Abstract
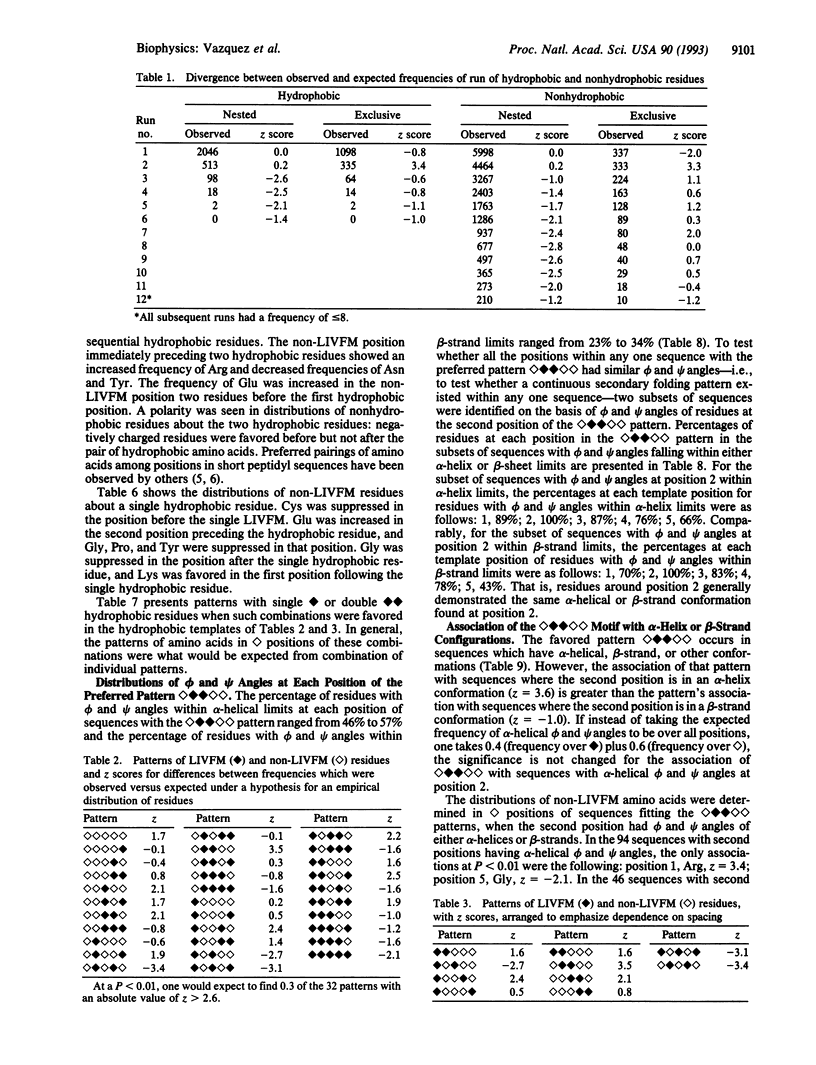
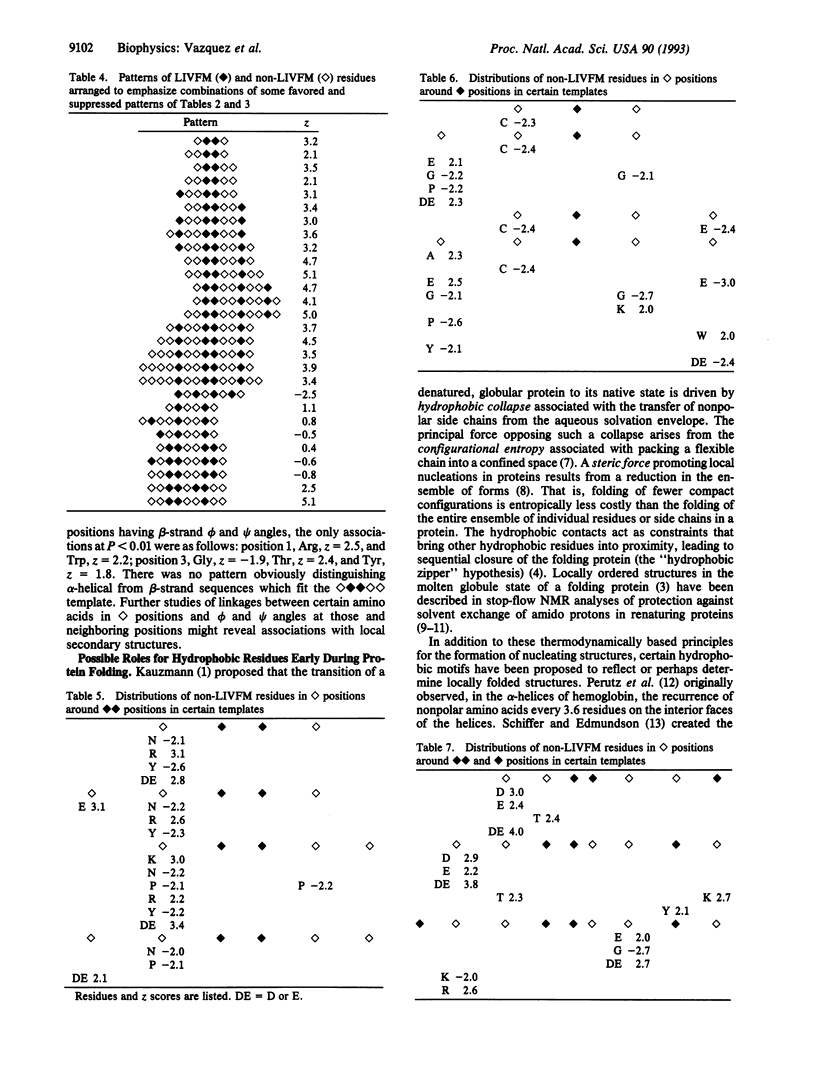
Hydrophobic amino acids of the group Leu, Ile, Val, Phe, and Met (LIVFM) are distributed in favored or suppressed patterns within protein sequences. The frequencies of all five-position combinations of [symbol: see text] = LIVFM and [symbol: see text] = non-LIVFM residues were analyzed in 48 proteins of known crystallographic structure. Some motifs were strongly preferred or suppressed; e.g., [symbol: see text] was favored (z = 3.5), while [symbol: see text] was suppressed (z = -3.4). In longer patterns, [symbol: see text] followed by [symbol: see text] and one [symbol: see text] was favored ([symbol: see text], z = 5.1), while conversion of the single hydrophobic residue to a pair was not ([symbol: see text], z = 0.8). Distributions of certain non-LIVFM amino acids around [symbol: see text] positions in strongly favored patterns were also favored or disfavored (Asp, Glu, Lys, Arg, Asn, Cys, Tyr, and Pro; for each magnitude of z > 2.0). While the strongly favored pattern [symbol: see text] was found in both alpha-helical and beta-strand sequences, it associated significantly with alpha-helices (z = 3.6 for the second-position alpha-helical phi and psi angles) but not with beta-strands (z = -1.1). Certain motifs of LIVFM and non-LIVFM residues might be selected if they lead efficiently to the local nucleations hypothesized to characterize molten globule intermediates in the folding of proteins.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bowie J. U., Clarke N. D., Pabo C. O., Sauer R. T. Identification of protein folds: matching hydrophobicity patterns of sequence sets with solvent accessibility patterns of known structures. Proteins. 1990;7(3):257–264. doi: 10.1002/prot.340070307. [DOI] [PubMed] [Google Scholar]
- Brendel V., Bucher P., Nourbakhsh I. R., Blaisdell B. E., Karlin S. Methods and algorithms for statistical analysis of protein sequences. Proc Natl Acad Sci U S A. 1992 Mar 15;89(6):2002–2006. doi: 10.1073/pnas.89.6.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan H. S., Dill K. A. Origins of structure in globular proteins. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6388–6392. doi: 10.1073/pnas.87.16.6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cserzö M., Simon I. Regularities in the primary structure of proteins. Int J Pept Protein Res. 1989 Sep;34(3):184–195. doi: 10.1111/j.1399-3011.1989.tb00229.x. [DOI] [PubMed] [Google Scholar]
- Dill K. A. Dominant forces in protein folding. Biochemistry. 1990 Aug 7;29(31):7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- Dill K. A., Fiebig K. M., Chan H. S. Cooperativity in protein-folding kinetics. Proc Natl Acad Sci U S A. 1993 Mar 1;90(5):1942–1946. doi: 10.1073/pnas.90.5.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill K. A. Theory for the folding and stability of globular proteins. Biochemistry. 1985 Mar 12;24(6):1501–1509. doi: 10.1021/bi00327a032. [DOI] [PubMed] [Google Scholar]
- Eisenberg D., Weiss R. M., Terwilliger T. C. The hydrophobic moment detects periodicity in protein hydrophobicity. Proc Natl Acad Sci U S A. 1984 Jan;81(1):140–144. doi: 10.1073/pnas.81.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finer-Moore J., Stroud R. M. Amphipathic analysis and possible formation of the ion channel in an acetylcholine receptor. Proc Natl Acad Sci U S A. 1984 Jan;81(1):155–159. doi: 10.1073/pnas.81.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAUZMANN W. Some factors in the interpretation of protein denaturation. Adv Protein Chem. 1959;14:1–63. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- Klapper M. H. The independent distribution of amino acid near neighbor pairs into polypeptides. Biochem Biophys Res Commun. 1977 Oct 10;78(3):1018–1024. doi: 10.1016/0006-291x(77)90523-x. [DOI] [PubMed] [Google Scholar]
- Kuwajima K. The molten globule state as a clue for understanding the folding and cooperativity of globular-protein structure. Proteins. 1989;6(2):87–103. doi: 10.1002/prot.340060202. [DOI] [PubMed] [Google Scholar]
- McCaldon P., Argos P. Oligopeptide biases in protein sequences and their use in predicting protein coding regions in nucleotide sequences. Proteins. 1988;4(2):99–122. doi: 10.1002/prot.340040204. [DOI] [PubMed] [Google Scholar]
- Muthusamy R., Ponnuswamy P. K. Variation of amino acid properties in protein secondary structures, alpha-helices and beta-strands. Int J Pept Protein Res. 1990 May;35(5):378–395. doi: 10.1111/j.1399-3011.1990.tb00064.x. [DOI] [PubMed] [Google Scholar]
- Neri D., Billeter M., Wider G., Wüthrich K. NMR determination of residual structure in a urea-denatured protein, the 434-repressor. Science. 1992 Sep 11;257(5076):1559–1563. doi: 10.1126/science.1523410. [DOI] [PubMed] [Google Scholar]
- Regan L., DeGrado W. F. Characterization of a helical protein designed from first principles. Science. 1988 Aug 19;241(4868):976–978. doi: 10.1126/science.3043666. [DOI] [PubMed] [Google Scholar]
- Rennell D., Poteete A. R., Beaulieu M., Kuo D. Z., Lew R. A., Humphreys R. E. Critical functional role of the COOH-terminal ends of longitudinal hydrophobic strips in alpha-helices of T4 lysozyme. J Biol Chem. 1992 Sep 5;267(25):17748–17752. [PubMed] [Google Scholar]
- Roder H., Elöve G. A., Englander S. W. Structural characterization of folding intermediates in cytochrome c by H-exchange labelling and proton NMR. Nature. 1988 Oct 20;335(6192):700–704. doi: 10.1038/335700a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer M., Edmundson A. B. Use of helical wheels to represent the structures of proteins and to identify segments with helical potential. Biophys J. 1967 Mar;7(2):121–135. doi: 10.1016/S0006-3495(67)86579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. W., Kaiser E. T. Structure-function analysis of proteins through the design, synthesis, and study of peptide models. Methods Enzymol. 1987;154:473–498. doi: 10.1016/0076-6879(87)54091-5. [DOI] [PubMed] [Google Scholar]
- Torgerson R. R., Lew R. A., Reyes V. E., Hardy L., Humphreys R. E. Highly restricted distributions of hydrophobic and charged amino acids in longitudinal quadrants of alpha-helices. J Biol Chem. 1991 Mar 25;266(9):5521–5524. [PubMed] [Google Scholar]
- Udgaonkar J. B., Baldwin R. L. NMR evidence for an early framework intermediate on the folding pathway of ribonuclease A. Nature. 1988 Oct 20;335(6192):694–699. doi: 10.1038/335694a0. [DOI] [PubMed] [Google Scholar]
- Vazquez S. R., Kuo D. Z., Bositis C. M., Hardy L. W., Lew R. A., Humphreys R. E. Residues in the longitudinal, hydrophobic strip-of-helix relate to terminations and crossings of alpha-helices. J Biol Chem. 1992 Apr 15;267(11):7406–7410. [PubMed] [Google Scholar]
- Vazquez S. R., Kuo D. Z., Salomon M., Hardy L., Lew R. A., Humphreys R. E. Prediction of alpha-helices in proteins with the hydrophobic strip-of-helix template and distributions of other amino acids around the hydrophobic strip. Arch Biochem Biophys. 1993 Sep;305(2):448–453. doi: 10.1006/abbi.1993.1445. [DOI] [PubMed] [Google Scholar]



