Abstract
Structure has traditionally been interrelated with sequence, usually in the framework of comparing sequences across species sharing a common structural fold. However, the nature of information within the sequence and structure databases is evolving, changing the type of comparisons possible. In particular, we now have a vast amount of personal genome sequences from human populations and a larger fraction of new structures contain interacting proteins within large complexes. Consequently, we have to recast our conception of sequence conservation and its relation to structure – e.g. focusing more on selection within the human population. Moreover, within structural biology there is less emphasis on the discovery of novel folds and more on relating structures to networks of protein interactions. We cover this changing mindset here.
Introduction
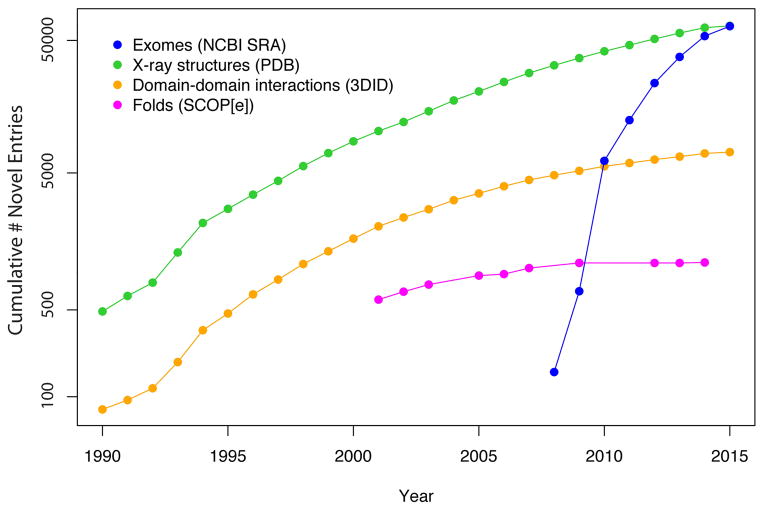
The amount of personal genomic information is growing at a rapid pace leading to a vast change in the nature of information stored within biological databases (Figure 1) [1]. In particular, before the completion of the human genome project in 2003, we had a large amount of genomic sequence information from different species and structural data in the databases. Due to the technological advances in next-generation sequencing, the amount of human sequence information has grown at an unprecedented pace. Meanwhile, even though the number of protein structures in the PDB database [2] has also increased, the pace of identifying new folds has slowed down indicating that few new folds remain undiscovered. However, a large number of novel domain-domain interactions are detected in the newly deposited structures indicating that the complexity of the structures in the PDB database continues to grow (Figure 1). This trend illustrates an increasing emphasis among structural biologists to treat biomolecules not as individual folds but rather as complex molecular machines that interact and regulate each another as they function within the cellular environment. Together, these trends suggest that the stage is set to integrate sequence and structural information to rationalize the effect of variants on protein function.
Figure 1.
The pace of novel fold discovery has begun to saturate, while the volume of X-Ray crystal structures and structurally-resolved protein-protein interactions has continued to grow. However, the pace with which personal genomic sequencing databases are growing is considerably greater than the pace at which structure databases are growing.
The identification and characterization of pathological disease-associated variants is an essential goal of genomic sequencing efforts [3,4]. A large number of medically-relevant mutations occur within proteins, some of which are available through databases such as the Online Database of Mendelian Inheritance in Man (OMIM) [5], the Human Gene Mutation Database (HGMD) [6], Humsavar [7], and ClinVar [8]. It is essential to utilize structural information for rationalizing the evolutionary pressure acting on these proteins as well as for developing drugs to combat the effects of disease-causing variants. However, it remains challenging to annotate the physical effects of these mutations on proteins and protein complexes, as the nature of functional constraints is highly multifaceted. A protein-coding variant may cause local or global changes in structure, or it may have a substantial impact on the protein-protein interaction (PPI) network, and each type of change adds a different layer of functional constraints on the protein. Such analyses are further complicated by the fact that we currently have incomplete knowledge of these constraints, and also by the fact that specific combinations of individually benign variants may cause disease.
While structural data provides an invaluable guide for rationalizing disease-associated variants, we also expect the growing genomic information to be a valuable resource for structural biologists. In particular, as the amount of genomic data continues to grow, we envision a future in which biologists will utilize genetic variation within human population(s) to help interpret their structural data [9,10]. Population genetic analysis within human proteins has already been used to identify novel species-specific functional constraints within a protein family [11]. In addition, a number of fundamental insights about biological pathways can be garnered by analyzing newly discovered loci associated with a disease [12].
In this review article, we initially explain how genomic information is used to identify pathological disease associated variants as well as variants that are harmful to protein function even within healthy individuals. We later describe how structural information is utilized to understand the harmful effects of different variants. Finally, we discuss the need to integrate sequence and structural data with a holistic system or network perspective before predicting phenotypic effects of the variants.
Classical Sequence Comparison
Typically, structural biologists identify functionally constrained regions within a protein family by comparing homologous sequences from different species (Figure 2a) [13,14]. They focus on changes that take place over longer evolutionary timescales by comparing the reference (or dominant) sequence within each species rather than focusing on intra-species changes. Nucleotides that do not change across different species are conserved over millions of years and are hence considered to be functionally important. Due to redundancy within the genetic code, some of the changes in the coding regions are silent as they occur without a corresponding change in the protein sequence (synonymous changes). With rare exceptions, all synonymous changes and a majority of the nonsynonymous changes are expected to be neutral or harmful to the protein function. A small fraction of the nonsynonymous changes can, however, be beneficial to the fitness of the species.
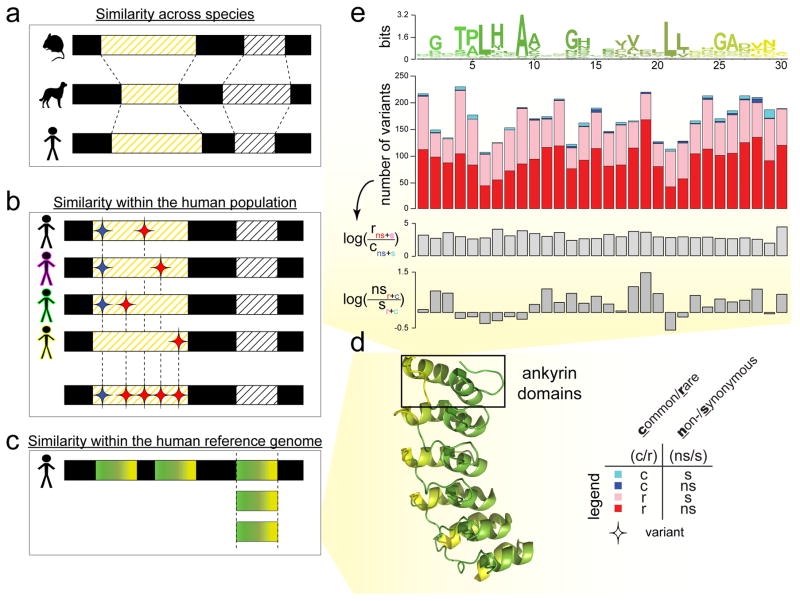
Figure 2. Evolutionary conservation in different contexts.
Evolutionary conservation can be inferred via sequence comparison in different contexts. (A) The examination of sequence conservation in orthologous sequences across multiple species looks at a longer evolutionary timescale. (B) The examination of the enrichment of rare variants (or depletion of common variants) in the same genomic element across multiple individuals within a single species or population looks at a shorter evolutionary timescale. Here, the red diamonds denote variants that are rare in a single human population (found in only one or a small number of individuals) and the blue diamonds denote variants that are commonly found in multiple individuals in the population. (C) The examination of sequence conservation in similar protein domain sequences within a single genome can reveal species- and domain-specific conservation that might be important to the structure or function of the domain family. (D) To illustrate (C), we use ankyrin protein domains as an example. We translate the DNA sequence of each ankyrin domain into its amino acid sequence. In order to relate the positions of the linear sequence of an ankyrin repeat domain to their structural locations, we then specifically paint each of the six ankyrin domains found in the structure of the human Notch 1 ankyrin domain (PDB ID: 1YYH) similar to the sequence profile in (E). (E) The top plot in this panel is the sequence profile of an ankyrin repeat domain with 30 amino acids, colored by position left to right, from green to yellow, corresponding to the coloring of the motifs of the human Notch 1 PDB structure in (D). In the sequence profile, the height of the amino acid letters connotes the degree of conservation of a particular residue at a specific location along the ankyrin repeat; the degree of conservation is computed using relative entropy in bits of information. To examine evolutionary conservation in more detail, the sequence profile can be further analysed with genomic variant profiles. For example, for each of the position along the ankyrin motif, the second plot shows the absolute numbers of variants binned into four categories: cyan bars show the number of variants that are common (c) and synonymous (s); blue bars for variants that are common and non-synonymous (ns); pink bars, rare (r) and synonymous; red bars, rare and non-synonymous. Subsequently, we can derive log ratios from these numbers to demonstrate an enrichment (or depletion) of categories of variants, in order to gain further biological insights. Here, the third subplot displays a general enrichment of rare variants relative to common variants across the entire motif, suggesting a uniform evolutionary importance of the ankyrin domain in the human population. However, the fourth subplot exhibits a depletion of nonsynonymous variants relative to the synonymous variants at more conserved motif positions (in the sequence profile), hinting at only a subset of positions being of particular functional importance to the ankyrin domain family.
The ratio of nonsynonymous to synonymous variants (dN/dS) is commonly utilized to characterize the selection pressure on the coding regions of the genome (Figure 2) [15]. If the dN/dS ratio for a coding region is substantially less than 1, it indicates that a few of these mutations are harmful or deleterious and that the protein is under negative selection. On the other hand, a dN/dS ratio substantially exceeding unity indicates that evolution is promoting a change in the protein sequence and that this protein (or protein region) is under positive selection [11]. Proteins undergoing positive selection may improve the fitness of an organism to different environments.
Introduction to Population Sequencing
The vast amounts of genomic and exome sequences available are providing unique opportunities to characterize genetic variation within the human population (Table 1). The exome comprises the coding sequences of all protein-coding genes and constitutes approximately 1% of the total genomic sequence [16]. Due to the reduced cost of exome sequencing and better-characterized clinical relevance of variation within the coding regions of the genome, it is more widely used for genetic diagnosis. Variants within an individual’s genome are either acquired at birth (germline mutations) or during the person’s lifetime (somatic mutations) as a consequence of errors during cell division. While germline mutations are typically present in every cell of the person, somatic mutations only affect certain cells and are typically not passed on to the next generation. There are approximately 74 de novo (new) variants that occur during each generation [17]. As only germline mutations are passed on to the next generation, somatic mutations are not under conventional evolutionary selection.
Table 1.
Some existing and ongoing human genome sequencing projects.
| Dataset | Number of individuals | Healthy/Diseases (H/D) | Exome/Genome (E, G, E+G) | Ref |
|---|---|---|---|---|
| Complete Genomics Data | 69 | H | G | 1 |
| Singapore Sequencing Malay Project | 100 | H | G | 2 |
| Genome of the Netherlands | 767 | D | G | 3 |
| 1000 Genome Project Phase 3 | 2,504 | H | E+G | 4 |
| Personal Genome Project | 4,419* | H | G | 5 |
| Exome Sequencing Project (ESP) | 6,515 | D | E | 6 |
| UK10K project | 10,000 | D | E+G | 7 |
| The Cancer Genome Atlas (TCGA) | 11,080 | H+D | E+G | 8 |
| Exome Aggregation Consortium (ExAC) | 60,706 | H+D | E | 9 |
| TOTAL | 82,772^ |
The numbers in the table are correct as of July 28th 2015.
The Personal Genome Project sets a target of sequencing 100,000 personal genomes.
This total excludes 1,851 individuals from 1000 Genomes Project Phase 3, 3,936 from the ESP and 7,601 from TCGA since they are also included in the ExAC dataset.
References
Complete Genomics: http://www.completegenomics.com/public-data/69-Genomes/
Wong, L.-P., Ong, R. T.-H., Poh, W.-T., Liu, X., Chen, P., Li, R., Lam, K. K.-Y., Pillai, N. E., Sim, K.-S., Xu, H., Sim, N.-L., Teo, S.-M., Foo, J.-N., Tan, L. W.-L., Lim, Y., Koo, S.-H., Gan, L. S.-H., Cheng, C.-Y., Wee, S., Yap, E. P.-H., Ng, P. C., Lim, W.-Y., Soong, R., Wenk, M. R., Aung, T., Wong, T.-Y., Khor, C.-C., Little, P., Chia, K.-S. & Teo, Y.-Y. Deep whole-genome sequencing of 100 southeast Asian Malays. Am J Hum Genet 92, 52–66 (2013).
Genome of the Netherlands: http://www.genoomvannederland.nl/?page_id=9
The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature (2015) (in press)
Personal Genome Project: https://my.personalgenomes.org/users
Tennessen, J. A., Bigham, A. W., O’Connor, T. D., Fu, W., Kenny, E. E., Gravel, S., McGee, S., Do, R., Liu, X., Jun, G., Kang, H. M., Jordan, D., Leal, S. M., Gabriel, S., Rieder, M. J., Abecasis, G., Altshuler, D., Nickerson, D. A., Boerwinkle, E., Sunyaev, S., Bustamante, C. D., Bamshad, M. J. & Akey, J. M. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science (New York, N.Y.) 337, 64–9 (2012).
UK10K: http://www.uk10k.org/
The Cancer Genome Atlas Portal: https://dcc.icgc.org/
Exome Aggregation Consortium: http://exac.broadinstitute.org/faq
The human genome exhibits extensive variation [18–22]. On average, any individual genome contains 20,000–25,000 coding variants (Table 2), of which 9,000–11,000 are nonsynonymous. The frequency with which a particular variant or allele occurs within a population is used to characterize the evolutionary pressure acting on it as common variants (minor allele frequency greater > 5%) are expected to be benign. However, rare variants (minor allele frequency < 0.5%) are rare either because they are harmful (deleterious) to a protein’s function or because the variant has been introduced recently into the population. The ratio of common to rare variants is often used as a proxy to characterize the evolutionary pressure acting on a locus. Although most of the variants within any particular individual are common, most coding variants manifest as distinct single nucleotide variants (SNVs), each of which occurs very rarely within the human population. About 25–50% of the rare nonsynonymous variants within healthy individuals are estimated to be deleterious, suggesting that the human proteome is highly robust to a large number of non-specific perturbations and because most rare deleterious variants are heterozygous implying that the cell also contains a functional copy of the gene [20,21].
Table 2.
Per-individual whole exome SNV load in the 1000 Genomes Project Phase 1 data.
| SYNONYMOUS | NON-SYNONYMOUS | |||||||
|---|---|---|---|---|---|---|---|---|
| DAF <0.5% | DAF 0.5–5% | DAF >5% | Total | DAF <0.5% | DAF 0.5–5% | DAF >5% | Total | |
| AVERAGE | 295 | 1,014 | 12,892 | 14,201 | 434 | 1,055 | 10,816 | 12,305 |
| YRI | 547 | 2,468 | 12,190 | 15,205 | 691 | 2,377 | 10,056 | 13,130 |
| CEU | 175 | 593 | 13,237 | 14,006 | 298 | 709 | 11,173 | 12,180 |
| CHB | 218 | 497 | 13,077 | 13,792 | 355 | 563 | 11,026 | 11,944 |
| JPT | 240 | 500 | 13,067 | 13,807 | 387 | 571 | 11,012 | 11,970 |
The number of synonymous and non-synonymous SNVs are categorized into three ranges of derived allele frequency (DAF; defined as the allele alternative to the ancestral allele). DAF<0.5% are considered ‘rare’. Ancestry legend, YRI: Yoruba in Ibadan, Nigeria; CEU: Utah Residents (CEPH) with Northwestern European ancestry; CHB: Han Chinese in Beijing; JPT: Japanese in Tokyo, Japan.
Despite the fact that new genomic data is still being produced, about 200,000–500,000 previously unobserved SNVs are still discovered after each personal genome is sequenced, suggesting that we have not yet reached a saturation in the extent of available human polymorphism data [20,21]. Indeed, the number of rare variants continues to grow even after the 1000 Genomes Consortium and Exome Aggregation Consortium data (60,706 individuals) [23] has become available. As deleterious mutations tend to occur at very low frequencies, we need to continue sequencing a large number of individuals to characterize and catalog these variants and their frequencies within the human population.
As such, we can turn to intra-human comparisons to uncover more human- or domain-specific features (Figure 2). There is, however, an important distinction between interpreting inter- and intra-species conservation due to the huge disparities in the associated evolutionary timescales (Figure 2a–c). While performing such an analysis, one can also align homologous coding regions not only between individuals (Figure 2b), but also within a single human genome (i.e., paralogs), such as proteins originating from the same structural domain family (Figure 2c). In particular, this can be used to elucidate domain-specific features.
Similar to the dN/dS ratio in cross-species comparisons, selective pressure on coding regions can be quantified using fraction of synonymous to nonsynonymous polymorphisms (pN/pS) at any site (Figure 2e). In addition, evolutionary pressure can also be quantified during intra-species comparison using the ratio of rare to common variants at each site as rare variants are under stronger negative selection (Figure 2e). A statistically significant depletion of common variants as compared to rare variants implies that the site is under stronger selective pressure. Furthermore, genomic variants that are increasing in frequency within a human population (positive selection) may help identify a novel gain-of-function event (such as a new protein-protein interaction). Some of these domain-specific events may be beneficial to the species. Comparative genetics/genomics studies have already uncovered a growing list of genes that might have experienced positive selection during the evolution of human and/or primates [11]. These genes offer valuable inroads into understanding the biological processes specific to humans, as well as the evolutionary forces that gave rise to them. It is also important to note that some variants occur in a correlated fashion within the population and these variants are said to be under linkage disequilibrium (LD). Note also that LD is statistically easier to observe for common variants than for rare ones.
Deleterious Effects of Variations on Protein Function
The patterns of conservation displayed by proteins are the product of a vast array of constraints active throughout its evolutionary history. In this regard, to understand the physical effects that cause a variant to be harmful, we need to consider the multitude of underlying constraints acting on the protein family. Such constraints are often intrinsic to the structure itself: they may include the need to maintain the integrity of functional hinge regions or interior packing geometry or the ability to regulate a protein through post-translational modifications at specific sites. They may also entail that residues at an interaction interface remain topologically compatible with those in the corresponding interface of an interaction partner. We can utilize the structural information in the PDB database to assess the effect of mutations on a protein’s stability as nonsynonymous changes that occur within the core of the protein or variants that disrupt the secondary structure of the protein could reduce its stability. Several computational tools based on sequence conservation (inter-species or intra-species) and/or several structural features (the physicochemical characteristics of the amino acid change, solvent accessibility, secondary structure, active site annotations, and protein-protein interfaces) were developed to predict the deleterious effect of sequence variations on a protein’s function [24–27]. Disease-associated mutations are highly enriched for residues in the interior of proteins (22% of all mutations in HGMD and OMIM), and active sites of proteins [18–21].
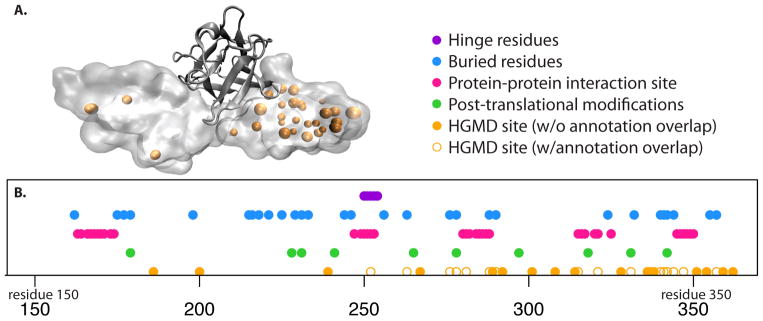
In terms of applying such a catalogue of rules as a means of understanding human disease-associated variants, the fibroblast growth factor receptor provides a case-in-point, several variants in which have been linked to craniofacial defects (Figure 3). The evolutionary constraints listed here provide sensible rationales for how many of these disease-associated variants may impart deleterious effects. Importantly, these constraints may act in synergistic ways rather than through isolated mechanisms [28,29]. However, the mechanisms for several other disease-associated variants fail to map to this catalogue, thereby underscoring the need to more comprehensively document sources of constraint. This more comprehensive documentation needs to transcend the native structure itself by including the folding pathways, allosteric regulation, and the functional roles of disordered regions or conformational transitions. Such mutations that affect the thermodynamic stability of different allosteric states of a protein [30] are typically ignored while predicting the deleteriousness of a putative variant. In addition, as discussed earlier, several deleterious mutations occur even in healthy individuals within the population, as discussed below, the network properties of a protein need to be integrated with this structural information before the phenotypic effect of any individual variant can be predicted.
Figure 3.
(A) The fibroblast growth factor receptor is shown in complex with FGF2 (PDB 1IIL), along with the loci of HGMD variants (orange spheres). (B) Various structural annotations (i.e., a “catalogue of constraints”) are shown in sequence space. Hinge residues are taken from HingeMaster [61], buried residues are identified using NACCESS [62], protein-protein interaction residues are defined to be those within 4.5 Angstroms of the co-crystallized growth factor, and post-translational modification sites are taken from UniProt. HGMD loci shown as holo circles coincide with the catalogue of constraints, and may thus likely be rationalized in light of such constraints. However, a large number of HGMD loci (shown in filled orange circles) fail to overlap with these annotations, highlighting the need to consider alternative sources of constraint.
Networks as a Framework for Understanding Deleterious Variants
While structural and sequence information are invaluable in providing a rationale for the deleterious effects of certain disease-causing and rare variations, it is often difficult to interpret the phenotypic effects of an individual variant without considering the broader cellular context. As proteins are extensively involved in protein-DNA interactions (gene regulatory network), protein-RNA interactions (post-transcriptional regulation), and protein-protein interactions (PPI) within the cellular milieu, variants that disrupt these interactions could potentially affect the viability of the cell. We refer the reader to comprehensive essays on the phenotypic effect of noncoding variation [31,32], and focus instead on deleterious effects of variants on the protein-protein interaction (PPI) network here.
Various experimental and computational approaches have been applied to characterize the PPI network in several model organisms and human beings [33,34] and these networks have been invaluable in interpreting the role of evolutionary constraints on a protein family. In the PPI network, a node represents a protein, while an edge represents an interaction between the two proteins connected by the edge. Proteins that are highly interconnected in PPI networks (hubs) are under strong negative selection while proteins under positive selection in humans tend to occur at the periphery of the network [35]. Proteins that are more central in an integrated “multinet” formed by integrating biological networks from different context (PPI, metabolic, post-translational modification, gene regulatory network, etc.) are under negative selection within human populations [36]. In agreement with this, perturbations to hub proteins are more likely to be associated with diseases than non-hub proteins [37].
The PPI networks are organized in a modular fashion as proteins associated with the same function are more likely to interact with one another [38] and proteins associated with similar diseases tend to occur within the same module [37]. The system properties of the network have also been useful in interpreting how the human proteome is robust even in the presence of a large number of deleterious variants within healthy individuals. Most deleterious variants observed in healthy individuals occur in peripheral regions of the interactome. Such limited effects may result as a consequence of compensatory mutations or functional redundancy [39]. On the other hand, cancer-associated somatic deleterious variations occur in the internal regions of the interactome and tend to have larger structural consequences on the PPI network.
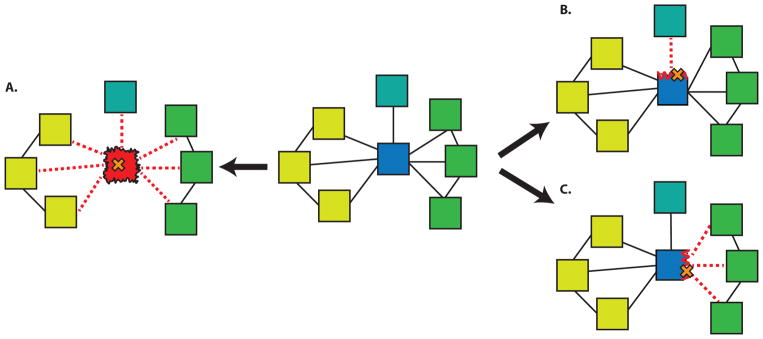
The interactome provides a convenient platform to measure the impact of a deleterious variant on the cell. As shown in Figure 4, a deleterious variant can either remove a protein (such a node effect would naturally also result in the removal of all the associated edges) from the PPI network by making a protein nonfunctional or it could lead to the loss of just one or more of its interactions (edgetic effects). Mutations at a PPI interface can have drastic effects on the biomolecular binding constant and several sequence and structure-based methods have been proposed to identify these interaction hotspots [40,41]. Even though we have incomplete information on the structures of protein complexes (Figure 1), it has been predicted that about 12% of all the HGMD and OMIM mutations occur at a PPI interface [42] while approximately 28% of experimentally-tested HGMD missense mutations affect one or more interactions, thus underscoring the importance of these interactions for annotating rare variants and disease-associated mutations [43].
Figure 4.
Various mechanisms of SNP-induced disruption in protein-protein interaction networks. A SNP that destabilizes a hub protein can ablate all associated interactions (A). SNPs disrupting different interfaces of the hub may interfere with interactions active in different tissues (B, C). Blue (hub protein), Yellow (nodes expressed in tissue1), Green (nodes expressed in tissue2), Turquoise (node expressed in tissue3). Mutation in cystathionine β-synthase (CBS) leads to metabolic disease called Homocystinuria. Among many HGMD SNPs impacting this protein, experimental evidence [63] suggest that I278T mutation leads to destabilization of CBS, which further disrupts of all three important interactions involving this protein and this is equivalent to removing a node from the PPI network. Mutation in EFHC1 gene, which has been implicated in epilepsy, presents a good example of edgetic effect [43]. This mutation perturbs interaction of EFHC1 with ZBED1 and TCF4. While the perturbed interaction between EFHC1 and ZBED1 interfere with cell proliferation [64], on the other hand disturbance in EFHC1 and TCF4 interaction influence the neuronal differentiation process [65].
In an effort to bridge the information gained from individual structures with network properties of the interactome, Kim, et al., [44] combined the experimentally determined interactome with structural information from the iPfam database to form the structural interaction network (SIN) and were able to obtain a higher-resolution understanding of the selection constraints on the hubs. Using structural information, the hubs were classified into different groups based on the number of distinct interfaces utilized for biomolecular complex formation and they showed that the number of distinct interfaces is a better proxy for evolutionary pressure acting on the hub rather than the number of edges in the PPI network. Consistent with this interpretation, hub proteins in the PPI network contain a higher fraction of disease-causing mutations on their solvent exposed surface, as compared to non-hub proteins suggesting that a larger fraction of a hub’s disease-associated mutations could affect its interactions [44].
Hub proteins interact with a large number of partners and tend to be more flexible and conformationally heterogenous than non-hub proteins [45]. Furthermore, the number of distinct interfaces in hub proteins is correlated with degrees of conformational heterogeneity [45]. To the extent that variants may enable or disable certain conformational states from being visited, such mutations could potentially affect protein complex formation and signaling pathways, and this has not yet been examined very closely. As deleterious mutations that affect hubs in networks tend to have a larger effect on the structures, they would also cause large changes in the PPI network. Proteins can utilize different interfaces for different (sets of) interactions, so multiple mutations on the same protein can be associated with drastically different diseases depending on the afflicted interface. Such mutations would have different edgetic effects on the protein’s interaction network - by breaking or weakening one of its interactions while the rest of its interactions remain intact - and a large proportion of HGMD and OMIM mutations are predicted to have edgetic effects on the PPI network [43,46].
It should also be noted that the hubs in PPI networks also tend to contain higher degrees of disordered regions (that display even higher amounts of conformational flexibility), and these regions typically become well-ordered upon ligand or protein binding [47,48]. Disease-associated mutations are enriched within disordered regions of the protein as they could affect post-translational modifications and/or protein-protein interaction sites [49,50]. The assessment of a mutation’s effect on the activity of an intrinsically disordered protein is even more challenging because it would be dependent upon the effects of these mutations upon the unfolded ensemble or the structure gained in the presence of its interaction partner. Due to their inherent flexibility, the unfolded ensembles of disordered proteins are especially difficult to characterize using either experimental or computational techniques [51,52], making variant annotation in the context of disordered proteins an uphill task. However, the phenotypic effect of mutations on the functional viability of a disordered protein is important because mutations to disordered regions tend to have large phenotypic effects as they could affect PPI interactions of hub proteins.
Ultimately, the goal is to develop an integrative framework to understand the effects of deleterious variants on the phenotype of the cell. However, a mutation typically displays tissue-specific phenotypic effects, hence an understanding of functional constraints on a protein should also incorporate tissue-specific information. While the gene regulatory network is being mapped out in a developmental time point and cell type-dependent fashion by several international consortia [53,54] the PPI network is largely treated in a static fashion. Recent works have tried to integrate proteome and gene expression profiles with PPI networks to create tissue-specific networks [55–57]. However, these studies typically neglect the protein isoform even though the protein’s interactions are dependent on its isoform [58,59]. A structural study on the effect of sequence variations on isoform-dependent PPI complexes has not been performed and would improve the prediction of phenotypic effects due to missense mutations. However, it is likely that the high costs in resources associated with studying isoform-specific assays in various cell types have impeded these types of studies. It should be noted that a number of proteins also change their interaction partners in a tissue-specific manner based upon the dominant isoform of the protein in that tissue [59]. Recent evidence suggests that many mutations occurring on these alternatively-spliced disordered motifs may drive cancer [60]. We anticipate that isoform-specific protein-protein interaction network annotation will become easier and more accessible in the near future, which will present new opportunities to better annotate such networks.
Conclusions
The exponential growth in genomic data has demonstrated that a large amount of genomic variation is present within the human population, and this data has also helped identify a vast number of rare variants and disease-associated variants. Though the motivation of developing methods to annotate the effects of variants that cause human disease is clear, it remains challenging to do so as it requires bridging disparate sources of information together to understand the functional constraints on a protein family. It is essential to utilize structural information to rationalize the effect of variants. The network properties of the protein in addition to sequence and structural information regarding the nonsynonymous amino acid changes need to be considered within a single framework before predicting the phenotypic impact of an amino acid change.
Highlights.
Increasing amounts of sequencing data add new dimension to study structural biology.
Next generation sequencing facilitates analysis of genetic variations.
Understanding evolutionary constraints acting on proteins remains elusive.
Essential to integrate sequences, structures, and interaction networks information to rationalize the phenotypic impact of these variations.
Acknowledgments
We acknowledge support from NIH and the AL Williams Professorship funds. DC acknowledges the support of the NIH Predoctoral Program in Biophysics (T32 GM008283-24).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1*.Stephens ZD, Lee SY, Faghri F, Campbell RH, Zhai C, Efron MJ, Iyer R, Schatz MC, Sinha S, Robinson GE. Big Data: Astronomical or Genomical? PLoS Biol. 2015;13:e1002195. doi: 10.1371/journal.pbio.1002195. This is an excellent perspective on genomics as a big data science and how new technologies will need to be developed to meet the computational challenges that genomics poses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Offit K. Personalized medicine: new genomics, old lessons. Hum Genet. 2011;130:3–14. doi: 10.1007/s00439-011-1028-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chin L, Andersen JN, Futreal PA. Cancer genomics: from discovery science to personalized medicine. Nat Med. 2011;17:297–303. doi: 10.1038/nm.2323. [DOI] [PubMed] [Google Scholar]
- 5.Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stenson PD, Mort M, Ball EV, Howells K, Phillips AD, Thomas NS, Cooper DN. The Human Gene Mutation Database: 2008 update. Genome Med. 2009;1:13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, Maglott DR. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sulkowska JI, Morcos F, Weigt M, Hwa T, Onuchic JN. Genomics-aided structure prediction. Proc Natl Acad Sci U S A. 2012;109:10340–10345. doi: 10.1073/pnas.1207864109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marks DS, Hopf TA, Sander C. Protein structure prediction from sequence variation. Nat Biotechnol. 2012;30:1072–1080. doi: 10.1038/nbt.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chothia C, Lesk AM. The relation between the divergence of sequence and structure in proteins. Embo j. 1986;5:823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durbin R, et al. Biological Sequence Analysis. Cambridge University Press; 1998. [Google Scholar]
- 15.Kryazhimskiy S, Plotkin JB. The population genetics of dN/dS. PLoS Genet. 2008;4:e1000304. doi: 10.1371/journal.pgen.1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler EE, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Veltman JA, Brunner HG. De novo mutations in human genetic disease. Nat Rev Genet. 2012;13:565–575. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- 18.Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khurana E, Fu Y, Colonna V, Mu XJ, Kang HM, Lappalainen T, Sboner A, Lochovsky L, Chen J, Harmanci A, et al. Integrative annotation of variants from 1092 humans: application to cancer genomics. Science. 2013;342:1235587. doi: 10.1126/science.1235587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22**.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. This is the final publication from the 1000 Genomes Consortium and contains the most up to date and comprehensive description and distribution of genetic variation within healthy human individuals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23*.Exome Aggregation Consortium (ExAC) on World Wide Web URL. http://exac.broadinstitute.orgThis is a resource from which people can download the common and rare variants found within exomes of healthy human populations.
- 24.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 25.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bromberg Y, Rost B. SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007;35:3823–3835. doi: 10.1093/nar/gkm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li B, Krishnan VG, Mort ME, Xin F, Kamati KK, Cooper DN, Mooney SD, Radivojac P. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics. 2009;25:2744–2750. doi: 10.1093/bioinformatics/btp528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishi H, Fong JH, Chang C, Teichmann SA, Panchenko AR. Regulation of protein-protein binding by coupling between phosphorylation and intrinsic disorder: analysis of human protein complexes. Mol Biosyst. 2013;9:1620–1626. doi: 10.1039/c3mb25514j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishi H, Hashimoto K, Panchenko AR. Phosphorylation in protein-protein binding: effect on stability and function. Structure. 2011;19:1807–1815. doi: 10.1016/j.str.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perica T, Kondo Y, Tiwari SP, McLaughlin SH, Kemplen KR, Zhang X, Steward A, Reuter N, Clarke J, Teichmann SA. Evolution of oligomeric state through allosteric pathways that mimic ligand binding. Science. 2014;346:1254346. doi: 10.1126/science.1254346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ward LD, Kellis M. Interpreting noncoding genetic variation in complex traits and human disease. Nat Biotechnol. 2012;30:1095–1106. doi: 10.1038/nbt.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Albert FW, Kruglyak L. The role of regulatory variation in complex traits and disease. Nat Rev Genet. 2015;16:197–212. doi: 10.1038/nrg3891. [DOI] [PubMed] [Google Scholar]
- 33.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 34.Rolland T, Tasan M, Charloteaux B, Pevzner SJ, Zhong Q, Sahni N, Yi S, Lemmens I, Fontanillo C, Mosca R, et al. A proteome-scale map of the human interactome network. Cell. 2014;159:1212–1226. doi: 10.1016/j.cell.2014.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim PM, Korbel JO, Gerstein MB. Positive selection at the protein network periphery: evaluation in terms of structural constraints and cellular context. Proc Natl Acad Sci U S A. 2007;104:20274–20279. doi: 10.1073/pnas.0710183104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36**.Khurana E, Fu Y, Chen J, Gerstein M. Interpretation of genomic variants using a unified biological network approach. PLoS Comput Biol. 2013;9:e1002886. doi: 10.1371/journal.pcbi.1002886. This study develops an integrative network framework and interprets human genetic variation within human populations using this network. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL. The human disease network. Proc Natl Acad Sci U S A. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharan R, Ulitsky I, Shamir R. Network-based prediction of protein function. Mol Syst Biol. 2007;3:88. doi: 10.1038/msb4100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia-Alonso L, Jimenez-Almazan J, Carbonell-Caballero J, Vela-Boza A, Santoyo-Lopez J, Antinolo G, Dopazo J. The role of the interactome in the maintenance of deleterious variability in human populations. Mol Syst Biol. 2014;10:752. doi: 10.15252/msb.20145222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ofran Y, Rost B. Protein-protein interaction hotspots carved into sequences. PLoS Comput Biol. 2007;3:e119. doi: 10.1371/journal.pcbi.0030119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aytuna AS, Gursoy A, Keskin O. Prediction of protein-protein interactions by combining structure and sequence conservation in protein interfaces. Bioinformatics. 2005;21:2850–2855. doi: 10.1093/bioinformatics/bti443. [DOI] [PubMed] [Google Scholar]
- 42*.Gao M, Zhou H, Skolnick J. Insights into Disease-Associated Mutations in the Human Proteome through Protein Structural Analysis. Structure. 2015;23:1362–1369. doi: 10.1016/j.str.2015.03.028. This article predicts the mechanistic effect of disease-associated mutations from the HGMD database using structures. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43**.Sahni N, Yi S, Taipale M, Fuxman Bass JI, Coulombe-Huntington J, Yang F, Peng J, Weile J, Karras GI, Wang Y, et al. Widespread macromolecular interaction perturbations in human genetic disorders. Cell. 2015;161:647–660. doi: 10.1016/j.cell.2015.04.013. This study tests the effect of a large subset of disease associated variants from the HGMD database on protein folding and protein-protein interactions and develops a framework to predict the phenotypic effect of mutations by measuring the perturbation in its interactions in yeast. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim PM, Lu LJ, Xia Y, Gerstein MB. Relating three-dimensional structures to protein networks provides evolutionary insights. Science. 2006;314:1938–1941. doi: 10.1126/science.1136174. [DOI] [PubMed] [Google Scholar]
- 45.Bhardwaj N, Abyzov A, Clarke D, Shou C, Gerstein MB. Integration of protein motions with molecular networks reveals different mechanisms for permanent and transient interactions. Protein Sci. 2011;20:1745–1754. doi: 10.1002/pro.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X, Wei X, Thijssen B, Das J, Lipkin SM, Yu H. Three-dimensional reconstruction of protein networks provides insight into human genetic disease. Nat Biotechnol. 2012;30:159–164. doi: 10.1038/nbt.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim PM, Sboner A, Xia Y, Gerstein M. The role of disorder in interaction networks: a structural analysis. Mol Syst Biol. 2008;4:179. doi: 10.1038/msb.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oldfield CJ, Dunker AK. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem. 2014;83:553–584. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- 49.Uversky VN, Dave V, Iakoucheva LM, Malaney P, Metallo SJ, Pathak RR, Joerger AC. Pathological unfoldomics of uncontrolled chaos: intrinsically disordered proteins and human diseases. Chem Rev. 2014;114:6844–6879. doi: 10.1021/cr400713r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vacic V, Iakoucheva LM. Disease mutations in disordered regions--exception to the rule? Mol Biosyst. 2012;8:27–32. doi: 10.1039/c1mb05251a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eliezer D. Biophysical characterization of intrinsically disordered proteins. Curr Opin Struct Biol. 2009;19:23–30. doi: 10.1016/j.sbi.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sethi A, Tian J, Vu DM, Gnanakaran S. Identification of minimally interacting modules in an intrinsically disordered protein. Biophys J. 2012;103:748–757. doi: 10.1016/j.bpj.2012.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Acuner Ozbabacan SE, Gursoy A, Nussinov R, Keskin O. The structural pathway of interleukin 1 (IL-1) initiated signaling reveals mechanisms of oncogenic mutations and SNPs in inflammation and cancer. PLoS Comput Biol. 2014;10:e1003470. doi: 10.1371/journal.pcbi.1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mosca R, Ceol A, Aloy P. Interactome3D: adding structural details to protein networks. Nat Methods. 2013;10:47–53. doi: 10.1038/nmeth.2289. [DOI] [PubMed] [Google Scholar]
- 57.Magger O, Waldman YY, Ruppin E, Sharan R. Enhancing the prioritization of disease-causing genes through tissue specific protein interaction networks. PLoS Comput Biol. 2012;8:e1002690. doi: 10.1371/journal.pcbi.1002690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ellis JD, Barrios-Rodiles M, Colak R, Irimia M, Kim T, Calarco JA, Wang X, Pan Q, O’Hanlon D, Kim PM, et al. Tissue-specific alternative splicing remodels protein-protein interaction networks. Mol Cell. 2012;46:884–892. doi: 10.1016/j.molcel.2012.05.037. [DOI] [PubMed] [Google Scholar]
- 59.Buljan M, Chalancon G, Eustermann S, Wagner GP, Fuxreiter M, Bateman A, Babu MM. Tissue-specific splicing of disordered segments that embed binding motifs rewires protein interaction networks. Mol Cell. 2012;46:871–883. doi: 10.1016/j.molcel.2012.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Colak R, Kim T, Michaut M, Sun M, Irimia M, Bellay J, Myers CL, Blencowe BJ, Kim PM. Distinct types of disorder in the human proteome: functional implications for alternative splicing. PLoS Comput Biol. 2013;9:e1003030. doi: 10.1371/journal.pcbi.1003030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Flores SC, Keating KS, Painter J, Morcos F, Nguyen K, Merritt EA, Kuhn LA, Gerstein MB. HingeMaster: normal mode hinge prediction approach and integration of complementary predictors. Proteins. 2008;73:299–319. doi: 10.1002/prot.22060. [DOI] [PubMed] [Google Scholar]
- 62.Hubbard S, Thornton J. Department of Biochemistry Molecular Biology, University College London, editor. NACCESS, Computer Program. 1993. [Google Scholar]
- 63.Zhong Q, Simonis N, Li QR, Charloteaux B, Heuze F, Klitgord N, Tam S, Yu H, Venkatesan K, Mou D, et al. Edgetic perturbation models of human inherited disorders. Mol Syst Biol. 2009;5:321. doi: 10.1038/msb.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamashita D, Sano Y, Adachi Y, Okamoto Y, Osada H, Takahashi T, Yamaguchi T, Osumi T, Hirose F. hDREF regulates cell proliferation and expression of ribosomal protein genes. Mol Cell Biol. 2007;27:2003–2013. doi: 10.1128/MCB.01462-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Flora A, Garcia JJ, Thaller C, Zoghbi HY. The E-protein Tcf4 interacts with Math1 to regulate differentiation of a specific subset of neuronal progenitors. Proc Natl Acad Sci U S A. 2007;104:15382–15387. doi: 10.1073/pnas.0707456104. [DOI] [PMC free article] [PubMed] [Google Scholar]