Long noncoding RNAs (lncRNAs) control diverse biological processes. Here the lncRNA SPRY4-IT1 is linked with TJ expression and subsequent gut permeability.
Abstract
Epithelial cells line the intestinal mucosa and form an important barrier to a wide array of noxious substances in the lumen. Disruption of the barrier integrity occurs commonly in various pathologies. Long noncoding RNAs (lncRNAs) control diverse biological processes, but little is known about the role of lncRNAs in regulation of the gut permeability. Here we show that the lncRNA SPRY4-IT1 regulates the intestinal epithelial barrier function by altering expression of tight junction (TJ) proteins. SPRY4-IT1 silencing led to dysfunction of the epithelial barrier in cultured cells by decreasing the stability of mRNAs encoding TJ proteins claudin-1, claudin-3, occludin, and JAM-1 and repressing their translation. In contrast, increasing the levels of SPRY4-IT1 in the intestinal mucosa protected the gut barrier in mice exposed to septic stress by increasing the abundance of TJ proteins. SPRY4-IT1 directly interacted with TJ mRNAs, and this process was enhanced through the association with the RNA-binding protein HuR. Of interest, the intestinal mucosa from patients with increased gut permeability exhibited a decrease in the levels of SPRY4-IT1. These findings highlight a novel role for SPRY4-IT1 in controlling the intestinal epithelial barrier and define a mechanism by which SPRY4-IT1 modulates TJ expression by altering the stability and translation of TJ mRNAs.
INTRODUCTION
Mammalian genomes transcribe a large number of noncoding RNAs with active roles in gene regulation (Ponting et al., 2009). Long noncoding RNAs (lncRNAs) are defined as transcribed RNAs spanning >200 nucleotides in length that lack protein-coding potential, although many of them display several mRNA-like properties, including transcription from multiexonic genes and the presence of a 5′ cap and a 3′ poly(A) tail (Mattick et al., 2006; Ulitsky et al., 2013). Some lncRNAs are ubiquitous, but others can be expressed with tissue-, differentiation stage–, and cell type–specific expression patterns (Batista et al., 2013). In addition, lncRNAs can regulate gene expression at different levels (Wang et al., 2011); for example, some lncRNAs can remodel chromatin, recruit transcriptional activators or repressors, assemble ribonucleoprotein (RNP) complexes, serve as decoys that titrate away RNA-binding proteins (RBPs), and function as scaffolds for protein assembly (Ponting et al., 2009; Batista et al., 2013; Ulitsky et al., 2013). Yet other lncRNAs can modulate gene expression by altering the stability and translation of mRNAs and working jointly with microRNAs (miRNAs) and RBPs (Wapinski et al., 2011; Yoon et al., 2012; Ulitsky et al., 2013). Collectively lncRNAs have been involved in a variety of cellular functions, physiological processes, and disease states (Tsai et al., 2011; Liu et al. 2012; Abdelmohsen et al., 2014; Takahashi et al., 2014; Uchida et al., 2015).
Epithelial cells line the intestinal mucosa and form an important barrier to a wide array of noxious substances in the lumen. The effectiveness and stability of the epithelial barrier depend on specialized structures composing different intercellular junctions, including tight junctions (TJs) and adherens junctions (AJs; Turner, 2009; Yang et al., 2014). The TJ is the apicalmost element of the junctional complex and seals intestinal epithelial cells (IECs) together in a way that prevents even small molecules from leaking between cells (Schneeberger et al., 2004; Turner, 2009). Four classes of TJ-transmembrane proteins and >30 TJ membrane–associated proteins have been identified in mammalian epithelial and endothelial cells (Schneeberger et al., 2004; Furuse et al., 2014). TJ complexes primarily consist of transmembrane proteins, such as occludin, tricellulin, and one or more members of the claudin family; these proteins also associate with a cytosolic plaque of TJ proteins such as ZO-1 that links tightly to the cortical cytoskeleton. TJs are highly dynamic, and their constituent proteins undergo continuous remodeling and turnover under tight regulation by numerous extracellular and intracellular factors. The dynamic maintenance of TJ protein levels is critical for normal function of the epithelial barrier, whereas disruption of TJ expression results in gut epithelial barrier dysfunction (Chen et al., 2008; Turner, 2009; Yang et al., 2014). Previous studies from our laboratory (Yu et al., 2011, 2013; Zhuang et al., 2013) and others (Ye et al., 2011; Sharma et al., 2013; Zhou et al., 2015) have shown that the RBP HuR and miRNAs (such as miR-29, miR-192, and miR-195) modulate the stability and translation of mRNAs encoding TJ proteins and play an important role in the control of intestinal epithelial TJ permeability. However, the exact roles of lncRNAs in the regulation of TJ expression and gut permeability have not been elucidated.
The lncRNA SPRY4-IT1 was originally identified as a 706–base pair transcript present in a large-scale study involving sequencing of adipose tissue cDNA (Qta et al., 2004) and was further shown to be broadly expressed in various human tissues, including the intestinal mucosa (Khaitan et al., 2011). SPRY4-IT1 is derived from the intronic region of the SPRY4 gene, but SPRY4 mRNA and SPRY4-IT1 are independent transcripts (Khaitan et al., 2011; Mazar et al., 2014). Expression of SPRY4-IT1 is up-regulated in human melanoma cells and predominantly localized in cytoplasmic polysomes or ribosomal clusters (Ingolia et al., 2012; Mazar et al., 2014). Inhibition of SPRY4-IT1 expression causes defects in cell proliferation and differentiation and induces apoptosis in melanoma cells, suggesting that it is implicated in melanocytic transformation (Khaitan et al., 2011; Mazar et al., 2014). Here we report a novel function for SPRY4-IT1 in protecting the intestinal epithelial barrier function by enhancing TJ expression posttranscriptionally. Because SPRY4-IT1 expression levels decrease in patients with increased gut permeability, our findings provide a strong rationale for developing new therapeutic strategies directed at SPRY4-IT1 to preserve the integrity of the gut epithelial barrier in various pathological conditions.
RESULTS
SPRY4-IT1 is essential for normal function of the intestinal epithelial barrier in vitro
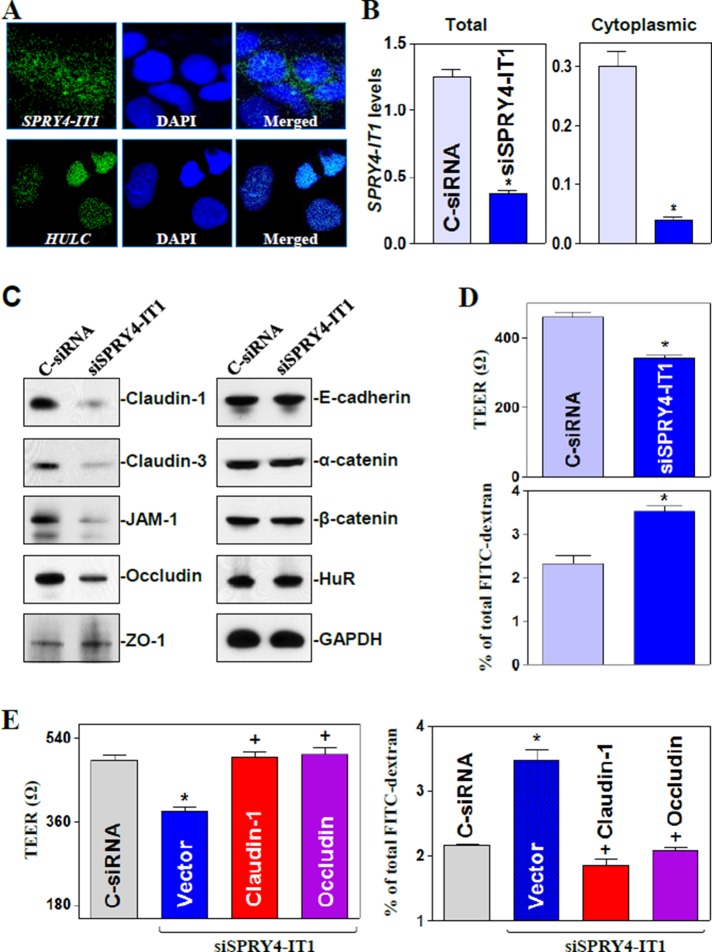
SPRY4-IT1 was highly expressed in IECs and distributed in both the cytoplasm and nucleus (Figure 1A), whereas the lncRNA HULC was localized only in the nucleus, as measured by confocal fluorescence analysis of optical sections and quantitative real-time PCR (Q-PCR) analysis (Supplemental Figure S1A). To identify the role of SPRY4-IT1 in the regulation of intestinal epithelial barrier function, we silenced expression of SPRY4-IT1 by transfecting Caco-2 cells with a specific small interfering RNA (siRNA) targeting SPRY4-IT1 (siSPRY4-IT1). As shown in Figure 1B, the levels of total and cytoplasmic SPRY4-IT1 decreased dramatically in siSPRY4-IT1-transfected cells compared with cells transfected with control siRNA (C-siRNA). Decreased levels of SPRY4-IT1 by siSPRY4-IT1 transfection specifically inhibited expression of TJ proteins claudin-1, claudin-3, JAM-1, and occludin but failed to alter the cellular abundance of TJ protein ZO-1, AJ proteins E-cadherin, α-catenin, or β-catenin, and RBP HuR (Figure 1C). The levels of claudin-1, claudin-3, JAM-1, and occludin proteins in SPRY4-IT1–silenced cells decreased by ∼95, ∼96, ∼93, and ∼85% (n = 3; p < 0.05), respectively, compared with those in cells transfected with C-siRNA. To exclude off-target effects, we tested another siRNA targeting SPRY4-IT1 (SPRY4-IT1-2), which showed a similarly repressive effect on the expression of SPRY4-IT1, as well as on this subset of TJ proteins (unpublished data). Consistent with this finding, SPRY4-IT1 silencing also disrupted the epithelial barrier function in an in vitro model, as evidenced by a decrease in transepithelial electrical resistance (TEER) values (Figure 1D, top) and an increase in the levels of paracellular flux of fluorescein isothiocyanate (FITC)–dextran (Figure 1D, bottom). Moreover, the barrier dysfunction induced by silencing SPRY4-IT1 was rescued by overexpression of TJ proteins, since decreased TEER and increased paracellular permeability were completely prevented when SPRY4-IT1–silenced cells were transfected with the claudin-1 or occludin expression vector (Figure 1E and Supplemental Figure S1). On the other hand, SPRY4-IT1 silencing did not affect cell viability, as measured by trypan blue staining (unpublished data), and failed to alter Caco-2 cell proliferation, as determined by the lack of significant differences in the expression levels of proliferation-associated proteins (CDK4, 14-3-3, and CUG-binding protein 1 [CUGBP1]) and the numbers of cells in SPRY4-IT1–silenced populations and C-siRNA cells (Supplemental Figure S2). We also examined changes in TJ expression after ectopic overexpression of SPRY4-IT1 and found that transfection of cells with the SPRY4-IT1 expression vector marginally increased expression levels of claudin-1 and occludin but did not affect claudin-3 or JAM-1 content (Supplemental Figure S3). In addition, neither TJ expression nor epithelial barrier function was affected by ectopic overexpression or silencing of lncRNA HULC (unpublished data). These data indicate that SPRY4-IT1 is necessary for normal expression of given TJ proteins and maintaining epithelial barrier function but not for increasing the basal levels of TJ proteins.
FIGURE 1:
SPRY4-IT1 silencing inhibits TJ expression and disrupts the epithelial barrier function. (A) Cellular distribution of lncRNAs SPRY4-IT1 (top) and HULC (bottom) in Caco-2 cells as measured by FISH assays. (B) Levels of total (left) and cytoplasmic (right) SPRY4-IT1 48 h after transfecting cells with siRNA targeting SPRY4-IT1 (siSPRY4-IT1) or control siRNA (C-siRNA). Values are means ± SEM from three separate experiments. *p < 0.05 compared with C-siRNA. (C) Representative immunoblots of tight junctions and adherens junctions in cells described in B. (D) Changes in epithelial barrier function as indicted by changes in TEER (top) and FITC-dextran paracellular permeability (bottom) after SPRY4-IT1 silencing. (E) Ectopic TJ overexpression rescues the barrier dysfunction in SPRY4-IT1–silenced cells. Cells were cotransfected with siSPRY4-IT1 and the claudin-1 or occludin expression vector; TEER and FITC-dextran permeability were examined 48 h thereafter. *,+p < 0.05 compared with C-siRNA or siSPRY4-IT1 alone, respectively.
Elevation of SPRY4-IT1 protects the gut barrier function in mice exposed to septic stress
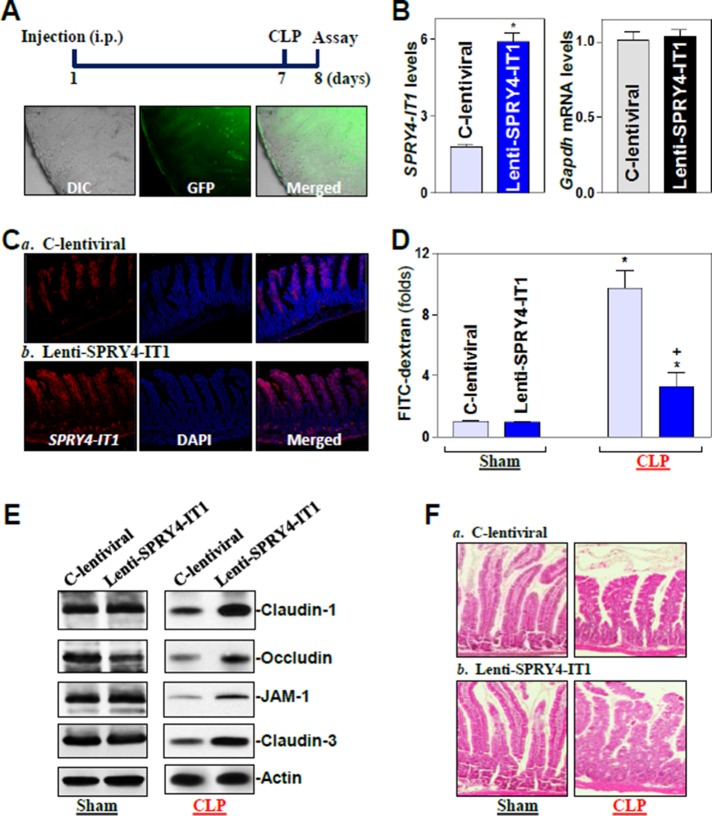
In an effort to define the in vivo importance of SPRY4-IT1 in regulating gut barrier function, we increased the levels of SPRY4-IT1 by infecting mice with the recombined SPRY4-IT1 lentiviral expression vector (lenti-SPRY4-IT1) as described previously (Scherr et al., 2007; Feng et al., 2012). Briefly, lenti-SPRY4-IT1 containing expressed SPRY4-IT1/GFP was under the control of the suCMV-promoter (AMSBIO, Cambridge, MA). As shown in Figure 2A, in situ detection in the mucosa of the small intestine showed a predominant accumulation of the lenti-SPRY4-IT1 in the villous area in mice infected with lenti-SPRY4-IT1 for 7 d. Minor FITC signal of the lenti-SPRY4-IT1 was also observed in the crypt area of the mucosa. By 7 d after infection with lenti-SPRY4-IT1, there was a sustained increase (∼3.2-fold) in SPRY4-IT1 in the intestinal mucosa (Figure 2B) compared with the levels in animals infected with the control lentiviral vector (C-lentiviral). RNA fluorescence in situ hybridization (RNA-FISH) analysis revealed that increased SPRY4-IT1 in the mucosa was localized at both villus and crypt areas (Figure 2C). To define the major cell type(s) present in the tissue extracted in these preparations, fluorescence-activated cell sorting analysis was performed as described previously (Terahara et al., 2008; Nagatake et al., 2014). As shown in Supplemental Figure S4, ∼90% of the cell population in the extracted intestinal mucosa scraped using a glass microscope slide were intestinal epithelial cells.
FIGURE 2:
Elevation of mucosal SPRY4-IT1 by infection with a SPRY4-IT1 lentiviral expression vector protects gut TJ barrier function in mice exposed to CLP. (A) Distribution of the lenti-SPRY4-IT1 (GFP) in the small intestinal mucosa 7 d after intraperitoneal (i.p.) injection. (B) Levels of SPRY4-IT1 in the small intestinal mucosa in mice described in A. Values are means ± SEM (n = 4). *p < 0.05 compared with C-control lentiviral vector (C-lentiviral). (C) Distribution of SPRY4-IT1 in the small intestine as measured by FISH in mice described in A. (D) Gut permeability in sham mice and mice exposed to CLP for 24 h. FITC-dextran was given orally, and blood samples were collected 4 h later. *,+p < 0.05 compared with sham or C-lentiviral-treated mice exposed to CLP, respectively. (E) Representative immunoblots of tight junctions in the small intestinal mucosa in mice described in D. (F) Hematoxylin/eosin staining of the intestinal mucosa.
Consistent with the findings observed in in vitro experiments, overexpression of SPRY4-IT1 by infection with lenti-SPRY4-IT1 did not alter basal expression levels of TJ proteins or gut permeability (Figure 2, D and E, left) in control mice (sham groups). To test whether SPRY4-IT1 overexpression enhanced the gut barrier function under critical pathological conditions, we subjected the mice to septic stress using the cecal ligation and puncture (CLP) model (Hubbard et al., 2005). Exposure to CLP for 24 h caused an acute gut barrier dysfunction in both C-lentiviral– and lenti-SPRY4-IT1–infected mice, as indicated by an increase in gut mucosal permeability to FITC-dextran. Of interest, however, gut permeability induced by CLP was significantly lower in mice infected with lenti-SPRY4-IT1 than in mice infected with C-lentiviral (Figure 2D, right). As expected, CLP stress also decreased the levels of TJ proteins claudin-1, claudin-3, occludin, and JAM-1 in the intestinal mucosa, but the inhibition of TJ protein expression by CLP was almost completely prevented or significantly reduced by increasing the levels of SPRY4-IT1 in lenti-SPRY4-IT1-infected mice (Figure 2E, right). On the other hand, there were no significant changes in histological features of the small intestinal mucosa between C-lentiviral–infected mice and mice infected with lenti-SPRY4-IT1 with or without CLP-induced stress (Figure 2F). These results indicate that increasing the levels of intestinal mucosal SPRY4-IT1 protects the barrier function against CLP-induced stress by derepressing TJ protein expression.
SPRY4-IT1 stabilizes TJ mRNAs and enhances their translation
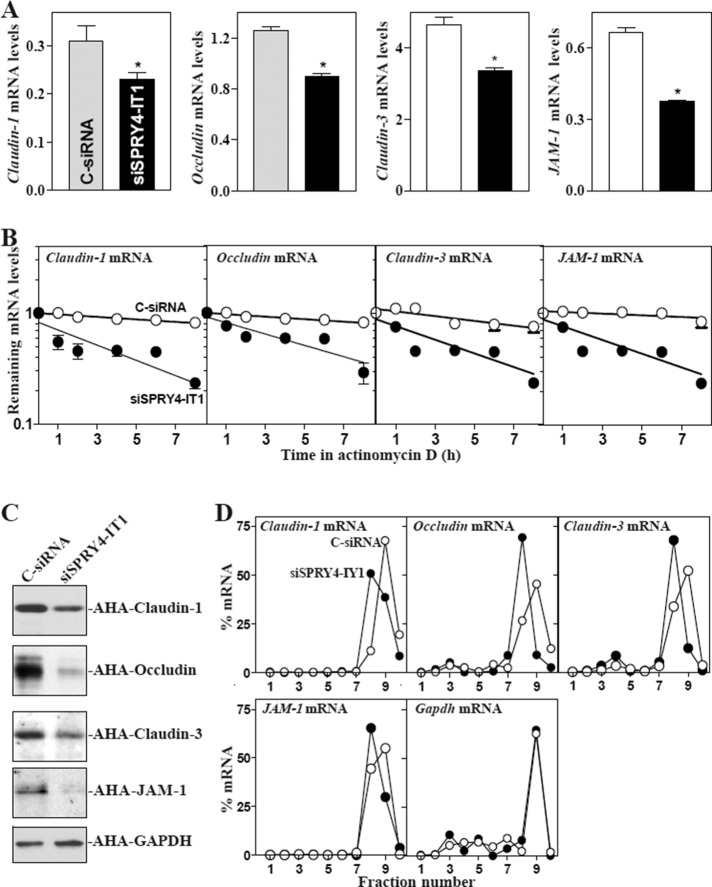
To investigate the mechanism underlying SPRY4-IT1 in regulating TJ expression, we examined changes in the stability and translation of TJ mRNAs after SPRY4-IT1 silencing. Knockdown of SPRY4-IT1 expression by transfection with siSPRY4-IT1 decreased the levels of claudin-1, occludin, claudin-3, and JAM-1 mRNAs (Figure 3A) by increasing their degradation rates. The half-lives of these TJ mRNAs in the SPRY4-IT1–silenced population of cells were decreased compared with those in cells transfected with C-siRNA (Figure 3B). However, quantitative analysis of these results indicated that the levels of TJ protein expression were decreased by >80% in SPRY4-IT1-silenced cells (Figure 1C), whereas the levels of the respective TJ mRNAs were reduced by only ∼25–40%.
FIGURE 3:
SPRY4-IT1 silencing enhances TJ mRNA decay and represses their translation. (A) Levels of claudin-1, occludin, claudin-3, and JAM-A mRNAs 48 h after transfection with siSPRY4-IT1 or C-siRNA. Values are means ± SEM from three separate experiments. *p < 0.05 compared with C-siRNA. (B) Stability of the TJ mRNAs in cells described in A. mRNA levels were examined at different times after administration with actinomycin D. (C) Newly synthesized TJ proteins in SPRY4-IT1–silenced cells. After cells were exposed to AHA, cell lysates were incubated with the reaction buffer containing biotin/alkyne reagent; the biotin-alkyne/azide–modified protein complex was pulled down by paramagnetic streptavidin-conjugated Dynabeads. (D) Distribution of claudin-1, occludin, claudin-3, and JAM-A mRNAs in each gradient fraction prepared from polysomal profile after SPRY4-IT1 silencing. Nuclei were pelleted, and the resulting supernatants were fractionated through a 10–50% linear sucrose gradient. Total RNA was isolated from different fractions, and the levels of TJ and Gapdh mRNAs were measured and plotted as a percentage of each of total TJ mRNAs and Gapdh mRNA levels in the samples. Three experiments were performed and showed similar results.
Therefore we investigated the involvement of SPRY4-IT1 in the regulation of TJ translation. De novo TJ protein synthesis after SPRY4-IT1 silencing was analyzed by measuring l-azidohomoalanine (AHA) incorporation. We found that the levels of newly synthesized claudin-1, clauidn-3, occludin, and JAM-1 proteins in siSPRY4-IT1–transfected cells decreased by ∼75, ∼90, ∼85, and ∼90% (n = 3; p < 0.05), respectively, compared with the levels in cells transfected with C-siRNA (Figure 3C). Inhibition of synthesis of these TJ proteins by SPRY4-IT1 silencing was specific, since there was no change in nascent glyceraldehyde-3-phosphate dehydrogenase (GAPDH) synthesis in siSPRY4-IT1–transfected cells. Changes in translation of these proteins were further studied by measuring the sizes of polysomes after silencing SPRY4-IT1. We fractionated cytoplasmic components through sucrose gradients and analyzed polysome distribution profiles. Although decreasing the levels of SPRY4-IT1 did not affect global polysomal profiles (unpublished data), the abundance of each of TJ mRNAs associated with actively translating fractions (peaking at fraction 9) decreased in siSPRY4-IT1–transfected cells, with a moderate leftward shift of the mRNAs toward lower-translating fractions (peaking at fraction 8; Figure 3D). In contrast, Gapdh mRNA, encoding a housekeeping protein, was distributed similarly in both groups. These results indicate that SPRY4-IT1 induces TJ levels by both stabilizing TJ mRNAs and enhancing their translation.
SPRY4-IT1 is required for HuR binding to and also directly interacts with TJ mRNAs
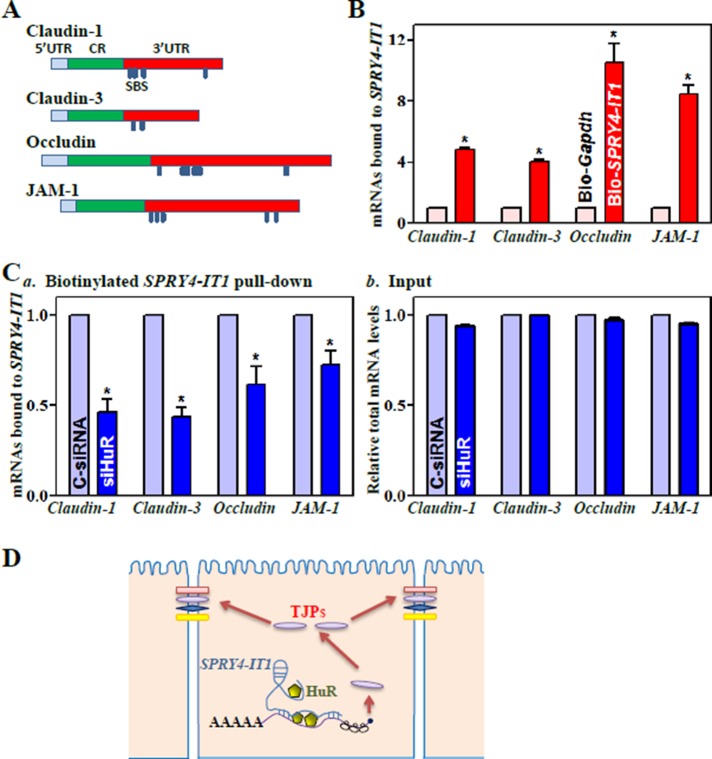
HuR bound the 3′-untranslated regions (UTRs) of claudin-1 and occludin mRNAs and enhanced their translation and stability (Yu et al., 2011; Sharma et al., 2013). The claudin-3 and JAM-1 mRNAs are also potential targets of HuR, as they contain computationally predicted HuR-binding sites in their 3′-UTRs. Because both HuR and SPRY4-IT1 regulate TJ expression at the posttranscriptional level, we tested the possibility that SPRY4-IT1 alters the association of HuR with the TJ mRNAs. To do so, we synthesized biotin-labeled SPRY4-IT1 and examined the direct interaction of SPRY4-IT1 with HuR in an in vitro system. Cytoplasmic lysates were initially incubated with biotinylated SPRY4-IT1 as described previously (Cao et al., 2014), and then the levels of HuR and other proteins in the pull-down material were assessed by Western blot analysis. As shown in Figure 4A, biotinylated SPRY4-IT1 specifically bound to HuR but did not bind other RBPs, such as CUGBP1 (also named as CELF1), TIAR, or AUF1, as determined by biotin pull-down analysis. SPRY4-IT1 also failed to directly interact with TJ proteins claudin-1, claudin-3, occludin, and JAM-1. To examine further the association of endogenous SPRY4-IT1 with endogenous HuR, we performed RNP immunoprecipitation (IP) using anti-HuR antibody and control immunoglobulin G (IgG), followed by isolation of bound RNA in both IP reactions. After reverse transcription (RT), Q-PCR analysis was used to measure the levels of SPRY4-IT1 enrichment in the HuR IP relative to IgG IP, as previously described (Zou et al., 2010). SPRY4-IT1 was highly enriched in HuR IP compared with control IgG IP (Figure 4Ba). In contrast, there were no significant changes in the levels of lncRNAs HULC and PTENP1 in HuR IP samples. As anticipated, the levels of TJ mRNAs, including claudin-1, claudin-3, occludin, and JAM-1 mRNAs but not ZO-1 mRNA, were also highly enriched in HuR IP samples (Figure 4Bb). HuR/SPRY4-IT1 association did not affect the stability of SPRY4-IT1, since HuR silencing by transfection with siRNA specifically targeting HuR (siHuR) did not alter cellular SPRY4-IT1 levels or its half-life (Supplemental Figure S5). Of interest, SPRY4-IT1 silencing by transfecting cells with siSPRY4-IT1 blocked HuR binding to claudin-1, claudin-3, occludin, and JAM-1 mRNAs (Figure 4C), although it did not affect total HuR abundance or its subcellular distribution (Supplemental Figure S6A). In contrast, silencing SPRY4-IT1 had no effect on HuR association with the Atf2 or Jund mRNAs (unpublished data).
FIGURE 4:
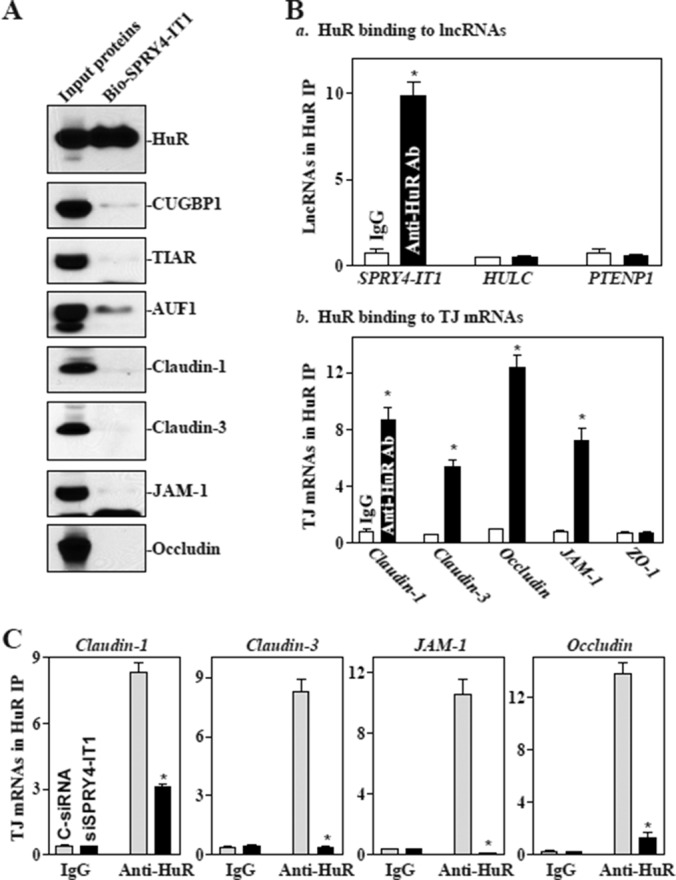
SPRY4-IT1 is essential for HuR interaction with TJ mRNAs. (A) SPRY4-IT1 directly binds to HuR but not to other RBPs and TJ proteins. After cytoplasmic lysates were incubated with biotinylated SPRY4-IT1, levels of HuR and other proteins in the pull-down material were assessed. (B) Association of endogenous HuR with endogenous SPRY4-IT1 (a) and TJ mRNAs (b). After IP of RNA–protein complexes from cell lysates using anti-HuR antibody (Ab) or control IgG1, RNA was isolated and measured by Q-PCR analysis. Values are means ± SEM from three separate experiments. *p < 0.05 compared with IgG. (C) SPRY4-IT1 silencing represses HuR association with TJ mRNAs. Forty-eight hours after cells were transfected with siSPRY4-IT1 and C-siRNA, the association of HuR with TJ mRNAs was measured. *p < 0.05 compared with C-siRNA.
There are multiple predicted SPRY4-IT1-binding sites in the 3′-UTRs of claudin-1, claudin-3, occludin, and JAM-1 (Figure 5A and Supplemental Table S1). RNA–RNA pull-down assays showed that the levels of claudin-1, claudin-3, occludin, and JAM-1 mRNAs were much higher in biotinylated SPRY4-IT1 pull-down samples than those in biotin-labeled Gapdh pull-down samples (Figure 5B). Of interest, transfection of cells with siHuR almost completely depleted HuR levels (Supplemental Figure S6B) but only decreased rather than totally prevented the binding of SPRY4-IT1 to claudin-1, claudin-3, occludin, and JAM-1 mRNAs (Figure 5C), indicating that SPRY4-IT1 also directly binds the TJ mRNAs and that this interaction is independent of its association with HuR. These findings strongly suggest a model in which SPRY4-IT1 controls intestinal epithelial TJ permeability (Figure 5D): 1) SPRY4-IT1 is essential for HuR binding to TJ mRNAs, 2) SPRY4-IT1 also directly interacts with TJ mRNAs, and 3) SPRY4-IT1/TJ mRNA associations stabilize and enhance TJ mRNA translation, thus increasing TJ protein production and promoting the function of the epithelial barrier.
FIGURE 5:
SPRY4-IT1 directly interacts with TJ mRNAs. (A) Schematic of TJ mRNAs depicting potential binding sites of SPRY4-IT1 in the 3′-UTRs of claudin-1, claudin-3, occludin, and JAM-1 mRNAs. SBS, potential SPRY4-IT1-binding site. (B) SPRY4-IT1 association with mRNAs encoding claudin-1, claudin-3, occludin, and JAM-1 as measured by using biotinylated SPRY4-IT1. Values are means ± SEM from three separate experiments. *p < 0.05 compared with biotin-labeled Gapdh. (C) Binding of biotinylated SPRY4-IT1 to TJ mRNAs after HuR silencing: (a) levels of TJ mRNAs in the materials pulled down by biotin-SPRY4-IT1, and (b) levels of total input mRNAs. Cells were transfected with siHuR or C-siRNA, and association of TJ mRNAs with SPRY4-IT1 was measured 48 h thereafter. *p < 0.05 compared with C-siRNA. (D) Schematic of proposed influence of SPRY4-IT1 on the epithelial TJ permeability. SPRY4-IT1 is essential for HuR binding to the TJ mRNAs and also directly interacts with the TJ mRNAs, thus enhancing TJ expression and barrier function.
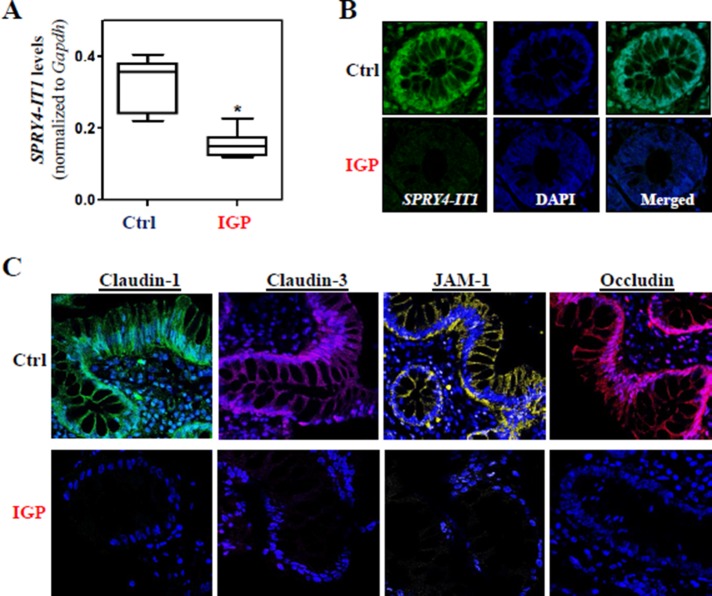
Decreased expression of SPRY4-IT1 in patients with increased gut permeability
To study the effect of SPRY4-IT1–regulated TJ expression on clinical conditions, we examined changes in the levels of SPRY4-IT1 in human intestinal mucosa from patients with increased gut permeability. The colonic mucosal tissues in patients with ulcerative colitis, who were diagnosed with increased gut permeability clinically, were collected for measurements of SPRY4-IT1 levels and TJ expression, with the mucosal samples from patients without significant changes in gut permeability serving as controls. As shown in Figure 6A, SPRY4-IT1 levels in the mucosa obtained from patients with increased gut permeability were significantly lower than those measured in control patients. The reduced SPRY4-IT1 expression levels in patients with increased gut permeability were further confirmed by RNA-FISH analysis, which revealed a decrease in SPRY4-IT1 fluorescence intensity in the mucosa (Figure 6B). Consistent with our findings from cultured IECs (Figure 1) and mice (Figure 2), decreased levels of SPRY4-IT1 in all of the patients we examined were correlated with an inhibition of TJ expression, as shown by a decrease in claudin-1, claudin-3, occludin, and JAM-1 immunostaining (Figure 6C). These results suggest that decreased SPRY4-IT1 and subsequent inhibition of TJ expression contribute to the pathogenesis of gut barrier dysfunction in patients.
FIGURE 6:
Association of decreased SPRY4-IT1 with reduction in the levels of TJs in patients with gut barrier dysfunction. (A) Levels of SPRY4-IT1 in the intestinal mucosa in patients with increased gut permeability (IGP) or controls (Ctrl) as measured by Q-PCR analysis and normalized to Gapdh levels. Values are means ± SEM (n = 7). *p < 0.05 compared with controls. (B) In situ hybridization of SPRY4-IT1 with fluorescent LNA RNA detection probe in the intestinal mucosa described in A. (C) Immunohistochemical staining of TJ proteins in the intestinal mucosa described in A.
DISCUSSION
Acute gut barrier dysfunction occurs commonly in certain pathological conditions. It can lead to the translocation of toxic substances and bacteria from the intestinal lumen to the bloodstream, and in extreme cases it can result in multiple organ dysfunction syndrome and death (Zhang et al., 2010; Carter et al., 2013). Because the exact mechanism responsible for increased gut permeability is obscure, effective therapies to preserve the integrity of the epithelial barrier are limited, especially in surgical intensive care patients supported with total parenteral nutrition (Mosenthal et al., 2002; Carter et al., 2013). Identification of the underlying causes and successful medical treatment are major challenges, and efforts to develop effective therapeutics to protect the barrier function are extremely important. Here we identify a novel mechanism by which the lncRNA SPRY4-IT1 controls intestinal epithelial barrier function by altering TJ expression and further show that SPRY4-IT1 regulates TJ levels primarily at the posttranscriptional level. Knockdown of SPRY4-IT1 expression caused epithelial barrier dysfunction as a result of inhibition of TJ expression in vitro, whereas increased levels of SPRY4-IT1 in the intestinal mucosa not only prevented CLP-induced TJ repression, but they also protected the gut epithelial barrier against septic stress in vivo. These findings are a significant conceptual advance by linking the lncRNA SPRY4-IT1 with TJ expression and subsequent gut permeability and underscore the effect of SPRY4-IT1 in the pathogenesis of acute gut barrier dysfunction.
SPRY4-IT1 is actively transcribed from an intron of the SPRY4 gene and undergoes maturation by cleavage of the 5′ region in the nucleus before transport to the cytoplasm (Mazar et al., 2014), and sequence analysis confirmed that SPRY4-IT1 does not show any coding potential. Although SPRY4-IT1 is expressed in different human tissues, our understanding of its biological function and involvement in pathology is limited. It has been reported that SPRY4-IT1 is implicated in the molecular etiology of human melanoma, because decreasing the levels of SPRY4-IT1 inhibits cell growth and induces apoptosis in melanoma cells (Khaitan et al., 2011). Further study demonstrated that SPRY4-IT1 binds to the lipid phosphatase lipin 2 and modulates apoptosis by altering lipin 2–mediated lipid metabolism (Mazar et al., 2014). A recent study also showed that SPRY4-IT1 stimulates proliferation of human breast cancer cells by up-regulating ZNF703 expression (Shi et al., 2015). Our present results provide new evidence that SPRY4-IT1 regulates the stability and translation of mRNAs encoding TJ proteins claudin-1, claudin-3, occludin, and JAM-1. Because SPRY4-IT1 is associated with polyribosomes (Ingolia et al., 2012; Mazar et al., 2014), it is likely that SPRY4-IT1 enhances TJ levels posttranscriptionally by increasing recruitment of these TJ mRNAs to polyribosomes. In support of this notion, our results show that SPRY4-IT1 was enriched in actively translating fractions prepared from polysomal gradients (Supplemental Figure S7) and that SPRY4-IT1 silencing caused a shift of TJ mRNAs from actively translating fractions to low-translating fractions of polyribosomes. Efforts are underway in our laboratory to understand the specific mechanisms by which SPRY4-IT1 regulates the translation of TJ proteins and whether SPRY4-IT1 might also modulate TJ gene transcription.
The results reported here also indicate that SPRY4-IT1 interacts with HuR and that SPRY4-IT1/HuR associations are essential for HuR binding to the TJ mRNAs. HuR has three RNA recognition motifs through which it directly interacts with numerous mRNAs to modulate their translation and/or stability (Mukherjee et al., 2011; Yoon et al., 2012; Liu et al., 2014). Through its effect on target mRNAs, HuR has been implicated in many aspects of cellular functions and human diseases (Srikantan et al., 2012a). Our previous studies revealed that HuR functions as a master regulator of TJ expression in the intestinal epithelium and that inhibition of HuR expression or disruption of its binding affinity causes intestinal epithelial barrier dysfunction in vitro as well as in vivo (Yu et al., 2011, 2013; Yang et al., 2014; Xiao et al., 2014). HuR also interacts with noncoding RNAs (ncRNAs) such as miRNAs and lncRNAs to jointly regulate target transcripts antagonistically or synergistically (Kim et al., 2009; Liu et al., 2009; Mukherjee et al., 2011; Srikantan et al., 2012b; Yoon et al., 2012; Abdelmohsen et al., 2014). For example, HuR binds lncRNA-p21 and induces the recruitment of let-7/Ago to lncRNA-p21, leading to destabilization of lncRNA-p21 (Yoon et al., 2012); HuR also competes with ncRNA 7SL to modulate p53 translation (Abdelmohsen et al., 2014). In this study, although SPRY4-IT1 silencing did not affect total HuR levels or subcellular distribution, it robustly decreased the levels of the TJ mRNAs encoding claudin-1, claudin-3, occludin, and JAM-1 in HuR IP materials (Figure 4C), indicating that SPRY4-IT1 is required for HuR binding to these TJ mRNAs. On the other hand, the 3′-UTRs of all of these TJ mRNAs contain multiple predicted SPRY4-IT1-binding sites, and HuR silencing decreases but does not completely block SPRY4-IT1 association with the TJ mRNAs, as measured by biotinylated SPRY4-IT1 pull-down assays (Figure 5C), suggesting that SPRY4-IT1 is also capable of interacting directly with TJ mRNAs independently of its interaction with HuR.
Because patients with increased gut permeability displayed decreased levels of SPRY4-IT1 in the gut mucosa, therapies aimed at systemically overexpressing SPRY4-IT1 might enhance intestinal epithelial barrier function in specific clinical settings. Although the complete sets of mRNAs controlled by SPRY4-IT1 and the underlying mechanism in the intestinal epithelium remain to be fully elucidated, the fact that overexpressing SPRY4-IT1 using a lentivirus protected the gut barrier function in mice exposed to CLP highlights the effect of an individual lncRNA in controlling gut permeability. These findings are particularly significant in patients with traumatic damage, thermal injury, shock, and recovery from major surgical operations, as acute gut barrier dysfunction occurs commonly in these critical surgical conditions (Zhang et al., 2010; Carter et al., 2013).
Unlike most traditional therapeutics, in which drugs have specific cellular targets, modulating lncRNAs or miRNAs regulates entire functional gene networks (Wapinski et al., 2011; Mendell et al., 2012). Of course, this broad spectrum of actions should be pursued with caution, since unintended off-target effects may also occur. In sum, our results indicate that SPRY4-IT1 is a potent biological regulator of the intestinal epithelial TJ permeability through its interaction with HuR. These findings can also potentially provide innovative molecular therapy to protect the gut epithelial barrier function in patients with various critical disorders.
MATERIALS AND METHODS
Cell culture and animals
The Caco-2 human colon carcinoma cell line was purchased from the American Type Culture Collection (Manassas, VA) and maintained in standard culture conditions (Chen et al., 2008). Tissue culture medium and dialyzed fetal bovine serum were obtained from Invitrogen (Carlsbad, CA), and biochemicals were obtained from Sigma-Aldrich (St. Louis, MO). Antibodies recognizing claudin-1, claudin-3, occludin, ZO-1, HuR, and β-actin were obtained from Santa Cruz Biotechnology (Santa Cruz, CA) and BD Biosciences (Sparks, MD), and the secondary antibody conjugated to horseradish peroxidase was obtained from Sigma-Aldrich.
C57BL/65 mice (male and female, 6–9 wk old) were purchased from the Jackson Laboratory (Bar Harbor, ME) and housed in specific pathogen-free animal facility at the Baltimore VA Medical Center. All animal experiments were conducted in accordance with National Institutes of Health guidelines and were approved by the Institutional Animal Care and Use Committee of the University of Maryland School of Medicine and Baltimore VA Hospital.
Plasmid construction and RNA interference
Lenti-SPRY4-IT1 was custom made by AMSBIO, in which SPRY4-IT1/GFP expression was under the control of the suCMV-promoter. Lenti-SPRY4-IT1 and C-lentiviral were packaged in lentiviral production cells, concentrated by ultracentrifugation, resuspended in phosphate-buffered saline (PBS), and used to increase SPRY4-IT1 in vivo as described (Scherr et al., 2007; Feng et al., 2012). An expression vector containing SPRY4-IT1 cDNA under control of pCMV-promoter was constructed (Liu et al., 2009) and used to increase SPRY4-IT1 in Caco-2 cells; claudin-1 and occludin expression vectors were obtained from Origene (Rockville, MD).
Expression of SPRY4-IT1 and HuR was silenced by transfection with specific siRNA as described (Cui et al., 2011; Liu et al., 2015). The siSPRY4-IT1, siHuR, and C-siRNA were purchased from Santa Cruz Biotechnology. For each 60-mm cell culture dish, 15 μl of the 20 μM stock duplex siSPRY4-IT1, siHuR, or C-siRNA was used. Forty-eight hours after transfection using Lipofectamine, cells were harvested for analysis.
Quantitative real-time PCR and immunoblotting analyses
Total RNA was isolated by using the RNeasy minikit (Qiagen, Valencia, CA) and used in RT and PCR amplification reactions as described (Zou et al., 2015). Q-PCR was performed using Step-one-plus Systems with specific primers, probes, and software (Applied Biosystems, Foster City, CA).
To examine protein levels, whole-cell lysates were prepared using 2% SDS, sonicated, and centrifuged at 4°C for 15 min. The supernatants were boiled for 5 min and size fractionated by SDS–PAGE. After transferal of proteins onto nitrocellulose filters, the blots were incubated with primary antibodies recognizing TJ proteins or RBPs; after incubations with secondary antibodies, immunocomplexes were visualized by using chemiluminescence.
Isolation of nuclear and cytoplasmic RNA
Nuclear and cytoplasmic RNA fractions from Caco-2 cells were isolated and purified with the SurePrep Nuclear/Cytoplasmic RNA Purification Kit from Fisher Bioreagents (Fair Lawn, NJ), following the manufacturer’s instructions. Briefly, after lysis solution was added, whole-cell lysate was transferred and centrifuged at 17,000 × g for 3 min. The supernatant containing cytoplasmic RNA and the pellet containing nuclear RNA were saved separately and then mixed with the binding solution. After addition of ethanol, the mixtures were applied onto a spin column and centrifuged at 17,000 × g for 1 min, and then RNA was eluted. Gapdh mRNA levels were measured as an internal control for normalization. To assess any cross-contamination between cytoplasmic and nuclear fractions, the levels of β-tubulin (a specific cytoplasmic protein marker) and lamin B (a nuclear protein marker) were examined in the two fractions (Zou et al., 2006).
Analysis of newly translated protein and polysome analysis
De novo synthesis of nascent TJ proteins was detected by a Click-IT Protein Analysis Detection Kit (Life Technologies, Grand Island, NY) and performed following the manufacturer’s instructions. Briefly, cells were incubated in methionine-free medium and then exposed to AHA. After mixing cell lysates with the reaction buffer containing biotin/alkyne reagent and CuSO4 for 20 min, the biotin-alkyne/azide–modified protein complex was pulled down using paramagnetic streptavidin-conjugated Dynabeads. The pull-down material was resolved by 10% SDS–PAGE and analyzed by Western immunoblotting analysis using the antibody against TJ proteins or GAPDH.
Polysome analysis was performed as described (Xiao et al., 2013). Briefly, cells at ∼70% confluence were incubated for 15 min in 0.1 mg/ml cycloheximide and then lifted by scraping in 1 ml of polysome extraction lysis buffer and lysed on ice for 10 min. Nuclei were pelleted, and the resulting supernatant was fractionated through a 15–60% linear sucrose gradient to fractionate cytoplasmic components according to their molecular weight. The eluted fractions were prepared with a fraction collector (Brandel, Gaithersburg, MD), and their quality was monitored at 254 nm using a UV-6 detector (ISCO, Louisville, KY). After RNA in each fraction was extracted, the levels of each individual mRNA were quantified by RT, followed by Q-PCR in each of the fractions.
Biotin-labeled SPRY4-IT1 pull-down and RNP-IP assays
After biotin-labeled SPRY4-IT1 was incubated with cytoplasmic proteins at room temperature for 1 h, the mixture was mixed with streptavidin-Dynal beads and incubated at 4°C on a rotator overnight. After the beads were washed thoroughly, the beads-bound RNA was isolated and subjected to RT, followed by Q-PCR analysis. To examine association of SPRY4-IT1 with RBPs, levels of HuR and other proteins in the pull-down material were assessed after cytoplasmic lysates were incubated with biotinylated SPRY4-IT1.
To assess the association of endogenous HuR with endogenous TJ mRNAs or SPRY4-IT1, IP of RNP complexes was performed as described (Yu et al., 2011). Twenty million cells were collected per sample, and lysates were used for IP for 4 h at room temperature in the presence of excess (30 μg) IP antibody (IgG, or anti-HuR). RNA in IP materials was used in RT, followed by PCR and Q-PCR analysis to detect the presence of TJ and Gapdh mRNAs.
RNA-FISH assay
The RNA-FISH assay was performed with the ISH Optimization Kit from Exiqon (Vedbaek, Denmark) as described (Elmen et al., 2008). Briefly, the mucosa was fixed with 4% fresh paraformaldehyde overnight and embedded in paraffin for sections. The slides were deparaffinized and then incubated with proteinase-K. After washes with PBS, the slides were dehydrated and hybridized with 25 nM fluorescent locked nucleic acid (LNA) probe for 1 h at 60°C. The slides were washed with saline sodium citrate buffer and PBS, covered with coverslides, and then processed using a Zeiss confocal microscope (Zeiss, Jena, Germany).
Measurements of gut epithelial barrier function
The epithelial barrier function in vitro was examined by paracellular tracer flux assays using a 12-well Transwell plate (surface area, 1.12 cm2) as described (Guo et al., 2003; Yu et al., 2013). FITC-dextran (70 kDa; Sigma-Aldrich), a membrane-impermeable molecule, served as the paracellular tracer and was added to the apical bathing wells. The basal bathing well had no added tracers and contained the same flux assay medium as in the apical compartment. All flux assays were performed at 37°C, and the basal medium was collected 2 h after addition of the FITC-dextran. The concentration of the FITC-dextran in the basal medium was determined using a fluorescence plate reader with an excitation wavelength at 490 nm and an emission wavelength of 530 nm. TEER was measured with an epithelial voltmeter under open-circuit conditions (WPI, Sarasota, FL) as described (Yu et al., 2013), and the TEER of all monolayers was normalized to that of control monolayers in the same experiment.
Gut permeability in vivo was determined by examining the appearance in blood of FITC-dextran administered by gavage as described (Furuta et al., 2001). Briefly, mice were gavaged with FITC-dextran at a dose of 60 mg/100 g wt at 4 h before harvest. Blood sample was collected by cardiac puncture. The serum concentration of the FITC-dextran was determined using a fluorescence plate reader as described.
Surgical procedures
CLP was performed as described previously (Hubbard et al., 2005). Mice were anesthetized by Nembutal (5.5 mg/100 g wt, intraperitoneally), and a midline abdominal incision was performed. The distal portion of the cecum (1 cm) was ligated with 5-0 silk suture. The ligated cecum was then punctured with a 25-gauge needle and slightly compressed with an applicator until a small amount of stool appeared. In sham-operated animals, the cecum was manipulated but without ligation and puncture and placed back in the peritoneum. The incision was closed using a two-layer procedure: 5-0 silk suture on the muscle layer and the skin, respectively. Mice received 1 ml of saline intraperitoneally for fluid resuscitation at the time of closure and 0.1 mg/100 g wt Buprenex subcutaneously four times at 12-h intervals to minimize distress.
Statistics
Values are means ± SEM from three to six samples. Autoradiographic results were repeated three times. The significance of the difference between means was determined by analysis of variance. Level of significance was determined by Duncan’s multiple-range test (Harter, 1960).
Supplementary Material
Acknowledgments
This work is supported by Merit Review Awards (to J.Y.W. and J.N.R.) from the U.S. Department of Veterans Affairs, grants from the National Institutes of Health (DK57819, DK61972, and DK68491 to J.Y.W.), and funding from the National Institute on Aging–Intramural Research Program, National Institutes of Health (to M.G.). J.Y.W. is a Senior Research Career Scientist, Biomedical Laboratory Research and Development Service, U.S. Department of Veterans Affairs.
Abbreviations used:
- AHA
l-azidohomoalanine
- CUGBP1
CUG-binding protein 1
- IEC
intestinal epithelial cell
- lncRNA
long noncoding RNA
- IP
immunoprecipitation
- miRNA
microRNA
- RBP
RNA-binding protein
- RNP
ribonucleoprotein
- siHuR
siRNA targeting HuR
- UTR
untranslated region.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-10-0703) on December 17, 2015.
REFERENCES
- Abdelmohsen K, Panda AC, Kang MJ, Guo R, Kim J, Grammatikakis I, Yoon JH, Dudekula DB, Noh JH, Yang X, et al. 7SL RNA represses p53 translation by competing with HuR. Nucleic Acids Res. 2014;42:10099–10111. doi: 10.1093/nar/gku686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S, Xiao L, Rao JN, Zou T, Liu L, Zhang D, Turner DJ, Gorospe M, Wang JY. Inhibition of Smurf2 translation by miR-322/503 modulates TGF-β/Smad2 signaling and intestinal epithelial homeostasis. Mol Biol Cell. 2014;25:1234–1243. doi: 10.1091/mbc.E13-09-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SR, Zahs A, Palmer JL, Wang L, Ramirez L, Gamelli RL, Kovacs EJ. Intestinal barrier disruption as a cause of mortality in combined radiation and burn injury. Shock. 2013;40:281–289. doi: 10.1097/SHK.0b013e3182a2c5b5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Xiao L, Rao JN, Zou T, Liu L, Bellavance E, Gorospe M, Wang JY. JunD represses transcription and translation of the tight junction protein zona occludens-1 modulating intestinal epithelial barrier function. Mol Biol Cell. 2008;19:3701–3712. doi: 10.1091/mbc.E08-02-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui YH, Xiao L, Rao JN, Zou T, Liu L, Chen Y, Turner DJ, Gorospe M, Wang JY. miR-503 represses CUG-binding protein 1 translation by recruiting CUGBP1 mRNA to processing bodies. Mol Biol Cell. 2011;23:151–162. doi: 10.1091/mbc.E11-05-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- Feng SY, Dong CG, Wu WK, Wang XJ, Qiao J, Shao JF. Lentiviral expression of anti-microRNAs targeting miR-27a inhibits proliferation and invasiveness of U87 glioma cells. Mol Med Rep. 2012;6:275–281. doi: 10.3892/mmr.2012.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M, Izumi Y, Oda Y, Higashi T, Iwamoto N. Molecular organization of tricellular tight junctions. Tissue Barriers. 2014;2:e28960. doi: 10.4161/tisb.28960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta GT, Turner JR, Taylor CT, Hershberg RM, Comerford K, Narravula S, Podolsky DK, Colgan SP. Hypoxia-inducible factor 1-dependent induction of intestinal trefoil factor protects barrier function during hypoxia. J Exp Med. 2001;193:1027–1034. doi: 10.1084/jem.193.9.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Rao JN, Liu L, Zou TT, Turner DJ, Bass BL, Wang JY. Regulation of adherens junctions and epithelial paracellular permeability: a novel function for polyamines. Am J Physiol. 2003;285:C1174–C1187. doi: 10.1152/ajpcell.00015.2003. [DOI] [PubMed] [Google Scholar]
- Harter JL. Critical values for Duncan’s new multiple range tests. Biometric. 1960;16:671–685. [Google Scholar]
- Hubbard WJ, Choudhry M, Schwacha MG, Kerby JD, Rue LW, 3rd, Bland KI, Chaudry IH. Cecal ligation and puncture. Shock. 2005;24(Suppl 1):52–57. doi: 10.1097/01.shk.0000191414.94461.7e. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS, Perera RJ. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 2011;71:3852–3862. doi: 10.1158/0008-5472.CAN-10-4460. [DOI] [PubMed] [Google Scholar]
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Christodoulou-Vafeiadou E, Rao JN, Zou T, Xiao L, Chung HK, Yang H, Gorospe M, Kontoyiannis D, Wang JY. RNA-binding protein HuR promotes growth of small intestinal mucosa by activating the Wnt signaling pathway. Mol Biol Cell. 2014;25:3308–3318. doi: 10.1091/mbc.E14-03-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Ouyang M, Rao JN, Zou T, Xiao L, Chung HK, Wu J, Donahue JM, Gorospe M, Wang JY. Competition between RNA-binding proteins CELF1 and HuR modulates MYC translation and intestinal epithelium renewal. Mol Biol Cell. 2015;26:1797–1810. doi: 10.1091/mbc.E14-11-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Rao JN, Zou T, Xiao L, Wang PY, Turner DJ, Gorospe M, Wang JY. Polyamines regulate c-Myc translation through Chk2-dependent HuR phosphorylation. Mol Biol Cell. 2009;20:4885–4898. doi: 10.1091/mbc.E09-07-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Li D, Zhang W, Guo M, Zhan Q. Long non-coding RNA gadd7 interacts with TDP-43 and regulates Cdk6 mRNA decay. EMBO J. 2012;31:4415–4427. doi: 10.1038/emboj.2012.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. (Spec No 1) [DOI] [PubMed] [Google Scholar]
- Mazar J, Zhao W, Khalil AM, Lee B, Shelley J, Govindarajan SS, Yamamoto F, Ratnam M, Aftab MN, Collins S, et al. The functional characterization of long noncoding RNA SPRY4-IT1 in human melanoma cells. Oncotarget. 2014;5:8959–8969. doi: 10.18632/oncotarget.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosenthal AC, Xu D, Deitch EA. Elemental and intravenous total parenteral nutrition diet-induced gut barrier failure is intestinal site specific and can be prevented by feeding nonfermentable fiber. Crit Care Med. 2002;30:396–402. doi: 10.1097/00003246-200202000-00022. [DOI] [PubMed] [Google Scholar]
- Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M, Jr, Tuschl T, Ohler U, Keene JD. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatake T, Fujita H, Minato N, Hamazaki Y. Enteroendocrine cells are specifically marked by cell surface expression of claudin-4 in mouse small intestine. PLoS One. 2014;9:e90638. doi: 10.1371/journal.pone.0090638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Scherr M, Venturini L, Battmer K, Schaller-Schoenitz M, Schaefer D, Dallmann I, Ganser A, Eder M. Lentivirus-mediated antagomir expression for specific inhibition of miRNA function. Nucleic Acids Res. 2007;35:e149. doi: 10.1093/nar/gkm971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger EE, Lynch RD. The tight junction: a multifunctional complex. Am J Physiol. 2004;286:C1213–C1228. doi: 10.1152/ajpcell.00558.2003. [DOI] [PubMed] [Google Scholar]
- Sharma A, Bhat AA, Krishnan M, Singh AB, Dhawan P. Trichostatin-A modulates claudin-1 mRNA stability through the modulation of Hu antigen R and tristetraprolin in colon cancer cells. Carcinogenesis. 2013;34:2610–2621. doi: 10.1093/carcin/bgt207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Li J, Liu Y, Ding J, Fan Y, Tian Y, Wang L, Lian Y, Wang K, Shu Y. The long noncoding RNA SPRY4-IT1 increases the proliferation of human breast cancer cells by upregulating ZNF703 expression. Mol Cancer. 2015;14:51. doi: 10.1186/s12943-015-0318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantan S, Gorospe M. HuR function in disease. Front Biosci. 2012;17:189–205. doi: 10.2741/3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantan S, Tominaga K, Gorospe M. Functional interplay between RNA-binding protein HuR and microRNAs. Curr Protein Pept Sci. 2012;13:372–379. doi: 10.2174/138920312801619394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yan I, Haga H, Patel T. Long noncoding RNA in liver diseases. Hepatology. 2014;60:744–753. doi: 10.1002/hep.27043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terahara K, Yoshida M, Igarashi O, Nochi T, Pontes GS, Hase K, Ohno H, Kurokawa S, Mejima M, Takayama N, et al. Comprehensive gene expression profiling of Peyer’s patch M cells, villous M-like cells, and intestinal epithelial cells. J Immunol. 2008;180:7840–7846. doi: 10.4049/jimmunol.180.12.7840. [DOI] [PubMed] [Google Scholar]
- Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71:3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9:799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- Uchida S, Dimmeler S. Long noncoding RNAs in cardiovascular diseases. Circ Res. 2015;116:737–750. doi: 10.1161/CIRCRESAHA.116.302521. [DOI] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Xiao L, Rao JN, Zou T, Liu L, Cao S, Martindale JL, Su W, Chung HK, Gorospe M, Wang JY. miR-29b represses intestinal mucosal growth by inhibiting translation of cyclin-dependent kinase 2. Mol Biol Cell. 2013;24:3038–3046. doi: 10.1091/mbc.E13-05-0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Wang JY. RNA-binding proteins and microRNAs in gastrointestinal epithelial homeostasis and diseases. Curr Opin Pharmacol. 2014;19:46–53. doi: 10.1016/j.coph.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Rao JN, Wang JY. Posttranscriptional regulation of intestinal epithelial tight junction barrier by RNA-binding proteins and microRNAs. Tissue Barriers. 2014;2:e28320. doi: 10.4161/tisb.28320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye D, Guo S, Al-Sadi R, Ma TY. MicroRNA regulation of intestinal epithelial tight junction permeability. Gastroenterology. 2011;141:1323–1333. doi: 10.1053/j.gastro.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TX, Rao JN, Zou T, Liu L, Xiao L, Ouyang M, Cao S, Gorospe M, Wang JY. Competitive binding of CUGBP1 and HuR to occludin mRNA controls its translation and modulates epithelial barrier function. Mol Biol Cell. 2013;24:85–99. doi: 10.1091/mbc.E12-07-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TX, Wang PY, Rao JN, Zou T, Liu L, Xiao L, Gorospe M, Wang JY. Chk2-dependent HuR phosphorylation regulates occludin mRNA translation and epithelial barrier function. Nucleic Acids Res. 2011;39:8472–8487. doi: 10.1093/nar/gkr567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yuan C, Hua G, Tong R, Luo X, Ying Z. Early gut barrier dysfunction in patients with severe acute pancreatitis: attenuated by continuous blood purification treatment. Int J Artif Organs. 2010;33:706–715. [PubMed] [Google Scholar]
- Zhou Q, Costinean S, Croce CM, Brasier AR, Merwat S, Larson SA, Basra S, Verne GN. microRNA-29 targets nuclear factor-κB-repressing factor and claudin 1 to increase intestinal permeability. Gastroenterology. 2015;148:158–169. doi: 10.1053/j.gastro.2014.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang R, Rao JN, Zou T, Liu L, Xiao L, Cao S, Hansraj NZ, Gorospe M, Wang JY. miR-195 competes with HuR to modulate stim1 mRNA stability and regulate cell migration. Nucleic Acids Res. 2013;41:7905–7919. doi: 10.1093/nar/gkt565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou T, Mazan-Mamczarz K, Rao JN, Liu L, Marasa BS, Zhang AH, Xiao L, Pullmann R, Gorospe M, Wang JY. Polyamine depletion increases cytoplasmic levels of RNA-binding protein HuR leading to stabilization of nucleophosmin and p53 mRNAs. J Biol Chem. 2006;281:19387–19394. doi: 10.1074/jbc.M602344200. [DOI] [PubMed] [Google Scholar]
- Zou T, Rao JN, Liu L, Xiao L, Chung HK, Li Y, Chen G, Gorospe M, Wang JY. JunD enhances miR-29b levels transcriptionally and post-transcriptionally to inhibit proliferation of intestinal epithelial cells. Am J Physiol. 2015;308:C813–C824. doi: 10.1152/ajpcell.00027.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou T, Rao JN, Liu L, Xiao L, Yu TX, Jiang P, Gorospe M, Wang JY. Polyamines regulate the stability of JunD mRNA by modulating the competitive binding of its 3’ untranslated region to HuR and AUF1. Mol Cell Biol. 2010;30:5021–5032. doi: 10.1128/MCB.00807-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.