The neuron-enriched Na+/H+ exchanger NHE5 is expressed in C6 glioma cells, acidifies recycling endosomes, and modulates cell surface abundance of receptor tyrosine kinases MET and EGFR. NHE5 depletion impairs MET recycling and facilitates degradation, thereby impairing cell migration and polarity.
Abstract
Increased recycling and elevated cell surface expression of receptors serve as a mechanism for persistent receptor-mediated signaling. We show that the neuron-enriched Na+/H+ exchanger NHE5 is abundantly expressed in C6 glioma cells and plays an important part in regulating cell surface expression of the receptor tyrosine kinases MET and EGF receptor. NHE5 is associated with transferrin receptor (TfR)- and Rab11-positive recycling endosomal membranes, and NHE5 knockdown by short hairpin RNA significantly elevates pH of TfR-positive recycling endosomes. We present evidence that NHE5 facilitates MET recycling to the plasma membrane, protects MET from degradation, and modulates HGF-induced phosphatidylinositol-3-kinase and mitogen-activated protein kinase signaling. Moreover, NHE5 depletion abrogates Rac1 and Cdc42 signaling and actin cytoskeletal remodeling. We further show that NHE5 knockdown impairs directed cell migration and causes loss of cell polarity. Our study highlights a possible role of recycling endosomal pH in regulating receptor-mediated signaling through vesicular trafficking.
INTRODUCTION
The MET protein is a receptor tyrosine kinase (RTK) and the receptor for hepatocyte growth factor (HGF)/scatter factor (Boccaccio and Comoglio, 2006). Binding of HGF to MET initiates various biological responses, including cell proliferation and survival, detachment from adjacent cells, epithelial to mesenchymal transition, and degradation of and migration through extracellular matrices (Trusolino et al., 2010). While HGF/MET signaling is important for organogenesis and other physiological processes, aberrant activation of HGF/MET signaling facilitates metastasis in malignant tumors (Gherardi et al., 2012), and elevated expression of MET is associated with poor prognosis of glioma (Nabeshima et al., 1997; Kong et al., 2009; Liu et al., 2011; Miyamoto et al., 2011). As activation of RTKs typically occurs on the cell surface, postendocytic degradation and recycling of RTKs play a crucial role in determining receptor availability and coordinating downstream signaling (Lemmon and Schlessinger, 2010). Endocytosis/recycling of oncogenic MET mutant proteins is required for oncogenicity (Joffre et al., 2011), whereas decreased MET recycling attenuates mitogen-activated protein kinase (MAPK) signaling and cell migration (Parachoniak et al., 2011). Moreover, endocytic recycling of MET facilitates sustained Rac1 signaling for optimal membrane ruffling and cell migration and invasion, and recruitment of the p85 regulatory subunit of phosphatidylinositol-3-kinase (PI3K) and a Rac GDP/GTP exchange factor Vav2 to recycling endosomes was suggested as an underlying mechanism (Menard et al., 2014). Thus MET targeting to and from recycling endosomes may account for a significant step linking HGF stimuli with actin cyoskeletal reorganization during cell migration. It was also reported that tumor-derived mutant p53 proteins enhance MET recycling and signaling, which drive invasion and scattering of human non–small cell lung carcinoma, H1299 cells, further signifying the role of MET recycling in metastatic progression (Muller et al., 2013). These observations collectively suggest that up-regulated MET recycling promotes tumor invasion and metastasis.
Increasing evidence points to the promigratory and proinvasive role of the acidic luminal pH of endosomes. Vacuolar-type H+-ATPases (V-ATPases) are expressed in most eukaryotic cells and play a pivotal role in establishing the acidic luminal pH of endosomal and secretory organelles by pumping protons into the lumen (Forgac, 2007). Treatment of human breast cancer cells with a V-ATPase inhibitor disrupted the leading-edge association of epidermal growth factor receptor (EGFR; Wiedmann et al., 2012). Moreover, inhibition of V-ATPases attenuates Rab5-mediated Rac1 activation in early endosomes, perturbs actin cytoskeletal remodeling, and impedes cell migration. Intravenous injection of an inhibitor against V-ATPases drastically reduced metastasis of mouse breast cancer cells (Wiedmann et al., 2012). Chloride intracellular channels (CLICs) residing in endosomal membranes are another group of acidifiers that shunt the proton current created by V-ATPases (Stauber et al., 2012). CLIC3, a proposed late endosomal acidifier, plays a proinvasive role in tumor metastasis by facilitating targeting of membrane-type matrix metalloproteinase (Macpherson et al., 2014) and integrins (Dozynkiewicz et al., 2012) to the plasma membrane. Recycling endosomes possess slightly acidic luminal pH (Presley et al., 1997; D’Souza et al., 1998), but the role of the acidic pH of recycling endosomes in tumor cell migration remains poorly understood. We now show that the neuron-enriched Na+/H+ exchanger NHE5 potently acidifies recycling endosomes of C6 rat glioma cells and demonstrate that NHE5 is required for MET recycling. Silencing NHE5 impairs MET targeting to the leading edge, facilitates its degradation, and suppresses downstream signaling and directed cell migration.
RESULTS
NHE5 is a potent endosomal acidifier in C6 glioma
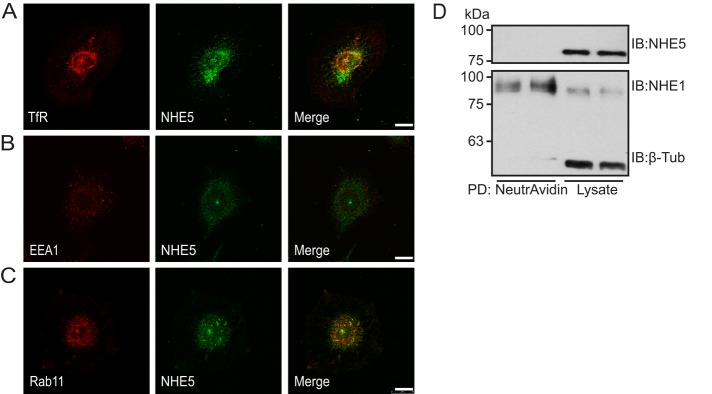
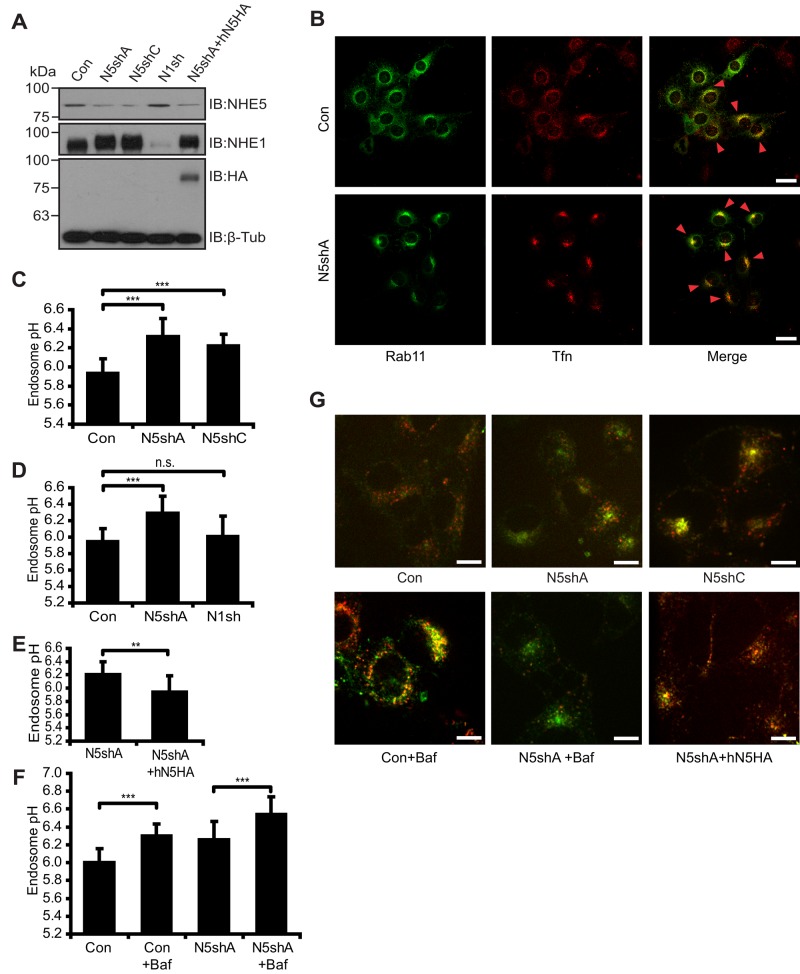
Whereas NHE5 protein (Diering et al., 2011) and mRNA (Baird et al., 1999) were not detected in glia-rich brain regions, we noted that C6 glioma cells abundantly express NHE5 protein. Immunofluorescence microscopy revealed that NHE5 is associated with Rab11-positive and transferrin (Tfn)-positive recycling endosomes (Figure 1, A–C). Cell surface biotinylation did not detect any appreciable association of NHE5 with the plasma membrane in C6 cells when cultured under regular conditions (Figure 1D). Plasmid-based short hairpin RNA (shRNA) shNHE5A, shNHE5B, and shNHE5C constructs (Diering et al., 2011, 2013) were individually expressed in C6 cells, and several independent clones stably expressing the shRNA were isolated for further experiments (Figure 2A). For determination of the endosomal pH, NHE5-knockdown cells and control cells were incubated with a mixture of two types of Tfn, one conjugated with fluorescein, a pH-sensitive green fluorophore, and another conjugated with Alexa Fluor 568, a pH-insensitive red fluorophore. Following the chase incubation at 37°C for 15 min, Tfn accumulated to the perinuclear Rab11-positive recycling endosomes (Figure 2B). Fluorescence signal was captured under a spinning-disk confocal microscope, and pH was determined by ratiometric measurement of the two fluorescence intensities (Diering et al., 2013). Recycling endosomal pH of C6 cells was close to 6.0, whereas C6 cell lines stably expressing shRNAs for NHE5 exhibited a significantly higher pH of around 6.2 (Figure 2C). In contrast, recycling endosomal pH of C6 cells expressing NHE1 shRNA was not significantly different from that of control cells (Figure 2D). The elevated recycling endosomal pH of NHE5-knockdown cells was rescued by stable expression of human NHE5 (Figure 2E). Treatment of C6 cells with the V-ATPase inhibitor bafilomycin A1 (Baf) alkalinized recycling endosomal pH to around 6.3, whereas recycling endosomes of Baf-treated NHE5-knockdown cells showed a pH of around 6.5 (Figure 2F). These results suggest that NHE5 and V-ATPases play major roles in generating acidic-recycling endosomal pH. Internalized Tfn appears to be more tightly packed in the juxtanuclear region of NHE5-knockdown cells (Figure 2B), a similar effect was observed when parental C6 cells were treated with 50 nM Baf for 16 h, but not when they were treated with up to 200 nM Baf for 45 min (Supplemental Figure 1).
FIGURE 1:
NHE5 is associated with recycling endosomes in C6 glioma. (A–C) C6 cells were grown on coverslips overnight and double stained for TfR (red in A), early endosome antigen-1 (EEA1, red in B), or Rab11 (red in C), and NHE5 (green). Scale bars: 20 μm. (D) Cell surface biotinylation detected NHE1, but not NHE5, associated with the plasma membrane. The cytosolic protein β-tubulin was not labeled by cell surface biotin, indicating that the membrane integrity was maintained during the experiments.
FIGURE 2:
NHE5 acidifies recycling endosomes. NHE5 expression in C6 cells stably expressing shRNA plasmids for NHE5 (N5shA, N5shC), scrambled control for N5shA (Con), NHE1shRNA (N1sh), and N5sh cells expressed with HA-tagged human NHE5 (hN5HA). (A) NHE5 and NHE1 protein expression was probed with anti-NHE5 and anti-NHE1 antibodies, respectively. Exogenous NHE5 expression was detected by an anti-HA antibody. An anti β-tubulin antibody was used as a loading control. (B) GFP-Rab11–transfected cells were serum-starved overnight and incubated with 30 μg/ml of Alexa Fluor 568–conjugated transferrin (Tfn-568) for 30 min at 37°C. Red arrowheads mark the endosomal compartments where Rab11 and Tfn colocalize. Scale bars: 25 μm. (C–F) Endosomal pH estimations. +Baf, treated with 200 nM Baf. The bar graph presents means ± SD of at least three experiments in which more than five live-cell images per cell line or treatment condition were used for fluorescence ratiometric analysis. p Values were obtained by Tukey-Kramer test following ANOVA: (C) F(2, 56) = 37.84, p < 0.0001, ***p < 0.001; (D) F(2,71) = 22.12, p < 0.0001, ***p < 0.001, n.s., not significant; (F) F(3,52) = 25.12, p < 0.0001, ***p < 0.001; (E) paired Student’s t test, **p < 0.01; (G) Representative confocal images of live cells for C–F. Scale bars: 10 μm. **p < 0.01, ***p < 0.001, n.s., not significant.
NHE5 regulates surface expression of MET and epidermal growth factor receptor
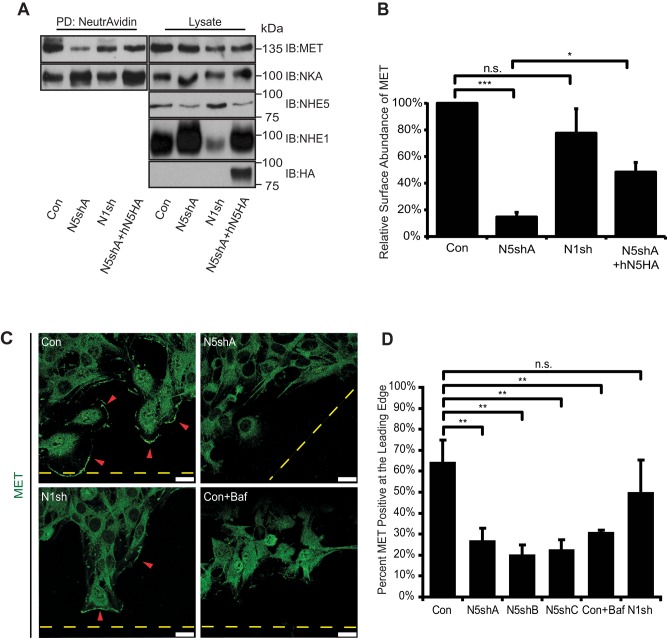
Our previous finding that NHE5 is required for endocytic recycling of the nerve growth factor (NGF) receptor tyrosine kinase TrkA in PC12 cells (Diering et al., 2013) led us to postulate that NHE5 might regulate RTK targeting and signaling. HGF receptor tyrosine kinase MET and epidermal growth factor receptor (EGFR) expression were detected in C6 cells. To investigate the role of recycling endosomal pH on MET trafficking and downstream signaling, we examined the abundance of MET populations residing in the plasma membrane. Knocking down NHE5 by shRNA led to a substantial decrease of cell surface–biotinylated MET to ∼15% of that of control cells, whereas NHE1-depleted cells showed no significant alterations (Figure 3, A and B). Reexpressing human NHE5 into NHE5-knockdown cells increased the cell surface expression of MET to approximately threefold of NHE5-knockdown cells, a significant (p < 0.05 by Tukey-Kramer test, N = 3) and yet not a complete rescue. Fluorescence microscopy revealed that a substantial population of control cells (∼65%) showed clear association of MET with the leading edge when directed migration was induced (Figure 3, C and D). Leading-edge association of MET was also apparent in NHE1-knockdown cells, in which more than half of the cells exhibited similar MET localization. In contrast, ∼20% of the cells stably expressing NHE5 shRNA exhibited a typical appearance of MET in association with the cell front. Similarly, MET did not localize to the leading edge when cells were treated with Baf, suggesting the potential importance of the acidic luminal pH of organelles in MET targeting. Reduced cell surface abundance of EGFR was also detected in NHE5-knockdown cells (Supplemental Figure 2, A and B).
FIGURE 3:
Cell surface expression and polarized targeting of MET are reduced in NHE5-deficient cells. (A and B) C6 cells expressing shRNA plasmids for NHE5 (N5shA), NHE1 (N1sh), N5shA cells expressing HA-tagged human NHE5 (N5shA +hN5HA), and control cells (Con) were treated with a membrane-impermeable biotinylating reagent (sulfo-NHS-SS-biotin), and biotinylated proteins were affinity purified by NeutrAvidin and detected by immunoblotting. A representative immunoblot is shown. Signal intensities were determined by densitometry, and relative levels of cell surface MET are presented in B. Data represent mean ± SEM of five experiments, except for N1sh with N = 3. p Values by Tukey-Kramer test following ANOVA: F(3,10) = 37.27, p < 0.0001; *p < 0.05, ***p < 0.001, n.s., not significant. (C and D) Intracellular localization of MET during directional migration of confluent monolayer cells toward an open space was investigated in C6 cells stably expressing shRNAs for NHE5 (A–C), NHE1 (N1sh), and scrambled shRNA control (Con). (C) Representative confocal images of Con, N5shA, N1sh, and Baf-treated cells are shown. Dashed lines mark the edge of open space introduced to the confluent monolayer cells, and arrowheads indicate the leading edge displaying positive membrane MET fluorescence. Scale bars: 20 μm. (D) Mean (± SD) percentages of cells positive for MET at the leading edge from three to five experiments are shown. On average, 150 cells per cell line were scored in each experiment. p Values were by Tukey-Kramer test following ANOVA: F(5,8) = 33.0, p < 0.0001, ** p < 0.01, n.s., not significant.
NHE5 knockdown limits MET recycling and accelerates HGF-induced degradation
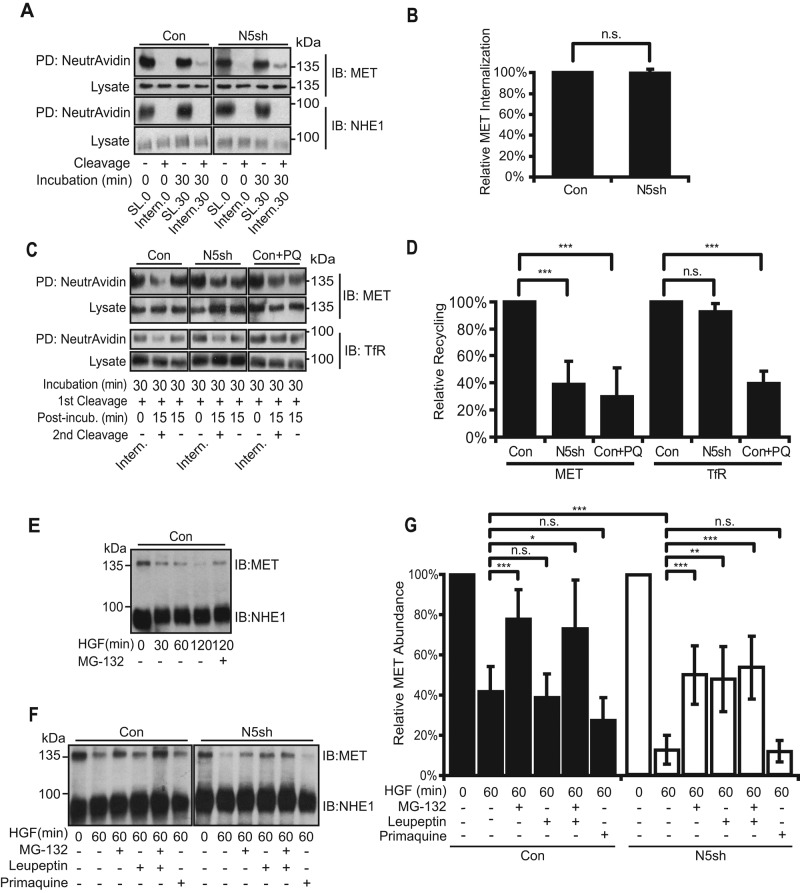
A decrease in cell surface population of MET may be caused by increased internalization from the plasma membrane, reduced recycling from endosomes to the plasma membrane, or both. For investigation of these possibilities, MET residing in the plasma membrane was labeled by biotinylation, and internalized proportions following a chase incubation were determined. No difference was observed in MET endocytosis between control and NHE5-knockdown cells (Figure 4, A and B). We next examined the effect of NHE5 knockdown on MET recycling by probing the cell surface population of biotinylated MET returning from the endosomal pool. After a 15-min chase incubation at 37°C, ∼40% of MET recycled back to the plasma membrane in NHE5-depleted cells relative to control cells (Figure 4, C and D). Recycling of transferrin receptor (TfR) was not affected by NHE5 depletion, while primaquine, an endosomal recycling blocker (Hiebsch et al., 1991; van Weert et al., 2000), inhibited MET and TfR recycling. As receptor internalization and subsequent degradation also halt cell surface ligand-mediated signaling, we next tested whether NHE5 depletion affects MET degradation. In agreement with the previous studies (Jeffers et al., 1997; Hammond et al., 2001; Petrelli et al., 2002), ligand-induced degradation of MET was significantly reduced by concomitant treatment with the proteasomal inhibitor MG-132 (Figure 4E). While 40% of MET remained intact in control cells after a 60-min treatment with HGF, more than 80% of MET was degraded in NHE5-knockdown cells after HGF treatment, which was substantially protected by MG-132 (Figure 4, F and G). Intriguingly, leupeptin, an inhibitor of serine and cysteine proteases, inhibited degradation of MET in NHE5-knockdown cells but not in the control cells. The endocytic recycling inhibitor primaquine had no effect on MET degradation in either type of cells.
FIGURE 4:
NHE5 depletion impairs MET recycling and enhances ligand-dependent degradation but has no effect on endocytosis. (A) Cells stably expressing shRNA for NHE5 (N5sh) and scrambled control (Con) were treated with sulfo-NHS-SS-biotin at 4°C and subjected to a chase incubation at 37°C for 0 (Intern.0) or 30 min (Intern.30) to let the biotinylated surface receptors internalize. Cells were then treated with glutathione to cleave biotin from surface proteins. Internalized proteins were protected from cleavage. Some samples were left untreated with glutathione (SL.0 and SL.30). Biotinylated proteins were affinity purified by NeutrAvidin and analyzed by immunoblotting. For MET cell surface biotinylation experiments, five times more total proteins from NHE5-knockdown cell lysates (250 μg) than control cells (50 μg) were used to obtain an equivalent signal of MET labeled on the cell surface (PD: NeutrAvidin, IB: MET), whereas equal amounts of biotinylated proteins (50 μg) were analyzed to probe NHE1 on the cell surface (PD: NeutrAvidin, IB: NHE1). Lysates (4 μg) were loaded to probe the amount of MET and NHE1 (Lysate, IB: MET and IB: NHE1). (B) The intensity of each band was determined by densitometry, and the internalized proportion relative to the control was calculated. A bar graph shows mean ± SD of five experiments. n.s. = not significant by paired Student’s t test. (C) MET recycling in control (Con), NHE5-shRNA–expressing cells (N5sh), and primaquine (PQ)-treated cells. Cells were treated with sulfo-NHS-SS-biotin, biotinylated receptors were internalized, and the biotin was removed from the proteins remaining on the plasma membrane (1st Cleavage). Some samples were subjected to an additional 15-min incubation in culture media at 37°C and an additional round of glutathione cleavage to remove biotin from proteins recycled back to the cell surface (2nd Cleavage). Biotinylated proteins were affinity-purified and analyzed by immunoblotting. Fivefold larger amounts of proteins from NHE5-knockdown cells (750 μg) than those from control cells (150 μg) were used for MET pull down, whereas equal amounts of lysates from each cell line (150 μg) were used for the TfR experiments. Lysates (4 μg) were loaded to probe the amount of MET and TfR. (D) Relative recycling was calculated by taking densitometry ratios of recycled populations ([1st Cleavage +, Post-incub.15, 2nd Cleavage–] – [1st Cleavage +, Post-incub.15, 2nd Cleavage +]) to internalized populations (1st Cleavage +, Post-incub.0, 2nd Cleavage–; Intern.) and expressed as percentages to the values of the control. Mean ± SD of three to five experiments. p Values were obtained by Tukey-Kramer test following ANOVA: F(2,14) = 193.9, p < 0.0001. (E) HGF-induced MET degradation in C6 cells. Serum-starved cells were stimulated with 50 ng/ml of HGF for up to 120 min, and the same amount of lysates (4 μg) was analyzed by immunoblotting. Treatment of 20 μM MG-132 significantly slowed down the MET degradation over time. (F) HGF-induced MET degradation in control and NHE5-depleted cells. A representative blot is shown. Where indicated, cells were treated with 20 μM MG-132, 100 μM leupeptin, or 100 μM primaquine. Four micrograms total protein was analyzed by immunoblotting. (G) Relative abundance of MET in each condition was determined by normalization to the unstimulated level. Mean ± SD of three to eight experiments. p Values by Tukey-Kramer test following ANOVA: F(6,85) = 24.6, p < 0.0001 for conditions; and F(1,85) = 25.6, p < 0.0001 for cell lines; *, p < 0.05, **, p < 0.01, ***, p < 0.001, n.s., not significant.
NHE5 is required for HGF signaling
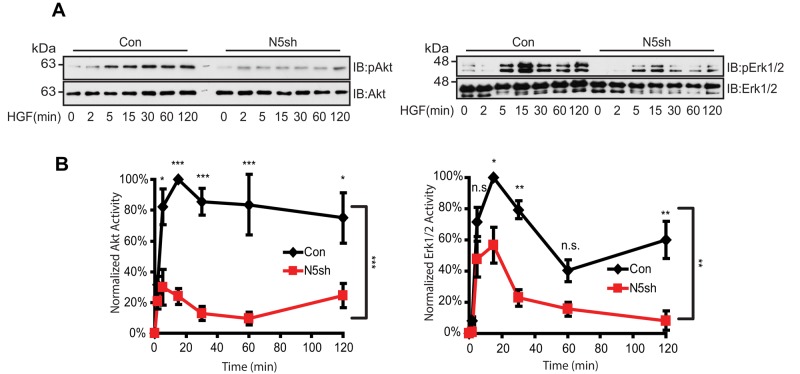
Having shown the critical role of NHE5 in modulating endocytic recycling, degradation, and surface abundance of MET, we next examined the impact of NHE5 depletion on HGF/MET signaling. C6 cells stably expressing shRNA for NHE5 and control cells were stimulated by HGF, and the phosphorylation status of Akt and Erk1/2 was assessed by immunoblotting to determine the PI3K and MAPK activities, respectively. Although both cell lines exhibited a steep peak for Akt phosphorylation within 15 min of stimulation, the peak intensity was substantially lower in NHE5-knockdown cells compared with control cells (Figure 5A). Densitometric analyses of three independent experiments revealed that the maximum Akt phosphorylation is reduced by approximately fourfold in NHE5-knockdown cells (Figure 5B). Down-regulation of PI3K-Akt signaling was confirmed by three different NHE5-silencing constructs (Supplemental Figure 3, A and B). The level of peak phosphorylation of Erk1/2 in NHE5-knockdown cells was ∼55% of the peak phosphorylation level in control cells (Figure 5, A and B).
FIGURE 5:
HGF-induced Akt/Erk phosphorylation is attenuated by NHE5 knockdown. Cells serum-starved overnight were stimulated with 50 ng/ml of HGF. The amount of phosphorylated and total Akt and Erk1/2 was detected by immunoblotting. (A) Representative immunoblots are shown. (B) The relative level of phospho-Akt and phospho-Erk are expressed as mean ± SEM of four experiments. p Values for pairwise comparisons at each time point were obtained following repeated-measures ANOVA with time as the within-subject factor. *, p < 0.05, **, p < 0.01, ***, p < 0.001, n.s., not significant.
Rac1 and Cdc42 activities are reduced in NHE5-knockdown cells
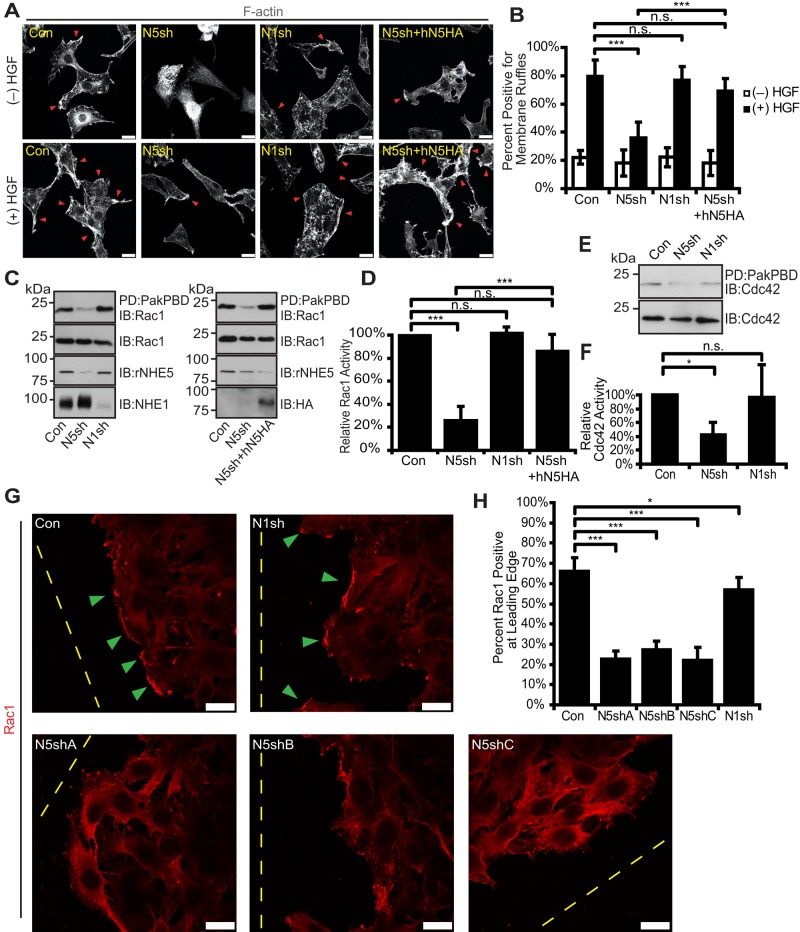
During the course of morphological characterization, we noticed that NHE5-knockdown cells exhibit limited F-actin structures. F-actin staining by fluorescent phalloidin revealed some patchy appearance on the plasma membrane as well as in the cytosol of C6 cells, and also the peripheral membrane ruffles became evident upon HGF treatment (Figure 6A). In contrast, the actin-filament network at the edge of the NHE5-knockdown cells was not apparent, even after HGF treatment. We counted the cells with and without membrane ruffles upon HGF stimulation; ∼80% of C6 cells exhibited membrane ruffles. In contrast, <40% of NHE5-knockdown cells showed these structures, whereas close to 80% of NHE1-knockdown cells formed HGF-induced membrane ruffles (Figure 6B). Close to 70% of NHE5-knockdown cells stably expressing human NHE5 displayed membrane ruffles upon HGF stimulation, representing a substantial restoration of the phenotype (Figure 6, A and B). We next asked whether NHE5 influences migration-induced membrane ruffles. The mesh-like structure of actin filaments formed at the edge of migrating front was also not apparent in NHE5-knockdown cells (Supplemental Figure 4, A and B). Approximately 75% of control cells had membrane ruffles at the leading edge, whereas ∼50% of NHE5-knockdown cells showed membrane ruffles. Expression of human NHE5 substantially rescued the phenotype in which more than 70% of cells showed membrane ruffles. The Rho-family small GTPase Rac1 is a downstream signaling molecule of HGF/MET that facilitates actin remodeling and cell migration in canine kidney epithelial MDCK cells (Ridley et al., 1995; Chianale et al., 2010), murine cortical neurons (Segarra et al., 2006), human glioma SNB19 cells (Kwiatkowska et al., 2012), breast cancer MDA-468 cells (Menard et al., 2014), and human pulmonary microvascular endothelial cells (Singleton et al., 2007; Usatyuk et al., 2014). We therefore examined Rac1 activity in NHE5-knockdown cells by using glutathione S-transferase (GST) pull down that captures only the GTP-bound active form of Rac1. Rac1 activity showed an approximately fourfold increase upon HGF treatment in control cells (Supplemental Figure 4C), which was abolished by applying NSC 23766, an inhibitor of Rac1 activation, to the cells (Supplemental Figure 4D). Remarkably, NHE5- but not NHE1-knockdown cells showed a marked reduction in Rac1 activity to ∼20% of that in control cells, and expression of hNHE5 to knockdown cells restored the Rac1 activity to the level close to that of control cells (Figure 6, C and D). As Cdc42 shares some common regulators and effectors with Rac1 and also promotes actin cytoskeleton remodeling (Ridley, 2011), we tested Cdc42 activity in NHE-knockdown cells. A significant reduction in Cdc42 activity was detected in NHE5-depleted but not in NHE1-depleted cells (Figure 6, E and F). Recruitment of Rac1 to the plasma membrane is critical in establishing spatially restricted signals for cell migration (Palamidessi et al., 2008). As endocytic trafficking of MET and EGFR has been implicated in spatial activation and localization of Rac1 (Joffre et al., 2011; Wiedmann et al., 2012), we postulated that NHE5 knockdown might affect the membrane association of Rac1. Membrane fractions and cytosolic fractions were isolated, and the same amount of proteins in each fraction was analyzed by immunoblotting to test this possibility. Less Rac1 was associated with membrane fractions in NHE5-knockdown cells than in control cells or NHE1-knockdown cells (Supplemental Figure 4E). For further examination of whether Rac1 is associated with morphologically distinguishable membrane fractions, Rac1 was visualized by immunofluorescence in cells migrating toward an open space (wound) introduced to a confluent monolayer sheet of cells. Association of Rac1 with the leading edge of the migration front was significantly impaired in NHE5-knockdown cells, whereas NHE1 knockdown showed only marginal effects on Rac1-localization (Figure 6, G and H).
FIGURE 6:
Rac1 and Cdc42 activities and Rac1 targeting to the leading edge were perturbed by NHE5 depletion. (A and B) HGF-induced membrane ruffle formation was impaired by silencing NHE5. Serum-starved C6 cells stably expressing shRNA for NHE5 (N5sh), NHE1 (N1sh), and N5sh cells expressing HA-tagged human NHE5 (N5sh+hN5HA) were left untreated or treated with 50 ng/ml HGF for 60 min. Cells were then fixed and stained with fluorescently labeled phalloidin. (A) Representative images are shown. Arrowheads point to the positive membrane ruffling observed as increased phalloidin staining associated with wavelike protrusions or tiny spicules. Scale bars: 20 μm. (B) The cells with membrane ruffles were counted. The bar graph presents mean ± SD of percentages of cells positive for membrane ruffles from a total of 8–14 images per condition for each cell line from three experiments. On average, 23 cells per image were counted. p Values were obtained by Tukey-Kramer test following ANOVA using images as a unit of analysis: F(3,42) = 40.31, p < 0.0001; *, p < 0.05, **, p < 0.01; ***, p < 0.001, n.s., not significant. (C and E) Rac1 and Cdc42 activities in control, N5sh, N1sh, and N5sh+hN5HA cells. GTP-bound Rac1 and Cdc42 were pulled down with PBD beads, and eluted samples were analyzed by immunoblotting. Two micrograms of total proteins were loaded as input. Representative immunoblots are shown. (D) Relative GTPase activities were calculated by taking densitometric ratio of the pulled-down (active form) and total proteins that were normalized to the value of control cells. The bars represent normalized mean ± SD of GTPase activities from three (N1sh) to nine experiments. p Values by Tukey-Kramer test following ANOVA: F(3, 22) = 85.27, p < 0.0001; ***, p < 0.001; n.s., not significant. (F) Mean ± SD with four experiments. p Values by Tukey-Kramer test following ANOVA: F(2, 8) = 85.27, p < 0.0001; *, p < 0.05, n.s., not significant. (G) Control cells and three independent N5shRNA clones were grown into a monolayer, and polarized targeting of Rac1 toward an open space was observed. Representative confocal images are shown. Rac1 associates with the leading edge of the majority of migratory control and NHE1-knockdown cells, whereas Rac1 association with the leading edge is markedly impaired in NHE5-knockdown cells. Arrowheads mark Rac1 associated with the leading edge. Dashed lines indicate edges of wounds. Scale bars: 25 μm. (H) Percentage of cells at the leading edge that had positive Rac1 staining. The bar chart represents mean ± SD of three experiments, consisting of, on average, a total of 82.5 cells per experiment. p Values were from Tukey-Kramer test following ANOVA: F(4, 35) = 114.40, p < 0.0001; *, p < 0.05, ***, p < 0.001.
NHE5 is required for cell migration and polarity establishment
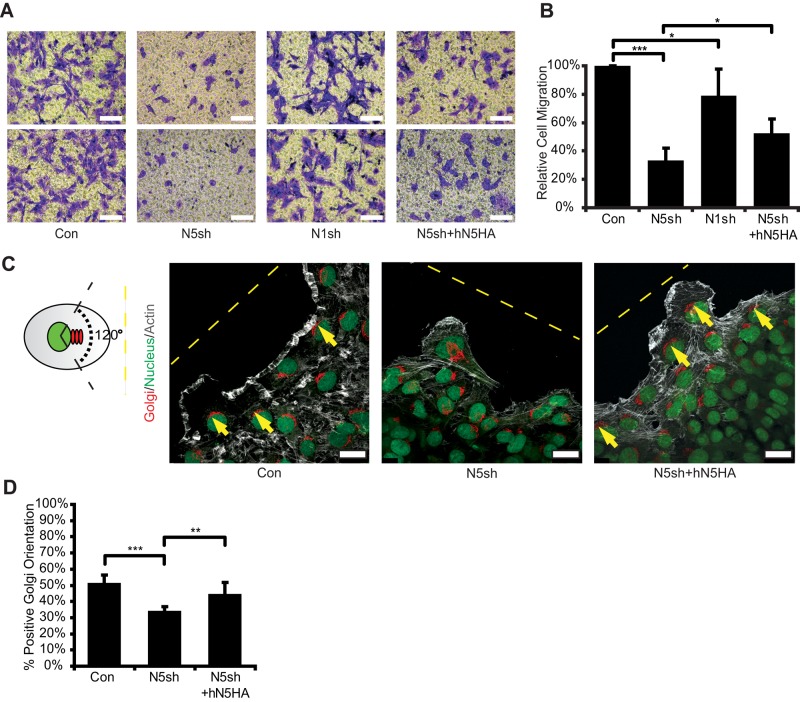
Having found a marked decline in Rac1 activity and severe impairment of actin cytoskeletal organization at the migrating edges in NHE5-knockdown cells, we next investigated an impact of NHE5 knockdown on cell migration. When serum-starved cells were placed in the upper chamber and rendered to move across the polycarbonate membrane toward the bottom chamber, which was filled with serum-containing media, considerably fewer NHE5-knockdown cells than control cells (∼30% of control) migrated to the bottom chamber (Figure 7, A and B). Expression of human NHE5 into NHE5-knockdown cells increased such chemotaxis by 1.5-fold, showing a significant and partial rescue of the phenotype. Activation of Rac1 and Cdc42 at the migrating front is crucial for cell polarity establishment during directional migration (Nelson, 2009; Ridley, 2011). We therefore asked whether NHE5 is needed for cell-polarity establishment. Cell polarity can be assessed by the position of Golgi stacks relative to the position of the nucleus and migrating edge (Bornens, 2008; Figure 7C). Approximately 50% of control cells at the wound edge showed the Golgi apparatus oriented within a 120° angle projected from the nucleus toward the leading edge, whereas 33% of NHE5-knockdown cells displayed correct alignment of the Golgi apparatus, representing a significant difference (Figure 7, C and D). A modest but significant rescue was observed by stable expression of hNHE5.
FIGURE 7:
NHE5 deficiency impairs cell migration and polarity. Cell migrations of C6 cells expressing shRNAs for scrambled (Con), NHE5 (N5sh) and NHE1 (N1sh), and N5sh cells expressing HA-tagged human NHE5 (N5sh+hN5HA) were analyzed by Transwell migration assays. Representative images of crystal violet staining of the Transwell membrane are shown in A. Scale bars: 500 μm. (B) Cells that migrated through the membrane were counted in five randomly selected microscopic fields and expressed as a percentage relative to those in control cells. Mean ± SD of 6–10 experiments. p Values were obtained by Tukey-Kremer test following ANOVA: F(3, 19) = 56.22, p < 0.0001; *, p < 0.05, ***, p < 0.001. (C) Control, N5sh, and N5sh+hN5HA cells migrating toward the open space introduced in the confluent monolayer were labeled with GM130 antibodies for Golgi (red), DRAQ5 for nucleus (green), and fluorescent phalloidin for F-actin (gray). Correct cell polarity is defined as the Golgi apparatus residing within a 120° angle of the migration axis. Representative confocal images are shown. Dashed lines indicate edges of wounds, and cells with correct orientation were marked. Scale bars: 25 μm. (D) Cells that displayed correct cell polarity were counted, and the number was divided by the total number of cells at the leading edge, using images as the unit of analysis. Bars represent mean (± SD) percentage of cells with correct polarity of a total of 13–18 images per cell line containing more than 50 cells in each image from two experiments. p Values were obtained by ANOVA: F(2, 2) = 11.19, p = 0.082, followed by Tukey-Kramer test; **, p < 0.01, ***, p < 0.001.
DISCUSSION
Most malignant tumor cells primarily use aerobic glycolysis rather than mitochondrial oxidative phosphorylation and produce large amounts of lactate and acidic metabolites (Vander Heiden et al., 2009). To overcome the cytotoxicity of metabolic acids, tumor cells have evolved sophisticated mechanisms to pump H+ out of the cell (Gatenby and Gillies, 2008; Parks et al., 2013). H+-extruding activity across the plasma membrane by Na+/H+ exchangers (NHEs) is among the best-studied mechanisms that create the unique pH environment of glioma (McLean et al., 2000; Cong et al., 2014). Since their discovery as electroneutral secondary active transporters that mobilize H+ across the membrane in response to growth factors (Sardet et al., 1989), the concept of NHEs has been broadened, and select isoforms are now known to associate with organelle membranes (Ohgaki et al., 2011; Orlowski and Grinstein, 2011; Donowitz et al., 2013; Fuster and Alexander, 2014). C6 glioma cells are equipped with elevated ion transporters, including NHE1, and have been commonly used as a model to study the role of NHEs in invasive migration (Honasoge and Sontheimer, 2013). However, the role of NHEs in recycling endosomal pH regulation and their potential contributions to RTK signaling, tumor invasion, and migration are largely unknown. We now show that the neuron-enriched NHE5 is abundantly expressed in C6 cells and plays a critical role in acidifying recycling endosomes. We have found that the recycling endosomal pH of C6 cells is around 6.0, a more acidic value than previously reported in CHO cells that do not express NHE5 (pH = 6.4–6.5; Presley et al., 1997; D’Souza et al., 1998). We showed that reduced expression of NHE5 elevates recycling endosomal pH to around 6.2 (Figure 2). Considering that inhibition of V-ATPase by Baf raises the recycling endosomal pH by ∼0.3, the impact of NHE5 silencing seems substantial. PC12 rat pheochromocytoma cells of adrenal medulla origin also have an acidic recycling endosomal pH of ∼6.2, and either NHE5 depletion or V-ATPase inhibition elevates recycling endosomal pH to 6.5 (Diering et al., 2013). These results collectively suggest a substantial role of NHE5 in recycling endosomal acidification, although its relative contribution of NHE5 may vary depending on cell type. Depletion of NHE5 abrogates MET recruitment to the leading edge of the migrating cells, down-regulates HGF-induced PI3K and Rac1/Cdc42 activity, and causes loss of cell polarity as well as perturbed cell migration. While HGF-induced phosphorylation of Erk and Akt are both decreased when NHE5 is silenced, it is interesting to note that the degree of suppression of Akt phosphorylation is greater than that of Erk phosphorylation; a similar tendency is observed in NHE5-depleted PC12 cells (Diering et al., 2013). Aside from receptor recycling, it is possible that NHE5 has an additional mechanism(s) that influences PI3K-Akt signaling. Rac1 is a downstream effector of PI3K-Akt signaling, and a fraction of Rac1 binds to endosomes and recycles through the ADP-ribosylation factor Arf6 when cells are stimulated with HGF (Palamidessi et al., 2008). We previously found that Arf6-mediated endocytic recycling also contributes to cell surface targeting of NHE5 heterologously expressed in CHO cells (Diering et al., 2009). Moreover, Arf6-activation by its guanine nucleotide exchange factor ARNO was considered to occur on the cytosolic face of endosomes in a luminal acidification–dependent manner (Hurtado-Lorenzo et al., 2006). Thus an intriguing possibility is that Rac1 recruited to the NHE5-positive endosomal membrane is activated in an Arf6-dependent manner when the lumen is acidified by NHE5. Upon human NHE5 expression to knockdown cells, Rac1 activity and HGF- or cell migration–induced actin remodeling (Figure 6, B and D, and Supplemental Figure 4B) appeared to be rescued more substantially than other phenotypes such as MET surface expression (Figure 3B) and cell migration (Figure 7B). It is tempting to speculate that NHE5 modulates Rac1 activity via different mechanisms in addition to the one mediated by MET-PI3K signaling.
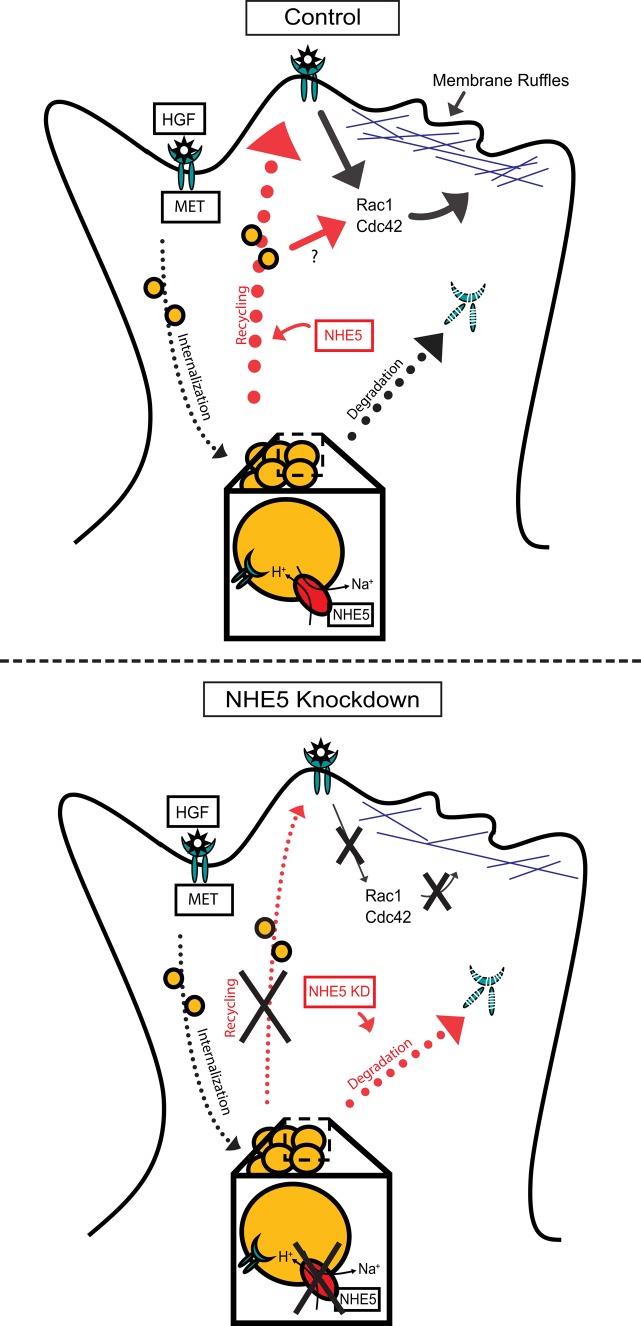
NHE5 influences endocytic recycling of multiple RTKs. In addition to MET, we found that surface abundance of EGFR is also affected by NHE5 knockdown (Supplemental Figure 2). Depletion of NHE5 in PC12 cells markedly reduces the cell surface targeting of TrkA and severely impairs NGF-TrkA signaling (Diering et al., 2013). V-ATPase inhibition also perturbs targeting of MET (Figure 3C) and EGFR (Wiedmann et al., 2012) to the leading edge of migratory cells similar to the impairment of NGF-induced TrkA recycling by Baf (Diering et al., 2013). Importantly, endocytic recycling of a non-RTK cargo TfR is not noticeably affected by NHE5 knockdown (Figure 4, C and D; Diering et al., 2013), although other processes of Tfn (and TfR) trafficking along the endocytic circuit may be affected by NHE5 depletion (Supplemental Figure 1). A recent study has revealed that a common clathrin-independent endocytic mechanism governs ligand-induced endocytosis for various growth factor receptors, thereby impacting on growth factor signaling, cell migration, and neurite extension (Boucrot et al., 2015). Thus it is possible that RTKs internalized by ligand-induced endocytosis follow a common recycling route that is regulated by a pH-dependent mechanism. We have further shown that NHE5 depletion significantly facilitates HGF-induced MET degradation (Figure 4, F and G). Consistent with a previous study (Jeffers et al., 1997), HGF-induced MET degradation in both control and NHE5-knockdown cells was blocked by the proteasomal inhibitor MG-132. The lysosomal protease inhibitor leupeptin also showed a protective effect on MET degradation in NHE5-knockdown cells but not in control cells. The effect of leupeptin may reflect differences in degradation kinetics of MET between the two cell lines. It is also possible that the enzyme contents in lysosomes and multiple vesicular bodies are influenced by NHE5-depletion. On the basis of these observations, we propose a model in which NHE5 depletion abrogates cell surface delivery of MET and promotes targeting of MET toward degradation (Figure 8). If neutralizing the acidic luminal pH of recycling endosomes decreases ligand-induced cell surface targeting and promotes down-regulation of RTKs, pharmacological interventions that elevate endosomal pH may become a new strategy to reduce the RTK signaling. Mildly acidic endosomal pH has been implicated in drug delivery to multidrug-resistant cancer cells (Lee et al., 2008; Gao et al., 2010; Wu et al., 2013). Developing new drugs that alter recycling endosomal pH may become an effective approach to treat malignant tumors.
FIGURE 8:
Proposed model of MET endocytic trafficking influenced by NHE5. On HGF stimulation, MET is internalized to endosomes, where a population of MET is degraded and the rest return to the plasma membrane. NHE5-mediated acidification of recycling endosomes facilitates plasma membrane targeting of MET, maintaining surface abundance of MET and downstream signaling. In the absence of NHE5, recycling of MET is halted and degradation is enhanced.
NHE7 was the first NHE shown to exhibit a unique cation-nonspecific (Na+, K+)/H+ exchanger activity across the trans-Golgi network (TGN) (Numata and Orlowski, 2001), and a similar cation-nonspecific organellar NHE (CNO-NHE) activity was later identified in NHE6, NHE8, and NHE9 in a liposome reconstitution system (Nakamura et al., 2005). CNO-NHEs are evolutionarily conserved from yeast to humans (Brett et al., 2005), and NHE6–NHE9 are ubiquitously expressed in most mammalian cell types (Nakamura et al., 2005). While NHE7 is predominantly associated with the TGN membrane (Lin et al., 2005), it shuttles along the endocytic-recycling trafficking pathway (Lin et al., 2007) and shares some common binding proteins with NHE5 (Lin et al., 2005; Kagami et al., 2008; Diering et al., 2009). CNO-NHEs were suggested “leaking” protons using the inwardly directed K+ concentration gradient across the organelle membrane as a driving force (Orlowski and Grinstein, 2007; Ohgaki et al., 2011). Interestingly, heterologous overexpression of NHE7 into breast cancer cells facilitates invasion, anchorage-independent growth, and tumor formation in nude mice (Onishi et al., 2012). A more recent study showed that heterologous overexpression of NHE9 in glioma cells alkalinizes the luminal pH of endosomes and suggested that increased cell surface expression and decreased degradation of EGFR facilitate tumor growth and invasion (Kondapalli et al., 2015). However, pharmacological manipulations that elevate the luminal pH of endosomes did not affect the endocytic behavior of TfR in agreement with our current findings. Moreover, depletion of NHE8 from HeLa cells was reported to facilitate EGF degradation, whereas NHE8 knockdown did not affect pH of late endosomes in which NHE8 was largely detected (Lawrence et al., 2010). Whether aberrant EGFR turnover and cell migration caused by altered CNO-NHE expression are mediated by changes in the degree of “proton leak” remains somewhat controversial, and future studies are needed to clarify this point. Regardless of the mechanism, it is important to note that NHE9 overexpression increases Tfn endocytosis (Kondapalli et al., 2015), whereas NHE5 depletion does not (Figures 4, C and D), suggesting that NHE5 and NHE9 target distinct trafficking pathways. These discrepancies can be partly explained by the difference in distinct intracellular localization between NHE9 (Kondapalli et al., 2015) and NHE5 (Figure 1). We propose a model in which acidification of confined recycling endosomal populations by NHE5 has a unique effect on RTK targeting, signaling, and degradation, while indiscriminate alkalinization of acidic organelles may also influence some (e.g., RTK recycling) but not all phenotypes detected in NHE5-knockdown cells. In summary, we identified an endosomal acidifier NHE5 as a regulator of MET recycling and degradation. This study lays the foundation for future studies exploring a role of NHE5 in tumor invasion and progression.
MATERIALS AND METHODS
Cell culture and transfection
C6 rat glioma cells were cultured in DMEM containing 10% fetal bovine serum (FBS). Stable knockdown clones were generated by transfecting the cells with the corresponding shRNA plasmid and maintaining them in DMEM + 10% FBS supplemented with either G418 or hygromycin. The target sequences for NHE5 are 1) GTTTGCTCTTGGTGAAACAGATGTTA, 2) ATAGTGGTGGCCACAAAGTAGTCCT, and 3) TTTGTGGTAATCACTCCTCTTCACC (Diering et al., 2011, 2013). C6 cells were transfected using the Bio-Rad GenePulser Xcell electroporation system (Bio-Rad, Hercules, CA) or the calcium phosphate method. For electroporation, cells were resuspended in ice-cold 1M HEPES buffer supplemented with 6 mM glucose, mixed with plasmid DNA, subjected to electric shock (250 V, 200 μF) in a 1-mm cuvette (1652089; Bio-Rad), and immediately placed into prewarmed culture media.
Antibodies, reagents, and DNA constructs
Rabbit anti-rat NHE5 antibody was generated in our laboratory and tested previously (Diering et al., 2011, 2013). Anti–phospho-Erk1/2 (pT185/pY187, 44-680G), TfR (clone H68.4, 13-6800), Alexa Fluor–conjugated goat anti-mouse (A-21235 and A-11004), anti-rabbit (A-11079 and A-21210) secondary antibodies and DRAQ5 (62251) were obtained from Invitrogen/Life Technologies/Thermo Fisher (Carlsbad, CA). Anti-EEA1 (clone 14/EEA1, 610456), Rab11 (clone 47/Rab11, 610656), and GM130 (clone 35/GM130, 610822) antibodies were obtained from BD Biosciences (Mississauga, ON, Canada). Anti-Rac1 (clone 23A8, 05-389) antibody was purchased from Millipore (Billerica, MA). Anti-Cdc42 (clone B-8, sc-8401), rabbit polyclonal anti-MET (SP260, sc-162), and EGFR (1005, sc-03) antibodies were obtained from Santa Cruz Biotechnology (Dallas, TX). Anti-hemagglutinin (HA; clone 16B12) antibody was obtained from Covance (Princeton, NJ). Anti-Na+/K+ ATPase (α5) and β-tubulin (clone E7) antibodies were obtained from the Developmental Study Hybridoma Bank (Iowa City, IA). Rabbit polyclonal anti phospho-Akt (pT308, 9275), Akt (9272), Erk1/2 (9102), and mouse monoclonal anti-MET (clone 25H2, 3127) antibodies were obtained from Cell Signaling Technology (Danvers, MA). Hoechst 33342 was obtained from Sigma-Aldrich (St. Louis, MO).
Baf (B-1080) was purchased from LC Laboratories (Woburn, MA). Leupeptin (LDJ691) was purchased from Bio Basic Canada (Markham, ON, Canada). Hepatocyte growth factor (CYT-090) was obtained from ProSpec-Tany TechnoGene (Rehovot, Israel). Nigericin (BML-CA421) was obtained from Enzo Life Sciences (Farmingdale, NY). Alexa Fluor 568– (T-23365) and fluorescein-conjugated Tfn (T-2871) were purchased from Invitrogen/Life Technologies. Primaquine bisphosphate (160393), MG132 (C2211), and sodium orthovanadate (S6508) were obtained from Sigma-Aldrich. EZ-Link sulfo-NHS-SS-biotin (21331) was purchased from Thermo Fischer Scientific (Waltham, MA). NSC-23766 was a generous gift from Ivan Robert Nabi (Department of Cellular and Physiological Sciences, University of British Columbia). Human NHE5 36HA and NHE5- and NHE1-specific shRNA constructs were previously described (Diering et al., 2011, 2013). YFP-Rac1 pEYFP-C1 (11391; Hoppe and Swanson, 2004) and GST-PBD pGEX2TK (12217) were purchased from Addgene (Cambridge, MA).
Immunofluorescence
Cells were grown on glass coverslips overnight and fixed with 3% paraformaldehyde (PFA) for 10 min at room temperature (RT); this was followed by permeabilization with 0.1% Triton X-100 in phosphate-buffered saline (PBS) for 10 min. For visualization of endogenous NHE5, cells were fixed and permeabilized in cold 1:1 methanol/acetone at −20°C for 10 min. The cells were then blocked with a 1% bovine serum albumin (BSA)-PBS solution, probed with primary antibodies diluted with 0.1% BSA-PBS at 4°C overnight, and incubated with Alexa Fluor–conjugated secondary antibodies in 0.1% BSA-PBS at RT for 45 min. After extensive washing, coverslips were mounted onto microscope slides using ProLong Gold Antifade Reagent (Invitrogen/Life Technologies/Thermo Fisher). Images were collected with a Leica (Wetzlar, Germany) TCS-SP8 laser scanning confocal microscope (63×/1.40 NA oil objective, Diode/Argon/HeNe lasers, and SP detector).
Immunoblotting
Cells were rinsed with PBS and lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH 7.2, 50 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Nonidet P-40) with protease inhibitor (PI) cocktail (Roche Diagnostic, Indianapolis, IN). Cell debris was cleared by centrifugation at 16,000 × g at 4°C for 15 min, and protein concentration was determined by Bradford assay (Bio-Rad Laboratories). After being denatured at 65°C for 15 min and then at RT for 45 min, proteins were resolved in an SDS–PAGE and electrotransferred onto polyvinylidene fluoride membranes. Membranes were blocked with 5% nonfat milk in PBS-Tw (0.075% Tween-20), incubated overnight with primary antibodies at 4°C, washed, incubated with horseradish peroxidase–conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA) at RT for 40 min, washed, and detected by chemiluminescence using Luminata Forte (Millipore). Quantifications of the blots were done by densitometry analysis with ImageJ software (National Institutes of Health, Bethesda, MD).
Endosomal pH measurement
Endosomal pH measurements were conducted as described previously (Diering et al., 2013). Cells were grown overnight and serum starved for 3 h before the experiment. Serum-starved cells were incubated with 30 μg/ml of fluorescein-conjugated Tfn and 25 μg/ml of Tfn-Alexa Fluor 568 at 37°C for 30 min; this was followed by a chase incubation for 15 min in the absence of Tfn. For some experiments, 200 nM of Baf was added during Tfn labeling and maintained throughout the experiments. Fluorescein was imaged by excitation with a 488-nm laser, and Alexa Fluor 568 was imaged using a 546-nm laser. Two-color live-cell imaging of perinuclear fluorescence was captured by a spinning-disk confocal microscopy using a 100×/1.45 NA oil objective with appropriate filters (Carl Zeiss Axiovert 200M, Jena, Germany). To convert the fluorescein/Alexa Fluor 568 fluorescence ratios to pH, we imaged fluorescein- or Alexa Fluor 568–conjugated Tfn-loaded cells under pH-clamp conditions (140 mM KCl, 10 mM glucose, 1 mM MgCl2, 2 mM CaCl2, 20 mM HEPES [pH 7.0]/PIPES [pH 6.5]/MES [pH 6.0, 5.5], 10 μM Nigericin).
HGF-induced Akt/Erk activation assay
Cells were grown at 37°C and serum starved overnight before HGF stimulation. Cells were incubated with 50 ng/ml of HGF in prewarmed serum-free DMEM for 0, 2, 5, 15, 30, 60, or 120 min at 37°C. They were then quickly washed with ice-cold PBS and lysed in 1% NP40-TBS containing 2 mM sodium orthovanadate and protease inhibitors, and quantified cell lysates were analyzed by immunoblotting. Signals were quantified by densitometric analysis. Relative Erk and Akt activities were obtained as ratios of phosphorylated and total corresponding proteins that are normalized to the peak intensity at 15 min of the control cells.
Cell surface biotinylation
Semiconfluent cells were rinsed twice with ice-cold PBS-CM (1 mM MgCl2, 0.1 mM CaCl2) and incubated with biotinylation solution (0.5 mg/ml sulfo-NHS-SS-biotin in PBS-CM) at 4°C for 45 min. Cells were then incubated with quenching buffer (20 mM glycine in PBS-CM) for 10 min, washed with ice-cold PBS, and lysed with RIPA buffer supplemented with PI, and 30 μg of total proteins were incubated overnight at 4°C with NeutrAvidin agarose beads (Thermo Fischer Scientific). After extensive washing, proteins were eluted from the beads and analyzed by immunoblotting. For examining internalization of cell surface biotinylated proteins, cells treated with sulfo-NHS-SS-biotin were subjected to a chase incubation at 37°C in prewarmed culture media; this was followed by an incubation with a cleavage buffer (50 mM glutathione, 90 mM NaCl, 1 mM MgCl2, 0.1 mM CaCl2, 0.2% BSA, pH 8.6) at 4°C for 15 min. Internalized proteins during the postbiotinylation incubation are protected from the cleavage reagent, whereas biotinylated proteins remaining on the plasma membrane are cleaved. For examination of the proportion of cell surface biotinylated proteins recycled, cells were first treated with sulfo-NHS-SS-biotin and incubated in the culture medium at 37°C for 30 min to facilitate internalization of cell surface biotinylated proteins. The subsequent incubation of the cells with the membrane-impermeable cleavage buffer at 4°C removed biotin from biotinylated proteins remaining on the cell surface. The cells were then subjected to a second round of incubation in the culture medium at 37°C for 15 min to facilitate recycling of biotinylated proteins. When cells were treated with the cleavage buffer, biotin was removed from the “recycled” proteins but not from proteins remaining inside the cell (“nonrecycled” proteins). For some experiments, cells were treated with 100 μM of primaquine before biotin labeling and maintained in the culturing media for the duration of the experiment.
Cell polarity assays
Equal-width wounds were generated with a pipette tip to a confluent monolayer of serum-starved cells. Following incubation at 37°C in culture media containing 10% FBS for 6 h, cells were fixed, and the positions of the Golgi apparatus and nuclei were visualized by anti-mouse GM130 antibody and Hoechst 33342, respectively. For some experiments, 50 nM of Baf was added 5 h before the induction of wounds and during the 6-h incubation period. Cell polarity was quantified by a percentage of cells exhibiting correct orientation at the leading edge (polarity index). Cell orientations were determined by placing an arbitrary rhombus on each cell with the tip of 120° angle at the center of nucleus and with the longer axis parallel to the wound. Correct orientation is scored if the Golgi apparatus resides within the plane. Ten to 15 confocal images per cell type were acquired, and more than 300 cells were scored per experiment.
Degradation assays
Cells seeded at a density of 5 × 105 into a 35-mm plastic dish were grown 1 d and serum starved for 16 h. Cells were then treated with 50 ng/ml of HGF for 0, 30, 60, and 120 min. For examination of the effect of specific degradation pathways, cells were treated with 20 μM of MG-132 for 3 h, 100 μM of primaquine for 3 h, 100 μM of leupeptin for 23 h, or MG-132 and leupeptin in conjunction before as well as during the 60-min HGF stimulation period. Cells were then lysed, and equal amounts of proteins were analyzed by immunoblotting. For every sample, the MET signal was first normalized to the NHE1 signal, and the resulting value was then normalized to the unstimulated level.
Rho GTPase activity assays
Pak-PBD (p21-binding domain of p21 activated protein kinase 1) inserted into pGEX-2T (GE Healthcare, Little Chalfont, UK) was transformed into BL21. GST-PBD fusion protein expression was induced by 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) for 4 h. Bacterial pellets were collected, lysed, and cleared by centrifugation, and the bacterial cell lysate obtained was incubated with glutathione beads (GE Healthcare) to affinity purify the GST-PBD fusion protein. Two micrograms of immobilized recombinant protein was incubated with either 25 μg or 100 μg C6 cell lysates for 1 h at 4°C to detect Rac1 and Cdc42, respectively. Beads were then washed with RIPA buffer, lysates were eluted in SDS sample buffer by heating at 65°C for 15 min in SDS sample buffer, and Rac1 or cdc42 bound to GST-PBD was detected by immunoblotting. The intensity of the signal was quantified by densitometric analysis. Relative GTPase activities were normalized to the value of the control cells. For some experiments, cells were treated with 100 μM of the Rac1 inhibitor NSC-23766 (Santa Cruz Biotechnology) for 24 h. For HGF activation assay, cells were serum starved overnight; this was followed by addition of 50 ng/ml HGF for 5 min.
Membrane fractionation
Cells were collected and resuspended in sucrose buffer (250 mM sucrose, 1 mM EDTA, 10 mM Tris, pH 7.4, PI). The cell suspension was passed through a 26.5-gauge needle on ice 20 times to shear cells, and the cell debris was cleared by centrifugation at 800 × g at 4°C for 15 min twice. The S-0.8 supernatant was subjected to ultracentrifugation at 98,000 × g at 4°C for 30 min, and the resulting S-98 supernatant was collected as a cytosolic fraction. The pellet was gently washed with ice-cold PBS and resuspended in RIPA buffer. Insoluble debris was cleared by centrifugation, and the supernatant was collected as a membrane fraction. Proteins in each fraction were quantified by Bradford assays and analyzed by immunoblotting.
Migration assays
Cells were seeded into the Transwell permeable support insert (Corning, NY) at 4 × 104 in 200 μl serum-free media per insert and incubated inside a 24-well plate containing 300 μl of DMEM with 20% FBS per well at 37°C for 6 h at 5% CO2. Cells were then fixed with 3% PFA for 15 min. Cells that remained on the upper side of the membrane were scraped off with a cotton swab, and those that migrated through the membrane pores were stained with 0.1% crystal violet for 20 min. The inserts were rinsed with water and air-dried. Images were taken using an inverted light microscope. Five randomly selected images of each cell line were scored, and experiments were repeated five to eight times depending on the cell line.
Statistical analysis
We conducted analysis of variance (ANOVA) when comparing more than two groups and obtained p values using the Tukey-Kramer test for pairwise comparisons. When necessary, the model included experiment dates as an explanatory variable. Student’s paired t test (two-tailed) was used for comparisons of two groups only. With the p-Akt or p-ERK time-course data for Figure 5B, repeated-measures ANOVA was conducted, with time as a within-subject factor. p Values for pairwise comparison were obtained at each time point following the ANOVA. All the analyses were conducted using Microsoft Excel (Microsoft, Redmond, WA) or Stata/IC 14.0 (StataCorp LP, College Station, TX).
Supplementary Material
Acknowledgments
We thank Ivan Robert Nabi, Wun-Chey Sin, Caroline Perronnet, Hakima Moukhles (Cellular and Physiological Sciences, University of British Columbia), and Pieter Cullis (Biochemistry and Molecular Biology, University of British Columbia) for providing reagents and Andy Yu-Ting Chen for technical assistance. This study was partly supported by Natural Sciences and Engineering Research Council of Canada (NSERC) discovery grants.
Abbreviations used:
- ANOVA
analysis of variance
- Baf
bafilomycin A1
- BSA
bovine serum albumin
- CLICs
chloride intracellular channels
- CNO-NHEs
cation nonspecific organelle-membrane bound NHEs
- EGFR
epidermal growth factor receptor
- FBS
fetal bovine serum
- GST
glutathione S-transferase
- HA
hemagglutinin
- HGF
hepatocyte growth factor
- MAPK
mitogen-activated protein kinase
- NGF
nerve growth factor
- NHEs
Na+/H+-exchangers
- PBD
p21-binding domain of p21 activated protein kinase 1
- PBS
phosphate-buffered saline
- PFA
paraformaldehyde
- PI
protease inhibitor
- PI3K
phosphatidylinositol-3-kinase
- RIPA
radioimmunoprecipitation assay
- RT
room temperature
- RTK
receptor tyrosine kinase
- shRNA
short hairpin RNA
- Tfn
transferrin
- TfR
transferrin receptor
- TGN
trans-Golgi network
- V-ATPase
vacuolar-type proton-translocating ATPase.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-04-0257) on December 23, 2015.
REFERENCES
- Baird NR, Orlowski J, Szabo EZ, Zaun HC, Schultheis PJ, Menon AG, Shull GE. Molecular cloning, genomic organization, and functional expression of Na+/H+ exchanger isoform 5 (NHE5) from human brain. J Biol Chem. 1999;274:4377–4382. doi: 10.1074/jbc.274.7.4377. [DOI] [PubMed] [Google Scholar]
- Boccaccio C, Comoglio PM. Invasive growth: a MET-driven genetic programme for cancer and stem cells. Nat Rev Cancer. 2006;6:637–645. doi: 10.1038/nrc1912. [DOI] [PubMed] [Google Scholar]
- Bornens M. Organelle positioning and cell polarity. Nat Rev Mol Cell Biol. 2008;9:874–886. doi: 10.1038/nrm2524. [DOI] [PubMed] [Google Scholar]
- Boucrot E, Ferreira APA, Almeida-Souza L, Debard S, Vallis Y, Howard G, Bertot L, Sauvonnet N, McMahon HT. Endophilin marks and controls a clathrin-independent endocytic pathway. Nature. 2015;517:460–465. doi: 10.1038/nature14067. [DOI] [PubMed] [Google Scholar]
- Brett CL, Donowitz M, Rao R. Evolutionary origins of eukaryotic sodium/proton exchangers. Am J Physiol Cell Physiol. 2005;288:C223–C239. doi: 10.1152/ajpcell.00360.2004. [DOI] [PubMed] [Google Scholar]
- Chianale F, Rainero E, Cianflone C, Bettio V, Pighini A, Porporato PE, Filigheddu N, Serini G, Sinigaglia F, Baldanzi G, Graziani A. Diacylglycerol kinase alpha mediates HGF-induced Rac activation and membrane ruffling by regulating atypical PKC and RhoGDI. Proc Natl Acad Sci USA. 2010;107:4182–4187. doi: 10.1073/pnas.0908326107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong D, Zhu W, Shi Y, Pointer KB, Clark PA, Shen H, Kuo JS, Hu S, Sun D. Upregulation of NHE1 protein expression enables glioblastoma cells to escape TMZ-mediated toxicity via increased H+ extrusion, cell migration and survival. Carcinogenesis. 2014;35:2014–2024. doi: 10.1093/carcin/bgu089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diering GH, Church J, Numata M. Secretory carrier membrane protein 2 regulates cell-surface targeting of brain-enriched Na+/H+ exchanger NHE5. J Biol Chem. 2009;284:13892–13903. doi: 10.1074/jbc.M807055200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diering GH, Mills F, Bamji SX, Numata M. Regulation of dendritic spine growth through activity-dependent recruitment of the brain-enriched Na+/H+ exchanger NHE5. Mol Biol Cell. 2011;22:2246–2257. doi: 10.1091/mbc.E11-01-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diering GH, Numata Y, Fan S, Church J, Numata M. Endosomal acidification by Na+/H+ exchanger NHE5 regulates TrkA cell-surface targeting and NGF-induced PI3K signaling. Mol Biol Cell. 2013;24:3435–3448. doi: 10.1091/mbc.E12-06-0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donowitz M, Ming Tse C, Fuster D. SLC9/NHE gene family, a plasma membrane and organellar family of Na+/H+ exchangers. Mol Aspects Med. 2013;34:236–251. doi: 10.1016/j.mam.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dozynkiewicz MA, Jamieson NB, Macpherson I, Grindlay J, van den Berghe PV, von Thun A, Morton JP, Gourley C, Timpson P, Nixon C, et al. Rab25 and CLIC3 collaborate to promote integrin recycling from late endosomes/lysosomes and drive cancer progression. Dev Cell. 2012;22:131–145. doi: 10.1016/j.devcel.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza S, Garcia-Cabado A, Yu F, Teter K, Lukacs G, Skorecki K, Moore HP, Orlowski J, Grinstein S. The epithelial sodium-hydrogen antiporter Na+/H+ exchanger 3 accumulates and is functional in recycling endosomes. J Biol Chem. 1998;273:2035–2043. doi: 10.1074/jbc.273.4.2035. [DOI] [PubMed] [Google Scholar]
- Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- Fuster DG, Alexander RT. Traditional and emerging roles for the SLC9 Na+/H+ exchangers. Pflügers Arch. 2014;466:1–16. doi: 10.1007/s00424-013-1408-8. [DOI] [PubMed] [Google Scholar]
- Gao W, Chan JM, Farokhzad OC. pH-responsive nanoparticles for drug delivery. Mol Pharmaceutics. 2010;7:1913–1920. doi: 10.1021/mp100253e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatenby RA, Gillies RJ. A microenvironmental model of carcinogenesis. Nat Rev Cancer. 2008;8:56–61. doi: 10.1038/nrc2255. [DOI] [PubMed] [Google Scholar]
- Gherardi E, Birchmeier W, Birchmeier C, Woude GV. Targeting MET in cancer: rationale and progress. Nat Rev Cancer. 2012;12:89–103. doi: 10.1038/nrc3205. [DOI] [PubMed] [Google Scholar]
- Hammond DE, Urbe S, Vande Woude GF, Clague MJ. Down-regulation of MET, the receptor for hepatocyte growth factor. Oncogene. 2001;20:2761–2770. doi: 10.1038/sj.onc.1204475. [DOI] [PubMed] [Google Scholar]
- Hiebsch RR, Raub TJ, Wattenberg BW. Primaquine blocks transport by inhibiting the formation of functional transport vesicles. Studies in a cell-free assay of protein transport through the Golgi apparatus. J Biol Chem. 1991;266:20323–20328. [PubMed] [Google Scholar]
- Honasoge A, Sontheimer H. Involvement of tumor acidification in brain cancer pathophysiology. Front Physiol. 2013;4:316. doi: 10.3389/fphys.2013.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe AD, Swanson JA. Cdc42, Rac1, and Rac2 display distinct patterns of activation during phagocytosis. Mol Biol Cell. 2004;15:3509–3519. doi: 10.1091/mbc.E03-11-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado-Lorenzo A, Skinner M, El Annan J, Futai M, Sun-Wada GH, Bourgoin S, Casanova J, Wildeman A, Bechoua S, Ausiello DA, et al. V-ATPase interacts with ARNO and Arf6 in early endosomes and regulates the protein degradative pathway. Nat Cell Biol. 2006;8:124–136. doi: 10.1038/ncb1348. [DOI] [PubMed] [Google Scholar]
- Jeffers M, Taylor GA, Weidner KM, Omura S, Vande Woude GF. Degradation of the Met tyrosine kinase receptor by the ubiquitin-proteasome pathway. Mol Cell Biol. 1997;17:799–808. doi: 10.1128/mcb.17.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joffre C, Barrow R, Menard L, Calleja V, Hart IR, Kermorgant S. A direct role for Met endocytosis in tumorigenesis. Nat Cell Biol. 2011;13:827–837. doi: 10.1038/ncb2257. [DOI] [PubMed] [Google Scholar]
- Kagami T, Chen S, Memar P, Choi M, Foster LJ, Numata M. Identification and biochemical characterization of the SLC9A7 interactome. Mol Membr Biol. 2008;25:436–447. doi: 10.1080/09687680802263046. [DOI] [PubMed] [Google Scholar]
- Kondapalli KC, Llongueras JP, Capilla-Gonzalez V, Prasad H, Hack A, Smith C, Guerrero-Cazares H, Quinones-Hinojosa A, Rao R. A leak pathway for luminal protons in endosomes drives oncogenic signalling in glioblastoma. Nat Commun. 2015;6:6289. doi: 10.1038/ncomms7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong DS, Song SY, Kim DH, Joo KM, Yoo JS, Koh JS, Dong SM, Suh YL, Lee JI, Park K, et al. Prognostic significance of c-Met expression in glioblastomas. Cancer. 2009;115:140–148. doi: 10.1002/cncr.23972. [DOI] [PubMed] [Google Scholar]
- Kwiatkowska A, Didier S, Fortin S, Chuang Y, White T, Berens ME, Rushing E, Eschbacher J, Tran NL, Chan A. The small GTPase RhoG mediates glioblastoma cell invasion. Mol Cancer. 2012;11:65–72. doi: 10.1186/1476-4598-11-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence SP, Bright NA, Luzio JP, Bowers K. The sodium/proton exchanger NHE8 regulates late endosomal morphology and function. Mol Biol Cell. 2010;21:3540–3551. doi: 10.1091/mbc.E09-12-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ES, Gao Z, Bae YH. Recent progress in tumor pH targeting nanotechnology. J Control Release. 2008;132:164–170. doi: 10.1016/j.jconrel.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PJ, Williams WP, Kobiljski J, Numata M. Caveolins bind to (Na+, K+)/H+ exchanger NHE7 by a novel binding module. Cell Signal. 2007;19:978–988. doi: 10.1016/j.cellsig.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Lin PJ, Williams WP, Luu Y, Molday RS, Orlowski J, Numata M. Secretory carrier membrane proteins interact and regulate trafficking of the organellar (Na+,K+)/H+ exchanger NHE7. J Cell Sci. 2005;118:1885–1897. doi: 10.1242/jcs.02315. [DOI] [PubMed] [Google Scholar]
- Liu W, Fu Y, Xu S, Ding F, Zhao G, Zhang K, Du C, Pang B, Pang Q. c-Met expression is associated with time to recurrence in patients with glioblastoma multiforme. J Clin Neurosci. 2011;18:119–121. doi: 10.1016/j.jocn.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Macpherson IR, Rainero E, Mitchell LE, van den Berghe PVE, Speirs C, Dozynkiewicz MA, Chaudhary S, Kalna G, Edwards J, Timpson P. CLIC3 controls recycling of late endosomal MT1-MMP and dictates invasion and metastasis in breast cancer. J Cell Sci. 2014;127:3893–3901. doi: 10.1242/jcs.135947. [DOI] [PubMed] [Google Scholar]
- McLean LA, Roscoe J, Jorgensen NK, Gorin FA, Cala PM. Malignant gliomas display altered pH regulation by NHE1 compared with nontransformed astrocytes. Am J Physiol Cell Physiol. 2000;278:C676–C688. doi: 10.1152/ajpcell.2000.278.4.C676. [DOI] [PubMed] [Google Scholar]
- Menard L, Parker PJ, Kermorgant S. Receptor tyrosine kinase c-Met controls the cytoskeleton from different endosomes via different pathways. Nat Commun. 2014;5:3907. doi: 10.1038/ncomms4907. [DOI] [PubMed] [Google Scholar]
- Miyamoto M, Ojima H, Iwasaki M, Shimizu H, Kokubu A, Hiraoka N, Kosuge T, Yoshikawa D, Kono T, Furukawa H. Prognostic significance of overexpression of c-Met oncoprotein in cholangiocarcinoma. Br J Cancer. 2011;105:131–138. doi: 10.1038/bjc.2011.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PA, Trinidad AG, Timpson P, Morton JP, Zanivan S, van den Berghe PV, Nixon C, Karim SA, Caswell PT, Noll JE, et al. Mutant p53 enhances MET trafficking and signalling to drive cell scattering and invasion. Oncogene. 2013;32:1252–1265. doi: 10.1038/onc.2012.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K, Shimao Y, Sato S, Kataoka H, Moriyama T, Kawano H, Wakisaka S, Koono M. Expression of c-Met correlates with grade of malignancy in human astrocytic tumours: an immunohistochemical study. Histopathology. 1997;31:436–443. doi: 10.1046/j.1365-2559.1997.3010889.x. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Tanaka S, Teko Y, Mitsui K, Kanazawa H. Four Na+/H+ exchanger isoforms are distributed to Golgi and post-Golgi compartments and are involved in organelle pH regulation. J Biol Chem. 2005;280:1561–1572. doi: 10.1074/jbc.M410041200. [DOI] [PubMed] [Google Scholar]
- Nelson WJ. Remodeling epithelial cell organization: transitions between front-rear and apical-basal polarity. Cold Spring Harb Perspect Biol. 2009;1:a000513. doi: 10.1101/cshperspect.a000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numata M, Orlowski J. Molecular cloning and characterization of a novel (Na+, K+)/H+ exchanger localized to the trans-Golgi network. J Biol Chem. 2001;276:17387–17394. doi: 10.1074/jbc.M101319200. [DOI] [PubMed] [Google Scholar]
- Ohgaki R, van ISC, Matsushita M, Hoekstra D, Kanazawa H. Organellar Na+/H+ exchangers: novel players in organelle pH regulation and their emerging functions. Biochemistry. 2011;50:443–450. doi: 10.1021/bi101082e. [DOI] [PubMed] [Google Scholar]
- Onishi I, Lin PJ, Numata Y, Austin P, Cipollone J, Roberge M, Roskelley CD, Numata M. Organellar (Na+, K+)/H+ exchanger NHE7 regulates cell adhesion, invasion and anchorage-independent growth of breast cancer MDA-MB-231 cells. Oncol Rep. 2012;27:311–317. doi: 10.3892/or.2011.1542. [DOI] [PubMed] [Google Scholar]
- Orlowski J, Grinstein S. Emerging roles of alkali cation/proton exchangers in organellar homeostasis. Curr Opin Cell Biol. 2007;19:483–492. doi: 10.1016/j.ceb.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlowski J, Grinstein S. Na+/H+ exchangers. Compr Physiol. 2011;1:2083–2100. doi: 10.1002/cphy.c110020. [DOI] [PubMed] [Google Scholar]
- Palamidessi A, Frittoli E, Garre M, Faretta M, Mione M, Testa I, Diaspro A, Lanzetti L, Scita G, Di Fiore PP. Endocytic trafficking of Rac is required for the spatial restriction of signaling in cell migration. Cell. 2008;134:135–147. doi: 10.1016/j.cell.2008.05.034. [DOI] [PubMed] [Google Scholar]
- Parachoniak CA, Luo Y, Abella JV, Keen JH, Park M. GGA3 functions as a switch to promote Met receptor recycling, essential for sustained ERK and cell migration. Dev Cell. 2011;20:751–763. doi: 10.1016/j.devcel.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks SK, Chiche J, Pouysségur J. Disrupting proton dynamics and energy metabolism for cancer therapy. Nat Rev Cancer. 2013;13:611–623. doi: 10.1038/nrc3579. [DOI] [PubMed] [Google Scholar]
- Petrelli A, Gilestro GF, Lanzardo S, Comoglio PM, Migone N, Giordano S. The endophilin-CIN85-Cbl complex mediates ligand-dependent downregulation of c-Met. Nature. 2002;416:187–190. doi: 10.1038/416187a. [DOI] [PubMed] [Google Scholar]
- Presley JF, Mayor S, McGraw TE, Dunn KW, Maxfield FR. Bafilomycin A1 treatment retards transferrin receptor recycling more than bulk membrane recycling. J Biol Chem. 1997;272:13929–13936. doi: 10.1074/jbc.272.21.13929. [DOI] [PubMed] [Google Scholar]
- Ridley AJ. Life at the leading edge. Cell. 2011;145:1012–1022. doi: 10.1016/j.cell.2011.06.010. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Comoglio PM, Hall A. Regulation of scatter factor/hepatocyte growth factor responses by Ras, Rac, and Rho in MDCK cells. Mol Cell Biol. 1995;15:1110–1122. doi: 10.1128/mcb.15.2.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardet C, Franchi A, Pouyssegur J. Molecular cloning, primary structure, and expression of the human growth factor-activatable Na+/H+ antiporter. Cell. 1989;56:271–280. doi: 10.1016/0092-8674(89)90901-x. [DOI] [PubMed] [Google Scholar]
- Segarra J, Balenci L, Drenth T, Maina F, Lamballe F. Combined signaling through ERK, PI3K/AKT, and RAC1/p38 is required for Met-triggered cortical neuron migration. J Biol Chem. 2006;281:4771–4778. doi: 10.1074/jbc.M508298200. [DOI] [PubMed] [Google Scholar]
- Singleton PA, Salgia R, Moreno-Vinasco L, Moitra J, Sammani S, Mirzapoiazova T, Garcia JG. CD44 regulates hepatocyte growth factor-mediated vascular integrity. Role of c-Met, Tiam1/Rac1, dynamin 2, and cortactin. J Biol Chem. 2007;282:30643–30657. doi: 10.1074/jbc.M702573200. [DOI] [PubMed] [Google Scholar]
- Stauber T, Weinert S, Jentsch TJ. Cell biology and physiology of CLC chloride channels and transporters. Compr Physiol. 2012;2:1701–1744. doi: 10.1002/cphy.c110038. [DOI] [PubMed] [Google Scholar]
- Trusolino L, Bertotti A, Comoglio PM. MET signalling: principles and functions in development, organ regeneration and cancer. Nat Rev Mol Cell Biol. 2010;11:834–848. doi: 10.1038/nrm3012. [DOI] [PubMed] [Google Scholar]
- Usatyuk PV, Fu P, Mohan V, Epshtein Y, Jacobson JR, Gomez-Cambronero J, Wary KK, Bindokas V, Dudek SM, Salgia R, et al. Role of c-Met/phosphatidylinositol 3-kinase (PI3k)/Akt signaling in hepatocyte growth factor (HGF)-mediated lamellipodia formation, reactive oxygen species (ROS) generation, and motility of lung endothelial cells. J Biol Chem. 2014;289:13476–13491. doi: 10.1074/jbc.M113.527556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Weert AW, Geuze HJ, Groothuis B, Stoorvogel W. Primaquine interferes with membrane recycling from endosomes to the plasma membrane through a direct interaction with endosomes which does not involve neutralisation of endosomal pH nor osmotic swelling of endosomes. Eur J Cell Biol. 2000;79:394–399. doi: 10.1078/0171-9335-00062. [DOI] [PubMed] [Google Scholar]
- Wiedmann RM, von Schwarzenberg K, Palamidessi A, Schreiner L, Kubisch R, Liebl J, Schempp C, Trauner D, Vereb G, Zahler S. The V-ATPase-inhibitor Archazolid abrogates tumor metastasis via inhibition of endocytic activation of the Rho-GTPase Rac1. Cancer Res. 2012;72:5976–5987. doi: 10.1158/0008-5472.CAN-12-1772. [DOI] [PubMed] [Google Scholar]
- Wu H, Zhu L, Torchilin VP. pH-sensitive poly(histidine)-PEG/DSPE-PEG co-polymer micelles for cytosolic drug delivery. Biomaterials. 2013;34:1213–1222. doi: 10.1016/j.biomaterials.2012.08.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.