The histone demethylase enzyme Kdm5b is identified as a regulator of neural stem cells (NSCs) from the subventricular zone (SVZ) of adult mice. shRNA knockdown and cell culture assays demonstrate that Kdm5b inhibition promotes neurogenesis and derepresses Reln (reelin) in SVZ NSCs.
Abstract
The role of epigenetic regulators in the control of adult neurogenesis is largely undefined. We show that the histone demethylase enzyme Kdm5b (Jarid1b) negatively regulates neurogenesis from adult subventricular zone (SVZ) neural stem cells (NSCs) in culture. shRNA-mediated depletion of Kdm5b in proliferating adult NSCs decreased proliferation rates and reduced neurosphere formation in culture. When transferred to differentiation culture conditions, Kdm5b-depleted adult NSCs migrated from neurospheres with increased velocity. Whole-genome expression screening revealed widespread transcriptional changes with Kdm5b depletion, notably the up-regulation of reelin (Reln), the inhibition of steroid biosynthetic pathway component genes and the activation of genes with intracellular transport functions in cultured adult NSCs. Kdm5b depletion increased extracellular reelin concentration in the culture medium and increased phosphorylation of the downstream reelin signaling target Disabled-1 (Dab1). Sequestration of extracellular reelin with CR-50 reelin-blocking antibodies suppressed the increase in migratory velocity of Kdm5b-depleted adult NSCs. Chromatin immunoprecipitation revealed that Kdm5b is present at the proximal promoter of Reln, and H3K4me3 methylation was increased at this locus with Kdm5b depletion in differentiating adult NSCs. Combined the data suggest Kdm5b negatively regulates neurogenesis and represses Reln in neural stem cells from the adult SVZ.
INTRODUCTION
Adult neurogenesis is a multistep process in which neural stem cell (NSC) lineages progress through a series of well-characterized cell stages to generate functional interneurons in the olfactory bulb and hippocampal dentate gyrus (Ming and Song, 2011; Lim and Alvarez-Buylla, 2014). In the subventricular zone (SVZ), quiescent radial glial-like (RGL) progenitor cells are the multipotent NSC population (Doetsch et al., 1999) and are capable of undergoing symmetrical cell division to maintain the stem cell pool and asymmetric cell division to generate rapidly dividing transit amplifying cells (also referred to as intermediate progenitors), as well as astrocytes and oligodendrocytes (Doetsch et al., 1997; Jackson et al., 2006; Bonaguidi et al., 2011). Transit amplifying cells differentiate into neuroblasts that migrate to the olfactory bulb through the rostral migratory stream (RMS) to settle as interneurons in the periglomerular and granule cell layers (Lois et al., 1996; Alvarez-Buylla and Garcia-Verdugo, 2002; Ming and Song, 2011). NSCs in the SVZ continually generate neurons throughout the adult life of mice, and stem cell pool maintenance and differentiation are tightly regulated.
Morphogens, growth factors, neurotrophins, cytokines, hormones, and neurotransmitters in the neurogenic niche regulate adult NSC (aNSC) biology (Zhao et al., 2008; Ming and Song, 2011; Berg et al., 2013) and interact with transcription factor and epigenetic programs to control NSC fate (Hsieh, 2012; Gonzales-Roybal and Lim, 2013). Epigenetic mechanisms regulating adult neurogenesis include chromatin remodeling by Ezh2, a component of the Polycomb repressive complex 2, which catalyzes the trimethylation of lysine 27 (K27) of histone H3 (H3K27), Mll1 (mixed-lineage leukemia 1), a trithorax group (trxG) H3K4 methyltransferase, and Jmjd3 (Kdm6b), a H3K27 demethylase. Ezh2 represses Ink4a/Arf and the basic helix-loop-helix transcription factor Olig2 to regulate NSC self-renewal and neurogenesis in the adult SVZ (Hwang et al., 2014). Mll1 and Jmjd3 are required for normal Dlx2 expression in neuroblasts and neurogenesis from the adult SVZ via mechanisms involving Mll1/Jmjd3 localization and H3K27me3 demethylation at the I12b enhancer of Dlx2 (Lim et al., 2009; Park et al., 2014). Combined these studies firmly establish histone methylation status as a key epigenetic determinant of adult NSC fate.
Kdm5b (Jarid1b) is a histone demethylase enzyme that catalyzes the removal of trimethyl groups at H3K4 (Xiang et al., 2007). Trimethylation of H3K4 (H3K4me3) occurs at transcriptional start sites, is enriched at actively transcribed genes (Barski et al., 2007), and is a binding site for the general transcription factor TFIID (Vermeulen et al., 2007). Accordingly, Kdm5b demethylation of H3K4me3 is primarily associated with transcriptional repression, and Kdm5b has been shown to be necessary for the silencing of stem and germ cell–specific genes in neuronal differentiation from mouse embryonic stem cells (Dey et al., 2008; Schmitz et al., 2011). Kdm5b-null mice predominantly die within the first day after birth and have disorganized cranial and spinal nerve projections (Albert et al., 2013), consistent with Kdm5b function in developmental neurogenesis. Of interest, differential Kdm5b expression status between embryonic and adult NSCs has been observed (Marei and Ahmed, 2013), suggesting potential differences in its activity between the two developmental states. However, the role of Kdm5b in adult neurogenesis has not been determined.
We investigated the function of Kdm5b in adult SVZ NSCs using short hairpin RNA (shRNA)–mediated depletion of Kdm5b. We demonstrate Kdm5b depletion impairs adult NSC proliferation and accelerates neuroblast migration in cell culture assays. These effects on adult NSC biology were correlated with gene expression changes consistent with cell differentiation, including increased transcription of the secreted extracellular matrix glycoprotein reelin (Reln). We show that increases in Reln transcription are associated with increased H3K4me3 at the Reln promoter in Kdm5b-depleted adult NSCs stimulated to differentiate. On the basis of these results, we propose that Kdm5b negatively regulates neurogenesis and represses Reln in NSCs from the adult SVZ.
RESULTS
Kdm5b is expressed in the adult SVZ
Kdm5b expression has been detected by Western blot and quantitative reverse transcription PCR (qRT-PCR) in discrete tissues in adult mice, including the brain (Schmitz et al., 2011). In situ hybridization (ISH) data from the Allen Brain Atlas (http://mouse.brain-map.org/) show Kdm5b mRNA expression throughout gray matter areas in the adult mouse brain, including neurogenic niches at the SVZ and subgranular zone of the dentate gyrus (DG). Kdm5b ISH signal is noticeably reduced in white compared with gray matter areas in the Allen Brain Atlas data set. To verify Kdm5b expression in neurogenic niches, we compared Kdm5b fluorescence in situ hybridization (FISH) signals in the SVZ and DG with those in the corpus callosum (CC) in adult mouse brain tissue sections. We used coimmunohistochemical labeling of glial fibrillary acidic protein (GFAP) to identify RGL stem cells (in the SVZ and DG) and astrocytes. FISH labeling resulted in concentrated, dense signal in the SVZ and DG, with FISH present in GFAP-labeled RGL stem cells in both neurogenic niches (Figure 1). In contrast, FISH labeling in the CC was sparse and diffuse (Figure 1). We quantitatively compared Kdm5b FISH signal in the SVZ and the adjacent CC to determine the relative levels of Kdm5b expression in these two structures. The data showed significantly greater numbers of FISH-labeled cells in the SVZ (91.4 ± 1.5 per 135 μm2) than in the CC (63.4 ± 1.2 per 135 μm2) in coronal sections of the adult mouse brain (p < 0.001, t test, n = 3). To take into account differences in cell density between these two structures, we also quantified the number of FISH foci per cell. SVZ cells contained 9.0 ± 0.6 foci/cell compared with 1.8 ± 0.5 foci/cell in the CC, a statistically significant difference (p < 0.001, t test, n = 3). We conclude that Kdm5b expression is highly expressed in the SVZ of the adult mouse compared with the adjacent corpus callosum.
FIGURE 1:
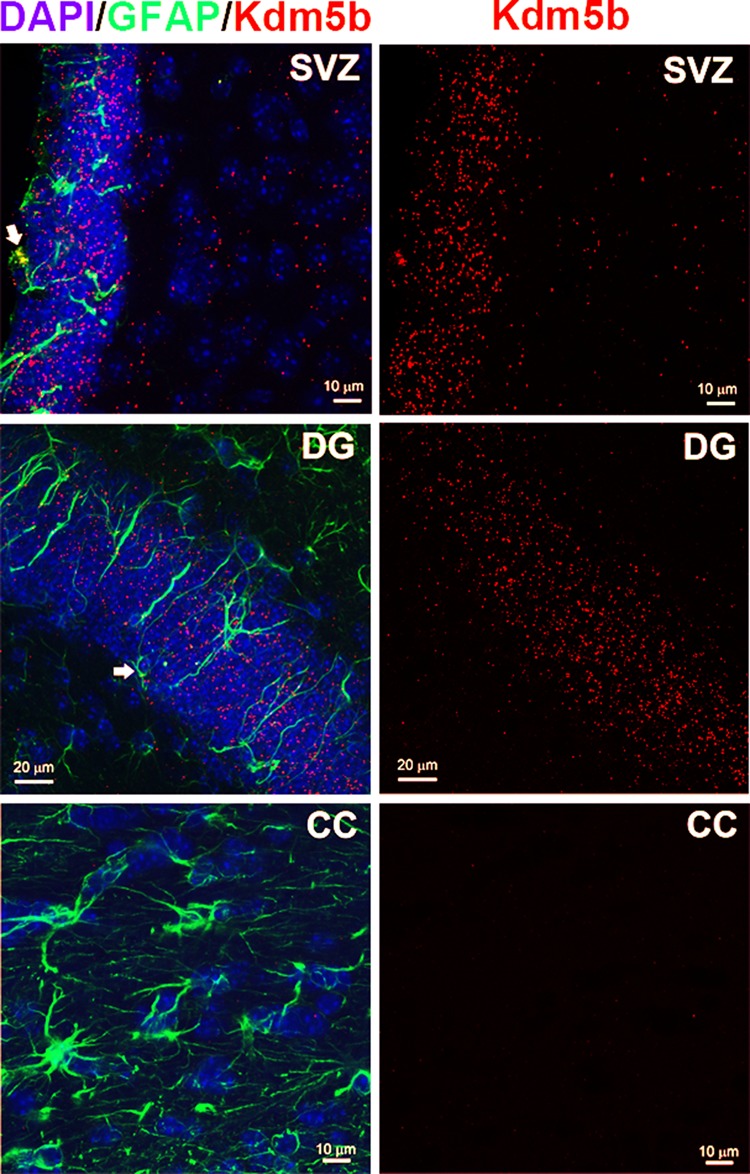
Kdm5b is highly expressed in neurogenic regions of the adult mouse brain. High-resolution confocal images of the SVZ, DG, and, for comparison, the CC taken from coronal sections of the adult mouse forebrain processed for FISH, anti-GFAP immunohistochemistry, and DAPI counterstain. Left, merged images. Right, Kdm5b FISH–only images. Note the high density of ISH signal in the SVZ and DG. Arrows denote Kdm5b FISH in GFAP-immunolabeled RGL progenitor cells.
shRNA-mediated knockdown of Kdm5b decreases aNSC proliferation and reduces neurosphere formation in culture
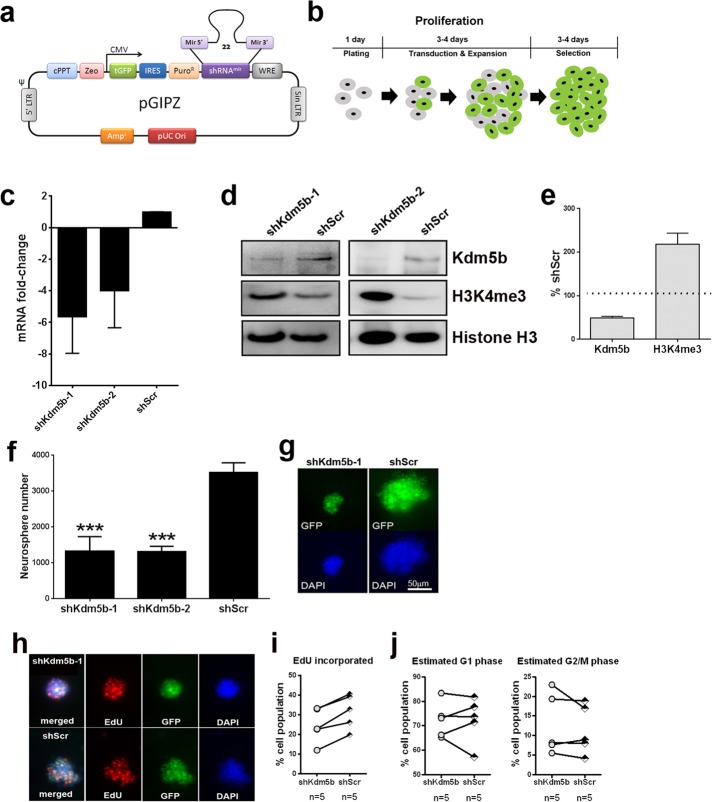
To investigate the function of Kdm5b in adult SVZ NSCs, we used GIPZ lentiviral shRNA vectors to knock down Kdm5b expression in adult SVZ neurosphere cultures. GIPZ lentiviral vectors express a turbo green fluorescent protein (tGFP) reporter for visual tracking of transduced cells (Figure 2a). Preliminary screening in mouse NIH3T3 of two GIPZ shRNA lentiviral vectors (referred to as shKdm5b-1 and shKdm5b-2) showed that both shRNAs depleted Kdm5b at the mRNA and protein levels compared with cells transduced with scrambled nonsilencing shRNA (shScr) control lentivirus (mRNA, 3.5- to 3.8-fold lower by qRT-PCR; protein, 31–46% of control shScr by Western blot). Next we determined shKdm5b effects in aNSCs. Total passage number 5–10 aNSCs from the SVZ were transduced with shKdm5b or shScr lentivirus 24 h after plating in NeuroCult medium containing NSC Proliferation Supplement, epidermal growth factor (EGF), and basic fibroblast growth factor (bFGF; referred to as proliferation medium). aNSCs were maintained in proliferation medium containing lentivirus for 3-4 d before the media was refreshed with proliferation media containing 5 μg/ml puromycin. aNSCs were cultured for a further 3–4 d in the presence of puromycin before analysis (Figure 2b). qRT-PCR data for Kdm5b confirmed mRNA depletion in aNSCs with shKdm5b-1 (5.6 ± 2.3–fold lower, n = 4) and shKdm5b-2 (4.0 ± 2.3–fold lower, n = 3) transduction (Figure 2c). Similarly, Western blot revealed depleted Kdm5b protein levels in cells transduced with shKdm5b-1 and -2 (Figure 2d). Correspondingly, Western blot levels of the Kdm5b substrate H3K4me3 were enhanced with Kdm5b knockdown (Figure 2d), indicating global suppression of H3K4me3 demethylation in aNSCs with shKdm5b lentivirus. Indeed, a quantitative comparison of Kdm5b protein and H3K4me3 levels in cells transduced with shKdm5b-1 or shScr shows that Kdm5b protein levels were 49.1 ± 3.1% (n = 9) and H3K4me3 signal 218.0 ± 25.7% (n = 4) of controls (Figure 2e).
FIGURE 2:
shRNA-mediated knockdown of Kdm5b in culture elevates global H3K4me3 levels in aNSCs and reduces neurosphere formation. Schematic representations of (a) the pGIPZ shRNA lentiviral vectors used to target mouse Kdm5b for knockdown and (b) the lentiviral transduction and selection protocol used to enrich for shRNA-expressing aNSCs. shKdm5b-1 and shKdm5b-2 vectors deplete Kdm5b mRNA (c) and protein (d) levels and elevate H3K4me3 (d) levels in cultured aNSCs. (e) Kdm5b protein is depleted by 49.1 ± 3.1% (n = 9) and H3K4me3 signal elevated 218.0 ± 25.7% (n = 4) by shKdm5b-1 vectors. (f) shKdm5b-1 and shKdm5b-2 transduction significantly reduces aNSC neurosphere size in culture (***p < 0.001, one-way ANOVA with Tukey’s multiple comparisons test, n > 4). (g) Representative tGFP-epifluorescent and DAPI-stained neurospheres generated from aNSCs transduced with shKdm5b-1 or shScr. (h) aNSCs transduced with shKdm5b-1 and pulsed with EdU to label proliferating cells during S phase. (i) Lower EdU incorporation rates in aNSCs transduced with shKdm5b-1 than with shScr at each experimental trial. (j) There is no consistent difference in the estimated percentage of cells in G1 or G2/M phase.
We noticed that transduction with shKdm5b appeared to impede aNSC growth in culture, as shKdm5b-transduced cells produced smaller neurospheres than with shScr controls. This was confirmed by quantification of the number of neurospheres >50 μm in diameter after 3–4 d of lentiviral exposure followed by 3–4 d of puromycin selection. Counts revealed a significant reduction in neurospheres with shKdm5b-1 (p < 0.001) or shKdm5b-2 (p < 0.001) transduction compared with shScr controls (Figure 2, f and g). We next quantified the effects of shKdm5b on aNSC cell cycle dynamics using 5-ethynyl-2´-deoxyuridine (EdU) incorporation, DNA staining, and flow cytometry (Figure 2, h–j). For these experiments, we used sh-Kdm5b-1 lentivirus. The shKdm5b-1 and shScr transduced aNSCs were exposed to EdU (10 μM) for 4 h in proliferation medium after 3–4 d of 5 μg/ml puromycin selection. Cell viability was measured by LIVE/DEAD stain and EdU incorporation compared in live-cell gated populations. When analyzed as a whole, this data set did not reveal statistical differences in EdU incorporation rates between shKdm5b-1– and shScr-transduced controls (unpublished data). However, EdU incorporation rates varied substantially between trials, and the SDs were large. When we plotted line plots of EdU incorporation rates in shKdm5b-1– compared with shScr–transduced cells from the same aNSC, the data revealed a consistent reduction in EdU incorporation rates in shKdm5b-1 compared with shScr controls within the same trial (Figure 2i). In contrast, line plots revealed no consistent difference in the percentage of cells estimated to be in G1 or G2/M phases of the cell cycle, using the Dean–Jett–Fox model to compute the percentage of events in each phase (Figure 2j). Combined these data show that shKdm5b lentiviral vectors deplete Kdm5b in aNSCs, resulting in reduced H3K4me3 levels and decreased neurosphere formation in culture, possibly via disruptions in cell cycle S phase.
Kdm5b knockdown results in increased transcription of Reln
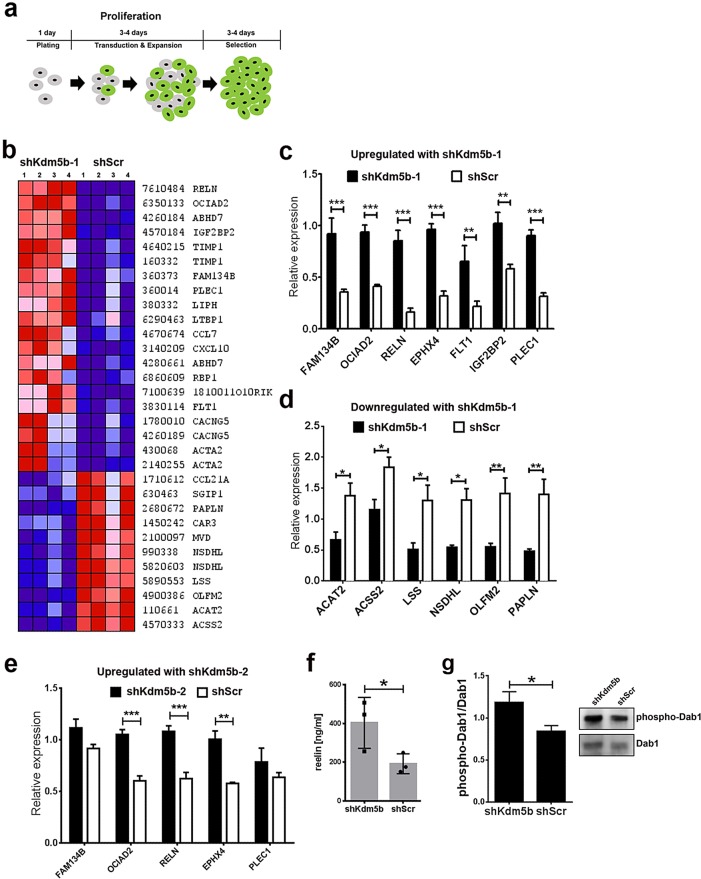
To determine the transcriptional effects of Kdm5b depletion in aNSCs, we conducted RNA expression analysis of shKdm5b-1– and shScr-transduced aNSCs. As before, shKdm5b-1or shScr lentivirus was added to proliferation medium 1 d after plating, the medium was replaced with proliferation medium containing 5 μg/ml puromycin after 3–4 d of exposure, and aNSCs were maintained in this medium for 3–4 d before mRNA harvest (Figure 3a). We profiled aNSC RNA samples using MouseRef-8 v2.0 Expression BeadChips on the HiScan Array Scanner to perform a genome-wide gene expression analysis of four independent aNSC culture samples (passage numbers 5–10) transduced with shKdm5b-1 or shScr lentivirus. Gene expression analysis of these eight samples resulted in the detection of 11,785 expressed genes across all samples. There was no significant difference in the number of expressed genes between shKdm5b-1– and shScr-transduced aNSC samples (p = 0.23). However, there were 163 differentially expressed genes (107 up-regulated and 56 down-regulated in shKdm5b-1 samples as compared with shScr controls) with expression level difference greater than twofold. Next we conducted gene annotation relationship analysis on the differentially expressed gene set determined by comparative marker selection. Gene annotation relationship analysis based on biological functions revealed significant enrichment (Table 1) for functions in steroid metabolism (9 genes, p < 0.0001), intracellular transport (26 genes, p = 0.001), response to immune stimuli (14 genes, p = 0.02), and regulation of transcription (10 genes, p = 0.03). Of note, the shKdm5b-1 transduced aNSCs were associated with a significant number of down-regulated transcripts for steroid metabolism activities (eight of nine genes). Of interest, Reln (reelin) was the highest differentially expressed gene in shKdm5b-1 versus shScr control samples (p = 0.03), but reelin did not have a relationship with any of the other differentially expressed genes in these functional subsets. Hierarchical clustering of the highest differentially expressed genes (p < 0.05, fold change ≥3, average signal >30) results in robust stratification of the samples based on shKdm5b-1 or shScr transduction (Figure 3b).
FIGURE 3:
shRNA-mediated knockdown of Kdm5b in culture results in large-scale gene expression changes in aNSCs. (a) Schematic representation of the lentiviral transduction and selection protocol used to enrich for shRNA-expressing aNSCs for analysis. (b) Hierarchical cluster heatmap of gene expression features identified by BeadChip genome-wide RNA expression screening of four independent aNSC culture samples (passage numbers 5–10) transduced with shKdm5b-1 or shScr lentivirus. Red and blue depict higher and lower gene expressions, respectively. Color intensity depicts magnitude of expression differences. (c, d) qRT-PCR validation of seven genes identified as up-regulated (c) and six down-regulated (d) in shKdm5b-1 transduced aNSCs by BeadChip expression screening. ***p < 0.001, **p < 0.01, and *p < 0.05, two-way ANOVA with Bonferroni’s multiple comparison test, n = 4. (e) qRT-PCR analysis of aNSCs transduced with shKdm5b-2 and shScr demonstrates five genes identified as up-regulated in shKdm5b-1 aNSCs by BeadChip expression screening are up-regulated by shKdm5b-2 in aNSCs . ***p < 0.001 and **p < 0.01, two-way ANOVA with Bonferroni’s multiple comparison test, n = 3. (f) ELISA confirms the shKdm5b-mediated increase in reelin expression in aNSCs. Secreted reelin protein concentrations are significantly higher in shKdm5b aNSC compared with control shScr aNSC–treated medium. *p < 0.05, one-tailed unpaired t test based on a priori sequence data of increased Reln expression in shKdm5b aNSCs, n = 3. (g) Elevated phospho-Dab1 levels in aNSCs transduced with shKdm5b compared with shScr lentivirus. *p < 0.05, two-tailed unpaired t test, n = 5.
TABLE 1:
Differentially expressed genes enriched for annotated relationships in aNSCs with basal versus depleted Kdm5b activity.
| Up-regulated in: | Gene symbol | Fold ratio | p |
|---|---|---|---|
| Function: steroid metabolism | |||
| shScr | Dhcr24 | 2.09 | <0.001 |
| shScr | Dhcr7 | 2.16 | <0.001 |
| shScr | Fdps | 2.27 | <0.001 |
| shScr | Ldlr | 2.83 | <0.001 |
| shScr | Lss | 3.10 | <0.001 |
| shScr | Mvd | 2.82 | <0.001 |
| shScr | Nsdhl | 2.99 | <0.001 |
| shKdm5b | Sqrl1 | 3.64 | <0.05 |
| shScr | Sqle | 2.00 | <0.001 |
| Function: regulation of transcription | |||
| shKdm5b | Cebpb | 2.14 | <0.05 |
| shScr | Ebf1 | 2.58 | <0.001 |
| shScr | Gtf2a1 | 2.74 | <0.001 |
| shKdm5b | Hmga1 | 2.06 | <0.05 |
| shScr | Irx2 | 3.76 | <0.001 |
| shScr | Mta1 | 2.35 | <0.05 |
| shKdm5b | Relb | 2.29 | <0.05 |
| shKdm5b | Tcea3 | 2.47 | <0.05 |
| Function: cellular transport | |||
| shKdm5b | Cacna2d1 | 2.57 | <0.05 |
| shKdm5b | Cacng5 | 2.70 | <0.05 |
| shKdm5b | Col4a6 | 3.35 | <0.05 |
| shKdm5b | Col6a1 | 22.27 | <0.05 |
| shKdm5b | Copz2 | 2.60 | <0.05 |
| shKdm5b | Ecm1 | 2.35 | <0.05 |
| shKdm5b | Gria1 | 2.08 | <0.05 |
| shKdm5b | Hmga1 | 2.06 | <0.05 |
| shScr | Kcnk13 | 2.84 | <0.001 |
| shKdm5b | Kdelr3 | 2.54 | <0.05 |
| shScr | Ldlr | 2.83 | <0.001 |
| shKdm5b | Rab15 | 2.46 | <0.05 |
| shKdm5b | Rab38 | 2.44 | <0.05 |
| shKdm5b | Rac3 | 2.06 | <0.05 |
| shKdm5b | Rasl11A | 2.31 | <0.05 |
| shKdm5b | Rbp1 | 2.68 | <0.05 |
| shKdm5b | Scara3 | 2.37 | <0.05 |
| shKdm5b | Slc16a6 | 2.47 | <0.05 |
| shScr | Slc2a6 | 2.12 | <0.001 |
| shScr | Slc39a12 | 2.19 | <0.001 |
| shKdm5b | Slc6a17 | 2.24 | <0.05 |
| shKdm5b | Sorl1 | 3.64 | <0.05 |
| shKdm5b | Syt4 | 8.91 | <0.05 |
| shScr | Wnk1 | 2.08 | <0.001 |
| Function: response to immune stimulus | |||
| shKdm5b | Anxa11 | 2.06 | <0.05 |
| shScr | C2 | 2.47 | <0.001 |
| shKdm5b | Ccl7 | 4.61 | <0.05 |
| shKdm5b | Cebpb | 2.14 | <0.05 |
| shKdm5b | Cxcl10 | 3.93 | <0.05 |
| shKdm5b | Gbp2 | 2.52 | <0.05 |
| shKdm5b | Hspb1 | 2.23 | <0.05 |
| shKdm5b | Hspb8 | 2.25 | <0.05 |
| shKdm5b | Psmb9 | 2.01 | <0.05 |
| shKdm5b | Scara3 | 2.37 | <0.05 |
| shKdm5b | Tap1 | 2.41 | <0.05 |
| shKdm5b | Tlr2 | 3.95 | <0.05 |
Subsets of genes are shown from the complete differentially expressed set that are significantly (p < 0.05) supported with a cluster of associations for an annotated relationship or function. Individual differentially expressed genes associated with an annotation are listed with the condition for up-regulated expression, the fold ratio of expression, and p for statistical test of sample expression values compared with the other condition.
To qualify our array data, we selected seven genes up-regulated and six genes down-regulated in aNSCs with shKdm5b-1 when compared with shScr controls. In the case of each of the 13 genes, the directionality of qPCR values between shKdm5b-1 and shScr sample sets matched the original BeadArray data, and two-way analysis of variance (ANOVA) with Bonferroni post tests revealed that all of these differences were significant (p < 0.05 to p < 0.001; Figure 3, c and d). In general, the qPCR data revealed a consistently greater difference between genes up-regulated with Kdm5b knockdown (p < 0.01–0.001) compared with genes down-regulated (p < 0.05–0.01). To independently validate our array data, we performed the qPCR analysis using the distinct shKdm5b-2 versus control shScr lentivirus. aNSCs were transduced with shKdm5b-2 lentivirus as depicted in Figure 3a for validation. We chose five of the seven genes qualified for up-regulation in shKdm5b-1 aNSCs. The data revealed higher mRNA expression levels for all five genes in shKdm5b-2 compared with shScr sample sets. Statistical comparisons revealed that three of the five genes (including Reln, p < 0.001) were significantly up-regulated in shKdm5b-2 compared with shScr-treated aNSCs (p < 0.01 to p < 0.001; Figure 3e). Taken together, the reduction in steroid biosynthetic gene expression, combined with increased expression of intracellular transport component genes, as well as the increased expression of Reln, suggests increased differentiation in shKdm5b-transduced aNSCs.
Kdm5b knockdown in cultured aNSCs results in increased extracellular reelin and increased phosphorylation of the downstream reelin signaling target Disabled-1
The most striking finding of our expression screen was the increase in Reln transcription with Kdm5b knockdown in aNSCs. Because reelin is a secreted glycoprotein, we developed a custom-made enzyme-linked immunosorbent assay (ELISA) to determine whether increased Reln transcription was associated with the increased presence of reelin in the medium of aNSCs transduced with shKdm5b. Two mouse monoclonal antibodies to reelin of differing immunoglobulin 1 (IgG1) and IgG2b isotypes (de Bergeyck et al., 1998; Jossin et al., 2007) were used to create a sandwich ELISA. Culture medium was harvested from shKdm5b- and shScr-transduced aNSCs following the same selection protocol used for the transcriptome analysis (Figure 3a). On the basis of the transcriptomics data, we predicted increased reelin in the media of aNSCs transduced with shKdm5b lentivirus. This was validated by our ELISA, which found 403.1 ± 76.0 ng/ml reelin in shKdm5b aNSC conditioned medium versus 191.8 ± 29.6 ng/ml in control shScr-conditioned samples (p < 0.05; Figure 3f). Combined these data demonstrate that shRNA-mediated knockdown of Kdm5b results in the up-regulation of reelin expression in aNSCs. Reelin binds to the apolipoprotein E receptor 2 and the very low density lipoprotein receptor to phosphorylate the intracellular adapter Disabled-1 (Dab1; D’Arcangelo et al., 1999; Hiesberger et al., 1999; Trommsdorff et al., 1999). We compared Western blot levels of phosphorylated Dab1 (pDab1) in aNSCs transduced with shKdm5b or shScr to determine the relative rates of reelin signaling. The data revealed a small but significant increase in the ratio of pDab1 to total Dab1 protein in aNSCs transduced with shKdm5b compared with shScr controls (p > 0.05; Figure 3g), consistent with increased reelin signaling in aNSC cultures transduced with shKdm5b lentivirus.
Kdm5b knockdown results in accelerated aNSC migration
Reelin has well-established roles in neuronal migration and is required for normal laminar positioning in the developing brain (for a recent review, see Sekine et al., 2014). In adult mice, reelin has been shown to stimulate the detachment and dispersal of aNSCs from the RMS (Simo et al., 2007; Courtes et al., 2011). We were therefore interested in Kdm5b-knockdown effects on aNSC migration. To investigate this, we used time-lapse videomicroscopy to measure the migration rates of aNSCs from neurospheres in culture. Adult NSCs were transduced with shKdm5b or shScr lentivirus in proliferation medium for 3–4 d, followed by 3–4 d of puromycin selection. Neurospheres were then seeded at low density in Matrigel-coated six-well plates in NeuroCult medium supplemented with mouse NSC Differentiation Supplement (referred to as differentiation medium; Figure 4a). The cultures were maintained in a microscope incubation chamber, and tGFP epifluorescence images of neurospheres were captured from preset coordinates after plating. In all, a total of 875 individual cells were traced for 17 h postplating to calculate cell velocity. Time-lapse images revealed a consistently greater cell movement velocity in aNSCs transduced with shKdm5b than with shScr control lentivirus (p < 0.0001, two-way ANOVA of shScr versus shKdm5b; Figure 4, b and c). To determine whether increased reelin activity in cultures transduced with shKdm5b lentivirus accounted for the increase in migration, we also recorded aNSC velocity in the presence of the reelin-blocking antibody CR-50 (Utsunomiya-Tate et al., 2000). CR-50 antibodies (20 μg/ml) were added to the proliferation medium for 24 h before neurospheres were transferred to differentiation and to the differentiation medium during time-lapse imaging (Figure 4a). CR-50 antiserum incubation significantly reduced the migration velocity of shKdm5b-transduced aNSCs (p < 0.0001, two-way ANOVA of shKdm5b vs. shKdm5b + CR-50), such that Kdm5b-transduced aNSCs were restored to migration rates comparable to those for shScr controls (Figure 4, b and c). Combined the results demonstrate that Kdm5b depletion increases aNSC migratory velocity by a reelin-dependent mechanism in culture.
FIGURE 4:
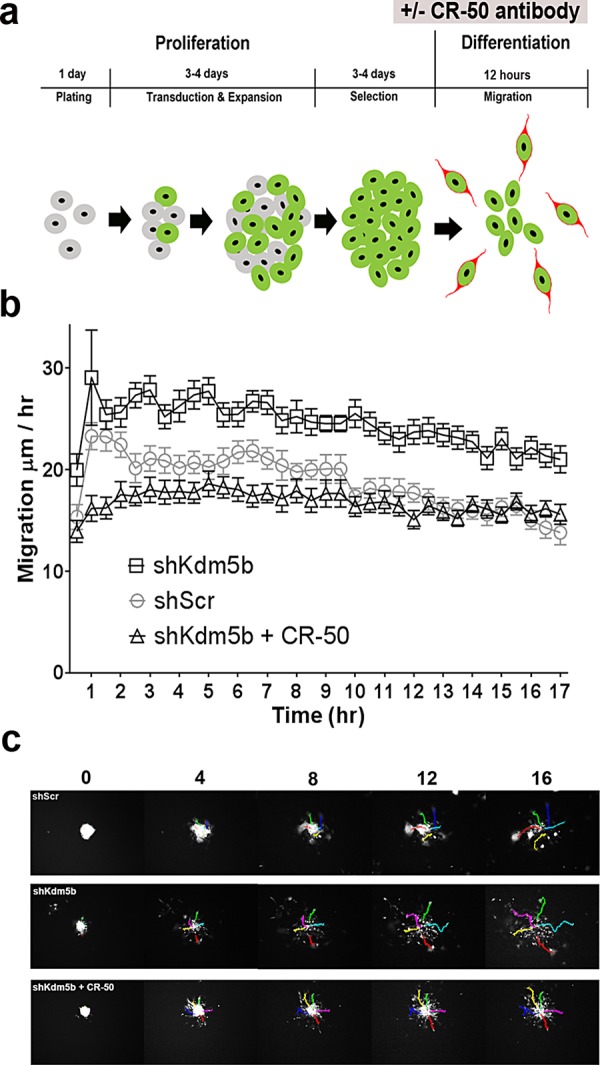
Kdm5b knockdown results in accelerated aNSC migration and increased neuronal differentiation from neurospheres in culture. (a) Schematic representation of the lentiviral transduction/selection and differentiation protocol used to measure aNSC dispersal, showing the timeline of CR-50 antibody incubation. (b) Time-lapse videomicroscopy reveals accelerated migration velocities of aNSCs transduced with shKdm5b compared with control shScr lentivirus (p < 0.0001), which is blocked by the addition of CR-50 reelin block to the culture medium (p < 0.0001, two-way ANOVA). (c) Representative images from time-lapse fluorescence microscopy of aNSCs transduced with shKdm5b–, shScr–, and shKdm5b–transduced aNSCs cultured in the presence of CR-50 antisera. See Supplemental Data QuickTime movies: shKdm5b, shScr, and shKdm5b + CR-50.
shKdm5b-mediated increases in Reln transcription are associated with increased H3K4me3 methylation at the Reln proximal promoter in aNSCs stimulated to differentiate
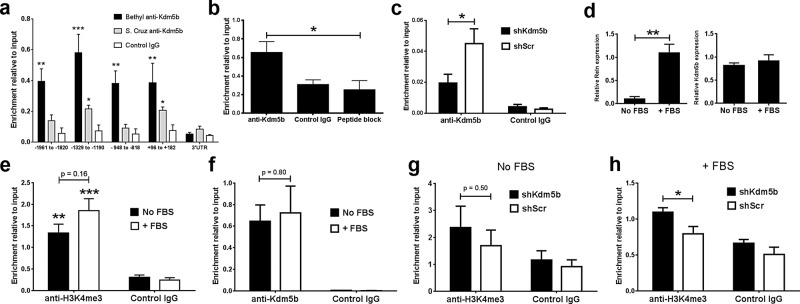
Bioinformatic searches of chromatin immunoprecipitation (ChIP) data indicate the Kdm5b substrate H3K4me3 is highly enriched at the proximal promoter and 5’ open reading regions of Reln (Supplemental Figure S1). We examined Kdm5b occupancy at the Reln loci by ChIP, using two rabbit polyclonal antibodies to Kdm5b. Adult NSCs were grown for ∼7 d in proliferation medium to generate neurospheres for chromatin harvest and ChIP. Real-time PCR analysis of the ChIP samples indicated enriched anti-Kdm5b binding at genomic regions proximal to the Reln transcriptional start site (TSS) but not at the 3’-untranslated region of the gene (Figure 5a). Of the two antisera tested for ChIP, the Bethyl polyclonal antisera resulted in the greater enrichment relative to input, and this antibody was validated in more detail. We measured the extent of nonspecific binding by the Bethyl antisera in our ChIP assay by preincubation with the immunizing peptide (peptide block). With primers directed to −1329 to −1199 to the Reln TSS, peptide block significantly decreased (p < 0.05) ChIP enrichment (Figure 5b). To further corroborate our ChIP assay, we performed ChIP on aNSC transduced with shKdm5b or shScr as controls. The data revealed a significant reduction in Kdm5b ChIP signal with shRNA-mediated knockdown of Kdm5b (Figure 5c). We conclude that our ChIP assay detects Kdm5b binding using Bethyl rabbit polyclonal anti-Kdm5b antisera and that the data reveal Kdm5b enrichment at proximal promoter regions of Reln.
FIGURE 5:
H3K4me3 is enriched at the Reln proximal promoter with Kdm5b depletion. (a) ChIP assay with two rabbit polyclonal antisera reveals Kdm5b enrichment at regions proximal to the Reln TSS. ***p < 0.001, **p < 0.01, and *p < 0.05, two-way ANOVA with Bonferroni’s multiple comparison tests vs. IgG control, n = 3. (b) Preincubation with the immunizing peptide (peptide block) significantly decreases Bethyl anti-Kdm5b ChIP at −1329 to −1190 to the Reln TSS. *p < 0.05, two-way ANOVA with Bonferroni’s multiple comparison test, n = 4. (c) Kdm5b knockdown significantly decreases Bethyl anti-Kdm5b ChIP at −1329 to −1190 to the Reln TSS. *p < 0.05, two-way ANOVA with Bonferroni’s multiple comparison test, n = 3. (d) FBS (5%) addition to induce neurosphere attachment and cell differentiation significantly increases Reln (**p < 0.01) but not Kdm5b transcription (two-tailed unpaired t test, both n = 3). (e) Anti-H3K4me3 ChIP reveals significant enrichment of H3K4me3 compared with IgG controls at −1329 to −1190 to the Reln TSS (***p < 0.001 and **p < 0.01, two-way ANOVA with Bonferroni’s multiple comparison test, n = 6), but H3K4me3 is not significantly increased by FBS-induced differentiation (p = 0.16). (f) FBS addition does not alter Bethyl anti-Kdm5b ChIP at −1329 to −1190 to the Reln TSS. p = 0.80, n = 3. (g) shKdm5b knockdown does not significantly increase H3K4me3 levels at −1329 to −1190 to the Reln TSS in aNSCs grown in proliferation medium. p = 0.50, n = 9. (h) In contrast, shKdm5b knockdown significantly increases H3K4me3 levels at this Reln locus after 5% FBS treatment to stimulate Reln transcription. *p < 0.05, two-way ANOVA with Bonferroni’s multiple comparison test, n = 6.
We next examined H3K4me3 levels at the −1329 to −1199 Reln locus using a rabbit polyclonal anti-H3K4me3 antiserum previously validated for ChIP (Kolasinska-Zwierz et al., 2009). Two culture conditions were used to prepare the aNSCs for ChIP: 1) aNSCs were cultured in proliferation medium for ∼7 d as before; 2) aNSCs were cultured in proliferation medium for ∼7 d, followed by the addition of 5% fetal bovine serum (FBS) for 48 h to induce neurosphere attachment and differentiation. Real-time PCR showed that FBS addition significantly increased aNSC Reln mRNA levels ∼10-fold from basal levels in proliferation medium (p < 0.01) without changing Kdm5b transcription (Figure 5d). ChIP assays of aNSCs showed significant H3K4me3 enrichment at the −1329 to −1199 Reln locus in aNSCs cultured in both conditions compared with IgG controls (p < 0.01 no FBS, p < 0.001 + FBS; Figure 5e). However, although there is a small increase in H3K4me3 enrichment with the addition of FBS, this increase is not significant (p = 0.16 no FBS vs. +FBS, n = 6; Figure 5e). Furthermore, ChIP showed that Kdm5b association at the −1329 to −1199 Reln locus is not significantly altered by the addition of FBS (Figure 5f). We next examined the effects of Kdm5b depletion on H3K4me3 levels at the −1329 to −1199 Reln locus in the two culture conditions. shRNA-mediated depletion of Kdm5b did not significantly increase H3K4me3 at the Reln promoter in aNSCs grown in proliferation medium alone (Figure 5g). In contrast, Kdm5b depletion significantly increased H3K4me3 levels at the proximal Reln promoter when differentiation was stimulated by the addition of 5% FBS to the medium (p < 0.05; Figure 5h). Taken together, the data indicate that shKdm5b-mediated (and FBS-treated) increases in Reln transcription occur independently of significant increases in H3K4me3 at the −1329 to −1199 Reln locus in aNSCs. The finding that increased H3K4me3 at this locus requires cotreatment with shKdm5b and FBS suggests a synergistic response of histone methylation to Kdm5b depletion and FBS-induced differentiation in aNSCs.
2-4(4-Methylphenyl)-1,2-benzisothiazol-3(2H)-one, a small-molecule inhibitor of Kdm5b, can mimic the shRNA-mediated increases in Reln transcription in aNSCs
Structure–function studies of Kdm5b catalytic activity have identified the small molecules 2,4-pyridinedicarboxylic acid (2,4-PDCA) and 2-4(4-methylphenyl)-1,2-benzisothiazol-3(2H)-one (PBIT) as inhibitors of H3K4me3 demethylation by Kdm5b (Kristensen et al., 2012; Sayegh et al., 2013). We tested the effects of these molecules on aNSCs grown in proliferation medium with 5% FBS to stimulate Reln transcription. We added 2,4-PDCA or PBIT with 5% FBS for 24 h before analysis. Dose-response data suggested that PBIT but not 2,4-PDCA administration increased Reln transcription (Figure 6a). Increasing PBIT concentration increased Reln mRNA levels, but the differences were not statistically significant due to toxicity effects at the highest, 100 μM dose. Accordingly, we replicated the analysis at a 50 μM dose. We added 50 μM PBIT with 5% FBS and analyzed aNSCs 24 and 48 h later. Real-time PCR revealed a significant increase in Reln transcription after 24- but not 48-h treatment compared with vehicle (dimethyl sulfoxide) controls (p < 0.05; Figure 6b). PBIT-mediated increases in Reln mRNA were not associated with significant changes in Kdm5b mRNA (p = 0.90, n = 3; Figure 6, c and d) or protein levels (p = 0.24, n = 3, Figure 6d), and, unlike shRNA depletion of Kdm5b, PBIT treatment did not result in changes in global H3K4me3 levels in aNSCs (Figure 6, c and d). Furthermore, H3K4me3 levels at the −1329 to −1199 Reln locus were not significantly different between aNSCs treated with PBIT and vehicle (Figure 6e). Combined the data demonstrate that treatment with PBIT, a small-molecule inhibitor of Kdm5b, up-regulates Reln transcription in aNSCs, but this effect does not result in significant global changes in H3K4me3 abundance or H3K4me3 enrichment at the −1329 to −1199 Reln locus.
FIGURE 6:
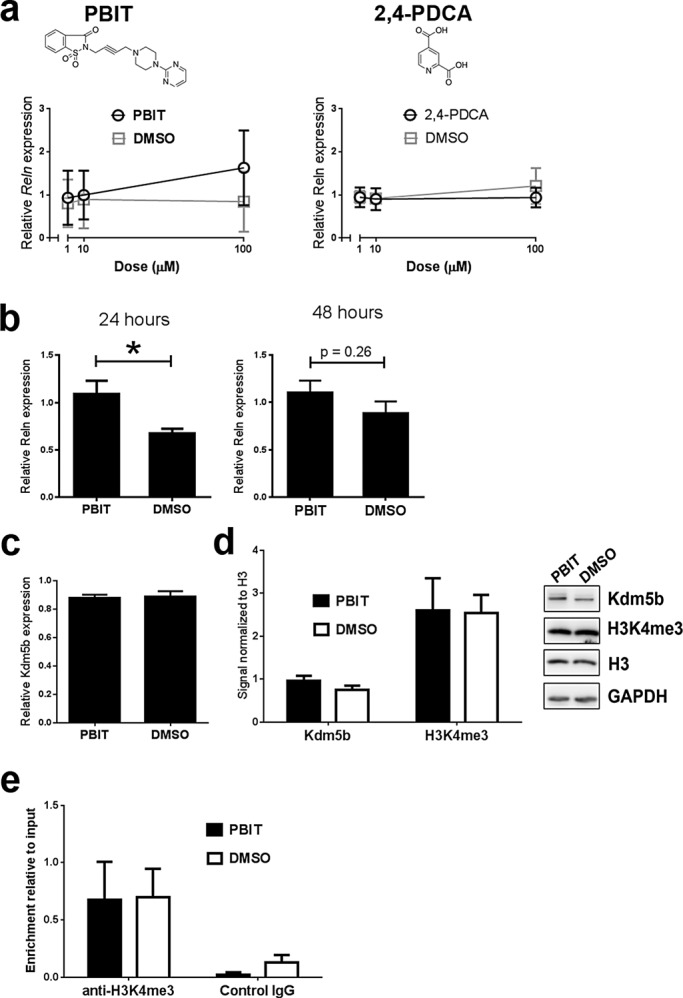
PBIT, a small-molecule inhibitor of Kdm5b, can mimic the shRNA-mediated increases in Reln transcription in aNSCs. (a) Dose-response curves suggest that PBIT but not 2,4-PCDA treatment affects Reln transcription in cultured aNSCs. (b) A 24-h but not 48-h treatment with 50 μM PBIT in culture significantly increases Reln transcription in aNSCs. *p < 0.05, unpaired t test, n = 6. (c) A 50 μM PBIT treatment does not inhibit (c) Kdmb transcription or (d) Kdm5b expression or H3K4me3 abundance (n = 3) in aNSCs. (e) ChIP reveals no significant change in H3k4me3 enrichment at −1329 to −1190 to the Reln TSS in aNSCs with 50 μM PBIT treatment (n = 3).
DISCUSSION
Our data demonstrate that Kdm5b depletion decreases proliferation and accelerates migration in cultured aNSCs. shRNA-mediated depletion of Kdm5b resulted in global increases in H3K4me3 levels in aNSCs, consistent with H3K4me3 demethylase activity for Kdm5b (Yamane et al., 2007). Gene expression screening revealed widespread transcriptional changes with Kdm5b depletion, notably the up-regulation of reelin (Reln), the inhibition of steroid biosynthetic pathway component genes, and the activation of genes with intracellular transport functions in cultured aNSCs. Kdm5b depletion resulted in accelerated aNSC migration in vitro, consistent with increased reelin activity, and treatment with reelin-blocking antibodies reversed this phenotype. ChIP assays demonstrated that Kdm5b binds to H3K4me3-rich regions immediately upstream of the Reln TSS, and shRNA depletion of Kdm5b leads to increased H3K4me3 levels at this locus in aNSCs stimulated to differentiate. Furthermore, treatment with the small-molecule inhibitor PBIT increased Reln transcription in aNSCs, mimicking the effects of shRNA-mediated Kdm5b knockdown. Combined the data suggest that Kdm5b negatively regulates neurogenesis and represses Reln in neural stem cells from the adult SVZ.
Kdm5b depletion decreases aNSC proliferation and induces widespread changes in gene transcription
shRNA-mediated knockdown of Kdm5b increased global H3K4me3 levels and decreased proliferation rates in aNSCs grown in proliferation culture conditions. Decreases in aNSC proliferation were associated with gene expression changes consistent with increased differentiation in Kdm5b-depleted aNSCs. Gene pathway analysis revealed inhibition of steroid biosynthetic pathway gene components and up-regulation of genes with functions in intracellular transport. Proliferating stem cell populations have high anabolic rates of metabolism to meet the demands of constant cell replication. As progenitors differentiate, cell metabolism shifts from glycolysis to the more ATP-efficient oxidative metabolism to supply the increased energy required for cellular homeostasis (Yanes et al., 2010). Thus down-regulation of steroid biosynthesis genes with Kdm5b depletion is consistent with a metabolic shift away from anabolism. Conversely, up-regulation of intracellular transport genes with shKdm5b knockdown has clear links with increased parcellation of cell structure and function in differentiating cells.
Of interest, increased aNSC proliferation with Kdm5b depletion is in contrast to the lack of proliferation effects in SVZ aNSCs after targeted deletion of the trxG H3K4 methyltransferase Mll1 (Lim et al., 2009). Using transgenic hGFAP-Cre mice to delete Mll1 in multipotent RGL progenitor cells in the E13.5 mouse, the authors found that SVZ cell proliferation rates were unaffected by Mll1 status. Instead, neurogenesis but not gliogenesis was severely impaired in Mll1-deficient SVZ aNSCs. These differences suggest divergent requirements for Kdm5b and Mll1 activity in establishing the chromatin environment for “proliferation” gene expression states in aNSCs. Microarray analysis showed that the expression of cell cycle regulatory genes is unaffected in Mll1-deficient aNSCs (Potts et al., 2014). A similar lack of expression changes in cell cycle–associated genes after Kdm5b depletion suggests that direct disruption of cell cycle regulation is not responsible for decreased proliferation. Because transcriptional control of genes central to lipid metabolism are necessary for precise cell cycle progression (Stumpf et al., 2013), the significant number of down-regulated steroid metabolism genes associated with Kdm5b depletion may explain the atypical cell cycle delay within S phase in our data.
Kdm5b depletion promotes aNSC migration and differentiation
Phenotypic effects of Kdm5b depletion in aNSCs persisted when these cells were induced to differentiate in culture. We used time-lapse videomicroscopy to track aNSC dispersal in culture to measure the effects of Kdm5b depletion on cell migration. The results revealed accelerated migration of aNSC dispersal from adherent neurospheres with Kdm5b knockdown. Gene expression, ELISA, Western blot, and antibody-blocking experiments provide evidence that shKdm5b-mediated increases in reelin expression and signaling in aNSCs accelerate the movement of these cells from neurospheres. Reelin is classically known for its role during embryonic development of the brain but has more recently been implicated in adult brain function (Levenson et al., 2008). Reelin expression switches from Cajal Retzius cells during development to widespread GABAergic interneuron populations in the adult brain, including the cortex and olfactory bulb (Ogawa et al., 1995; Pesold et al., 1998; Ramos-Moreno et al., 2006). In the olfactory bulb, reelin immunoreactive cells are present in the periglomerular and granule cell layers (Ramos-Moreno et al., 2006), sites of adult neurogenesis from SVZ progenitors (Ming and Song, 2011). In the cortex, reelin has been shown to stimulate the ectopic recruitment of SVZ-derived progenitors to perilesional sites in the adult mouse cortex in response to injury, and reelin treatment of adult forebrain explant cultures increases both migration velocity and the rate of detachment of neuroblasts from the SVZ (Courtes et al., 2011). Extracellular reelin is therefore present throughout the SVZ-RMS-olfactory bulb pathway, with the potential to regulate aNSC lineage progression from SVZ progenitors to olfactory bulb interneurons.
Kdm5b regulates Reln expression in aNSCs
Combined our gene expression and ChIP data identify Kdm5b as a regulator of Reln transcription in adult SVZ NSCs. Reln transcription is regulated at both genetic and epigenetic levels. Transcription factor mouse mutants have identified Ascl1, Pax6, NPAS1/3, and Tbr1 regulation of reelin expression in Cajal Retzius cells in the developing cortex (Hevner et al., 2001; Erbel-Sieler et al., 2004; Swanson et al., 2005; Dixit et al., 2011). Transcription factor–binding sites are located within a large CpG island at the Reln promoter, and loss of methylation is associated with increased reelin expression in cell and animal models (Chen et al., 2002, 2007; Tremolizzo et al., 2002, 2005; Dong et al., 2005; Kundakovic et al., 2009). Furthermore, studies with histone deacetylase (HDAC) inhibitors reveal a functional link between DNA methylation status and histone acetylation in Reln repression through the binding of MeCP2 to hypermethylated DNA and MeCP2 recruitment of HDACs (Kundakovic et al., 2007). Accordingly, treatment with HDAC inhibitors increases Reln transcription in neurons (Costa et al., 2003; Tremolizzo et al., 2005; Dong et al., 2007; Kundakovic et al., 2009). Through shRNA-mediated depletion and treatment with PBIT (a recently identified inhibitor of Kdm5b activity; Sayegh et al., 2013), we identified Kdm5b as a coregulator of Reln transcription in aNSCs. Of interest, although genome-wide screening revealed increased Reln transcription with Kdm5b depletion in proliferating aNSCs, increased H3K4me3 levels at the Reln promoter were detected by ChIP in differentiating (FBS-treated) but not proliferating aNSCs with Kdm5b depletion. Furthermore, FBS addition alone, although it increased Reln transcription ∼10-fold, did not lead to a significant enrichment of H3K4me3 levels at the Reln promoter in our ChIP assay (Figure 5e). Although these findings are predicated on the sensitivity of the antibody for ChIP and a short locus as readout, the data support a loose association between H3K4me3 levels and Reln transcription in aNSCs. The fact that Kdm5b depletion increased H3K4me3 levels at the Reln promoter in differentiating (FBS-treated) but not proliferating aNSCs indicates that Kdm5b demethylation of H3K4me3 is regulated by factors associated with aNSC differentiation state.
MATERIALS AND METHODS
Animals
We purchased 8- to 10-wk-old male and female (equal numbers) C57BL/6J mice from the Jackson Laboratory (Bar Harbor, ME) and housed them in the Uniformed Services University’s Center for Laboratory Animal Medicine before experimental use. Animals were handled in accordance with procedures approved by the Uniformed Services University of the Health Sciences Institutional Animal Care and Use Committee. All research complied with Department of Defense (DoD) regulations as published in DoD Directive 3216.1. The University’s Center for Laboratory Animal Medicine is a fully accredited institution with the Association for Assessment and Accreditation of Laboratory Animal Care.
Fluorescent in situ hybridization
Ten-week-old C57BL/6J mice were fixed by intracardiac perfusion of 4% paraformaldehyde in phosphate-buffered saline (PBS). The brain was cryoprotected in sucrose, and frozen sections of the forebrain were cut on a cryostat and postfixed in fresh fixative. FISH was performed on forebrain sections using an RNAscope probe for mouse Kdm5b according to the instructions from the manufacturer (Advanced Cell Diagnostics, Hayward, CA). After FISH, sections were processed for fluorescence immunohistochemistry with anti-GFAP (DAKO, Carpinteria, CA) antibodies. Tissue sections were 4′,6-diamidino-2-phenylindole (DAPI) counterstained and mounted under coverslips, and images were collected on a Zeiss 710 laser scanning confocal microscope.
Adult neural stem cell culture
aNSCs were harvested from the SVZ of adult C57BL/6J mice for culture. Mice were deeply anesthetized with vaporized isoflurane and decapitated, the whole brain was dissected out, and the forebrain was cut in serial, coronal sections 1 mm thick using a Sorvall tissue chopper. The SVZ was dissected out from coronal sections and dissociated to a single-cell suspension using the Neural Tissue Dissociation Kit (Mitenyi Biotech) according to the manufacturer’s protocol. Cells were seeded at a cell density of 1 × 105 cells/ml in mouse NeuroCult NSC Basal Medium supplemented with mouse NeuroCult NSC Proliferation Supplement, 20 ng/ml recombinant human (rh) EGF, 10 ng/ml rh bFGF, and 2 μg/ml heparin (all StemCell Technologies, Vancouver, Canada). Cells were cultured at 37°C and 5% CO2, and cultures were passaged every 5–7 d. Cells were dissociated to a single-cell suspension in 0.025% trypsin-EDTA (Life technologies) and reseeded in culture medium at 1 × 105 cells/ml. Cells were passaged a minimum of five times before experimental analysis to ensure a high enrichment of multipotent stem cells.
Lentiviral production and transduction
GIPZ lentiviral shRNA (shKdm5b) and GIPZ lentiviral nonsilencing shRNA control (shScr) plasmids were purchased from Open Biosystems/GE Dharmacon (Lafayette, CO). To generate replication-incompetent lentiviral particles, a Trans-Lentiviral Packaging plasmid mixture (GE Dharmacon) containing five packaging plasmids (pTLA1-Pak, pTLA1-Enz, pTLA1-Env, pTLA1-Rev, and pTLA1-TOFF) was cotransfected with shKdm5b and control shScr plasmids into HEK293T producer cells in normal serum medium (10% FBS, high-glucose DMEM, 2 mM l-glutamine) by calcium phosphate transfection. Medium was changed to reduced serum medium (5% FBS, high-glucose DMEM, 2 mM l-glutamine) 12–16 h after the transfection, and viral particles were harvested from the medium of transfected cells 48 h later. The medium was centrifuged at 1600 × g at 4°C for 10 min to remove cell debris and then filtered through a 0.45-μm filter. Viral particles were concentrated by ultracentrifugation at 25,000 × g for 2 h and resuspended in sterile DMEM. Viral titer was determined by analysis of GFP expression in HEK293T transduced cells. For lentiviral transduction, adult NSCs were plated at a density of 1 × 105 cells/ml and viral particles added to the culture medium at a concentration of (2–4) × 105 titers/ml (multiplicity of infection [MOI] of 2–4). The medium was replaced 24 h later and cells expanded for 2–4 d before selection with 5 μg/ml puromycin. Medium containing puromycin was replaced every 48 h. NIH3T3 cells were plated in DMEM with 10% FBS. Cells were seeded at (1–5) × 105 cells/ml in six-well culture plates. After 24 h, shKdm5b or shScr lentivirus was added to the medium at a concentration of (2–10) × 105 viral particles/ml (MOI, 2–10). NIH3T3 cells were expanded for 2 d before selection with 5 μg/ml puromycin before analysis.
Neurosphere counts
Neurospheres 50 μm in diameter were counted after shKdm5b/shScr lentiviral transduction, followed by 3–4 d of 5 μg/ml puromycin selection. The total number of neurospheres per 100 mm cell culture plate was estimated from 10 randomly selected 2.1025-mm2 counting areas using an optical graticule on a Zeiss inverted A1 microscope. Counting area selection and cell counting were performed by blinded investigators.
Flow cytometry, Click-iT EdU, and cell cycle DNA labeling
aNSCs were pulsed with 10 μM EdU in cell culture medium for 4 h at 37°C and 5% CO2 after shKdm5b/shScr lentiviral transduction and 3–4 d of 5 μg/ml puromycin selection. aNSCs were harvested and labeled with Alexa Fluor 555 dye by Click-iT (Life Technologies, Carlsbad, CA). For combined cell cycle analysis, cells were colabeled with CellCycle 488-red (7-AAD) and LIVE/DEAD Fixable Violet stains (Life Technologies). DNA content and proliferation status data acquisition was performed on a LRS II Flow Cytometer System at low flow rate with standard compensation and double discrimination parameters. Data were analyzed using FlowJo 9.0.1 (Ashland, OR). Briefly, events were first gated based on FSC and SSC parameters for the prominent cell population and then gated based on LIVE/DEAD signal. Live cells were analyzed for EdU incorporation for proliferation and also DNA content. Cell cycle analysis of the live population was performed in FlowJo using a Dean–Jett–Fox model to compute the percentage of events in each phase.
RNA isolation
Total RNA was extracted from aNSCs and stored at –80°C until use. Approximately 5 × 106 aNSCs were used for RNA extraction in TRIzol (Life Technologies), followed by chloroform extraction and precipitation with isopropyl alcohol (0.5 ml per 1.0 ml of TRIzol). Qiagen RNeasy Mini columns were used to isolate total RNA precipitated from aNSCs. Genomic DNA contaminant was removed by treating 0.5–10 μg of total RNA with 2 U of DNase for 30 min at 37°C (TURBO DNA-free; Life Technologies). RNA concentration and purity were evaluated by absorbance spectrophotometry (NanoDrop 1000C; ThermoFisher Scientific, Waltham, MA).
Genome-wide gene expression microarray analysis
Genome-wide gene expression profiling was conducted using MouseRef-8 v2.0 Expression BeadChips (Illumina, San Diego, CA), which contain ∼25,600 well-annotated RefSeq transcript features. Total RNA integrity was determined using the Experion RNA HighSens Analysis Chip (Bio-Rad, Hercules, CA) on the Experion Automated Electrophoresis System. RNA quality indicator values were >9.0 for all samples. Total RNA was labeled using TotalPrep RNA Labeling Kit (Ambion) before incubation on MouseRef-8 v2.0 BeadChips and imaging by the HiScan Array Scanner (Illumina, San Diego, CA) at the University of Chicago Genomics Facility (Chicago, IL). Four experimental replicates of aNSCs transduced with shKdm5b or control shScr lentiviral vector were simultaneously profiled on a single MouseRef-8 BeadChip. Raw data produced from HiScan imaging were outputted with the GenomeStudio software package and GEX module (Illumina).
Gene expression data analysis
Raw data were preprocessed by offset background correction and quantile normalization. Non–accurately detected BeadArray features were removed from preprocessed data for features with a p cutoff of >0.1 in 75% (six of eight) samples. Differentially expressed BeadArray features of transcript abundance values for accurately detected transcripts between shKdm5b and shScr control samples were determined using the ComparativeMarkerSelection module found in the GenePattern 3.2.3 software platform (Broad Institute, MIT, Cambridge, MA). Differentially expressed BeadArray features were filtered for features with changes greater than twofold with p < 0.05 between groups. Differentially expressed features were hierarchically clustered using the HierarchicalCluster module in GenePattern and analyzed for network connectivity using GATHER (Chang and Nevins, 2006) and Ingenuity Pathway Analysis (Qiagen, Valencia, CA).
Real-time quantitative reverse transcription PCR
First-strand cDNA was synthesized from 1 μg of total RNA using random primer/oligo(dT) primer according to the manufacturer’s instructions (ABI, Carlsbad, CA). Synthesized cDNA was diluted to a final concentration of 10 ng/μl for qPCR using SYBR Green Master Mix (ABI, ThermoFisher Scientific) on a thermal cycler C1000 with CFX384 real-time system. Optimum primers were designed using the National Center for Biotechnology Information’s Primer BLAST (www.ncbi.nlm.nih.gov/tools/primer-blast/). Primer sequence pairs used are Reln, CCA GAA AAC ATG GTC CGG TGC and GGC ACT GCT GCC CAA TGT AAA; Fam134b, AGC CTC ACA GTT GCT GCC AA and GGC CGG TCA AGA TCG TCA GAA; Ociad2, TGG TCC TGA GCA TAA CAG GCA C and TGG TTG TGA CCC TGC CTT CT; Ephx4, CGC ATC AAG GAC TCC GGG TTA and AGC AGC ATG AGC GGT TTG C; Flt1, CCA AAG AAA GGT ATG ACT TCT GCA C and CGT CAT GCG CTG AGA GAT GC; Igf2bp2, CGG GAA AAC CGT GAA CGA GC and TCT CGT CTG GCG TTT GGT CA; Plec1, CTC TGC GCT CGG AGT TGG AA and TGA TGG TCT TGA GTT TCT CCA GGT; Acat2, AGC GAC AAG ATG AAT GCG GG and GGC ACC ATT GAA GGA GCC TAT; Acss2, CAA TGA GCC AGG GGA GAC CA and TCA CCC TTC TGA ATG CCC TGT T; Lss, GGG CCC TGA ATG GAG TAA CCT and TAA TCA GGA GAC CTG GCA AGA GG; Nsdhl, TCC CCT CCG CCG TAC AGT AA and ATG AGT TTC TGA ACT CCG GCC TC; Olfm2, CCT GAT GGC AAG TGC GTC TG and TGG GAC ACA TTC TGG ACC TTC TC; and Papln, GGG TGA TGG CTC ATC CTG CT and GTA CTC GCC ACG GAT GCT CT.
Primer specificity was confirmed by verifying that a single PCR product had been generated by using ultraviolet gel electrophoresis, as well as by confirming that the melting temperature of the product had a single value on dissociation plots. Gene of interest (GOI) Ct values were normalized to internal control Actb and/or Hprt1 to calculate ∆Ct values. Relative expression levels were calculated using the Bio-Rad CFX Manager 3.1 software. GOI fold change (F) was estimated using the ∆∆Ct method: F = 2−∆∆Ct, where ∆∆Ct = GOI ∆Ct shScr control − GOI ∆Ct shKdm5b. Ct values were measured in triplicate and recorded if the SD < 0.3. All values represent a minimum of three biological replicates.
Western blot
Whole-cell protein extracts were obtained from aNSCs using NE-PER reagents (Thermo Scientific) according to the manufacturer’s instructions. Protein fractions were stored at −20ºC in 1× Halt Protease Inhibitor Cocktail (ThermoFisher Scientific) until use. For Dab1 only, 1× Halt Phosphatase Inhibitor Cocktail was added to fresh extract, and 50 μg of total protein was immunoprecipitated overnight at 4°C with 2 μg of rabbit polyclonal anti-Dab1 antibodies (Sigma-Aldrich, St. Louis, MO) conjugated to 25 μl of Dynabeads Protein G. Immunoprecipitation samples were washed and stored in SDS loading dye until use at −20ºC in 1× protease and phosphatase inhibitor cocktails until use. All samples were separated by SDS–PAGE and electrotransferred to polyvinylidene fluoride membranes. Except for immunoprecipitates, 20 μg of protein was loaded per lane. All washes and antiserum incubations were performed in 5% skim milk or 5% bovine serum albumin in Tris-buffered saline with 0.1% Tween-20. Blots were incubated with primary antiserum overnight at 4ºC on a rocking platform. Primary antisera and dilutions used were 1:1000 rabbit monoclonal anti-Dab1 (Millipore, Billerica, MA); 1:2000 rabbit polyclonal anti-Kdm5b (Bethyl Laboratories, Montgomery, TX); 1:1000 rabbit polyclonal anti-H3k4me3 (Active Motif, Carlsbad, CA); 1:1000 rabbit polyclonal anti-H3 (Cell Signaling Technology, Danvers, MA); 1:1000 rabbit polyclonal anti–trimethyl-histone H3 (Lys-4; Cell Signaling); and 1:800 rabbit polyclonal anti–phosphotyrosine 198 Dab1 (Millipore); 1:1000 mouse monoclonal anti-reelin (Millipore). The next day, blots were washed three times and incubated with a 1:5000 dilution of peroxidase-conjugated goat anti-mouse or anti-rabbit IgG secondary antisera (Cell Signaling Technologies) for 1 h at room temperature. Blots were developed in Immobilon chemiluminescent ECL substrate (Millipore) for 5 min at room temperature, and the fluorescence signal was captured using a Fujifilm LAS-3000 phosphoimager (Fujifilm, Valhalla, NY). Immunosignal intensities (protein density) were measured using Fujifilm Multi Gauge software and input normalized to Actb or H3 signal. Images were optimized for brightness and contrast for publication.
Enzyme-linked immunosorbent assay for reelin measurement
Mouse reelin protein concentration in media samples was measured using indirect ELISA methodology. High-binding wells of EIA/RIA 96-well plates (Costar; Corning, Glendale, AZ) were coated with monoclonal anti-mouse reelin (E4) IgG1 capture antibody (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA) before blocking nonspecific sites with 1% BSA in PBS plus 0.1% Tween. Standards with recombinant Reelin (R&D Systems, Minneapolis, MN) or unknown samples in 100 μl of medium were added to wells overnight at 4ºC. Incubation with a monoclonal anti-mouse reelin (R4b) IgG2 detection antibody (Developmental Studies Hybridoma Bank) was performed for 2 h at room temperature before the addition of anti-mouse IgG2b biotin-conjugated purified monoclonal antibody (Mabtech, Cincinnati, OH) and horseradish peroxidase–conjugated streptavidin (ThermoFisher Scientific). TMB ELISA Substrate (BD Biosciences) was used to develop wells before spectrophotometric measurement at 650 nm. All washes were conducted using PBS plus 0.1% Tween. Values were background subtracted from no-capture-antibody wells before fitting values on the within-plate standard curve. Values are averaged from technical triplicates.
In vitro neurosphere cell dispersal assay
After shKdm5b or shScr lentiviral transduction and 3–4 d of selection in 5 μg/ml puromycin, neurospheres were manually transferred at low density to six-well culture plates coated with Matrigel basement membrane matrix (BD Biosciences, San Jose, CA) and cultured in NeuroCult NSC Basal Medium supplemented with mouse NeuroCult NSC Differentiation Supplement (StemCell Technologies). Turbo GFP fluorescence images were recorded using a Leica AF6000 inverted microscope fitted with an incubation chamber and a holder for 24-well culture plates. Cultures were maintained with 70% relative humidity at 37°C and 5% CO2, and time-lapse serial image stacks were acquired through the full thickness of the Matrigel culture using a 10× objective. Neurospheres were first recorded at ∼+1 h after plating and every 0.5 h until +17 h after plating. Cells migrating from the periphery of the neurosphere (and distinguishable from the neurosphere) were identified and tracked through the image series by their x, y, and z positional coordinates. The distance moved by individual cells and their migration velocity were measured using the MTrackJ Java Plugin (Meijering et al., 2012) with FIJI image processing package. For reelin-blocking experiments, monoclonal CR-50 anti-reelin antibodies (Medical & Biological Laboratories, Des Plaines, IL) were added to Stem Cell proliferation medium containing shKdm5b and shScr lentiviral–transduced neurospheres at a concentration of 20 μg/ml IgG. Neurospheres were cultured in proliferation medium plus CR-50 antiserum for 24 h before manual transfer to six-well culture plates coated with Matrigel basement membrane matrix (BD Biosciences). Transferred neurospheres were cultured in NeuroCult NSC Basal Medium supplemented with mouse NeuroCult NSC Differentiation Supplement in the presence of 20 μg/ml IgG of CR-50 antiserum during time-lapse video fluorescence imaging. A total of 875 cells were tracked using this approach from three replicate experiments.
Chromatin immunoprecipitation
ShKdm5b/shScr-transduced aNSCs were treated for 10 min with 1% formaldehyde in PBS at room temperature. The cells were lysed in cell lysis buffer (5 mM 1,4-piperazinediethanesulfonic acid, pH 8.0, 85 mM KCl, 0.5% NP-40, and 1× protease inhibitors [Pierce, ThermoFisher Scientific]) and the crude cell lysate transferred to a sonication buffer (1× PBS, 1% NP-40, 0.5% sodium deoxycholate, and 1× protease inhibitors). The lysate was sonicated under conditions yielding fragments ranging from 500 to 1000 base pairs. Samples were subsequently precleared at 4°C with recombinant protein G agarose beads (Roche, Indianapolis, IN). Precleared lysate (100 μl) diluted in immunoprecipitation buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris⋅HCl, pH 8.0, 167 mM NaCl) was used for a 16-h overnight immunoprecipitation. Lysate was immunoprecipitated with 5 μg of polyclonal rabbit anti-Kdm5b (Bethyl Laboratories), 5 μg of polyclonal rabbit anti-H3K4me3 (Active Motif, Carlsbad, CA), or 5 μg of ChIP rabbit IgG control (Abcam, Cambridge, MA) at 4°C. Immunoprecipitated complexes were collected by incubation with recombinant protein G agarose beads (Roche, Indianapolis, IN) for 1–4 h at 4°C. After washing and elution, formaldehyde cross-linking was reversed with a 16-h overnight incubation at 65°C. Samples were purified using PCR purification kit columns (Qiagen, Valencia, CA) and used as a template for qPCR.
Enrichment relative to input was calculated from qPCR values from ChIP templates as follows. Standard curves were generated from serial 10-fold dilutions of 10% of input DNA and plotted on a log 10 scale to obtain the linear relationship between Ct value and template concentration. Using this equation, we normalized qPCR values to input, compared enrichment with IgG control samples, and statistically determined differences.
Reln primers used for ChIP were TTGGCCAGAGCAGCTGTTTA and TGGCTAGACCTCACATTGGC (−1961 to −1820 5’ of the Reln precursor start site); CAGAGACGGGAGAACAGAGC and TGGATCTTGCCTTCTGACGC (−1329 to −1190 5’ of the Reln precursor start site; AAAGGGAGATTGGGTGACGG and TCCGAGGAATCCCCTAGCAA (−948 to −818 5’ of the Reln precursor start site); TGCAGGGAAATGAGCACCTC and GCGCTTCTCGCCTTTCTTTT (+96 to +182 3’ of the Reln precursor start site); and TGC CAT TTC GTG TCT GTG GT and GTG TTG GCT CCA CCT CAT CA (+780 to + 886 3’ of the Reln precursor stop signal).
Data and statistics
Histograms are plotted as mean ± SEM error bar. Line graphs are plotted as mean ± SEM error bar above/below.
Unpaired t tests were used to compare the means of unmatched groups. All t tests were two tailed unless otherwise stated. Multiple comparisons were analyzed using one-way ANOVA with Tukey’s multiple comparisons test or two-way analysis of variance (ANOVA) with post hoc Bonferroni’s post test unless otherwise stated. p < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We are grateful for assistance with time-lapse video microscopy from Joshua Udovic (Zeiss), members of the laboratory of Sharon Juliano, and Dennis McDaniel of the Bio-instrumentation Center at the Uniformed Services University of the Health Sciences. This work was supported by a grant from the Center for Neuroscience and Regenerative Medicine.
Abbreviations used:
- aNSC
adult neural stem cell
- CC
corpus callosum
- ChIP
chromatin immunoprecipitation
- DG
dentate gyrus
- H3
histone 3
- H3K4me3
tri-methylated lysine 4 of histone H3
- HDM
histone demethylase
- HDMi
histone demethylase inhibitor
- PBIT
2-4(4-methylphenyl)-1,2-benzisothiazol-3(2H)-one
- 2,4-PDCA
2,4-pyridinedicarboxylic acid
- RGL
radial glial-like
- RMS
rostral migratory stream
- SVZ
subventricular zone.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-07-0513) on January 6, 2016.
REFERENCES
- Albert M, Schmitz SU, Kooistra SM, Malatesta M, Morales Torres C, Rekling JC, Johansen JV, Abarrategui I, Helin K. The histone demethylase Jarid1b ensures faithful mouse development by protecting developmental genes from aberrant H3K4me3. PLoS Genet. 2013;9:e1003461. doi: 10.1371/journal.pgen.1003461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Buylla A, Garcia-Verdugo JM. Neurogenesis in adult subventricular zone. J Neurosci. 2002;22:629–634. doi: 10.1523/JNEUROSCI.22-03-00629.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Berg DA, Belnoue L, Song H, Simon A. Neurotransmitter-mediated control of neurogenesis in the adult vertebrate brain. Development. 2013;140:2548–2561. doi: 10.1242/dev.088005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaguidi MA, Wheeler MA, Shapiro JS, Stadel RP, Sun GJ, Ming GL, Song H. In vivo clonal analysis reveals self-renewing and multipotent adult neural stem cell characteristics. Cell. 2011;145:1142–1155. doi: 10.1016/j.cell.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JT, Nevins JR. GATHER: a systems approach to interpreting genomic signatures. Bioinformatics. 2006;22:2926–2933. doi: 10.1093/bioinformatics/btl483. [DOI] [PubMed] [Google Scholar]
- Chen Y, Kundakovic M, Agis-Balboa RC, Pinna G, Grayson DR. Induction of the reelin promoter by retinoic acid is mediated by Sp1. J Neurochem. 2007;103:650–665. doi: 10.1111/j.1471-4159.2007.04797.x. [DOI] [PubMed] [Google Scholar]
- Chen Y, Sharma RP, Costa RH, Costa E, Grayson DR. On the epigenetic regulation of the human reelin promoter. Nucleic Acids Res. 2002;30:2930–2939. doi: 10.1093/nar/gkf401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa E, Grayson DR, Mitchell CP, Tremolizzo L, Veldic M, Guidotti A. GABAergic cortical neuron chromatin as a putative target to treat schizophrenia vulnerability. Crit Rev Neurobiol. 2003;15:121–142. doi: 10.1615/critrevneurobiol.v15.i2.20. [DOI] [PubMed] [Google Scholar]
- Courtes S, Vernerey J, Pujadas L, Magalon K, Cremer H, Soriano E, Durbec P, Cayre M. Reelin controls progenitor cell migration in the healthy and pathological adult mouse brain. PLoS One. 2011;6:e20430. doi: 10.1371/journal.pone.0020430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Arcangelo G, Homayouni R, Keshvara L, Rice DS, Sheldon M, Curran T. Reelin is a ligand for lipoprotein receptors. Neuron. 1999;24:471–479. doi: 10.1016/s0896-6273(00)80860-0. [DOI] [PubMed] [Google Scholar]
- de Bergeyck V, Naerhuyzen B, Goffinet AM, Lambert de Rouvroit C. A panel of monoclonal antibodies against reelin, the extracellular matrix protein defective in reeler mutant mice. J Neurosci Methods. 1998;82:17–24. doi: 10.1016/s0165-0270(98)00024-7. [DOI] [PubMed] [Google Scholar]
- Dey BK, Stalker L, Schnerch A, Bhatia M, Taylor-Papidimitriou J, Wynder C. The histone demethylase KDM5b/JARID1b plays a role in cell fate decisions by blocking terminal differentiation. Mol Cell Biol. 2008;28:5312–5327. doi: 10.1128/MCB.00128-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R, Zimmer C, Waclaw RR, Mattar P, Shaker T, Kovach C, Logan C, Campbell K, Guillemot F, Schuurmans C. Ascl1 participates in Cajal-Retzius cell development in the neocortex. Cereb Cortex. 2011;21:2599–2611. doi: 10.1093/cercor/bhr046. [DOI] [PubMed] [Google Scholar]
- Doetsch F, Caille I, Lim DA, Garcia-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- Doetsch F, Garcia-Verdugo JM, Alvarez-Buylla A. Cellular composition and three-dimensional organization of the subventricular germinal zone in the adult mammalian brain. J Neurosci. 1997;17:5046–5061. doi: 10.1523/JNEUROSCI.17-13-05046.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E, Agis-Balboa RC, Simonini MV, Grayson DR, Costa E, Guidotti A. Reelin and glutamic acid decarboxylase67 promoter remodeling in an epigenetic methionine-induced mouse model of schizophrenia. Proc Natl Acad Sci USA. 2005;102:12578–12583. doi: 10.1073/pnas.0505394102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E, Guidotti A, Grayson DR, Costa E. Histone hyperacetylation induces demethylation of reelin and 67-kDa glutamic acid decarboxylase promoters. Proc Natl Acad Sci USA. 2007;104:4676–4681. doi: 10.1073/pnas.0700529104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbel-Sieler C, Dudley C, Zhou Y, Wu X, Estill SJ, Han T, Diaz-Arrastia R, Brunskill EW, Potter SS, McKnight SL. Behavioral and regulatory abnormalities in mice deficient in the NPAS1 and NPAS3 transcription factors. Proc Natl Acad Sci USA. 2004;101:13648–13653. doi: 10.1073/pnas.0405310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales-Roybal G, Lim DA. Chromatin-based epigenetics of adult subventricular zone neural stem cells. Front Genet. 2013;4:194. doi: 10.3389/fgene.2013.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevner RF, Shi L, Justice N, Hsueh Y, Sheng M, Smiga S, Bulfone A, Goffinet AM, Campagnoni AT, Rubenstein JL. Tbr1 regulates differentiation of the preplate and layer 6. Neuron. 2001;29:353–366. doi: 10.1016/s0896-6273(01)00211-2. [DOI] [PubMed] [Google Scholar]
- Hiesberger T, Trommsdorff M, Howell BW, Goffinet A, Mumby MC, Cooper JA, Herz J. Direct binding of Reelin to VLDL receptor and ApoE receptor 2 induces tyrosine phosphorylation of disabled-1 and modulates tau phosphorylation. Neuron. 1999;24:481–489. doi: 10.1016/s0896-6273(00)80861-2. [DOI] [PubMed] [Google Scholar]
- Hsieh J. Orchestrating transcriptional control of adult neurogenesis. Genes Dev. 2012;26:1010–1021. doi: 10.1101/gad.187336.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WW, Salinas RD, Siu JJ, Kelley KW, Delgado RN, Paredes MF, Alvarez-Buylla A, Oldham MC, Lim DA. Distinct and separable roles for EZH2 in neurogenic astroglia. Elife. 2014;3:e02439. doi: 10.7554/eLife.02439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Garcia-Verdugo JM, Gil-Perotin S, Roy M, Quinones-Hinojosa A, VandenBerg S, Alvarez-Buylla A. PDGFR alpha-positive B cells are neural stem cells in the adult SVZ that form glioma-like growths in response to increased PDGF signaling. Neuron. 2006;51:187–199. doi: 10.1016/j.neuron.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Jossin Y, Gui L, Goffinet AM. Processing of Reelin by embryonic neurons is important for function in tissue but not in dissociated cultured neurons. J Neurosci. 2007;27:4243–4252. doi: 10.1523/JNEUROSCI.0023-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen LH, Nielsen AL, Helgstrand C, Lees M, Cloos P, Kastrup JS, Helin K, Olsen L, Gajhede M. Studies of H3K4me3 demethylation by KDM5B/Jarid1B/PLU1 reveals strong substrate recognition in vitro and identifies 2,4-pyridine-dicarboxylic acid as an in vitro and in cell inhibitor. FEBS J. 2012;279:1905–1914. doi: 10.1111/j.1742-4658.2012.08567.x. [DOI] [PubMed] [Google Scholar]
- Kundakovic M, Chen Y, Costa E, Grayson DR. DNA methyltransferase inhibitors coordinately induce expression of the human reelin and glutamic acid decarboxylase 67 genes. Mol Pharmacol. 2007;71:644–653. doi: 10.1124/mol.106.030635. [DOI] [PubMed] [Google Scholar]
- Kundakovic M, Chen Y, Guidotti A, Grayson DR. The reelin and GAD67 promoters are activated by epigenetic drugs that facilitate the disruption of local repressor complexes. Mol Pharmacol. 2009;75:342–354. doi: 10.1124/mol.108.051763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson JM, Qiu S, Weeber EJ. The role of reelin in adult synaptic function and the genetic and epigenetic regulation of the reelin gene. Biochim Biophys Acta. 2008;1779:422–431. doi: 10.1016/j.bbagrm.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Lim DA, Alvarez-Buylla A. Adult neural stem cells stake their ground. Trends Neurosci. 2014;37:563–571. doi: 10.1016/j.tins.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DA, Huang YC, Swigut T, Mirick AL, Garcia-Verdugo JM, Wysocka J, Ernst P, Alvarez-Buylla A. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature. 2009;458:529–533. doi: 10.1038/nature07726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois C, Garcia-Verdugo JM, Alvarez-Buylla A. Chain migration of neuronal precursors. Science. 1996;271:978–981. doi: 10.1126/science.271.5251.978. [DOI] [PubMed] [Google Scholar]
- Marei HE, Ahmed AE. Transcription factors expressed in embryonic and adult olfactory bulb neural stem cells reveal distinct proliferation, differentiation and epigenetic control. Genomics. 2013;101:12–19. doi: 10.1016/j.ygeno.2012.09.006. [DOI] [PubMed] [Google Scholar]
- Meijering E, Dzyubachyk O, Smal I. Methods for cell and particle tracking. Methods Enzymol. 2012;504:183–200. doi: 10.1016/B978-0-12-391857-4.00009-4. [DOI] [PubMed] [Google Scholar]
- Ming GL, Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron. 2011;70:687–702. doi: 10.1016/j.neuron.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Miyata T, Nakajima K, Yagyu K, Seike M, Ikenaka K, Yamamoto H, Mikoshiba K. The reeler gene-associated antigen on Cajal-Retzius neurons is a crucial molecule for laminar organization of cortical neurons. Neuron. 1995;14:899–912. doi: 10.1016/0896-6273(95)90329-1. [DOI] [PubMed] [Google Scholar]
- Park DH, Hong SJ, Salinas RD, Liu SJ, Sun SW, Sgualdino J, Testa G, Matzuk MM, Iwamori N, Lim DA. Activation of neuronal gene expression by the JMJD3 demethylase is required for postnatal and adult brain neurogenesis. Cell Rep. 2014;8:1290–1299. doi: 10.1016/j.celrep.2014.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesold C, Impagnatiello F, Pisu MG, Uzunov DP, Costa E, Guidotti A, Caruncho HJ. Reelin is preferentially expressed in neurons synthesizing gamma-aminobutyric acid in cortex and hippocampus of adult rats. Proc Natl Acad Sci USA. 1998;95:3221–3226. doi: 10.1073/pnas.95.6.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts MB, Siu JJ, Price JD, Salinas RD, Cho MJ, Ramos AD, Hahn J, Margeta M, Oldham MC, Lim DA. Analysis of Mll1 deficiency identifies neurogenic transcriptional modules and Brn4 as a factor for direct astrocyte-to-neuron reprogramming. Neurosurgery. 2014;75:472–482. doi: 10.1227/NEU.0000000000000452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Moreno T, Galazo MJ, Porrero C, Martinez-Cerdeno V, Clasca F. Extracellular matrix molecules and synaptic plasticity: immunomapping of intracellular and secreted Reelin in the adult rat brain. Eur J Neurosci. 2006;23:401–422. doi: 10.1111/j.1460-9568.2005.04567.x. [DOI] [PubMed] [Google Scholar]
- Sayegh J, Cao J, Zou MR, Morales A, Blair LP, Norcia M, Hoyer D, Tackett AJ, Merkel JS, Yan Q. Identification of small molecule inhibitors of Jumonji AT-rich interactive domain 1B (JARID1B) histone demethylase by a sensitive high throughput screen. J Biol Chem. 2013;288:9408–9417. doi: 10.1074/jbc.M112.419861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz SU, Albert M, Malatesta M, Morey L, Johansen JV, Bak M, Tommerup N, Abarrategui I, Helin K. Jarid1b targets genes regulating development and is involved in neural differentiation. EMBO J. 2011;30:4586–4600. doi: 10.1038/emboj.2011.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine K, Kubo KI, Nakajima K. How does Reelin control neuronal migration and layer formation in the developing mammalian neocortex. Neurosci Res. 2014;86C:50–58. doi: 10.1016/j.neures.2014.06.004. [DOI] [PubMed] [Google Scholar]
- Simo S, Pujadas L, Segura MF, La Torre A, Del Rio JA, Urena JM, Comella JX, Soriano E. Reelin induces the detachment of postnatal subventricular zone cells and the expression of the Egr-1 through Erk1/2 activation. Cereb Cortex. 2007;17:294–303. doi: 10.1093/cercor/bhj147. [DOI] [PubMed] [Google Scholar]
- Stumpf CR, Moreno MV, Olshen AB, Taylor BS, Ruggero D. The translational landscape of the mammalian cell cycle. Mol Cell. 2013;52:574–582. doi: 10.1016/j.molcel.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson DJ, Tong Y, Goldowitz D. Disruption of cerebellar granule cell development in the Pax6 mutant, Sey mouse. Brain Res Dev Brain Res. 2005;160:176–193. doi: 10.1016/j.devbrainres.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Tremolizzo L, Carboni G, Ruzicka WB, Mitchell CP, Sugaya I, Tueting P, Sharma R, Grayson DR, Costa E, Guidotti A. An epigenetic mouse model for molecular and behavioral neuropathologies related to schizophrenia vulnerability. Proc Natl Acad Sci USA. 2002;99:17095–17100. doi: 10.1073/pnas.262658999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremolizzo L, Doueiri MS, Dong E, Grayson DR, Davis J, Pinna G, Tueting P, Rodriguez-Menendez V, Costa E, Guidotti A. Valproate corrects the schizophrenia-like epigenetic behavioral modifications induced by methionine in mice. Biol Psychiatry. 2005;57:500–509. doi: 10.1016/j.biopsych.2004.11.046. [DOI] [PubMed] [Google Scholar]
- Trommsdorff M, Gotthardt M, Hiesberger T, Shelton J, Stockinger W, Nimpf J, Hammer RE, Richardson JA, Herz J. Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell. 1999;97:689–701. doi: 10.1016/s0092-8674(00)80782-5. [DOI] [PubMed] [Google Scholar]
- Utsunomiya-Tate N, Kubo K, Tate S, Kainosho M, Katayama E, Nakajima K, Mikoshiba K. Reelin molecules assemble together to form a large protein complex, which is inhibited by the function-blocking CR-50 antibody. Proc Natl Acad Sci USA. 2000;97:9729–9734. doi: 10.1073/pnas.160272497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Zhu Z, Han G, Ye X, Xu B, Peng Z, Ma Y, Yu Y, Lin H, Chen AP, Chen CD. JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc Natl Acad Sci USA. 2007;104:19226–19231. doi: 10.1073/pnas.0700735104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H, Taylor-Papadimitriou J, Tempst P, Zhang Y. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell. 2007;25:801–812. doi: 10.1016/j.molcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Yanes O, Clark J, Wong DM, Patti GJ, Sanchez-Ruiz A, Benton HP, Trauger SA, Desponts C, Ding S, Siuzdak G. Metabolic oxidation regulates embryonic stem cell differentiation. Nat Chem Biol. 2010;6:411–417. doi: 10.1038/nchembio.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.