crystal structure of mitochondrial respiratory membrane protein complex II. Cell 121:1043–1057. doi: 10.1016/j.cell.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 31.Faergemann J, Baran R. 2003. Epidemiology, clinical presentation and diagnosis of onychomycosis. Br J Dermatol 149(Suppl 65):1–4. [DOI] [PubMed] [Google Scholar]