Abstract
Avibactam is a novel non-β-lactam β-lactamase inhibitor that covalently acylates a variety of β-lactamases, causing inhibition. Although avibactam presents limited antibacterial activity, its acylation ability toward bacterial penicillin-binding proteins (PBPs) was investigated. Staphylococcus aureus was of particular interest due to the reported β-lactamase activity of PBP4. The binding of avibactam to PBPs was measured by adding increasing concentrations to membrane preparations of a variety of Gram-positive and Gram-negative bacteria prior to addition of the fluorescent reagent Bocillin FL. Relative binding (measured here as the 50% inhibitory concentration [IC50]) to PBPs was estimated by quantification of fluorescence after gel electrophoresis. Avibactam was found to selectively bind to some PBPs. In Escherichia coli, Pseudomonas aeruginosa, Haemophilus influenzae, and S. aureus, avibactam primarily bound to PBP2, with IC50s of 0.92, 1.1, 3.0, and 51 μg/ml, respectively, whereas binding to PBP3 was observed in Streptococcus pneumoniae (IC50, 8.1 μg/ml). Interestingly, avibactam was able to significantly enhance labeling of S. aureus PBP4 by Bocillin FL. In PBP competition assays with S. aureus, where avibactam was used at a fixed concentration in combination with varied amounts of ceftazidime, the apparent IC50 of ceftazidime was found to be very similar to that determined for ceftazidime when used alone. In conclusion, avibactam is able to covalently bind to some bacterial PBPs. Identification of those PBP targets may allow the development of new diazabicyclooctane derivatives with improved affinity for PBPs or new combination therapies that act on multiple PBP targets.
INTRODUCTION
The development and spread of antibacterial resistance in Gram-negative bacteria drives the critical medical need for new antibacterial agents (1, 2). A particular problem is resistance to β-lactam antibiotics resulting from a continuously expanding plethora of variants of β-lactamases (3) and dissemination of β-lactamase-producing clones, such as those that were found to occur within a population of clinical isolates of Klebsiella pneumoniae (4). Avibactam, a potent inhibitor of Ambler classes A, C, and some D serine β-lactamases, including TEM-1, CTX-M-15, and P99 β-lactamases, as well as K. pneumoniae carbapenemases (5–7), is a compound that can potentially address part of this medical need. It is a novel non-β-lactam β-lactamase inhibitor that is currently in advanced clinical development for use in combination with ceftazidime (8), aztreonam (9), and ceftaroline fosamil (10). The combination with ceftazidime was recently approved by the U.S. Food and Drug Administration (FDA) for certain infections in adults for which there are limited or no alternative treatment options (11). Avibactam does not contain a β-lactam ring, but it can covalently acylate a variety of serine β-lactamases to cause inhibition (5, 6) and thereby restore the microbiological activity of ceftazidime (12–17). Avibactam displays a broader spectrum of activity than the currently employed β-lactamase inhibitors clavulanic acid, sulbactam, and tazobactam.
Although avibactam possesses limited antibacterial activity (18, 19), the structural relationship between β-lactamases and bacterial penicillin-binding proteins (PBPs) (20) suggests that avibactam may also interact with PBPs. Indeed, the fact that the diazabicyclooctanes, a class of compounds of which avibactam is a member, were originally proposed as potential β-lactam mimics (21) led to the hypothesis that avibactam might bind to PBPs. Consequently, some measurable MICs for avibactam against at least some enteric bacteria have been reported (18, 19). In order to further understand the biological properties of avibactam as a member of the diazabicyclooctane class of compounds, the hypothesis regarding avibactam interactions with PBPs was tested in several Gram-negative and -positive species in the work reported here. Morinaka et al. (22) also recently explored this same hypothesis, obtaining quantitatively comparable results in one of these species, Escherichia coli.
PBPs are central to bacterial cell wall biosynthesis. Most PBPs have either transpeptidase or carboxypeptidase activities, and some also demonstrate transglycosylase activity. Inhibition of PBPs by β-lactam antibiotics occurs via competition with the peptidoglycan pentapeptide precursor for binding to the active site serine of the transpeptidase domain (23).
The affinities of avibactam for the PBPs of Escherichia coli, Pseudomonas aeruginosa, Haemophilus influenzae, Streptococcus pneumoniae, and Staphylococcus aureus were experimentally evaluated via inhibition of PBP acylation by the fluorescent penicillin marker Bocillin FL (24). Of particular interest was S. aureus PBP4, which appears to display some β-lactamase activity (25).
(Part of this work was presented in summary form at the 24th European Congress of Clinical Microbiology and Infectious Diseases, May 2014 [26].)
MATERIALS AND METHODS
Antibiotics.
Avibactam was provided by AstraZeneca Pharmaceuticals (Wilmington, NC, USA). Ceftazidime was obtained from Lab Express International (Fairfield, NJ, USA). Oxacillin and amdinocillin were from Sigma-Aldrich Canada (Oakville, ON, Canada). PBPs were labeled using the fluorescent reporter molecule Bocillin FL (Invitrogen-Molecular Probes, Eugene, OR, USA).
Antibiotic susceptibility testing.
Susceptibility testing was performed using the broth microdilution technique according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (27). MICs were determined in cation-adjusted Mueller-Hinton broth (CAMHB; Becton Dickinson Canada Inc., Mississauga, ON, Canada). MICs for S. pneumoniae were determined in CAMHB supplemented with 2.5% lysed horse blood.
Bacterial strains and growth conditions.
E. coli K-12 (250HT11) and P. aeruginosa 1771 (391HT2) were grown in brain heart infusion (BHI) broth at 35°C. For bacterial membrane preparation, an overnight broth culture was diluted in fresh medium and grown to an A600 of ∼1.5. H. influenzae strain Rd was grown in BHI broth supplemented with Haemophilus test medium supplement (Oxoid, Nepean, ON, Canada), which includes hemin and β-NAD (both at 15 μg/ml) to reach an A600 of ∼0.6 to 0.7 at 35°C. S. aureus ATCC 29213 was used as a typical methicillin-susceptible strain and was grown to reach an A600 of ∼0.6 to 0.7 in BHI broth at 35°C. S. pneumoniae R6 is a typical penicillin-susceptible strain (28). It was grown on blood agar plates at 37°C in 5% CO2. For bacterial membrane preparation, cells from the overnight blood agar plates were suspended in BHI and further incubated under agitation to reach an A600 of ∼1.0.
Preparation of bacterial membranes.
Bacterial cells were grown as described above. The cells from a 1.5-liter culture were harvested by centrifugation at 6,000 × g for 10 min at 4°C and washed, and E. coli and P. aeruginosa cells were suspended in 50 mM phosphate buffer (pH 7.0). For H. influenzae, S. aureus, and S. pneumoniae, KPN buffer (20 mM potassium phosphate, 140 mM NaCl [pH 7.5]) was used. Cells of S. aureus were first treated with lysostaphin (400 μg/ml) for 1 h at 37°C before addition of 3 μg/ml of a protease inhibitor cocktail (Sigma-Aldrich), DNase (6 μg/ml), and RNase (6 μg/ml). For S. pneumoniae and H. influenzae, lysozyme (400 μg/ml) was also added. After 30 min of treatment, cells were disrupted by a French press instrument, and the bacterial lysates were centrifuged at 6,000 × g for 30 min at 4°C to remove unbroken cells. The supernatant was then centrifuged at 150,000 × g for 40 min at 4°C using a fixed-angle rotor to collect the membranes. The membranes were suspended in a minimal volume of buffer (typically 500 μl) and stored at −80°C. The protein concentrations in the membrane preparations were estimated by using the Micro bicinchoninic acid protein assay kit (Thermo Fisher Scientific, Rockford, IL) with bovine serum albumin as a standard.
PBP binding and competition assays.
The relative binding of test compounds for bacterial PBPs was determined in a competition assay with the fluorescent reporter molecule Bocillin FL. An aliquot of bacterial membrane preparation was used for E. coli, P. aeruginosa, and S. aureus (10 to 20 μg of membrane proteins), S. pneumoniae (30 to 40 μg of membrane proteins), or H. influenzae (100 to 120 μg of membrane proteins) in each assay mixture. The amount of membrane preparation used for each species was selected so that the PBP banding patterns obtained on gels would reflect those previously published in the literature. In the sets of assay mixtures (∼7 μl of membrane preparation in a 15-μl final volume), increasing concentrations of each test compound were first added for 10 min at 30°C before Bocillin FL was added at a final concentration of 100 μM for an additional 20 min of incubation. Thereafter, electrophoresis loading buffer, containing sodium dodecyl sulfate (SDS) and fresh β-mercaptoethanol as a reducing reagent, was added to the mixture before heating for 3 min at 95°C. Samples were spun for 2 min in a microcentrifuge before loading for electrophoresis.
SDS-PAGE, PBP detection, and IC50 determinations.
Proteins from the PBP assay mixtures were separated by electrophoresis using an SDS-polyacrylamide discontinuous gel system (5% stacking and 10% separating gels). After electrophoresis, the gels were quickly rinsed in water and fixed in a 50% methanol–7% acetic acid solution for 30 min. Gels were scanned to collect the images of the PBP profiles by using either a Molecular Imager FX Pro instrument (Bio-Rad Laboratories, Mississauga, ON, Canada) or a Typhoon 9400 scanner (GE Healthcare) (excitation at 488 nm and emission at 520 to 530 nm). The PBP image was quantified using Quantity One 1-D analysis software (version 4.6.6; Bio-Rad Laboratories, Richmond, CA). Each individual labeled PBP band was selected, and its volume (surface × intensity) was measured. The relative binding or IC50, i.e., the concentration of test compound (in micrograms per milliliter) that was needed to reduce by 50% the binding of Bocillin FL to individual PBPs was then determined by plotting the PBP band volumes versus compound concentrations. The 100% binding of Bocillin FL was represented by the PBPs labeled with Bocillin FL but with no drug or avibactam added to the mixture. For drug combination experiments with S. aureus, the 100% value was represented by the PBPs labeled with Bocillin FL in the presence of avibactam at 4 μg/ml. The IC50 for each PBP of interest was calculated from at least eight test molecule concentrations (E. coli, P. aeruginosa) or three independent PBP binding assays using six different concentrations in each experiment (H. influenzae, S. aureus, S. pneumoniae).
RESULTS AND DISCUSSION
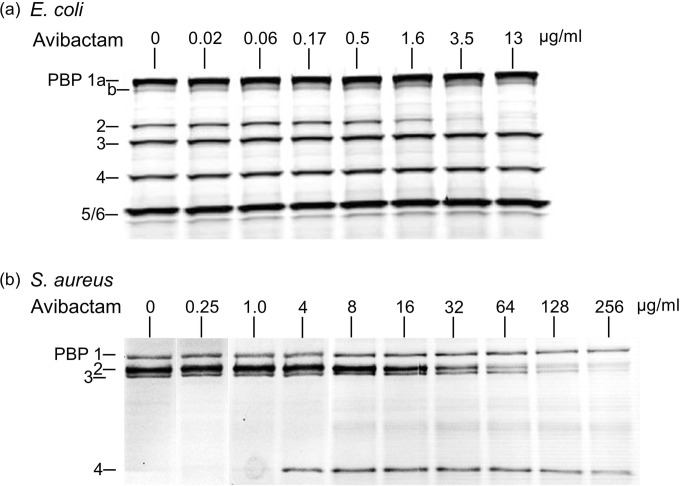
Figure 1 shows the pattern of avibactam binding to E. coli and S. aureus PBPs. Relative binding to PBPs (IC50s) are reported in Table 1 for those species and other Gram-negative and -positive bacteria. Avibactam binds moderately but very selectively to a few PBPs, notably to E. coli PBP2. Our results and IC50s for PBP2 (Table 1) confirmed the very recently reported E. coli PBP binding data for avibactam and another diazabicyclooctane derivative, namely, OP0595 (20). These results were very similar to those observed for amdinocillin (Table 1), as previously reported by Spratt and Pardee (29). Such selective PBP2 binding was also reported for the β-lactamase inhibitor clavulanic acid (30). The E. coli PBP banding profile we observed and the binding of the reference compounds amdinocillin and ceftazidime (Table 1) are consistent with published data (31, 32). PBP2 is essential for cell elongation and rod shape maintenance (33), and its inhibition causes rounding of the cells as reported after exposure of E. coli cells to amdinocillin (29) and OP0595 (22).
FIG 1.
Affinities of avibactam for E. coli (a) and S. aureus (b) PBPs. The indicated concentrations of avibactam were used in a PBP competition assay with Bocillin FL, a fluorescent reporter molecule that revealed the PBP profile.
TABLE 1.
Binding of test molecules to PBPs of Gram-negative and Gram-positive bacteria
| Species | Compounda | MIC (μg/ml) | Relative binding (IC50 [μg/ml]) for PBPb: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1a | 1b | 1a/b | 2 | 2X | 2a/b | 3 | 3a/b | 4 | 5 | 5/6 | |||
| E. coli | AVI | 8 | >13 | >13 | 0.92 | >13 | >13 | >13 | ||||||
| MEC | <0.06–0.1 | >13 | >13 | 0.02 | >13 | >13 | >13 | |||||||
| CAZ | 0.5 | 15 | 8.0 | >25 | 0.20 | >25 | >25 | |||||||
| P. aeruginosa | ||||||||||||||
| AVI | >128 | >13 | >13 | 1.1 | 1.8 | 11 | >13 | |||||||
| MEC | —c | >13 | >13 | 0.21 | >13 | >13 | >13 | |||||||
| CAZ | 4 | 0.19 | 3.9 | >25 | 0.04 | 1.6 | >25 | |||||||
| H. influenzae | ||||||||||||||
| AVI | 64 | >32 | >32 | 3.0 | >32 | >32 | >32 | |||||||
| S. pneumoniae | ||||||||||||||
| AVI | 256 | >64 | >64 | >64 | 8.1 | |||||||||
| S. aureus | ||||||||||||||
| AVI | 256 | >256 | 51.0 | 156 | >256 | |||||||||
| CAZ | 16 | 1.6 | 0.7 | >8 | >8 | |||||||||
| CAZ + AVId | 16 | 1.7 | 0.9 | 9.2 | 96 | |||||||||
AVI, avibactam; MEC, amdinocillin; CAZ, ceftazidime.
A > sign preceding a value indicates that the IC50 was greater than the highest dose tested.
—, the MIC was not measured.
The avibactam concentration was fixed at 4 μg/ml, while ceftazidime concentrations were varied in 2-fold increments.
The H. influenzae PBP banding profile generated by the fluorescent penicillin Bocillin FL appeared as previously reported for strain Rd (34, 35) and for strain ATCC 49766 (36) (data not shown). As for E. coli, the principal H. influenzae PBP target of avibactam was PBP2 (Table 1). Concerning P. aeruginosa, avibactam preferentially bound to PBP2 but also to PBP3, and even more moderately to PBP4. The relative binding levels of amdinocillin and ceftazidime for P. aeruginosa PBP2 and PBP3, respectively, were similar to those previously reported (32) and were higher than those measured for avibactam (Table 1).
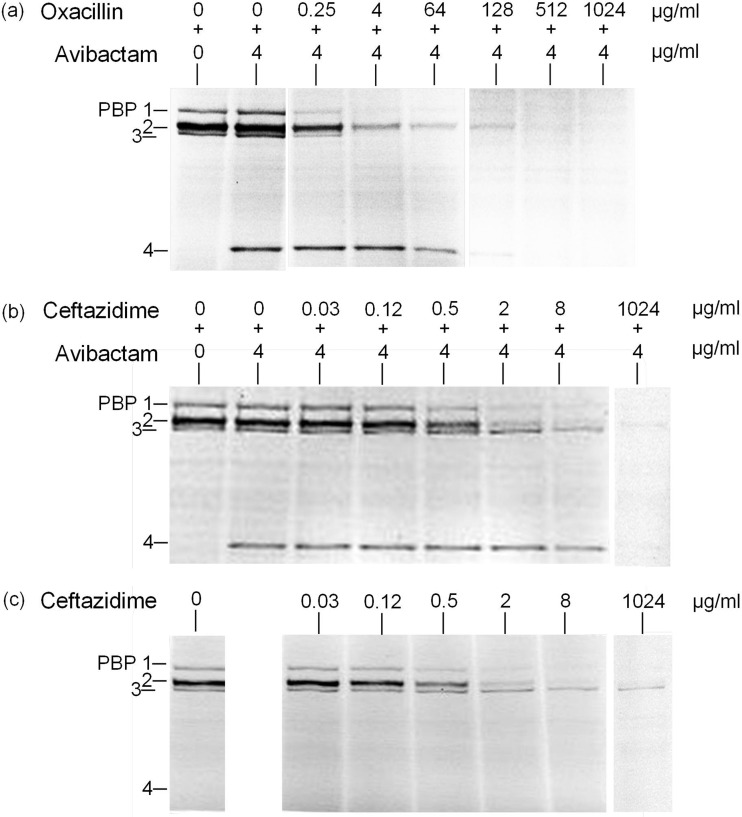
The PBP banding profile of S. aureus is shown in Fig. 1b. The principal PBP target of avibactam is PBP2 and to some extent PBP3 (Table 1). Using avibactam concentrations of 4 μg/ml and above, PBP4 became strongly labeled by the reporter molecule Bocillin FL (Fig. 1b). Accordingly, the addition of avibactam at a fixed concentration of 4 μg/ml allowed detection of oxacillin binding to PBP4; in other words, the binding of increasing concentrations of oxacillin to PBP4 were revealed by the reduction of PBP4 labeling by Bocillin FL (Fig. 2a). The same phenomenon was also observed with ceftazidime, although its affinity for PBP4 was much lower (Fig. 2b). The apparent IC50s of the combined agents (avibactam and ceftazidime) for each of the PBPs of S. aureus were found to be very similar to those determined for ceftazidime used alone (Fig. 2c and Table 1). The exception was PBP4, which was only sufficiently labeled by Bocillin FL in the presence of avibactam.
FIG 2.
Affinities of oxacillin and ceftazidime for S. aureus PBPs in the presence or absence of avibactam. The indicated concentrations of avibactam in combination with oxacillin (a) or ceftazidime (b), or of ceftazidime alone (c), were used in a PBP competition assay with Bocillin FL.
The strong labeling of S. aureus PBP4 by Bocillin FL in the presence of avibactam is intriguing, especially as this phenomenon was concentration dependent (Fig. 1b) and not observed for any other PBP of the other bacterial species tested. This phenomenon cannot only be due to the increased availability of Bocillin FL for PBP4 when a competing β-lactam binds to other PBP targets, since, for example, addition of ceftazidime (0.03 to 1,024 μg/ml) still did not increase labeling of PBP4 by Bocillin FL (Fig. 2c). Published crystal structures of PBP4–β-lactam complexes provide evidence of the β-lactamase activity of S. aureus PBP4 (25). Accordingly, PBP4 often has been difficult to detect by use of various β-lactam-based reporter molecules (34, 37). The observed binding of Bocillin FL to S. aureus PBP4 in the presence of avibactam may thus tentatively be explained by an inhibition of the PBP4 β-lactamase activity by avibactam. Some β-lactamases have been shown to release intact avibactam after being acylated (5), and hence avibactam can be qualified as a slowly reversible non-β-lactam inhibitor. In the PBP assay, a combined effect of avibactam binding to S. aureus PBP2 and PBP3 as well as inhibition of PBP4 β-lactamase activity may increase available amounts of Bocillin FL for PBP4 labeling. The putative inhibition of PBP4 β-lactamase activity by avibactam and the possibility of an alternate binding site for avibactam on PBP4, however, remain to be demonstrated biochemically.
S. aureus PBP4 activity determines the level of peptidoglycan cross-linking and together with PBP2 participates in methicillin-resistant S. aureus (MRSA) PBP2a function (38). Depending on the genetic background of the studied strains, PBP4 is either considered nonessential (39) or an important resistance determinant of community-acquired MRSA (CA-MRSA) (40). To that effect, the anti-PBP4 β-lactam cefoxitin has been shown to enhance oxacillin activity against CA-MRSA but not against hospital-acquired MRSA (40). It would be interesting to see if avibactam could help in tackling specific MRSA strains by helping binding of β-lactams to PBP4.
The structure of S. aureus PBP4 has many similarities to that of E. coli PBP5 (25). However, the Gram-positive functional homolog of E. coli PBP2 is more difficult to identify, especially in cocci that lack cylindrical elongation (41). While the class B-type PBP2 seems to be the preferred target of avibactam in E. coli, the class C PBP3 is the predominant target of avibactam in S. pneumoniae (Table 1). The S. pneumoniae PBP banding profile generated with Bocillin FL appeared as previously reported for strain R6 by Paik et al. (28) (data not shown). The class C type 5 PBP group includes not only S. pneumoniae PBP3 but also E. coli PBP5 and S. aureus PBP4 (33).
In conclusion, the non-β-lactam β-lactamase inhibitor avibactam is able to covalently bind to some bacterial PBPs, notably to E. coli and H. influenzae PBP2, to PBPs 2 and 3 of P. aeruginosa and S. aureus, and to PBP3 of S. pneumoniae, which may explain its moderate antibacterial activity against some bacterial strains and species. In addition to the ability of avibactam to inhibit several β-lactamase types, identification of those PBP targets may allow the development of new derivatives with improved affinity for PBPs or new combination therapies that act on multiple PBP targets. The possible inhibition of S. aureus PBP4 β-lactamase activity by avibactam also warrants further investigation.
ACKNOWLEDGMENTS
This work was supported by a research grant from AstraZeneca Pharmaceuticals.
We thank J.-M. Bruneau, M. T. Black, and P. Levasseur for providing previously unpublished data. We also thank E. Tremblay (CHUS, Université de Sherbrooke) for help with the molecular imager instrument.
Funding Statement
The funders had no role in data collection and interpretation.
REFERENCES
- 1.WHO. 2014. Antimicrobial resistance: global report on surveillance. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Boucher HW, Talbot GH, Benjamin DK, Bradley J, Guidos RJ, Jones RN, Murray BE, Bonomo RA, Gilbert D. 2013. 10 × '20 progress: development of new drugs active against Gram-negative bacilli. An update from the Infectious Diseases Society of America. Clin Infect Dis 56:1685–1694. doi: 10.1093/cid/cit152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bush K, Fisher JF. 2011. Epidemiological expansion, structural studies, and clinical challenges of new β-lactamases from Gram-negative bacteria. Annu Rev Microbiol 65:455–478. doi: 10.1146/annurev-micro-090110-102911. [DOI] [PubMed] [Google Scholar]
- 4.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, Brisse S, Cao H, Wilksch J, Gorrie C, Schultz MB, Edwards DJ, Nguyen KV, Nguyen TV, Dao TT, Mensink M, Minh VL, Nhu NT, Schultsz C, Kuntaman K, Newton PN, Moore CE, Strugnell RA, Thomson NR. 2015. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A 112:E3574–E3581. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ehmann DE, Jahić H, Ross PL, Gu RF, Hu J, Kern G, Walkup GK, Fisher SL. 2012. Avibactam is a covalent reversible non-β-lactam β-lactamase inhibitor. Proc Natl Acad Sci U S A 109:11663–11168. doi: 10.1073/pnas.1205073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ehmann DE, Jahić H, Ross PL, Gu RF, Hu J, Durand-Réville TF, Lahiri S, Thresher J, Livchak S, Gao N, Palmer T, Walkup GK, Fisher SL. 2013. Kinetics of avibactam inhibition against class A, C, and D β-lactamases. J Biol Chem 288:27960–27971. doi: 10.1074/jbc.M113.485979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stachyra T, Pèchereau M-C, Bruneau J-M, Claudon M, Frère JM, Miossec C, Coleman K, Black MT. 2010. Mechanistic studies of the inactivation of TEM-1 and P99 by NXL104, a novel non-β-lactam β-lactamase inhibitor. Antimicrob Agents Chemother 54:5132–5138. doi: 10.1128/AAC.00568-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lagacé-Wiens PRS, Walkty A, Karlowsky JA. 2014. Ceftazidime-avibactam: an evidence-based review of its pharmacology and potential use in the treatment of Gram-negative bacterial infections. Core Evid 9:13–25. doi: 10.2147/CE.S40698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.ClinicalTrials.gov. 2015. NCT01689207. To investigate the safety and tolerability of aztreonam-avibactam (ATM-AVI). https://clinicaltrials.gov/ct2/show/NCT01689207? term=%22aztreonam%22+AND+%22avibactam%22&rank=1 Accessed 27 October 2015.
- 10.Riccobene TA, Su SF, Rank D. 2013. Single- and multiple-dose study to determine the safety, tolerability, and pharmacokinetics of ceftaroline fosamil in combination with avibactam in healthy subjects. Antimicrob Agents Chemother 57:1496–1504. doi: 10.1128/AAC.02134-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forest Pharmaceuticals, Inc. AVYCAZ™ (ceftazidime-avibactam) for injection. Package insert. U.S. Food and Drug Administration, Rockville, MD: http://www.accessdata.fda.gov/drugsatfda_docs/label/2015/206494s000lbl.pdf Accessed 26 May 2015. [Google Scholar]
- 12.Aktaş Z, Kayacan C, Oncul O. 2012. In vitro activity of avibactam (NXL 104) in combination with β-lactams against Gram-negative bacteria, including OXA-48 β-lactamase-producing Klebsiella pneumoniae. Int J Antimicrob Agents 39:86–89. doi: 10.1016/j.ijantimicag.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 13.Flamm RK, Stone GG, Sader HS, Jones RN, Nichols WW. 2014. Avibactam reverts the ceftazidime MIC90 of European Gram-negative bacterial clinical isolates to the epidemiological cut off value. J Chemother 26:333–338. doi: 10.1179/1973947813Y.0000000145. [DOI] [PubMed] [Google Scholar]
- 14.Keepers TR, Gomez M, Celeri C, Nichols WW, Krause KM. 2014. Bactericidal activity, absence of serum effect, and time-kill kinetics of ceftazidime-avibactam against β-lactamase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother 58:5297–5305. doi: 10.1128/AAC.02894-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sader HS, Castanheira M, Flamm RK, Farrell DJ, Jones RN. 2014. Antimicrobial activity of ceftazidime-avibactam against Gram-negative organisms collected from U.S. medical centers in 2012. Antimicrob Agents Chemother 58:1684–1692. doi: 10.1128/AAC.02429-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Zhang F, Zhao C, Wang Z, Nichols WW, Testa R, Li H, Chen H, He W, Wang Q, Wang H. 2014. In vitro activities of ceftazidime-avibactam and aztreonam-avibactam against 372 Gram-negative bacilli collected in 2011 and 2012 from 11 teaching hospitals in China. Antimicrob Agents Chemother 58:1774–1778. doi: 10.1128/AAC.02123-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhanel GG, Lawson CD, Adam H, Schweizer F, Zelenitsky S, Lagacé-Wiens PR, Denisuik A, Rubinstein E, Gin AS, Hoban DJ, Lynch JP III, Karlowsky JA. 2013. Ceftazidime-avibactam: a novel cephalosporin/β-lactamase inhibitor combination. Drugs 73:159–177. doi: 10.1007/s40265-013-0013-7. [DOI] [PubMed] [Google Scholar]
- 18.Lagacé-Wiens PRS, Tailor F, Simner P, DeCorby M, Karlowsky JA, Walkty A, Hoban DJ, Zhanel GG. 2011. Activity of NXL 104 in combination with β-lactams against genetically characterized Escherichia coli and Klebsiella pneumoniae isolates producing class A extended spectrum β-lactamases and class C β-lactamases. Antimicrob Agents Chemother 55:2434–2437. doi: 10.1128/AAC.01722-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berkhout J, Melchers MJ, van Mil AC, Nichols WW, Mouton JW. 2015. In vitro activity of ceftazidime-avibactam combination in in vitro checkerboard assays. Antimicrob Agents Chemother 59:1138–1144. doi: 10.1128/AAC.04146-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Massova I, Mobashery S. 1998. Kinship and diversification of bacterial penicillin-binding-proteins and β-lactamases. Antimicrob Agents Chemother 42:1–17. doi: 10.1093/jac/42.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coleman K. 2011. Diazabicyclooctanes (DBOs): a potent new class of non-β-lactam β-lactamase inhibitors. Curr Opin Microbiol 14:550–556. doi: 10.1016/j.mib.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 22.Morinaka A, Tsutsumi Y, Yamada M, Suzuki K, Watanabe T, Abe T, Furuuchi T, Inamura S, Sakamaki Y, Mitsuhashi N, Ida T, Livermore DM. 2015. OP0595, a new diazabicyclooctane: mode of action as a serine β-lactamase inhibitor, antibiotic and β-lactam ‘enhancer’. J Antimicrob Chemother 70:2779–2786. 10.1093/jac/dkv166. [DOI] [PubMed] [Google Scholar]
- 23.Ghuysen JM. 1991. Serine beta-lactamases and penicillin-binding proteins. Annu Rev Microbiol 45:37–67. doi: 10.1146/annurev.mi.45.100191.000345. [DOI] [PubMed] [Google Scholar]
- 24.Zhao G, Meier TI, Kahl SD, Gee KR, Blaszczak LC. 1999. Bocillin FL: a sensitive and commercially available reagent for detection of penicillin-binding proteins. Antimicrob Agents Chemother 43:1124–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Navratna V, Nadig S, Sood V, Prasad K, Arakere G, Gopal B. 2010. Molecular basis for the role of Staphylococcus aureus penicillin binding protein 4 in antimicrobial resistance. J Bacteriol 192:134–144. doi: 10.1128/JB.00822-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asli A, Brouillette E, Malouin F. 2014. Binding of avibactam to penicillin-binding proteins of Staphylococcus aureus, Streptococcus pneumoniae and Haemophilus influenzae, poster P1564. 24th European Congress of Clinical Microbiology and Infectious Diseases, Barcelona, Spain, 10 to 13 May 2014. [Google Scholar]
- 27.Clinical and Laboratory Standards Institute. 2011. Performance standards for antimicrobial susceptibility testing: twenty-first informational supplement. Document M100-S21. CLSI, Wayne, PA. [Google Scholar]
- 28.Paik J, Kern I, Lurz R, Hakenbeck R. 1999. Mutational analysis of the Streptococcus pneumoniae bimodular class A penicillin-binding proteins. J Bacteriol 181:3852–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spratt BG, Pardee AB. 1975. Penicillin-binding proteins and cell shape in E. coli. Nature 254:516–517. doi: 10.1038/254516a0. [DOI] [PubMed] [Google Scholar]
- 30.Moosdeen F, Williams JD, Yamabe S. 1988. Antibacterial characteristics of YTR 830, a sulfone beta-lactamase inhibitor, compared with those of clavulanic acid and sulbactam. Antimicrob Agents Chemother 32:925–927. doi: 10.1128/AAC.32.6.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gutmann L, Vincent S, Billot-Klein D, Acar JF, Mrèna E, Williamson R. 1986. Involvement of penicillin-binding protein 2 with other penicillin-binding proteins in lysis of Escherichia coli by some beta-lactam antibiotics alone and in synergistic lytic effect of amdinocillin (mecillinam). Antimicrob Agents Chemother 30:906–912. doi: 10.1128/AAC.30.6.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davies TA, Page MG, Shang W, Andrew T, Kania M, Bush K. 2007. Binding of ceftobiprole and comparators to the penicillin-binding proteins of Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus, and Streptococcus pneumoniae. Antimicrob Agents Chemother 51:2621–2624. doi: 10.1128/AAC.00029-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev 32:234–258. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- 34.Dargis M, Malouin F. 1994. Use of biotinylated β-lactams and chemiluminescence for study and purification of penicillin-binding proteins in bacteria. Antimicrob Agents Chemother 38:973–980. doi: 10.1128/AAC.38.5.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Picard M, Malouin F. 1992. Molecular basis of the efficacy of cefaclor against Haemophilus influenzae. Antimicrob Agents Chemother 36:2569–2572. doi: 10.1128/AAC.36.11.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kubota T, Higa F, Kusano N, Nakasone I, Haranage S, Tateyama M, Yamane N, Fujita J. 2006. Genetic analysis of beta-lactamase-negative ampicillin-resistant Haemophilus influenzae strains isolated in Okinawa, Japan. Jpn J Infect Dis 59:36–51. [PubMed] [Google Scholar]
- 37.Moisan H, Pruneau M, Malouin F. 2010. Binding of ceftaroline to penicillin-binding proteins of Staphylococcus aureus and Streptococcus pneumoniae. J Antimicrob Chemother 65:713–716. doi: 10.1093/jac/dkp503. [DOI] [PubMed] [Google Scholar]
- 38.Leski TA, Tomasz A. 2005. Role of penicillin-binding protein 2 (PBP2) in the antibiotic susceptibility and cell wall cross-linking of Staphylococcus aureus: evidence for the cooperative functioning of PBP2, PBP4, and PBP2A. J Bacteriol 187:1815–1824. doi: 10.1128/JB.187.5.1815-1824.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katayama Y, Zhang HZ, Chambers HF. 2003. Effect of disruption of Staphylococcus aureus PBP4 gene on resistance to beta-lactam antibiotics. Microb Drug Resist 9:329–336. doi: 10.1089/107662903322762752. [DOI] [PubMed] [Google Scholar]
- 40.Memmi G, Filipe SR, Pinho MG, Fu Z, Cheung A. 2008. Staphylococcus aureus PBP4 is essential for β-lactam resistance in community-acquired methicillin-resistant strains. Antimicrob Agents Chemother 52:3955–3966. doi: 10.1128/AAC.00049-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zapun A, Vernet T, Pinho MG. 2008. The different shapes of cocci. FEMS Microbiol Rev 32:345–360. doi: 10.1111/j.1574-6976.2007.00098.x. [DOI] [PubMed] [Google Scholar]