Abstract
Background
The aim of this study was to determine the effect on proliferation, migration, and invasion after silencing HOTAIR in ovarian cancer SKOV3 cells, and to elucidate the potential mechanism.
Material/Methods
We analyzed the mRNA expression level of HOTAIR and PIK3R3 in ovarian cancer SKOV3, OVCAR3, and A2780 cell lines. We analyzed the mRNA expression level of HOTAIR and PIK3R3 in ovarian SKOV3 after transection with miR-214 or miR-217. We analyzed the mRNA and protein expression level of PIK3R3 when silencing HOTAIR. We analyzed the expression of HOTAIR when silencing PIK3R3. We analyzed the proliferation, migration and invasion in ovarian cancer SKOV3 after silencing HOTAIR or PIK3R3.
Results
The expression of HOTAIR and PIK3R3 in ovarian SKOV3 and OVCAR3 was increased compared with A2780 cells (P<0.05). The mRNA level of HOTAIR and PIK3R3 in ovarian cancer SKOV3 cells was decreased when transected with miR-214 or miR-217 compared to negative control (p<0.05). The mRNA and protein level of PIK3R3 was decreased when HOTAIR was silenced and the mRNA level of HOTAIR was decreased when PIK3R3 was silenced (p<0.05). The proliferation, migration and invasion was decreased in ovarian SKOV3 when HOTAIR or PIK3R3 was silenced (p<0.05).
Conclusions
HOTAIR can promote proliferation, migration, and invasion in ovarian SKOV3 cells as a competing endogenous RNA.
MeSH Keywords: Cell Migration Assays, Leukocyte; MicroRNAs; Ovarian Neoplasms
Background
Epithelial ovarian cancer (EOC) is among the leading causes of cancer death in women. Because most ovarian cancers become drug-resistant, the survival rates are still low [1]. Thus, it is important to understand the potential molecular mechanisms involved in EOC metastasis. It is crucial to find biomarkers of EOC for diagnosis and predict the clinical outcome of the patient.
Long non-coding RNAs (LincRNA Long intergenic non-coding RNA) have been reported to regulate gene expression at various levels, including in chromatin modification, transcription, and posttranscriptional processing [2,3]. LincRNA-p21 and GAS5 act as tumor-suppressor molecules [4]. Expression of lincRNA-p21 is regulated by the p53 and upon transcription, it suppresses expression of the genes transcriptionally regulated through p53 by binding to the hnRNP-K complex [5]. GAS5 plays an important role in the induction of apoptosis. It suppresses some antiapoptotic genes through binding to the glucocorticoid receptor (GR) and hence prevents GR from binding to the glucocorticoid response elements on the target DNA molecule [6].
Long noncoding RNA HOTAIR (HOX antisense intergenic RNA) has been considered as a pro-oncogene in multiple cancers [7]. HOTAIR expression was obviously increased in leukemic cell lines and primary AML blasts. It modulated c-KIT expression by competitively binding miR-193a [8]. The up-regulation of HOTAIR has been found in both estrogen receptor-positive and triple-negative breast cancer. It has been reported that HOTAIR is regulated by estrogen via ERs in ER-positive breast cancer [9]. HOTAIR expression was increased in oral squamous cell carcinoma compared to non-tumor tissue and is associated with metastasis, stage, and histological differentiation [10]. However, the underlying molecular mechanism of HOTAIR in ovarian cancer remains under unclear.
The PI3K (phosphoinositide-3-kinase, 3)/AKT/mammalian target of rapamycin (mTOR) pathway plays a critical role in the malignant transformation of human tumors and their subsequent growth, proliferation, and metastasis. Preclinical investigations have suggested that the PI3K/AKT/mTOR pathway is frequently activated in ovarian cancer [11,12]. PIK3R3 is regarded as a potential therapeutic target of ovarian cancer [13]. However, there is no report about the relation between PIK3R3 and HOTAIR.
In this study, we investigated the function of HOTAIR in ovarian cancer SKOV3 cells, then elucidated the mechanism.
Material and Methods
Immunohistochemistry (IHC)
The patient and control groups had no significantly difference in terms of age and parity (p>0.05). The tissue samples were fixed in formalin, embedded in paraffin, sectioned (5 μm thick), deparaffinized in xylene, dehydrated in a graded series of ethanol, subjected to antigen retrieval in citrate buffer for 30 min in a steamer, and washed in phosphate-buffered saline (PBS). PBS was used for all subsequent washes and for antiserum dilution. Tissue sections were quenched sequentially in 3% hydrogen peroxide and blocked with PBS containing 10% goat serum (Sigma) for 1 h at 37ºC. Next, slides were incubated with the primary antibody overnight at 4ºC, which included Rabbit monoclonal (1:100; ab186612; Abcam). Negative controls included omission of primary antibody and use of irrelevant primary antibodies. After several washes (3×3 min) to remove excess antibody, the slides were incubated with diluted (1:300) anti-rabbit biotinylated antibodies (Beijing Biosynthesis Biotechnology CO., LTD, China) for 1 h. All the slides were washed in PBS and were incubated in avidin biotin peroxidase complex (ABC, Beijing Zhongshan Golden Bridge Biotechnology Co. Ltd., Beijing, China) diluted 1:300 in PBS for 30 min in humidified chambers at 37ºC. DAB (Beijing Biosynthesis Biotechnology CO., Ltd., China) was used as a chromogen and hematoxylin was used as a nuclear counter-stain.
Slides were evaluated independently by 3 pathologists for distribution and intensity of signal as described by De Falco et al. [14] Intensity was scored from 0 to 3: 0 (absent immunopositivity); 1 (low immunopositivity); 2 (moderate immunopositivity); and 3 (intense immunopositivity). An average of 22 fields was observed for each tissue.
Cell lines and culture conditions
Ovarian cancer cell lines (SKOV3, OVCAR3, and A2780) were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (10% FBS), 100 U/ml penicillin, and 100 mg/ml streptomycin (Invitrogen) in humidified air at 37ºC with 5% CO2. The miRNA mimics were purchased from Genepharma Co., Ltd. (Shanghai, China). The negative control was non-specifically transfected siRNA.
Cell proliferation assay
Cell proliferation was determined using the CCK-8 assay, as described previously [15], and EdU assay was performed using the Cell-Light TM EdU imaging detecting kit according to the instructions in the kit (Ruibo Biotechnology, Guangzhou, China). EdU is a thymidine analog that can be used to label cells undergoing DNA replication [16].
Quantitative real-time polymerase chain reaction
Total RNA was isolated using an RNA pure High-purity Total RNA Rapid Extraction Kit (BioTeke, RP1201, China), as per the instructions provided in the kit. cDNA was synthesized using the iSCRIPT cDNA synthesis kit (Bio-Rad). The primers used for amplifying HOTAIR, PIK3R3, and GAPDH were synthesized by Guangzhou Funeng Co., Ltd. The real-time PCR kit was purchased from Guangzhou Funeng Co., Ltd. PCR conditions were 95ºC for 10 s, 60ºC for 20 s, and 72ºC for 10 s. Each sample was analyzed in triplicate. Relative quantification of mRNA was performed using the comparative threshold cycles (CT) method. This value was used to plot the gene expression using the formula 2−ΔΔCT.
Western blot analysis
Expression of MAPK1 protein was analyzed by Western blotting, as previously described [17]. The primary antibodies used were polyclonal goat anti-PIK3R3 (1:1000; ab186612; Abcam) and monoclonal rabbit anti-GAPDH (1:5000; ab181602; Abcam). The bands were detected via enhanced chemiluminescence (ECL) reagent (Beyotime Institute of Biotechnology, Jiangsu, China) and quantified by ImageQuant 3.3 software (Molecular Dynamics).
Statistical evaluation
All values are expressed as mean ±SEM. The Mann-Whitney U test was used. A p value of <0.05 was considered statistically significant.
Results
The expression of PIK3R3 in ovarian cancer tissues
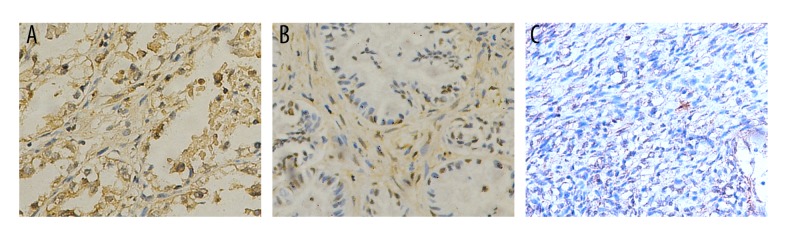
The expression of PIK3R3 was significantly higher in ovarian carcinoma samples than in normal ovarian samples (Figure 1; P<0.05). PIK3R3 was predominantly localized on the plasma membrane and cytoplasm (Figure 1A, 1B).
Figure 1.
The expression of PIK3R3 in ovarian cancer tissues. (A) Ovarian serous adenocarcinoma; (B) Mucinous adenocarcinoma; (C) Normal ovarian tissues. ×200 magnification.
The expression of HOTAIR and PIK3R3 in ovarian cancer cells
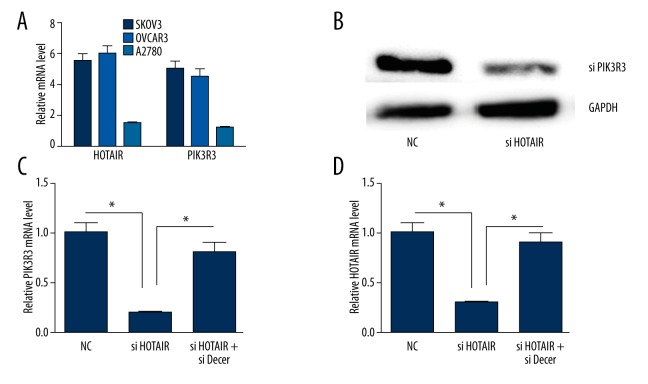
To investigate the expression of HOTAIR and PIK3R3 in ovarian cancer cells, we detected the mRNA expression of HOTAIR and PIK3R3 in SKOV3, OVCAR3, and A2780 cells. The mRNA expression of HOTAIR and PIK3R3 in ovarian cancer SKOV3 and OVCAR3 was increased compared with A2780 cells (Figure 2A) (P<0.05).
Figure 2.
The relative expression of mRNA and protein of HOTAIR or PIK3R3. * p<0.05 (versus control group); error bars, SEM. (A) HOTAIR or PIK3R3 mRNA was detected in A2780, OVCAR3 and SKOV3 cells. (B) silencing HOTAIR down-regulated the protein expression of PIK3R3. (C) silencing HOTAIR down-regulated the mRNA expression of PIK3R3. (D) Silencing PIK3R3 down-regulated the expression of HOTAIR.
The interaction between HOTAIR and PIK3R3 in ovarian cancer SKOV3 cells
We selected SKOV3 cells to determine the high-level expression of HOTAIR and PIK3R3. The protein and mRNA level of PIK3R3 were decreased when silencing HOTAIR. The mRNA level of HOTAIR was decreased when silencing PIK3R3 (P<0.05). When silencing dicer, the interaction between HOTAIR and PIK3R3 was abolished (Figure 2B–2D).
HOTAIR and PIK3R3 are targets of miR-214 or miR-217
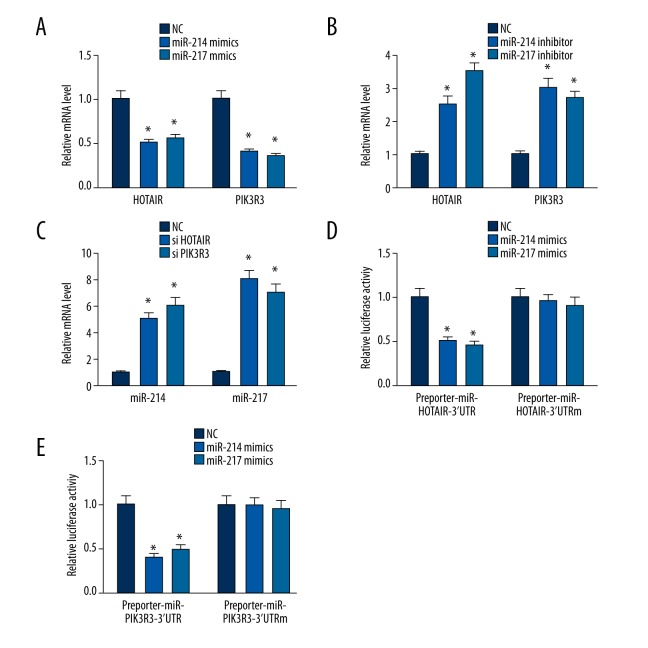
We investigated whether HOTAIR and PIK3R3 are targets of miR-214 or miR-217. MiR-214 or miR-217 mimics and inhibitors were transfected into ovarian cancer SKOV3 cells. The expression of HOTAIR and PIK3R3 was decreased when transfected with miR-214 or miR-217 mimics (Figure 3A; P<0.05). In contrast, the expression of HOTAIR and PIK3R3 was increased when transfected with miR-214 or miR-217 inhibitors (Figure 3B) (P<0.05).
Figure 3.
The expression of mRNA and luciferase. * p<0.05 (versus control group); error bars, SEM. (A) miR-214 and miR-217 mimics regulated the expression of HOTAIR or PIK3R3. (B) miR-214 and miR-217 inhibitors regulated the expression of HOTAIR or PIK3R3. (C) silencing HOTAIR or PIK3R3 regulated miR-214 and miR-217. (D, E) miR-214 and miR-217 regulated luciferase.
Silencing HOTAIR or PIK3R3 increased the expression of miR-214 or miR-217
We investigated whether HOTAIR and PIK3R3 regulates miR-214 or miR-217. The expression of miR-214 or miR-217 was increased when HOTAIR or PIK3R3 was silenced (Figure 3C) (P<0.05). The present work provides the first evidence for a positive HOTAIR/PIK3R3 correlation and the crosstalk between miR-214 or miR-217, HOTAIR, and PIK3R3. Luciferase assays confirmed the existence of specific crosstalk between the HOTAIR and PIK3R3 mRNA through competition for miR-214 or miR-217 binding (Figure 3D, 3E).
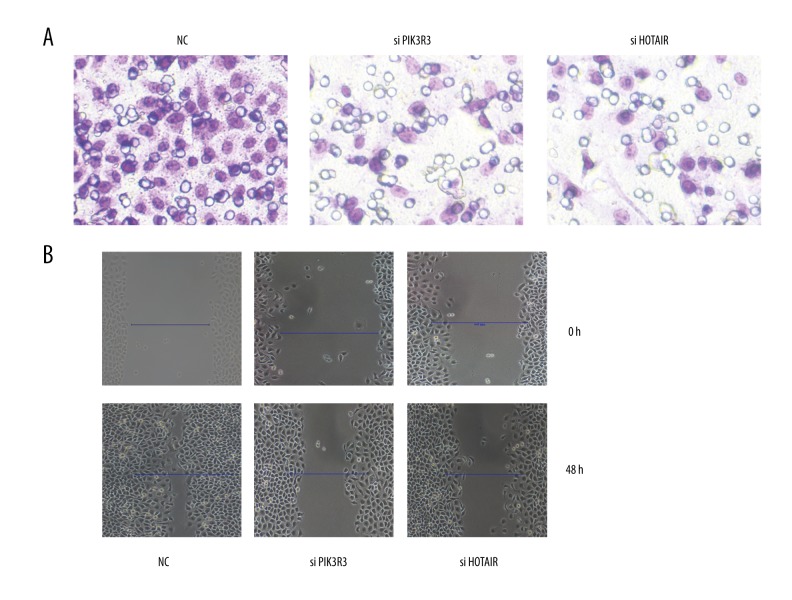
Silencing HOTAIR or PIK3R3 inhibited the proliferation, migration and invasion in SKOV3 cell
We studied the effects of HOTAIR or PIK3R3 on SKOV3 cells. The proliferation, migration, and invasion of SKOV3 was inhibited when HOTAIR or PIK3R3 was silenced (Figure 4 and 5)(P<0.05).
Figure 4.
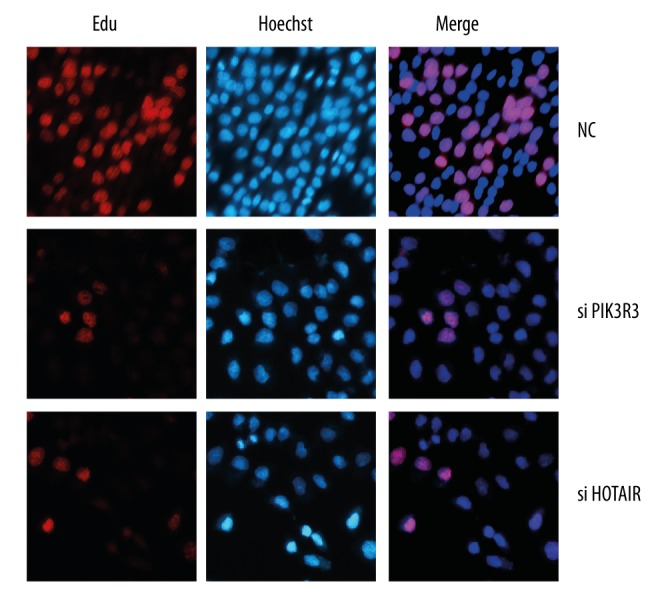
Cell proliferation was detected by EdU assay.
Figure 5.
The migration and invasion was detected. NC represents negative control.
Discussion
Long noncoding RNAs are a recently reported class of non-protein coding RNAs, which have now increasingly been shown to be involved in a wide variety of biological processes as regulatory molecules. Recent reports have suggested that lncRNAs could potentially interact with miRNAs and regulated their regulatory role by interactions [18].
In this study, we found that the expression of HOTAIR and PIK3R3 in ovarian SKOV3 and OVCAR3 was increased compared to that in A2780 cells. The repression of HOTAIR and PIK3R3 were strongly correlated. Furthermore, we found that HOTAIR interacted with PIK3R3 through miR-214 or miR-217. The proliferation, migration, and invasion were inhibited in ovarian SKOV3 after silencing HOTAIR or PIK3R3.
Emerging evidence suggests that lncRNAs may participate in this regulatory circuitry. We hypothesized that HOTAIR may also serve as a ceRNA. Bioinformation analysis (http://starbase.sysu.edu.cn/) indicated that HOTAIR is a putative ceRNA of PIK3R3. Firstly, we investigated the expression of PIK3R3 and HOTAIR in ovarian cancer SKOV3, OVCAR3, and A2780. We found that the expression of HOTAIR and PIK3R3 in ovarian SKOV3 and OVCAR3 was increased compared with A2780 cells (P<0.05) suggesting that HOTAIR may be related to PIK3R3. Silencing HOTAIR down-regulated the protein and mRNA level of PIK3R3. Silencing PIK3R3 downregulated the expression of HOTAIR, indicating that HOTAIR may interact with PIK3R3. When silencing dicer, the interaction between HOTAIR and PIK3R3 was abolished. Therefore, we hypothesized that the miRNA may be involved in this process.
Bioinformation analysis predicts that miR-214 and miR-217 are shared by HOTAIR and PIK3R3 [19,20]. Therefore, we determined whether the interaction of HOTAIR and PIK3R3 is regulated by miR-214 and miR-217. As expected, miR-214 and miR-217 mimics can inhibit the expression of HOTAIR and PIK3R3. When silencing HOTAIR, the expression of miR-214 and miR-217 was increased, suggesting that the interaction of HOTAIR and PIK3R3 is mediated through miR-214 and miR-217.
Phosphatidylinositol 3-kinase (PI3K) is involved in a wide range of cancer-associated signaling pathways [21]. The role of PI3K in ovarian cancer will undoubtedly provide new insights for pharmacologic intervention in this malignancy. Molecular cloning of PI3Ks reveals a large and complex family that contains 3 classes of multiple subunits and isoforms [22]. PIK3R3 had increased expression in ovarian cancer and knockdown of PIK3R3 mRNA expression increases cell apoptosis in ovarian cancer in vitro [13]. Our study revealed that silencing PIK3R3 can inhibit the proliferation, migration, and invasion in ovarian cancer SKOV3 cells. This is consistent with a previous report [13] that HOTAIR is overexpressed in males in colorectal cancer, hepatocellular carcinoma, pancreatic cancer, and gastrointestinal stromal tumors, and in females in ovarian cancer [23–37]. Down-regulation of HOTAIR can inhibit the malignant behavior of cancer [28]. In this study, silencing HOTAIR inhibited the proliferation, migration, and invasion of ovarian cancer SKOV3 cells.
Conclusions
Our study determined that HOTAIR interacting with PIK3R3 regulates ovarian cancer skov3 cells proliferation, migration, and invasion through miR-214 and miR-217. It can serve as a therapy target of ovarian cancer. This study gives new insight into ovarian cancer research, pathogenesis, and treatment. In the future, we will study the interaction between HOTAIR and PIK3R3 in vivo.
Footnotes
Source of support: Departmental sources
References
- 1.Rustin G, van der Burg M, Griffin C, et al. Early versus delayed treatment of relapsed ovarian cancer. Lancet. 2011;377(9763):380–81. doi: 10.1016/S0140-6736(11)60126-8. [DOI] [PubMed] [Google Scholar]
- 2.Shi Y, Zhou Y. The role of surgery in the treatment of gastric cancer. J Surg Oncol. 2010;101(8):687–92. doi: 10.1002/jso.21455. [DOI] [PubMed] [Google Scholar]
- 3.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneider C, King RM, Philipson L. Genes specifically expressed at growth arrest of mammalian cells. Cell. 1988;54(6):787–93. doi: 10.1016/s0092-8674(88)91065-3. [DOI] [PubMed] [Google Scholar]
- 5.Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–19. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kino T, Hurt DE, Ichijo T, et al. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3(107):ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu WM, Zhu X, Wang WM, et al. Hotair Mediates Hepatocarcinogenesis through Suppressing MiRNA-218 Expression and Activating P14 and P16 Signaling. J Hepatol. 2015;63(4):886–95. doi: 10.1016/j.jhep.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 8.Xing CY, Hu XQ, Xie FY, et al. Long non-coding RNA HOTAIR modulates c-KIT expression through sponging miR-193a in acute myeloid leukemia. FEBS Lett. 2015;589(15):1981–87. doi: 10.1016/j.febslet.2015.04.061. [DOI] [PubMed] [Google Scholar]
- 9.Tao S, He H, Chen Q. Estradiol induces HOTAIR levels via GPER-mediated miR-148a inhibition in breast cancer. J Transl Med. 2015;13:131. doi: 10.1186/s12967-015-0489-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Y, Zhang L, Zhang L, et al. Long non-coding RNA HOTAIR promotes tumor cell invasion and metastasis by recruiting EZH2 and repressing E-cadherin in oral squamous cell carcinoma. Int J Oncol. 2015;46(6):2586–94. doi: 10.3892/ijo.2015.2976. [DOI] [PubMed] [Google Scholar]
- 11.Mabuchi S, Kuroda H, Takahashi R, Sasano T. The PI3K/AKT/mTOR pathway as a therapeutic target in ovarian cancer. Gynecol Oncol. 2015;137(1):173–79. doi: 10.1016/j.ygyno.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 12.Li H, Zeng J, Shen K. PI3K/AKT/mTOR signaling pathway as a therapeutic target for ovarian cancer. Arch Gynecol Obstet. 2014;290(6):1067–78. doi: 10.1007/s00404-014-3377-3. [DOI] [PubMed] [Google Scholar]
- 13.Zhang L, Huang J, Yang N, et al. Integrative genomic analysis of phosphatidylinositol 3′-kinase family identifies PIK3R3 as a potential therapeutic target in epithelial ovarian cancer. Clin Cancer Res. 2007;13(18 Pt 1):5314–21. doi: 10.1158/1078-0432.CCR-06-2660. [DOI] [PubMed] [Google Scholar]
- 14.De Falco M, Fedele V, Cobellis L, et al. Pattern of expression of cyclin D1/CDK4 complex in human placenta during gestation. Cell Tissue Res. 2004;317(2):187–94. doi: 10.1007/s00441-004-0880-z. [DOI] [PubMed] [Google Scholar]
- 15.Zhang BG, Li JF, Yu BQ, et al. microRNA-21 promotes tumor proliferation and invasion in gastric cancer by targeting PTEN. Oncol Rep. 2012;27:1019–26. doi: 10.3892/or.2012.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci USA. 2008;105:2415–20. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He HJ, Zhu TN, Xie Y, et al. Pyrrolidine dithiocarbamate inhibits interleukin-6 signaling through impaired STAT3 activation and association with transcriptional coactivators in hepatocytes. J Biol Chem. 2006;281(42):31369–79. doi: 10.1074/jbc.M603762200. [DOI] [PubMed] [Google Scholar]
- 18.Jalali S, Bhartiya D, Lalwani MK, et al. Systematic transcriptome wide analysis of lncRNA-miRNA interactions. PLoS One. 2013;8(2):e53823. doi: 10.1371/journal.pone.0053823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang JH, Li JH, Shao P, et al. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39(Database issue):D202–9. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7(8):606–19. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 22.Nakayama K, Nakayama N, Kurman RJ, et al. Sequence mutations and amplification of PIK3CA and AKT2 genes in purified ovarian serous neoplasms. Cancer Biol Ther. 2006;5(7):779–85. doi: 10.4161/cbt.5.7.2751. [DOI] [PubMed] [Google Scholar]
- 23.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–76. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kogo R, Shimamura T, Mimori K, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71(20):6320–26. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 25.Geng YJ, Xie SL, Li Q, et al. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J Int Med Res. 2011;39(6):2119–28. doi: 10.1177/147323001103900608. [DOI] [PubMed] [Google Scholar]
- 26.Niinuma T, Suzuki H, Nojima M, et al. Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res. 2012;72(5):1126–36. doi: 10.1158/0008-5472.CAN-11-1803. [DOI] [PubMed] [Google Scholar]
- 27.Qiu JJ, Lin YY, Ye LC, et al. Overexpression of long non-coding RNA HOTAIR predicts poor patient prognosis and promotes tumor metastasis in epithelial ovarian cancer. Gynecol Oncol. 2014;134(1):121–28. doi: 10.1016/j.ygyno.2014.03.556. [DOI] [PubMed] [Google Scholar]
- 28.Kim HJ, Lee DW, Yim GW, et al. Long non-coding RNA HOTAIR is associated with human cervical cancer progression. Int J Oncol. 2015;46(2):521–30. doi: 10.3892/ijo.2014.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]