Abstract
Background
Rickettsia species are strictly intracellular bacteria that have undergone a reductive genomic evolution. Despite their allopatric lifestyle, almost half of the 26 currently validated Rickettsia species have plasmids. In order to study the origin, evolutionary history and putative roles of rickettsial plasmids, we investigated the evolutionary processes that have shaped 20 plasmids belonging to 11 species, using comparative genomics and phylogenetic analysis between rickettsial, microbial and non-microbial genomes.
Results
Plasmids were differentially present among Rickettsia species. The 11 species had 1 to 4 plasmid (s) with a size ranging from 12 kb to 83 kb. We reconstructed pRICO, the last common ancestor of the current rickettsial plasmids. pRICO was vertically inherited mainly from Rickettsia/Orientia chromosomes and diverged vertically into a single or multiple plasmid(s) in each species. These plasmids also underwent a reductive evolution by progressive gene loss, similar to that observed in rickettsial chromosomes, possibly leading to cryptic plasmids or complete plasmid loss. Moreover, rickettsial plasmids exhibited ORFans, recent gene duplications and evidence of horizontal gene transfer events with rickettsial and non-rickettsial genomes mainly from the α/γ-proteobacteria lineages. Genes related to maintenance and plasticity of plasmids, and to adaptation and resistance to stress mostly evolved under vertical and/or horizontal processes. Those involved in nucleotide/carbohydrate transport and metabolism were under the influence of vertical evolution only, whereas genes involved in cell wall/membrane/envelope biogenesis, cycle control, amino acid/lipid/coenzyme and secondary metabolites biosynthesis, transport and metabolism underwent mainly horizontal transfer events.
Conclusion
Rickettsial plasmids had a complex evolution, starting with a vertical inheritance followed by a reductive evolution associated with increased complexity via horizontal gene transfer as well as gene duplication and genesis. The plasmids are plastic and mosaic structures that may play biological roles similar to or distinct from their co-residing chromosomes in an obligate intracellular lifestyle.
Introduction
Rickettsia species (Order Rickettsiales, Family Rickettsiaceae) are obligate intracellular α-proteobacteria associated with diverse eukaryotic hosts. The genus Rickettsia emerged approximately 150 million years ago after several transitions from a presumably free-living ancestor of Rickettsiales to an intracellular life and then to primarily infecting arthropod lineages approx. 525–425 million years ago [1, 2]. Rickettsiae can infect humans or animals, mostly through arthropod bites, and can cause a range of mild to fatal diseases such as epidemic typhus and Rocky Mountain spotted fever. Twenty-six species currently have standing in the nomenclature, but many isolates await taxonomic classification [3]. Rickettsia species diverged into three phylogenetic groups: the spotted fever group (SFG) associated with ticks, fleas and mites and causing spotted fevers, the typhus group (TG) including R. prowazekii and R. typhii associated with body lice and rat fleas, respectively, and causing typhus, and the so-called ancestral group (AG) containing R. bellii and R. canadensis, associated with ticks [4,5]. However, recent studies have showed that other Rickettsia lineages exist, notably associated with protists, leeches or arthropods [1, 2].
Deciphering rickettsial genomes revealed unexpected genetic and evolutionary features. These bacteria have small genomes (1.1–1.8 Mbp) that have undergone a reductive evolution, possibly in relation to their strict intracellular lifestyle in which some metabolic pathways are progressively lost as host cells provide the missing metabolites. Consequences of this progressive genomic reduction included: i) an enrichment in A+T content, polyA/T homopolymers, conjugative clusters and mechanisms of adhesion to host and motility; ii) a high degree of inter-species genomic synteny; iii) an increased virulence; and iv) a variable distribution of plasmids [2, 6–11]. Although initially thought to be devoid of plasmids, such mobile genetic elements (MGEs) have now been identified in at least 11 Rickettsia species. In addition, these plasmids may be diversely distributed among strains of a species, some strains having no, one or several plasmids [12–22]. Finally, the presence of plasmids may also vary according to the passage history or intra-specific variability, as was observed in R. akarii and R. bellii [21, 22]. However, the mechanisms underlying the observed differences in plasmid content among species and strains remain unknown.
Rickettsial plasmids (RPs) were first detected in R. felis in which two isoforms of the putative conjugative pRf plasmid were identified [14]. Based on phylogenomic analysis, the genes in the R. felis pRf plasmid were suggested to have been acquired through a single horizontal gene transfer event in R. felis or in an ancestor before the divergence of the R. felis/ R. akari clade [14, 17]. Furthermore, on the basis of the presence of the pRf plasmid and one unpublished sequence of a putative plasmid in R. bellii OSU85-389, it was hypothesized that the primitive rickettsial ancestor harbored a plasmid system that was lost in certain lineages [23]. Moreover, Baldridge et al. [21, 22] suggested multiple possible origins of RPs as well as likely horizontal transfers from plasmids to chromosomes and vice versa using phylogenetic analyses of the parA, hsp1 and hsp2 genes.
However, to date, most of the studies on RPs have been based on a limited number of chromosomic and/or plasmidic sequences. Thus, the putative origin(s) and evolutionary processes of RPs remain mostly unknown. As recent studies have demonstrated that plasmids, not virus, are key vectors of genetic exchanges between bacterial chromosomes, even between distant phylogenetic groups [24, 25], we accepted the opportunity provided by the expanded number of publicly available genomes from arthropod-associated Rickettsia species to study the RPs by comparative genomic analysis and to reconstruct the evolutionary scenario of forces that shaped their structures and predicted functions. We inferred the origin, evolution and putative roles of RPs.
Material and Methods
Genomic sequences and annotation
Genomic sequences of plasmids and chromosomes (100% and 94% of which are both complete and circular, respectively) belonging to 26 species and 35 strains of Rickettsia and Orientia genera were downloaded from the NCBI ftp server (ftp://ftp.ncbi.nih.gov/Genome/). These species were collected and identified in diverse arthropod hosts (ticks, insects and fleas) or clinical patients (humans) from Africa, Europe, America, Azia and Australia from 1941 to 2002, and they are members of either spotted fever group (SFG), typhus group (TG) and ancestral groups (AG) rickettsiaes. To avoid potential biases across the originally published and unpublished data, that were generated by different gene identification and annotations, all genomes were subjected to CDSs (CoDing Sequences) predictions with the same AMIgene software [26] and automatic functional re-annotation against the RickBase [17] and non-redundant NR databases using pipRick (an in-house annotation pipeline written in Perl language) including the BLASTp algorithm [27]. The re-annotated plasmids were then manually curated and either complete (coverage > = 80% of the longest homolog), split (gene with at least two CDSs), fragment (coverage <80% of the longest homolog) or chimeric genes were distinguished [14, 17]. Functional classification of gene families (COG ID and Letters) was searched using COGnitor and COGsoft against COG database [28, 29]. Thus, a standardized database named RickettsiaDB was constructed for further investigations.
Comparative genomics and phylogenetics
In order to examine evolutionary relationships between Rickettsia plasmids, proteins were subjected to a reciprocical best BLAST hit (BBH) algorithm with all-against-all search (coverage of the query length > = 60% and E-value < 10-5) using COGsoft software [28, 29]. Each putative orthologous groups of plasmidic rickettsial genes was named pRIGs, and then a manual curation was performed to detect false negatives. A gene content microarray data matrix (pRIGs x plasmids) based on the presence and absence of genes including the degradation levels (complete: red, split: yellow, fragment: blue, chimeric: green, and absent or remnant: violet) was then constructed using TMeV software (http://sigenae.org/index.php?id=88). Sequence datasets of each pRIG and COGID were extracted and packaged into single fasta files for further analysis.
Evolutionary relationships between rickettsial plasmids, Rickettsia and Orientia chromosomes as well as microbial and non-microbial genomes were, first, examined using BLASTp and tBLASTn algorithms (Cutoffs: aa sequence identity > = 25%, coverage query> = 60%, E-value<10-5), search against RickettsiaDB (excluding plasmid sequences), non-redundant NR and RefSeq_genomic databases to identify their corresponding best homologs. The genes which did not exhibit, any significant hit neither with rickettsial chromosomes nor with other genomes from NCBI databases and having an E-value>0.01, were considered as ORFans. New visually inspected datasets that gathered sequences of rickettsial plasmids (each pRIG and COGID) and their corresponding homologs were downloaded and packaged into single FASTA files to perform phylogenetic analysis. Split genes were concatenated while highly degraded sequences (either very short fragments or remnants) were removed. Moreover, redundant sequences of the same gene from different BLAST analysis were removed to retain some representatives. Multiple sequence alignments were carried out using CLUSTALX and/or MUSCLE applications [30, 31]. Phylogenetic trees were computed with MEGA version 6 [32]. Both Neighbor-Joining (NJ) and Maximum Likelihood (ML) methods, respectively, under the JTT amino acid substitution matrix and the WAG model plus the Nearest-Neighbor-Interchange (NNI) were examined. Each tree was constructed by examining the following parameters: uniform sites, gamma (Γ) distribution of parameter α to account for substitution rate heterogeneity among sites and deletion either complete or partial (> = 90%). The robustness of the tree nodes was estimated by Bootstrap Percentage (BP) using 1000 and 100 replicates for NJ and ML, respectively. For each pRIG, either single or multiple phylogeny (ies) were examined to retain a consistent dataset for both evolutionary methods.
The evolutionary events including vertical and horizontal gene transfers (VGT and HGT) were identified using distinct cutoffs from best BLAST hits and/or their BP confidence in each phylogenetic trees as follows: VGT (proteins with 28> = aa identity < = 100%), evident HGT (aa identity > = 50% and BP> = 60) and probable HGT (25> = aa identity < = 80%). The recent duplications (i.e., inparalogs after the divergence of each species) were identified in phylogenetic trees or in case of ORFans by BLASTp with an identity cutoff > = 80%. To perform plasmid grouping and their putative evolutionary origins, the supertree algorithm based on the subtree prune-and-regraft distance [33] was also used from NJ trees of 10 selected genes present in most plasmids. The phylogenetic results were summarized into evolutionary networks between the rickettsial plasmids and microbial as well as non-microbial genomes using CYTOSCAPE software [34]. In the networks, the nodes represent the genes while the edges represent the links connecting them. Last, complete sequences of all plasmids were started from the dnaA-like gene and ended with the parA and some unknown genes, and then multiple pairwise alignments (cutoff: Evalue<10-5) were performed according to plasmid grouping using BLASTn and viewed under genoPlotR tool [35].
Statistical analysis
The correlation between Rickettsia plasmids and chromosomes sizes was performed using the correlation coefficient R of Pearson and the coefficient of determination R². Statistical analyses were performed using the R Commander software package (http://r-forge.r-project.org).
Results/Discussion
RPs are in a process of reductive evolution
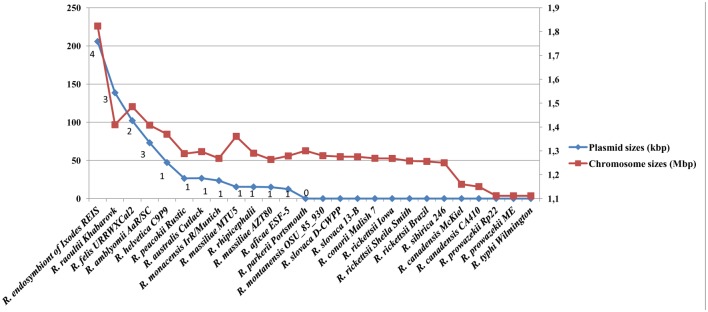
A total of 20 plasmids occurred in 11 SFG Rickettsia species, represented by 13 strains collected from various geographical locations worldwide (Table 1). In contrast, 9 SFG and TG species, represented by 15 strains, had no plasmids (Table 1). These included R. conorii, R. montanensis, R. parkeri, R. rickettsii, R sibirica, R. slovaca, R. prowazekii, R. typhi, and R. canadensis in which the absence of plasmid was confirmed in at least two strains by whole-genome sequencing (sequences available in GenBank) and/or pulsed-field gel electrophoresis (PFGE) [21,22]. In another three species (R. japonica, R. philipii and R. heilongjiangensis) the absence of plasmid needs to be confirmed in more than one strain. In R. akari and R. bellii (two SFG and AG species, respectively), Baldridge et al. [21, 22] detected plasmids in some strains by PFGE, but the currently available genomes (Rak strain Hartford and Rbe strains RML369-C and OSU-89-389) had none. This may be explained by strain differences (i. e., plasmid loss in nature), as observed in R. africae [9] or their passage history in cell culture, as observed in R. felis [36] or other bacteria such as Borrelia burgdorferi [37]. Among the other 5 genera within the order Rickettsiales (Anaplasma, Ehrlichia, Neorickettsia, Orientia, Wolbachia), all available genome sequences were devoid of plasmids [23]. RPs ranged in number and size per species from 1 to 4 plasmids and from 12 kb to 83 kb, respectively, and contained 15 to 85 genes (Table A in S1 File). A decrease in number and pooled plasmid size correlated with the decrease in chromosome size in rickettsiae (Fig 1, Figure A in S2 File). Such results had previously been observed in prokaryotes [38]. We did not find any evident correlation between presence, number and/or size of plasmids, and rickettsial host range (Table 1).
Table 1. Rickettsia and Orientia genomes extracted from the National Center for Biotechnology Information (NCBI) database.
| Species | Strain | Plasmid (s) | Host | Origin | Year | Rickettsiosis | Accession. No. |
|---|---|---|---|---|---|---|---|
| R. africae | ESF-5 | pRaf | Tick: Amblyomma variegatum | Ethiopia | 1966 | African tick bite fever | NC_012633–34 |
| R. massiliae | MTU5 | pRma | Tick: Rhipicephalus turanicus | France | 1990 | Unnamed rickettsiosis | NC_009897–900 |
| AZT80 | pRmaB | Tick: Rhipicephalus sanguineus | USA | 2004 | CP003319-20 | ||
| R. peacokii | Rustic | pRpe | Tick: Dermacentor andersoni | USA | Unknown | Not pathogenic | NC_012730–32 |
| R. monacensis | IrR/Munich | pRmo | Tick: Ixodes ricinus | Germany | 1998 | Unknown pathogenesis | NC_010927 |
| R. Helvetica | C9P9 | pRhe | Tick: Ixodes ricinus | Switzeland | 1979 | Unnamed rickettsiosis | CM001467-68 |
| R. felis | URRWXCal2 | pRfe/pdRfe | Flea: Ctenocephalides felis | USA | 1990 | Flea spotted fever | NC_007109–11 |
| RfeI1 | pRfeI1 | Insect: Liposcelis botrychophila | Australia | 1997 | GQ329881 | ||
| C. R. amblyomii | AaR/SC | pRam18/23/32 | Tick: Amblyomma americanus | South Carolina | 1999 | Unknown pathogenesis | NC_013937–38 |
| R. endosy. of Ixodes | REIS | pReis1/2/3/4 | Tick: Ixodes scapularis | USA | 2008 | Unknown pathogenesis | CM000770-73 GG688301-16 |
| R. raoultii | Khabarovsk | pRra1/2/3 | Tick: Dermacentor silvarum | Russia | 2005 | SENLAT | CP010969-72 |
| R. australis | Cutlack | pRau | Unknown | Unknown | Unknown | Queensland tick typhus | CP003338-39 |
| R. rhipicephalii | 3-7-female6-CWPP | pRrh | Unknown | Unknown | Unknown | Unknown pathogenesis | CP003342-43 |
| R. conorii | Malish 7 | - | Tick: Rhipicephalus sp. | South Africa | 1946 | Mediterranean spotted fever | NC_003103 |
| R. slovaca | 13-B | - | Tick: Dermacentor sp. | Slovakia | 1968 | Tick-borne lymphadenitis | CP002428 |
| D-CWPP | - | Unknown | Unknown | Unknown | CP003375 | ||
| R. sibirica | 246 | - | Unknown | Russia | Unknown | Siberian tick typhus | NZ_AABW00000000 |
| R. philipii | 364D | - | Unknown | Unknown | Unknown | Unknown pathogenesis | CP003308 |
| R. japonica | YH | - | Human: Homo sapiens sapiens | Japan | 1985 | Japanese spotted fever | AP011533 |
| R. parkerii | Portsmouth | - | Human: Homo sapiens sapiens | USA | 2002 | Unnamed rickettsiosis | CP003341 |
| R. montanensis | OSU_85_930 | - | Unknown | Unknown | Unknown | Unknown pathogenesis | CP003340 |
| R. heilongjiangensis | 054 | - | Unknown | Unknown | Unknown | Far-Eastern tick-borne rickett. | CP002912 |
| R. rickettsia | Sheila Smith | - | Human: Homo sapiens sapiens | Unknown | Unknown | Rocky mountain spotted fever | NC_009882 |
| Brazil | - | Unknown | Unknown | Unknown | NC_010263 | ||
| Iowa | - | Unknown | Unknown | Unknown | Not pathogenic mutant | CP003305 | |
| R. akari | Hartford | - | Mite:? | USA | Unknown | Rickettsial pox | NC_009881 |
| R. prowazekii | Rp22 | - | Human: Homo sapiens sapiens | Algeria | Unknown | Epidemic typhus | CP001584 |
| Madrid E | - | Human: Homo sapiens sapiens | Spain | 1941 | Not pathogenic mutant | NC_000963 | |
| R. typhi | Wilmington | - | Unknown | Unknown | Unknown | Murine typhus | NC_006142 |
| R. canadensis | McKiel | - | Tick: Haemaph. leporispalustris | Canada | Unknown | Unknown pathogenesis | NC_009879 |
| CA410 | - | Unknown | Unknown | Unknown | CP003304 | ||
| R. bellii | RML369-C | - | Tick: Dermacentor varabilis | USA | 1966 | Unknown pathogenesis | NC_007940 |
| OSU-89-389 | u. s. | Unknown | Unknown | Unknown | NC_009883 | ||
| O. tsutsugamushi | Boryong | - | Human: Homo sapiens sapiens | Japan | 1995 | Scrub typhus | NC_009488.1 |
| Ikeda | - | Human: Homo sapiens sapiens | Japan | 1979 | NC_010793.1 |
Data were reported from [2, 3] as well as PATRIC (http://patricbrc.org) and GENBANK (http://www.ncbi.nih.gov) databases. -, species without any plasmid. u.s., unavailabale sequences.
Fig 1. Rickettsial plasmid sizes (pooled) and numbers plotted with their chromosome sizes.
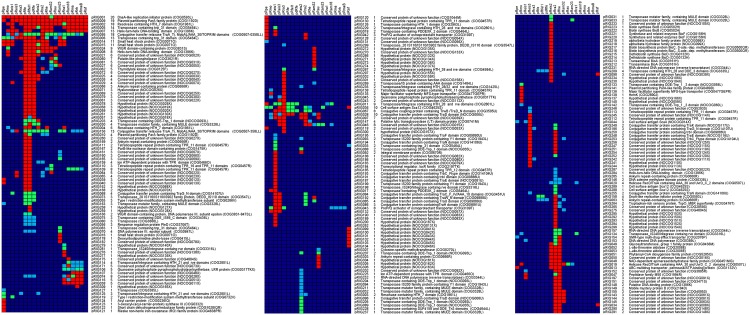
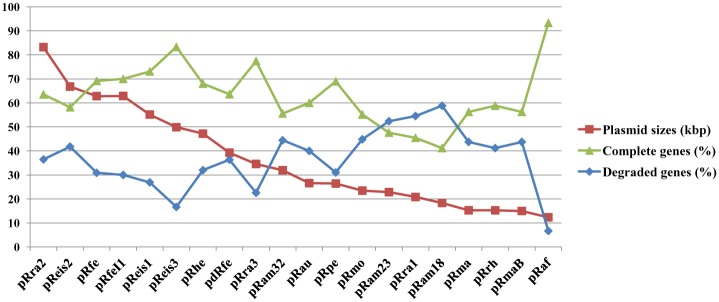
The plasmidic gene content (plasmidome) counted 260 pRIGs (i.e., putative orthologous groups of plasmidic rickettsial genes) constructed from 747 protein-coding genes (Fig 2). Of these genes, 65% were full-length genes, whereas 35% were partially degraded (i.e., either split, fragment or chimerical). These genes can also be either highly degraded remnants, absent or lost in several plasmids. Overall, the degradation levels varied among plasmids, ranging from 16 to 40% in larger plasmids (size >47 kbp) to 44 to 59% in smaller plasmids (<40 kbp), suggesting that RPs are in a progressive degradation process and size reduction (Fig 3). This may also contribute to variations in the coding capacity of the plasmids which ranged from 75 to 92% (Table A in S1 File).
Fig 2. Gene content array of Rickettsia plasmidome obtained from BBH analysis.
The right three columns showed the pRIG references of putative ortholog gene groups, the total number of genes for each pRIG and its corresponding annotation including its COGID form COG database. Each member of the pRIG was annotated as either complete (red), split (yellow), fragment (blue), chimeric (green) and absent (or remnant, violet).
Fig 3. Frequencies of complete and degraded (split, fragment or chimeric) genes identified in the plasmidome of Rickettsia.
Overall, these data suggest that modern RPs are in a process of reductive evolution similar to that observed in rickettsial chromosomes [17]. Such a phenomenon is typical of symbiotic/parasitic lifestyles [7, 12, 13, 17, 39]. However, this finding is in contrast with the megaplasmids of the free-living Bacillus cereus group which are a fusion of smaller plasmids [40]. RPs also exhibited G+C contents ranging from 32 to 35% (Table A in S1 File). Such values are similar to those of the rickettsial chromosomes (Table A in S1 File), suggesting that similar evolutionary forces such as vertical and horizontal gene transfers (VGT and HGT), duplications and selection pressure, impacted both genomic components.
Origin and divergence of RPs
We performed phylogenetic reconstructions of all 260 pRIGs to quantify the evolutionary processes that may have shaped the 20 current RPs, and to reconstruct the gene content of their last common ancestor and their evolutionary scenarios. The phylogenetic analysis revealed that some pRIGs contained sub-clusters of orthologous genes from multiple origins (see below e.g., parA gene); These sub-clusters were subsequently considered as distinct pRIGs, thus increasing the plasmidome pRIG content from 260 to 275. A total of 136 phylogenetic trees of 184 (67%) pRIGs were retained, for which both neighbor-joining (NJ) and maximum likelihood (ML) methods displayed similar topologies. For the remaining 91 (33%) pRIGs no phylogeny could be inferred.
Vertical evolution
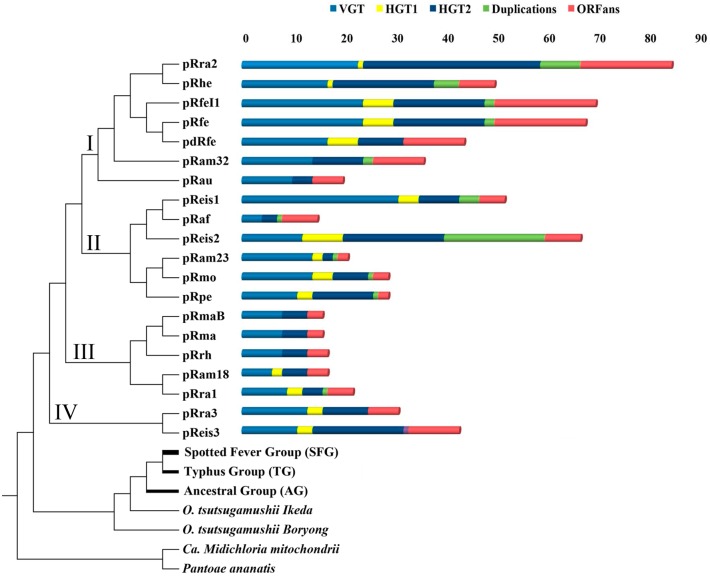
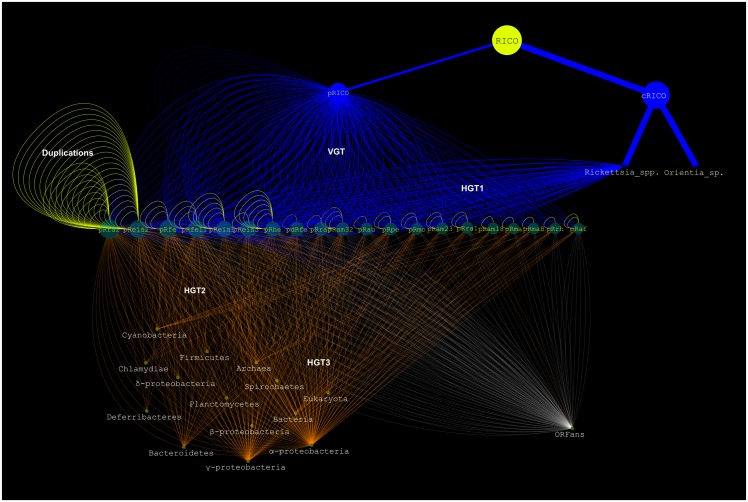
Phylogenetic trees of 12 pRIGs (4.4%), conserved in most plasmids, exhibited two phylogenetically-related clades, one including 5 to 20 RPs and the other made of 1 to 26 chromosomes from Rickettsia and/or Orientia (Figures A1 to A9 in S3 File). Moreover, the supertree constructed from 10 of the 12 pRIGs confirmed these two phylogenetically-related clades, one containing all the 20 RPs and the other all Rickettsia/Orientia chromosomes (Fig 4).
Fig 4. Supertree obtained from 10 genes of Rickettsia plasmids as well as Rickettsia and Orientia chromosomes using subtree prune-and-regraft distance (D = 64).
Candidatus Midichloria mitochondrii (YP_004679220.1) and Pantoea ananatis (YP_005193440.1) were used as outgroups. The list of genes used are two transposases, patatin-like phospholipase, thymidylate kinase, heat shock protein, dnaA-like replication initiator proteins, helix-turn-helix DNA-binding domain, cell surface antigen Sca12, conjugative transfer protein containing traD domain, and leucine rich-repeat containing protein. In the right, summary of evolutionary events that shaped rickettsial plasmids.
This supertree was used as a reference for all following analyses. Phylogenetic analysis of a second set of 41 (15%) pRIGs showed the same two related clusters in each tree, one containing 1 to 11 RPs and the other 1 to 30 Rickettsia/Orientia chromosomes (Figures A10 to A49 in S3 File). Moreover, the analysis of another four (1.4%) pRIGs (mostly present as fragments), did not result in any reliable phylogeny, but they best matched with homologs in rickettsial chromosomes (data not shown). In all, our results suggest that the current RPs have a vertical origin (VGT) from a last common plasmidic ancestor that we named “pRICO” (Fig 5).
Fig 5. Evolutionary scenario of rickettsial plasmids.
Both “pRICO” and “cRICO” (i.e., the last common ancestor of the current Rickettsia/Orientia chromosomes) may have been vertically inherited from a last common chromosomal ancestor, “RICO” (Figs 4 and 5). However, we observed that the internal phylogenetic clades inferred from plasmids and chromosomes were not often identical. This may be due to the facts that i) each species can harbor 1 to 4 plasmid(s) with similar or distinct evolutionary history, ii) both plasmids and chromosomes are in a process of gene degradation and loss, and iii) the selection pressure shaping plasmids and chromosomes may be similar or different in each species and/or between species. Moreover, 8 (3%) pRIGs were also vertically transmitted to RPs but matched non-Rickettsia/Orientia species, suggesting that they might have been horizontally acquired by pRICO from unknown species (Figures A50 to A53 in S3 File). Thus, our data strongly support that pRICO harbored around 65 (23%) genes, suggesting that it might have had an estimated size around 60 to 70 kbp (Fig 5). The vertical inheritance is also corroborated by the presence of plasmids in two distinct rickettsial groups (SFG and AG). In Salmonella enterica serovars, virulence plasmids were also described to be vertically transmitted [41], although no gene phylogeny and evolutionary relationships were carried out between Salmonella plasmids and chromosomes as well as microbial and non-microbial sequenced genomes to infer their origin and evolution.
In the supertree, the pRICO ancestor diverged into a single or multiple plasmid(s) in rickettsial species, and that these plasmids clustered into four putative groups (I to IV) (Fig 4): group I included 4 large and 3 small plasmids of 5 species (pRra2/pRhe/pRfe/pRfeI1/pdRfe/pRam32/pRau); group II clustered 2 large and 4 small plasmids belonging to 5 species (pReis1/pRaf/pReis2/pRam23/pRmo/pRpe); group III contained 5 small plasmids of 4 species (pRam18/pRrh/pRra1/pRma/pRmaB); and group IV gathered one large and one small plasmids from two species (pReis3/pRra3). At the inter-species level, plasmids of the same group showed variable sequence conservations (Fig 4 and Figures BA to BD in S2 File). At the intra-species level, plasmids either belonged to the same group with high sequence conservation, as was the case for R. felis (pRfe, pdRfe and pRfeI1) and R. massiliae (pRma and pRmaB) strains (Fig 4 and e.g., Figure BA in S2 File). In contrast, the plasmids from either R. raoultii (pRra1/2/3), Rickettsia endosymbiont of I. scapularis (pReis1/3, except pReis1/2) or R. amblyomii (pRam18/23/32) strains were resolved into distinct phylogenetic groups with variable sequence conservation, although they lived in the same bacterial strain as well as the same host-species (Fig 4 and Figures BE to BG in S2 File).
The current RPs conserved highly variable numbers of genes acquired by VGT across the branches of the tree, ranging from 4 genes (6% out of the 65 pRICO genes) in the small pRaf plasmid to 31 (47% out of 65) in the large pReis1 plasmid (Fig 4). Therefore, the loss of ancestral genes in RPs ranged from 53% in pReis1 to 94% in pRaf. The latter was the compacted RP in which reduction was the greatest driving force.
Horizontal evolution
The plasmidome exhibited 23 (8%) pRIGs resulting from evident horizontal gene exchanges (HGT1, plasmid-chromosome and/or plasmid-plasmid) within a given Rickettsia species, and/or between distinct Rickettsia species (Fig 5, Figures A37 and A54 to A74 in S3 File). RPs contained variable numbers of pRIGs resulting from HGT1 across the tree ranging from 0 in the small pRaf plasmid to 7 in the large pReis2 plasmid (Fig 4). Unexpectedly, 4 pRIGs (1.4%, originally present in pRICO) of pRfe, pdRfe, pRfeI1, pRam32 and pRau exhibited close phylogenetic relationships with a gene cluster in R. prowazekii chromosomes (Rpr22_687 to Rpr22_698 and RprME_862 to RprME_879, Figures A1, A5, A57 and A100 in S3 File). Similarly, 6 pRIGs (out of the 34 pRIGs acquired by HGT2) in pRpe showed close phylogenetic relationships with a gene cluster in the R. raoultii chromosome (Rra_909 to Rra_916, Figure C in S2 File, Figures A87 to A92 in S3 File). These data suggest that both Rickettsia species may have had one (for R. prowazekii) or an additional (for R. raoultii) plasmid that was disrupted in the course of the reductive genomic evolution in the genus Rickettsia, with subsequent HGT1 integration of plasmidic genes in their chromosomes. Alternatively, HGT1 events may have occurred between the pRfe, pRam or pRau plasmids and the R. prowazekii chromosome, and between pRpe and the R. raoultii chromosome, respectively.
Thirty-four (12%) pRIGs exhibited evident horizontal gene transfers (HGT2, plasmid-genome) with known non-Rickettsia/Orientia species, mostly belonging to α/γ-proteobacteria lineages (Fig 5, Figure D in S2 File, Figures A75 to A111 in S3 File). Moreover, 55 (20%) pRIGs displayed putative horizontal gene exchanges (HGT2) of unknown origin, but matching homologs of non-Rickettsia/Orientia species, also mostly belonging to α/γ-proteobacteria lineages (Fig 5, Figure E in S2 File, Figures A39 to A44, A46, A47, A100 and A112 to A136 in S3 File). RPs harbored highly variable numbers of genes exchanged by HGT2 events ranging from 0 in the small pRam23 plasmid to 24 in the large pRra2 plasmid (Fig 4).
Finally, five pRIGs (1.8%, originally present in 65 pRIGs inherited by VGT) and one pRIG (0,4%, originally present in the 34 pRIGs exchanged by HGT2) displayed evidence of additional gene transfers (HGT3, plasmid-genome) between Rickettsia and distant lineages including Cardinium endosymbiont of Encarsia pergandiella, Candidatus Amoebophilus asiaticus (Bacteroidetes) as well as bacterial endosymbionts of either Trichoplax adhaerens or Hydra magnipapillata (Bacteria) (Fig 5, Figure D in S2 File, Figures A10, A11, A55, A57, A58 and A101 in S3 File) [42–44].
ORFans
The plasmidome contained a set of 64 pRIGs (23%, one pRIG in 1 to 8 plasmid(s)) classified as ORFans, including 40 that are specific for one given plasmid (Fig 5). RPs harbored highly variable numbers of ORFans ranging from 2 in the small pRaf plasmid to 18 in the large pRra2 plasmid (Fig 4). These ORFans may have been horizontally acquired from microbes or eukaryotes not yet identified or that have disappeared, or have the de novo origin (gene genesis) either from non-coding regions, through an overprinting phenomenon of genes and/or underwent gene degradation processes [45–47].
Duplications
Recent duplication events drove 34 pRIGs (12%, one inparalog can be present 1 to 5 times in 1 to 4 plasmid(s)) after the divergence of pRICO (Fig 5). Of these, 21, 9 and 4 pRIGs were duplicated from pRIGs acquired by VGT and HGT2 (Figures A1, A4, A6 to A8, A10, A38, A50, A55 to A58, A60, A61, A81 to A86, A97, A100 and A129 in S3 File) as well as from ORFan pRIGs, respectively. The plasmids contained highly variable numbers of inparalogs across the tree ranging from 0 in the small plasmid pRaf to 20 in the large plasmid pReis2 (Fig 4). Overall, the gene proliferation by recent duplications in plasmids was found to be similar to duplication phenomenon in the corresponding chromosomes (see phylogenetic trees).
In summary, the current RPs were vertically transmitted from their last common ancestor pRICO. They evolved dynamically under an obligate intracellular lifestyle in two main phases: i) reductive evolution including pseudogenisation as well as gene degradation and loss, and ii) gene gain and innovation via HGT and proliferation-duplication as well as gene genesis. This mode of evolution is described in parasitic and symbiotic organisms as a recurrent biphasic model dominated by longer phases of genome reduction and simplification, punctuated by shorter phases of episodic complexification [39]. Although the expansion via HGT and gene duplications occurred in RPs, the contraction via gene degradation and loss severely contributed to the reduction in gene content and plasmid size, and thus probably led to plasmid degradation and/or loss in several rickettsial species. This evolution of RPs seems to be in accordance with a recent study which revealed that the prevailing mode of evolution in bacteria is genome reduction, which is partially compensated by the gain of new gene families via HGT [48]. The HGT process conferred on RPs a potential to exchange and disseminate their gene pools with their co-residing chromosomes and genomes of closely-related and distant phylogenetic lineages, while duplication events enabled an internal genetic amplification. This indicates that RPs were able to overcome the genetic barriers to HGT between distant phylogenetic lineages such as α-, γ-proteobacteria and/or Bacteroidetes. Similar horizontal transfer events between phylogenetically-distant clades were previously described (e.g., between plasmids from α-proteobacteria and Deinococci), underlying the key role of plasmids in genetic exchange throughout the microbial world [25]. Furthermore, all the identified evolutionary processes variably occurred across RPs suggesting that rickettsiaes may have been adapted to distinct biotic and/or abiotic factors for the invasion, survival and/or defense in eukaryotic host cells. Snel et al. [49] suggested that gene loss is under negative selection, while the process that adds genes is under positive selection.
Impact of evolution on functional diversity of RPs
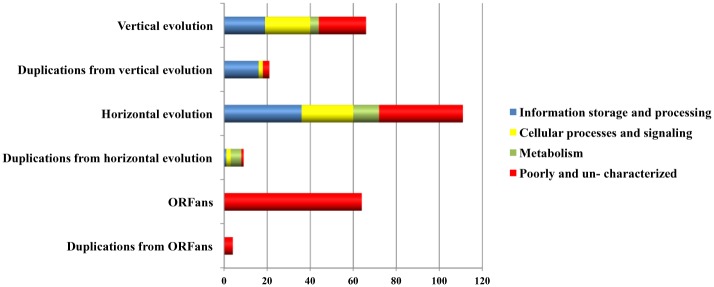
We examined the impact of the identified evolutionary forces (i. e., VGT, HGT, gene genesis, duplications and genome reduction) on the functional diversity of the RPs by a comparative analysis of the COG functional categories of the plasmidome (Fig 6). Overall, VGT had a weaker influence than HGT on pRIGs involved in information storage and processing (6% vs 14%, resp.), metabolism (1.4% vs 4%, resp.) and poorly- or un-characterized genes (8% vs 14%, resp.). In contrast, both forces had a similar impact on pRIGs related to cellular processes and signaling (7.6% vs 8.7%, resp.).
Fig 6. COG functional categories of Rickettsia plasmidome originated from vertical and horizontal evolutions as well as duplications.
Information storage and processing categories
In information storage and processing categories, both VGT and HGT forces drove high and distinct proportions of pRIGs associated with replication, recombination and repair (5 vs 13%, resp.), but less and similar proportions on those involved in transcription (0.7 vs 0.3%, resp.) and translation (0 vs 0.3%, resp.) (see details in Table B in S1 File). Moreover, duplication forces acted on pRIGs resulting more from VGT than HGT events, and involved in replication, recombination and repair (6 vs 0.3%, resp.) (see details in Table B in S1 File). The abundance of HGT pRIGs from rickettsial and non-rickettsial chromosomes and duplications from VGT pRIGs in the replication, recombination and repair category were mainly due to mobile transposable elements (i.e., transposases and integrases, 9 out of 13% and 5.1 out of 6%, resp.). These data from rickettsial plasmids corroborate a recent large-scale evolutionary study that showed that microbial plasmids share mainly genes encoding proteins involved in DNA transposition and recombination, some functions strongly linked to HGT process [25]. The single pRIG associated to translation was also exchanged with rickettsial chromosomes. Thus, VGT, HGT and duplication forces have driven pRIGs (e.g., dnaA-like, transposases, integrases, resolvases and relaxases) that may contribute to maintenance, rearrangements and plasticity of plasmids, making them self-replicative and simultaneously dynamic to gene exchange with various evolutionary lineages and consequent diversification of rickettsial biological activities and phenotypic traits.
Cellular processes and signaling categories
In cellular processes and signaling categories, both VGT and HGT forces acted in nearly equal proportions on pRIGs related to intracellular trafficking and secretion (tra operons) (4.3 vs 4%, resp.) (see details in Table B in S1 File). Based on the classification of plasmids by their mobility [38], our data predict that three (15%) large plasmids (pRra2/pReis1/pReis3) are self-transmissible or conjugative (i.e., having MOB or relaxases and TRA modules), whereas 8 (40%) small or large plasmids (pRam32/pRra3/pReis2/pRau/pdRfe/pRfe/pRfeI1/pRhe) are mobilisable (i.e., having MOB and/or degraded or no TRA modules). Some of these mobilisable replicons (e.g., pRra3) could be transmissible if using conjugative machineries of co-residing self-transmissible replicons (e.g., pRra2) [38, 50, 51]. The 9 (45%) remaining small plasmids (e.g., pRaf/pRam12/pRam23/pRrh/pRpe/pRmo/pRra1/pRma/pRmaB), lacked MOB and TRA modules and may not be transmissible. The three mobility types in RPs are common in various bacterial phyla including α-proteobacteria [38]. Overall, the presence of conjugative system in RPs may have strong implications in their evolutionary dynamics and history as well as in the acquisition and spread of ecologically important traits (novel and/or ancestral among various bacteria) that may be associated to rearrangement, adaptation and/or pathogenesis of rickettsiae. This is consistent with the analysis of gene sharing networks among 329 proteobacteria that suggested that gene transfer in this phylum is frequently mediated by conjugation [52].
Moreover, both VGT and HGT forces drove in a similar way pRIGs related to posttranslational modification and chaperones (0.7 vs 0.3%, resp.) and defense mechanisms (0.3 vs 0.7%, resp.), but not pRIGs associated to transduction mechanisms (e.g., 2 vs 0.3%, resp.) (see details in Table B in S1 File). Among these processes we can cite pRIGs associated to adaptation (e.g., hsp), defense (type I restriction-modification system methyltransferase subunit) and stress (e.g., spoT) that may help the survival of host bacteria in an obligate intracellular life style. However, HGT, but not VGT forces, drove 3.3% of pRIGs related to cell wall/membrane/envelope biogenesis and cycle control, including genes associated to plasmid segregation (e.g., parA/B) and regulator of cell division and wall (e.g., mobile mystery protein B, putative lytic transglycosylase, glycosyltransferase, group 1 family protein). In addition, duplication forces drove equal proportions of pRIGs resulting from both VGT and HGT events, and associated with intracellular trafficking and secretion, posttranslational modification and chaperones and cycle control (0.3 vs 0.3%, resp.).
Metabolism categories
In metabolism categories, both VGT and HGT forces drove equal proportions of pRIGs involved in inorganic ion transport and metabolism (0.3 vs 0.3%, resp.) (see details in Table B in S1 File). However, only VGT forces acted on 1.1% of pRIGs related to nucleotide/carbohydrate transport and metabolism (thymidylate kinase, major facilitator superfamily MFS-type transporter). In contrast, only HGT forces drove 4.4% of pRIGs associated with amino acid/lipid/coenzyme and secondary metabolites biosynthesis, transport and metabolism (e.g., L-allo-threonine aldolases, 3-oxoacyl-(acyl-carrier-protein) synthase III, bioA, B, C, D and F). Furthermore, duplication forces acted on 1.8% of pRIGs including bioA, B, C, D and F. Overall, VGT and duplication forces drove pRIGs common to RPs and Rickettsia/Orientia chromosomes which may increase the biological activities of some rickettsiae, whereas HGT forces enabled some RPs to acquire novel metabolic functions absent in Rickettsia/Orientia chromosomes that may complete host-metabolic gaps and confer host-adaptive phenotypes to obligate intracellular lifestyle.
Poorly and uncharacterized categories
In poorly and uncharacterized categories, VGT, HGT and de novo gene genesis drove distinct proportions of pRIGs (7.6, 13.8 and 23.3%, resp.) which have known or unknown functions and/or poorly or uncharacterized COG assignments and that remain to be elucidated for the biology of Rickettsia species (see details in Table B in S1 File). Moreover, duplication forces acted on similar proportions of pRIGs resulting from VGT, HGT and de novo genes genesis (1.1, 0.3 and 1.4%, resp.). Among the poorly and uncharacterized categories, VGT forces drove some pRIGs (2.5%) related to adhesion and/or infection such as those encoding the cell surface antigen proteins Sca and patatin-like phospholipase Pat [10, 53], whereas HGT forces acted on some pRIGs (5.8%) coding ankyrin repeat-containing proteins Ank, tetratricopeptide repeat-containing proteins Tpr known to play a role in protein-protein interactions in eukaryotes, and suspected to be associated to adaptation to hosts and/or to virulence [14].
In several human bacterial pathogens, such as Salmonella spp., Escherichia coli and Borrelia spp., virulence plasmids have been identified [37, 41, 54]. However, among the most pathogenic rickettsiae, R. prowazekii and R. conorii do not harbor any plasmid whereas R. australis contains one plasmid. These data suggest that the role of RPs in rickettsial virulence, remains uncertain and requires further investigations. In addition, the functional diversity of RPs observed in the present study suggests that some of them may be used as transformation vectors for rickettsiae, as was recently described using a shuttle vector system derived from R. amblyomii plasmids that enabled stable transformations of diverse rickettsiae [55, 56]. Thus, the genetic manipulation of rickettsiae using RPs may provide new insights in the pathogenesis of these obligate intracellular bacteria as well as the dissemination of their genotypic and phenotypic traits.
Conclusions
This study offers an overview of the evolution and functional diversity of rickettsial plasmids. The current RPs have been shaped by multiple evolutionary forces in a bottleneck ecological niche. Initially, they were vertically inherited and then may have evolved under a biphasic model including a strong reductive evolution as well as innovations by horizontal gene transfer and gene duplication and genesis. This study demonstrates for the first time that plasmids of obligate intracellular bacteria can evolve under both vertical and reductive forces similar to those shaping their co-residing chromosomes. The evolutionary processes that have shaped RPs suggest that they might be influenced by molecular interactions between rickettsiae and eukaroytic host cells including biological needs for adaptation and survival, as suggested for Buchnera aphidicola in which metabolic requirements could determine when genome reduction occurs [39]. RPs may play diverse functional roles that are either novel or similar to those of rickettsial chromosomes via various evolutionary mechanisms. Overall, this study provides insights into rickettsial plasmids and their biology and evolution.
Supporting Information
(PDF)
(PDF)
(PDF)
Acknowledgments
The study was funded by the Mediterranée-Infection foundation.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The study was funded by the Mediterranean-Infection foundation, http://en.mediterranee-infection.com/article.php?larub=83&titre=the-foundation.
References
- 1.Weinert LA, Werren JH, Aebi A, Stone GN, Jiggins FM. Evolution and diversity of Rickettsia bacteria. BMC Biol. 2009; 7: 6 10.1186/1741-7007-7-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merhej V, Raoult D. Rickettsial evolution in the light of comparative genomics. Biol Rev Camb Philos Soc. 2011; 86: 379–405. 10.1111/j.1469-185X.2010.00151.x [DOI] [PubMed] [Google Scholar]
- 3.Parola P, Paddock CD, Socolovschi C, Labruna MB, Mediannikov O, Kernif T, et al. Update on tick-borne rickettsioses around the world: a geographic approach. Clin Microbiol Rev. 2013; 26: 657–702. 10.1128/CMR.00032-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stothard DR, Clark JB, Fuerst PA. Ancestral divergence of Rickettsia bellii from the spotted fever and typhus groups of Rickettsia and antiquity of the genus Rickettsia. Int J Syst Bacteriol. 1994; 44: 798–804. [DOI] [PubMed] [Google Scholar]
- 5.Raoult D, Roux V. Rickettsioses as paradigms of new or emerging infectious diseases. Clin Microbiol Rev. 1997; 10: 694–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersson JO, Andersson SGE. Genome degradation is an ongoing process in Rickettsia. Mol Biol Evol. 1999; 16: 1178–1191. [DOI] [PubMed] [Google Scholar]
- 7.Andersson SGE, Zomorodipour A, Andersson JO, Sicheritz-Ponten T, Alsmark UCM, Podowski RM, et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998; 396: 133–140. [DOI] [PubMed] [Google Scholar]
- 8.Darby AC, Cho NH, Fuxelius HH, Westberg J, Andersson SG. Intracellular pathogens go extreme: genome evolution in the Rickettsiales. Trends Genet. 2007; 23: 511–520. [DOI] [PubMed] [Google Scholar]
- 9.Fournier PE, El Karkouri K, Leroy Q, Robert C, Giumelli B, Renesto P, et al. Analysis of the Rickettsia africae genome reveals that virulence acquisition in Rickettsia species may be explained by genome reduction. BMC Genomics. 2009; 10: 166 10.1186/1471-2164-10-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sahni SK, Narra HP, Sahni A, Walker DH. Recent molecular insights into rickettsial pathogenesis and immunity. Future Microbiol. 2013; 8: 1265–1288. 10.2217/fmb.13.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bechah Y, El Karkouri K, Mediannikov O, Leroy Q, Pelletier N, Robert C, et al. Genomic, proteomic, and transcriptomic analysis of virulent and avirulent Rickettsia prowazekii reveals its adaptive mutation capabilities. Genome Res. 2010; 20: 655–663. 10.1101/gr.103564.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McLeod MP, Qin X, Karpathy SE, Gioia J, Highlander SK, Fox GE, et al. Weinstock, Complete genome sequence of Rickettsia typhi and comparison with sequences of other rickettsiae. J Bacteriol. 2004; 186: 5842–5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogata H, Audic S, Renesto-Audiffren P, Fournier PE, Barbe V, Samson D, et al. Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science. 2001; 293: 2093–2098. [DOI] [PubMed] [Google Scholar]
- 14.Ogata H, La SB, Audic S, Renesto P, Blanc G, Robert C, et al. Genome sequence of Rickettsia bellii illuminates the role of amoebae in gene exchanges between intracellular pathogens. PLoS Genet. 2006; 2: e76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogata H, Renesto P, Audic S, Robert C, Blanc G, Fournier PE, et al. The genome sequence of Rickettsia felis identifies the first putative conjugative plasmid in an obligate intracellular parasite. PLoS Biol. 2005; 3: e248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blanc G, Ogata H, Robert C, Audic S, Claverie JM, Raoult D. Lateral gene transfer between obligate intracellular bacteria: evidence from the Rickettsia massiliae genome. Genome Res. 2007; 17: 1657–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blanc G, Ogata H, Robert C, Audic S, Suhre K, Vestris G, et al. Reductive genome evolution from the mother of Rickettsia. PLoS.Genet. 2007; 3: e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellison DW, Clark TR, Sturdevant DE, Virtaneva K, Porcella SF, Hackstadt T. Genomic comparison of virulent Rickettsia rickettsii Sheila Smith and avirulent Rickettsia rickettsii Iowa. Infect Immun. 2008; 76: 542–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fournier PE, Raoult D. Intraspecies diversity of Rickettsia conorii. J Infect Dis. 2009; 199: 1097–1098. 10.1086/597211 [DOI] [PubMed] [Google Scholar]
- 20.Felsheim RF, Kurtti TJ, Munderloh UG. Genome sequence of the endosymbiont Rickettsia peacockii and comparison with virulent Rickettsia rickettsii: identification of virulence factors. PLoS ONE. 2009; 4: e8361 10.1371/journal.pone.0008361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baldridge GD, Burkhardt NY, Felsheim RF, Kurtti TJ, Munderloh UG. Plasmids of the pRM/pRF family occur in diverse Rickettsia species. Appl Environ Microbiol. 2008; 74: 645–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baldridge GD, Burkhardt NY, Labruna MB, Pacheco RC, Paddock CD, Williamson PC, et al. Wide dispersal and possible multiple origins of low-copy-number plasmids in rickettsia species associated with blood-feeding arthropods. Appl Environ Microbiol, 2010; 76: 1718–1731. 10.1128/AEM.02988-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gillespie JJ, Beier MS, Rahman MS, Ammerman NC, Shallom JM, Purkayastha A, et al. Plasmids and rickettsial evolution: insight from Rickettsia felis. PLoS ONE. 2007; 2: e266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halary S, Leigh JW, Cheaib B, Lopez P, Bapteste E. Network analyses structure genetic diversity in independent genetic worlds. Proc Natl Acad Sci U S A. 2010; 107: 127–132. 10.1073/pnas.0908978107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamminen M, Virta M, Fani R, Fondi M. Large-scale analysis of plasmid relationships through gene-sharing networks. Mol Biol Evol. 2012; 29: 1225–1240. 10.1093/molbev/msr292 [DOI] [PubMed] [Google Scholar]
- 26.Bocs S, Cruveiller S, Vallenet D, Nuel G, Medigue C. AMIGene: Annotation of MIcrobial Genes. Nucleic Acids Res. 2003; 31: 3723–3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997; 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997; 278: 631–637. [DOI] [PubMed] [Google Scholar]
- 29.Kristensen DM, Kannan L, Coleman MK, Wolf YI, Sorokin A, Koonin EV, et al. A low-polynomial algorithm for assembling clusters of orthologous groups from intergenomic symmetric best matches. Bioinformatics. 2010; 26: 1481–1487. 10.1093/bioinformatics/btq229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007; 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- 31.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004; 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013; 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whidden C, Zeh N, Beiko RG. Supertrees Based on the Subtree Prune-and-Regraft Distance. Syst Biol. 2014; 63: 566–581. 10.1093/sysbio/syu023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guy L, Kultima JR, Andersson SG. genoPlotR: comparative gene and genome visualization in R. Bioinformatics. 2010; 26: 2334–2335. 10.1093/bioinformatics/btq413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fournier PE, Belghazi L, Robert C, El Karkouri K, Richards A, Greub G, et al. Variations of Plasmid Content in Rickettsia felis. PLoS ONE. 2008; 3: e2289 10.1371/journal.pone.0002289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chan K, Casjens S, Parveen N. Detection of established virulence genes and plasmids to differentiate Borrelia burgdorferi strains. Infect Immun. 2012; 80: 1519–1529. 10.1128/IAI.06326-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smillie C, Garcillan-Barcia MP, Francia MV, Rocha EP, de la Cruz F. Mobility of plasmids. Microbiol Mol Biol Rev. 2010; 74: 434–452. 10.1128/MMBR.00020-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolf YI, Koonin EV. Genome reduction as the dominant mode of evolution. Bioessays. 2013; 35: 829–837. 10.1002/bies.201300037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng J, Peng D, Ruan L, Sun M. Evolution and dynamics of megaplasmids with genome sizes larger than 100 kb in the Bacillus cereus group. BMC Evol Biol. 2013; 13: 262 10.1186/1471-2148-13-262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng Y, Liu J, Li YG, Cao FL, Johnston RN, Zhou J, et al. Inheritance of the Salmonella virulence plasmids: mostly vertical and rarely horizontal. Infect Genet Evol. 2012; 12: 1058–1063. [DOI] [PubMed] [Google Scholar]
- 42.Chapman JA, Kirkness E.F., Simakov O., Hampson S.E., Mitros T., Weinmaier T, et al. The dynamic genome of Hydra. Nature. 2010; 464: 592–596. 10.1038/nature08830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Driscoll T, Gillespie JJ, Nordberg EK, Azad AF, Sobral BW. Bacterial DNA sifted from the Trichoplax adhaerens (Animalia: Placozoa) genome project reveals a putative rickettsial endosymbiont. Genome Biol Evol. 2013; 5: 621–645. 10.1093/gbe/evt036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duron O. Lateral transfers of insertion sequences between Wolbachia, Cardinium and Rickettsia bacterial endosymbionts. Heredity. 2013; 111: 330–337. 10.1038/hdy.2013.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yin Y, Fischer D. On the origin of microbial ORFans: quantifying the strength of the evidence for viral lateral transfer. BMC Evol Biol. 2006; 6: 63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yomtovian I, Teerakulkittipong N, Lee B, Moult J, Unger R. Composition bias and the origin of ORFan genes. Bioinformatics. 2010; 26: 996–999. 10.1093/bioinformatics/btq093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tautz D, Domazet-Loso T. The evolutionary origin of orphan genes. Nat Rev Genet. 2011; 12: 692–702. [DOI] [PubMed] [Google Scholar]
- 48.Puigbò P, Lobkovsky AE, Kristensen DM, Wolf YI, Koonin EV. Genomes in turmoil: quantification of genome dynamics in prokaryote supergenomes. BMC Biol. 2014; 21: 12:66 10.1186/s12915-014-0066-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Snel B, Bork P. Genomes in flux: the evolution of archaeal and proteobacterial gene content. Genome Res. 2002; 12:17–25. [DOI] [PubMed] [Google Scholar]
- 50.Garcillan-Barcia MP, Francia MV, de la Cruz F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol Rev. 2009; 33: 657–687. [DOI] [PubMed] [Google Scholar]
- 51.Torres TG, Pistorio M, Althabegoiti MJ, Cervantes L, Wibberg D, Wibberg D, et al. Rhizobial plasmid pLPU83a is able to switch between different transfer machineries depending on its genomic background. FEMS Microbiol Ecol. 2014. 88: 565–578. 10.1111/1574-6941.12325 [DOI] [PubMed] [Google Scholar]
- 52.Kloesges T, Popa TO, Martin W, Dagan T. Networks of gene sharing among 329 proteobacterial genomes reveal differences in lateral gene transfer frequency at different phylogenetic depths. Mol Biol Evol. 2011. 28: 1057–1074. 10.1093/molbev/msq297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rahman MS, Gillespie JJ, Kaur SJ, Sears KT, Ceraul SM, Beier-Sexton M, et al. Rickettsia typhi Possesses Phospholipase A(2) Enzymes that Are Involved in Infection of Host Cells. Plos Pathog. 2013; 9: e1003399 10.1371/journal.ppat.1003399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson TJ, Nolan LK. Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol Mol Biol Rev. 2009; 73: 750–774. 10.1128/MMBR.00015-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burkhardt NY, Baldridge GD, Williamson PC, Billingsley PM, Heu CC, Felsheim RF, et al. Development of shuttle vectors for transformation of diverse Rickettsia species. PloS ONE. 2011; 6: e29511 10.1371/journal.pone.0029511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wood DO, Hines A, Tucker AM, Woodard A, Driskell LO, Burkhardt NY, et al. Establishment of a replicating plasmid in Rickettsia prowazekii. PloS ONE. 2012; 7: e34715 10.1371/journal.pone.0034715 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.