SUMMARY
Emerging evidence suggests that microRNAs can initiate asymmetric division, but whether microRNA and protein cell fate determinants coordinate with each other remains unclear. Here we show that miR-34a directly suppresses Numb in early-stage colon cancer stem cells (CCSCs), forming an incoherent feedforward loop (IFFL) targeting Notch to separate stem and non-stem cell fates robustly. Perturbation of the IFFL leads to a new intermediate cell population with plastic and ambiguous identity. Lgr5+ mouse intestinal/colon stem cells (ISCs) predominantly undergo symmetric division, but turn on asymmetric division to curb the number of ISCs when proinflammatory response causes excessive proliferation. Deletion of miR-34a inhibits asymmetric division and exacerbates Lgr5+ ISC proliferation under such stress. Collectively, our data indicate that microRNA and protein cell fate determinants coordinate to enhance robustness of cell fate decision, and they provide a safeguard mechanism against stem cell proliferation induced by inflammation or oncogenic mutation.
Graphical abstract
INTRODUCTION
Cells usually divide symmetrically, producing two identical daughter cells. However, there are prokaryotic and eukaryotic cells that can divide asymmetrically, giving rise to daughter cells with different characteristics (Li, 2013). In higher organisms, asymmetric division is a property associated with many types of stem and progenitor cells in embryo, nervous system, skin, mammary gland, blood, etc, in order to balance proliferation and differentiation as well as aging (Beckmann et al., 2007; Bultje et al., 2009; Inaba and Yamashita, 2012; Jackson et al., 2015; Katajisto et al., 2015; Knoblich, 2008; Neumuller and Knoblich, 2009; Williams et al., 2011). Asymmetric division manages differentiation and self-renewal simultaneously while keeping the number of stem cells constant, making it an attractive mechanism for tissue homeostasis. On the other hand, symmetric division expands the number of stem cells, and often occurs during early embryonic development, tissue regeneration and repair (Morrison and Kimble, 2006). These are certainly not fixed rules, because stem cells often rely on a spatial niche to regulate their number and behavior (Lander et al., 2012). For example, Lgr5+ crypt base columnar (CBC) cells in the intestine predominantly undergo symmetric division, and rely on a neutral drift process in the niche to stabilize their number (Lopez-Garcia et al., 2010; Snippert et al., 2010).
Cancer stem cells, or tumor initiating cells, of various cancer types, undergo both symmetric and asymmetric division (Bajaj et al., 2015; Cicalese et al., 2009; Dey-Guha et al., 2011; Lathia et al., 2011; O’Brien et al., 2012; Pece et al., 2010; Pine et al., 2010; Sugiarto et al., 2011). Loss of tumor suppressor genes often favors increased symmetric divisions of cancer stem cells, which promote proliferation and tumor growth.
Asymmetric cell division usually relies on imbalance of cell fate determinant proteins in the two cellular compartments to break symmetry, resulting in daughter cells with distinct cell fates. A canonical cell fate determinant in Drosophila neuroblasts and various mammalian stem cells, Numb targets membrane-bound Notch receptors for degradation (McGill and McGlade, 2003; Schweisguth, 2004). Furthermore, Numb is a cell fate determinant for various cancer stem cells, and has been used as a marker for distinguishing symmetric vs. asymmetric division (O’Brien et al., 2012).
Recently, emerging evidence suggests that asymmetric distribution of microRNAs can also give rise to asymmetric cell fates (Bu et al., 2013a; Hwang et al., 2014). For example, we have shown that miR-34a directly targets Notch to form a cell fate determination switch in colon cancer stem cells (CCSCs). A tumor suppressor in many cancer types, miR-34a regulates differentiation of embryonic and neural stem cell, somatic cell reprogramming, and cardiac aging (Boon et al., 2013; Choi et al., 2011; He et al., 2007; Liu et al., 2011). miR-34a mimics such as MRX34 are among the first microRNA mimics to reach clinical trial for cancer therapy (Bader, 2012; Bouchie, 2013).
However, this raises the question as to whether microRNA and protein cell fate determinants act independently or coordinate with each other to determine cell fate. The relationship between miR-34a and Numb is intriguing, because both target Notch in CCSCs. Here we show that miR-34a directly bind to the 3′UTR of Numb mRNA to suppress Numb expression, so that miR-34a, Numb, and Notch form an incoherent feedforward loop (IFFL). Combination of computational analysis and quantitative experiments revealed that the unique regulatory kinetics among miR-34a, Numb, and Notch enables a robust binary switch, so that Notch level is steady and insensitive to precise miR-34a level except for a sharp transition region. The switch enforces bimodality and cell fate bifurcation in the population. Subversion of this IFFL via Numb knockdown degrades Notch bimodality and gives rise to an intermediate subpopulation of cells with ambiguous and plastic cell fate. We further show that this cell fate determination switch plays a role in mouse intestinal stem cells (ISCs). Although Lgr5+ ISCs divide symmetrically in normal tissue homeostasis, we found that excessive proliferation caused by pro-inflammatory stress or APC deficiency triggers asymmetric division, which restrains the number of Lgr5+ ISCs. Silencing of the miR-34a-mediated switch inhibits ISC asymmetric division and contributes to CCSC-like proliferation in stressed tissue. Hence, the cell fate determinants provide a safeguard mechanism against excessive stem cell proliferation when normal homeostasis is disrupted by inflammation or oncogenic mutation.
RESULTS
miR-34a directly targets Numb
Using CCSCs derived from patient tumors as we have previously described and characterized (Bu et al., 2013a), we first examined whether miR-34a and Numb spatial distributions are independent or correlated in divided pairs by performing pair-cell assay with immunofluorescence (Bu et al., 2013a; Bultje et al., 2009) (Figure S1A). During asymmetric CCSC division, miR-34a and Numb are mostly present in the NotchlowALDH1low non-CCSC daughter cells (Figure S1B), consistent with their function as Notch suppressors (Figures S1C and S1D) (Bu et al., 2013a). According to co-immunofluorescence for mir-34a and Numb, miR-34a and Numb were present in the same daughter cells in 82% of the divided pairs, whereas they were present in different daughter cells in 18% of the divided pairs (Figures 1A and 1B). Expression of Numbl, a Numb homologue involved in neurogenesis, was not detectable in CCSC.
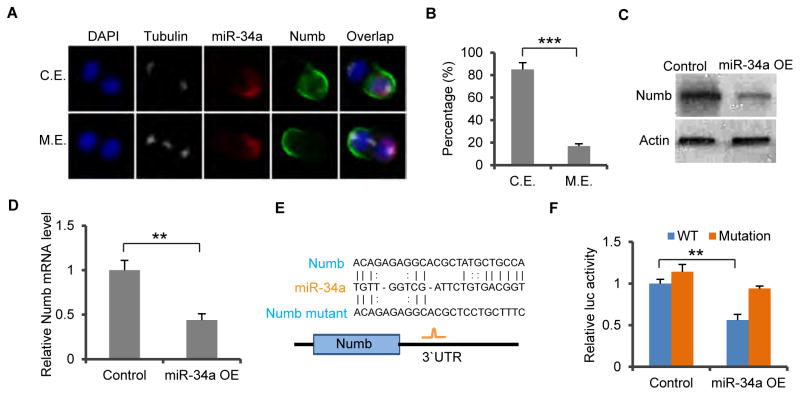
Figure 1. miR-34a directly targets Numb.
(A) Representative images of miR-34a (RNA FISH, red) and Numb (green) distribution during CCSC division. mir-34a and Numb can co-exist (C.E., top row) or be mutually exclusive (M.E., bottom row) in daughter cells. (B) Percentages of CCSC divisions wherein miR-34a and Numb are M.E. or C.E. (C and D) Western blot (C) and RT-qPCR (D) of Numb levels showing ectopic miR-34a expression (miR-34a OE) suppresses Numb expression compared to the control vector. (E) Schematic illustration of predicted binding between miR-34a and Numb 3′ UTR, and mutation introduced to the seed region. (F) Luciferase reporter assay confirming the miR-34a binding site in Numb 3′UTR. Numb 3′UTR sequences containing the wild-type (Wt) or mutated (Mut) putative miR-34a binding sites were cloned into the 3′UTR of firefly luciferase (Fluc). Fluc signals were normalized by Renillar luciferase (Rluc) signals. Mutation of the binding site attenuated suppression of Numb by ectopic miR-34a expression (miR-34a OE). Scale bar, 8μm. Error bars denote s.d. of triplicates. **, p<0.01; ***, p<0.001. p-value was calculated based on Student’s t-test. Also see Figure S1.
We then examined potential interaction between these two cell fate determinants, with the initial hypothesis that one might upregulate the other. We first expressed miR-34a in CCSCs using lentiviral infection and measured Numb expression levels by RT-qPCR and Western blot. Unexpectedly, ectopic miR-34a suppressed Numb expression (Figures 1C and 1D). To investigate whether miR-34a directly targets Numb, we used the microRNA target prediction tool RNA22 to analyze the 3′UTR sequence of Numb and found a putative miR-34a binding site (Figure 1E). The Numb 3′UTR was then cloned into a luciferase reporter, which showed that ectopic miR-34a expression suppressed firefly luciferase activity, whereas mutation in the putative miR-34a seed region in the Numb 3′UTRs abrogated the suppression by miR-34a (Figure 1F). Therefore, miR-34a directly targets Numb mRNA to silence its expression.
miR-34a, Numb, and Notch form an incoherent feedforward loop (IFFL)
It is counterintuitive that miR-34a targets Numb for suppression, considering that both cell fate determinants suppress Notch and promote differentiation. Why does miR-34a suppress Notch directly but upregulate Notch indirectly via Numb? Here, miR-34a, Numb, and Notch form a motif called incoherent feedforward loop (IFFL) (Figure 2A). miR-34a suppresses Notch1 and Numb translation by binding to the 3′UTRs of their mRNA, and Numb suppresses Notch1 by promoting its endocytosis and degradation.
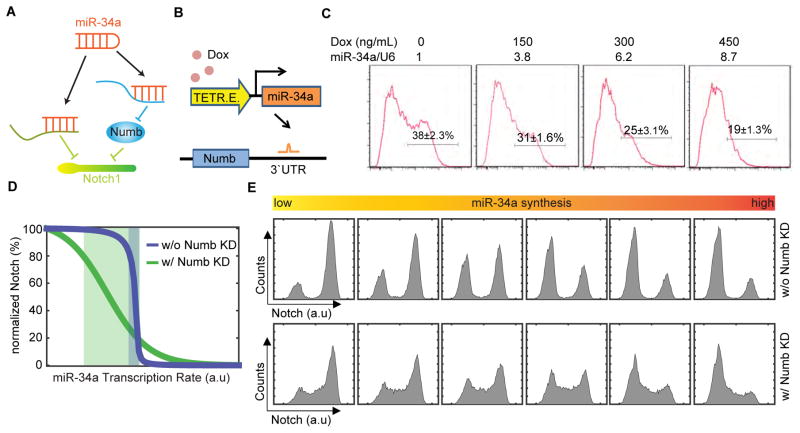
Figure 2. Computational analysis of the incoherent feedforward loop (IFFL).
(A) Schematic of the IFFL formed by miR-34a, Numb, and Notch1. (B) Schematic illustration of the inducible miR-34a construct used in the experiment shown in (C). (C) FACS analysis of Numb expression in CCSC sphere cells when miR-34a expression was incrementally induced by Doxycycline. (D) Simulated Notch1 vs. miR-34a levels from the ODE-based IFFL and Numb knockdown models. Shaded areas are transition regions (80% to 20% of peak Notch level). (E) Simulated Notch1 distributions with IFFL and Numb knockdown models. Also see Figure S2.
Previous studies have found that IFFL can generate non-monotonic, adaptive, or pulse-like responses in different contexts (Goentoro et al., 2009; Kaplan et al., 2008; Mangan et al., 2006), but none of these properties seemed to be particularly relevant to cell fate determination. There have also been computational analyses suggesting that microRNA may reduce noise in IFFL, but those referred to a different topology where the microRNA is suppressed by the protein (Osella et al., 2011).
To understand how miR-34a and Numb may synergize through this arrangement, we explored the quantitative aspects of this particular IFFL. We previously showed that miR-34a generates a threshold response from Notch due to mutual sequestration, while Numb regulates Notch in a graded, continuous way (Bu et al., 2013a; Levine et al., 2007; Mukherji et al., 2011), raising the prospect that this IFFL may possess unique properties. A similar setup was used to characterize the newly discovered miR-34a suppression of Numb. We incrementally increased ectopic miR-34a expression level using a Doxycycline-inducible promoter and performed Fluorescence Activated Cell Sorting (FACS) with antibody against Numb. FACS analysis revealed that incremental miR-34a induction gradually suppressed Numb levels in Numbhigh cells (Figures 2B and 2C).
A computational IFFL model was then constructed by expanding our previously published miR-34a/Notch model to include miR-34a suppression of Numb and Numb suppression of Notch (See Supplemental Information). The model assumes that miR-34a suppression of Notch1 is stronger than its suppression of Numb (which is more gradual), based on the experimental data. Simulation of the model over certain parameter ranges presented an interesting possibility that the IFFL could generate a more robust Notch switch than miR-34a alone (Figure 2D). With IFFL, ‘high’ and ‘low’ Notch levels are steady and insensitive to precise miR-34a level except for a narrow transition (threshold) region, which resembles a typical switch used in electronics. In contrast, Notch levels vary more with a wider transition region if there is only miR-34a but no Numb. Intuitively, when miR-34a level increases, Numb level is suppressed accordingly to offset, hence their combined suppression effect on Notch remains roughly constant until the mutual sequestration threshold is reached. Therefore, the IFFL buffers Notch level from miR-34a copy number variation and enforces a sharp transition only around the mutual sequestration threshold. Further simulations of the model suggested that the steepness of the transition is influenced by the relative strength between the direct and indirect paths (Figures S2A–S2B).
The model made a further prediction that IFFL produces better bimodality of Notch levels (and hence cell fate determination) in the population. Intuitively, the narrower transition region of the IFFL minimizes the number of cells with intermediate Notch levels (Figure 2D). Based on previous FACS measurements of Doxycycline-induced miR-34a level distributions in CCSC sphere cells (Bu et al., 2013a), we performed stochastic simulations of IFFL and miR-34a alone (Numb knockdown). The simulations suggested that, even though miR-34a alone could generate Notch bimodality due to mutual sequestration as previously demonstrated (Bu et al., 2013a), IFFL generates better Notch bimodality with more clearly defined peaks and fewer cells in between, thanks to its more robust switching behavior (Figure 2E).
miR-34a and Numb synergize for a robust Notch bimodal switch
Experiments were then designed to test whether the presence of Numb enhances miR-34a regulation of Notch as a cell fate switch. First, we measured the response of Notch1 level to incremental miR-34a levels with and without Numb. As previously demonstrated, we used CCSCs stably integrated with a lentiviral vector that drives ectopic miR-34a expression with a Doxycycline-inducible promoter(Bu et al., 2013a) (Figure 3A). CCSCs were then infected with a lentiviral vector expressing short hairpin RNAs (shRNAs) against Numb to knock down Numb. The efficiency of Numb knockdown in CCSCs was verified by western blot (Figure S2C).
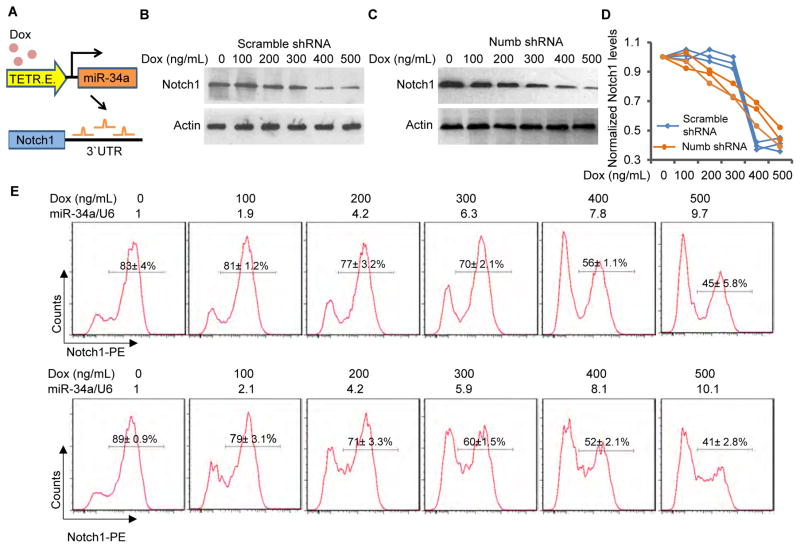
Figure 3. IFFL generates a robust Notch switch.
(A) Schematic illustration of the inducible miR-34a construct used in the experiments shown in (B to E). (B and C) Western blots of Notch levels in scramble shRNA (B) and Numb shRNA (C) infected CCSC spheres with incremental mir-34a induction by Doxycycline. (D) Quantification of Western blots in three independent repeats. (E) FACS analysis of Notch1 bimodality with incremental miR-34a induction by Doxycycline. Top row, intact IFFL; bottom row, Numb knockdown. miR-34a levels were measured by RT-qPCR and shown on top of the FACs plots. Also see Figure S2.
CCSCs in separate wells were then treated with incremental dosages (0, 100, 200, 300, 400, and 500 ng/ml) of Doxycycline. RT-qPCR verified that induced miR-34a expression level increased linearly with Doxycycline dosage in CCSCs with or without Numb knockdown (Figures S2D and S2E). Time-series measurements indicated that it took approximately 42 hours for Notch levels to stabilize after doxycycline induction (Figures S2F–S2I), so steady-state measurements were performed 48 hours post induction. With Numb, Notch levels remained largely steady until being abruptly turned off by 400 ng/ml Doxycycline induction of miR-34a (Figures 3B, 3D and S3J). In contrast, Notch levels gradually decreased and slowly turned off in response to increasing miR-34a levels when Numb was knocked down (Figures 3C, 3D and S3K). These measurements support the computational hypothesis in Figure 2D that miR-34a and Numb work in synergy to generate a more robust switch. Without Numb, Notch level is more sensitive to miR-34a variation.
We then tested whether the presence of Numb enhances miR-34a regulation of Notch to be more bimodal as the computational analysis predicted. Again, we induced miR-34a at different levels and measured Notch protein levels in individual cells using flow cytometry, with antibody against Notch. In CCSCs with Numb, Notch displayed clear bimodality and individual cells were clustered around the Notchhigh or Notchlow peaks. In contrast, in CCSCs with Numb knockdown, even though Notch level distribution was still overall bimodal due to mutual sequestration, bimodality was degraded by a subpopulation of cells with intermediate Notch levels between high and low (Figure 3E). This result is consistent with the computational prediction in Figure 2E that the IFFL improves Notch bimodality.
Intermediate Notch level leads to ambiguous and plastic cell fate
The implication of Notch bimodality on cell fate determination was then investigated. We isolated the Notchhigh, Notchlow and Notchinter cells by FACS (Figures 4A and S3A) and immediately performed immunofluorescence for the CCSC marker ALDH1 and differentiation marker CK20 (Figure 4B). Consistent with previous reports, Notchhigh cells are ALDH1+CK20− stem cells and Notchlow cells are ALDH1-CK20+ differentiated cells. Interestingly, the cells with intermediate Notch levels (Notchinter) expressed both ALDH1 and CK20, reflecting an intermediate state between stem cell and differentiation. RNA-seq transcriptome profiling revealed that Notchinter cells have a distinct gene expression signature between those of Notchhigh and Notchlow cells (Figures 4C and S3B). Notchinter cells express intermediate levels of stem cell and differentiation makers, while Notchhigh cells express high levels of stem cell markers and Notchlow cells express high levels of differentiation markers (Figures S3C). Gene Set Enrichment Analysis (GSEA) show that pathways commonly associated with CCSCs, such as Notch, Wnt, and MAPK signaling pathways, are upregulated in Notchhigh cells (Figure S3D).
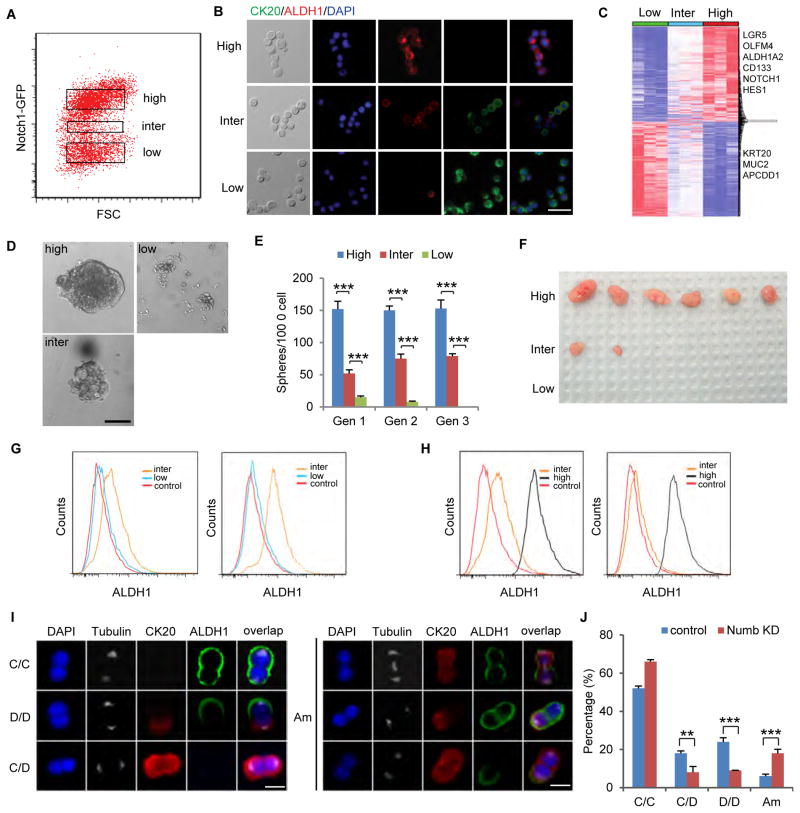
Figure 4. Numb knockdown gives rise to an intermediate population.
(A) FACS plot showing Notchhigh, Notchinter, and Notchlow subpopulations of Numb knockdown sphere cells, treated with 200ng/ml Doxycycline. (B) Immunofluorescence of Notchhigh, Notchinter, and Notchlow cells for CK20 (green) and ALDH1 (red). Scale bar, 20μm. (C) Heat-map of transcriptomes of Notchhigh, Notchinter, and Notchlow cells measured by RNA-seq. (D) Representative images of spheres grown from Notchhigh, Notchinter, and Notchlow cells. Scale bar, 50μm. (E) Serial Sphere propagation of Notchhigh, Notchinter, and Notchlow cells isolated from Numb knockdown sphere cells. Gen, generation. (F) Tumor images showing tumorigenic capability of transplanted Notchhigh, Notchinter, and Notchlow cells. (G) FACS analysis of Notchinter and Notchlow cells before (left) and after (right) being under stem cell culture condition for 7 days. Notchinter cells turned on ALDH1 expression under stem cell culture condition, whereas Notchlow cells did not. (H) FACS analysis of Notchinter and Notchhigh cells before (left) and after (right) being in FBS-containing medium for 24 hours. Notchinter cells lost ALDH1 expression, whereas Notchhigh cells did not. (I) Representative immunofluorescence images for ALDH1 (red) and CK20 (green) illustrating four types of division: CCSC/CCSC (C/C), CCSC/non-CCSC (C/D), non-CCSC/non-CCSC (D/D) and ambiguous (Am). Scale bar, 8μm. (J) Numb knockdown significantly increased Am divisions besides reducing C/D and D/D. Error bars denote s.d. of triplicates. **, p<0.01; ***, p<0.001. p-value was calculated based on Student’s t-test. Also see Figure S3.
We then performed serial sphere propagation assay to test these cells’ self-renewal ability, which is a measure of their stemness (Figures 4D, 4E, S3E). Notchhigh cells efficiently formed spheres in 3D Matrigel culture and maintained their sphere formation capability, whereas Notchlow cells formed few spheres in the first generation and lost their sphere formation capability after serial propagation. Notchinter cells could also form spheres, but the spheres were far fewer and smaller than those formed by Notchhigh cells. We then compared tumorigenic capability by subcutaneously injecting 1×104 Notchhigh, Notchinter and Notchlow cells respectively into Nude mice. During the observed period (6 weeks), all 6 mice injected with Notchhigh cells grew tumors, only 2 mice injected with Notchinter cells grew small tumors, and none of the mice injected with Notchlow cells grew tumors (Figure 4F). Similar results were observed in mice injected with Notchhigh, Notchinter and Notchlow cells sorted from a second CCSC (CCSC2) line (Figure S3F). Therefore, Notchinter cells have intermediate self-renewal and tumorigenic capability compared to Notchhigh and Notchlow cells, consistent with their intermediate gene expression signature.
Since Notchinter cells seem to occupy a state between Notchhigh CCSC and Notchlow non-CCSC, we next examined their plasticity, or ability to convert into CCSC or non-CCSC. When cultured in FBS-free stem cell medium and low-attachment flask, Notchinter cells upregulated the CCSC marker ALDH1, while Notchlow cells did not express ALDH1 after 7 days (Figure 4G). This suggests that Notchinter cells may possess the plasticity to dedifferentiate back into stem cells, in contrast to Notchlow cells. On the other hand, Notchinter cells are more ready to differentiate than Notchhigh cells. When cultured in differentiation medium, Notchinter cells lost ALDH1 expression within 24 hours, whereas Notchhigh cells still retained ALDH1 expression (Figure 4H). It took 10 days for most Notchhigh cells to lose ALDH1 and express CK20. Collectively, these data suggest that Notchinter cells are in an intermediate state that can dedifferentiate into CCSCs or readily commit to differentiation.
We then examined how the presence of the Notchinter cells affects cell division. Pair-cell assay followed by immunofluorescence for ALDH1 and CK20 revealed that Numb knockdown reduced asymmetric division and gave rise to significantly more ambiguous cell division outcomes, wherein one or both daughter cells co-expressed ALDH1 and CK20 (Figures 4I and 4J).
Altogether, the computational analysis and experimental data combined suggest that miR-34a suppresses Numb to form an IFFL, which acts as a robust switch to generate Notch bimodality. Undermining this switch by Numb knockdown results in a subpopulation of cells with intermediate Notch levels. These cells express both stem cell and differentiation markers, and show greater plasticity than Notchhigh and Notchlow cells.
miR-34a and Numb are associated with differentiation of mouse intestinal stem cells
We have previously shown that miR-34a mediated asymmetric cell fate determination is mostly active in CCSCs isolated from early-stage CRC patient specimens, and tends to be silenced in CCSCs isolated from late-stage CRC specimens. CCSCs from early-stage specimens form xenograft tumors in mice that maintain histopathology of their primary human CRCs, which still retain reminiscent features of original colon tissue (Bu et al., 2013a). This raised the possibility that miR-34a and Numb perform cell fate-related functions in normal tissues, which was initially inherited by early-stage CCSCs but eventually subverted in late-stage CCSCs.
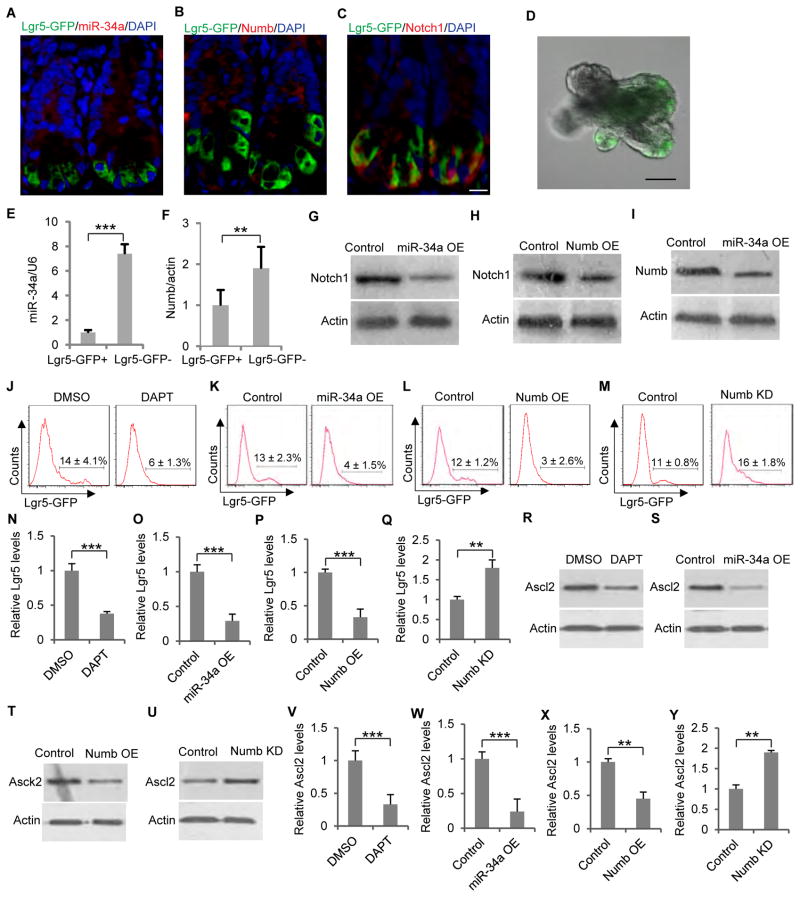
To test this possibility, we first performed immunofluorescence for miR-34a and Numb in cryosectioned mouse intestinal crypts harvested from Lgr5-EGFP-IRES-CreERT2 transgenic mice (Sato et al., 2009). miR-34a and Numb expression are low in GFP-labeled Lgr5+ intestinal stem cells (ISCs), but becomes higher in more differentiated cells above the stem cell niche (Figures 5A and 5B). On the other hand, Notch1, the target of miR-34a and Numb suppression, was more expressed in Lgr5+ ISCs (Figure 5C), consistent with previous reports that Notch is expressed in ISC and essential for ISC self-renewal (Fre et al., 2011; VanDussen et al., 2012).
Figure 5. miR-34a and Numb expression in mouse intestinal cells.
(A to C) Immunofluorescence images of intestinal crypts from an Lgr5-EGFP-CreERT2 transgenic mouse. Scale bar, 20μm. (D) A representative image of an intestinal organoid with Lgr5-GFP labeled ISCs. Scale bar, 50μm. (E and F) miR-34a (E) and Numb (F) expression levels in Lgr5-GFP+ and Lgr5-GFP− cells isolated from Lgr5-EGFP-CreERT2 intestinal organoids, measured by RT-qPCR. (G and H) Western blot showing that ectopic miR-34a (G) or Numb (H) expression decreased Notch1 level in organoid cells. (I) Western blot showing that ectopic miR-34a expression decreased Numb level in organoid cells. (J) DAPT treatment decreased the Lgr5-GFP cell population in Lgr5-EGFP-CreERT2 organoids. (K and L) Ectopic miR-34a (K) or Numb (L) expression decreased the Lgr5-GFP cell population in Lgr5-EGFP-CreERT2 organoids. (M) Numb knockdown gave rise to a subpopulation with intermediate Lgr5-GFP expression. (N to Q) RT-qPCR showing Lgr5 levels in conditions corresponding to J to M. (R to U) Western blot showing Ascl2 levels in conditions corresponding to J to M. (V to Y) RT-qPCR showing Ascl2 levels in conditions corresponding to J to M. Error bars denote s.d. of triplicates. **, p<0.01; ***, p<0.001. p-value was calculated based on Student’s t-test. Also see Figure S4.
To compare Numb and miR-34a expression levels between ISCs and more differentiated cells, we cultured mouse intestinal cells from Lgr5-EGFP-IRES-CreERT2 mice in 3D Matrigel, where they grew into crypt-villus like organoids (Sato et al., 2009) (Figure 5D). The Lgr5+ ISCs (also called CBCs) are capable of both self-renewal and generating other intestinal cell lineages in these organoids. RT-qPCR showed that both Numb and miR-34a expression levels are lower in Lgr5-GFP+ cells than in Lgr5-GFP− cells (Figures 5E and 5F). The difference in expression levels between Lgr5-GFP+ and Lgr5-GFP− cells is greater for miR-34a than for Numb. Flow analysis with RNA FISH probes confirmed low miR-34a expression in Lgr5-GFP+ cells (Figure S4A). Together, the immunofluorescence and RT-qPCR data suggest that miR-34a and Numb expression are associated with more differentiated cells, whereas Notch1 is associated with Lgr5+ ISC.
To validate whether miR-34a and Numb suppress Notch1 in intestinal cells, we infected organoids with lentiviral vectors that express miR-34a or Numb. Transduction and knockdown efficiency was validated by RT-qPCR and Western blot (Figures S4B–S4C). Western blot confirmed that ectopic miR-34a and Numb suppressed Notch1 expression in organoid cells (Figures 5G and 5H). Moreover, ectopic miR-34a expression also downregulated Numb expression, consistent with the IFFL (Figure 5I).
We then investigated how miR-34a, Numb and Notch impact ISC cell fate decision. Inhibition of Notch by treating the organoids with the γ-secretase inhibitor DAPT significantly reduced the Lgr5-GFP+ ISC population in the organoids (Figures 5J, 5N and S4D). Ectopic expression of miR-34a or Numb via lentiviral infection of organoid cells had a similar effect of reducing Lgr5-GFP+ ISCs, consistent with their role of Notch suppression (Figures 5K–5L and 5O–5P). Next, we used a lentiviral vector to express shRNA against Numb in organoids in order to examine whether knockdown of Numb would impact intestinal cell fate bimodality as it does to early-stage CCSCs. The efficiency of Numb knockdown was validated by Western blot (Figure S4E). Indeed, a subpopulation of cells with intermediate Lgr5 expression levels between Lgr5high ISCs and Lgr5low non-ISCs emerged, and the Lgr5-GFP distribution was no longer bimodal (Figures 5M and 5Q). The effects on ISCs were further validated by measuring the levels of Ascl2, an alternative marker for Lgr5+ ISC (van der Flier et al., 2009; VanDussen et al., 2012). Consistently, inhibition of Notch signaling by DAPT, ectopic miR-34a or Numb expression reduced Ascl2 levels, whereas Numb knockdown increased Ascl2 levels in organoids (Figures 5R–5Y). Notch inhibition by DAPT, ectopic miR-34a, or Numb expression also increased apoptotic cells shed into the lumen, a process reminiscent of the shedding of terminally differentiated cells in vivo (Figures S5F–S5I) (Sato et al., 2009).
Inflammatory stress induced miR-34a-dependent asymmetric division
To investigate how loss of the miR-34a-mediated switch may specifically impact ISC cell fate decision, we crossed miR-34aflox/flox mice (Concepcion et al., 2012) with Lgr5-EGFP-IRES-CreERT2 mice and then intraperitoneally administered Tamoxifen, which activated Cre to knock out miR-34a in Lgr5+ ISC specifically. miR-34a knockout in Lgr5+ ISCs did not cause noticeable changes to the crypt morphology or the number of Lgr5-GFP ISCs in vivo or in derived organoids (Figures S5A–S5B). To confirm that miR-34a is not essential for crypt homeostasis, we examined the intestinal crypts from a constitutive miR-34a knockout (miR-34a−/−) model (Choi et al., 2011). The crypt morphology again seems normal (Figures S5C). This suggests that the miR-34a-mediated switch is not essential for Lgr5+ ISC mediated intestinal homeostasis under normal physiological conditions.
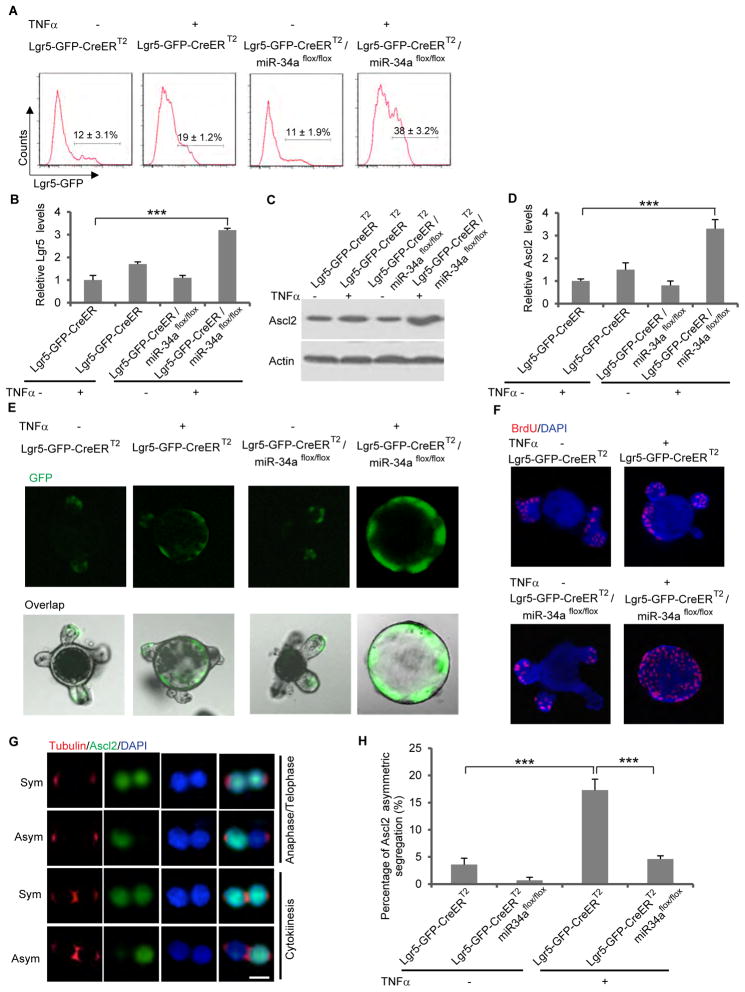
TNFα, a pro-inflammatory cytokine associated with chronic colitis, has been linked to risk of colorectal carcinogenesis (Coussens and Werb, 2002; Popivanova et al., 2008). A low dosage (10ng/ml) treatment of TNFα for 3 days caused modest proliferation of Lgr5-GFP+ ISCs, increasing their number from 12% to 19% of the total organoid cell population. The effect of TNFα treatment was amplified by miR-34a knockout. TNFα-induced ISC proliferation became more excessive in organoids derived from miR-34aflox/flox mice/Lgr5-GFP-IRES-CreERT2 mice after miR-34a knockout was induced, causing proliferating Lgr5-GFP+ ISCs to comprise 38% of the organoid cell population (Figure 6A). Consistent with the flow analyses, TNFα and loss of miR-34a greatly increased the expression of Lgr5 and Ascl2, the marker for Lgr5+ ISCs (Figures 6B–6D). Moreover, miR-34a knockout caused TNFα–treated organoids to grow into undifferentiated spheres that resemble CCSC spheres, with enrichment of Lgr5-GFP+ ISCs (Figure 6E). BrdU incorporation assay showed that loss of miR-34a led to excessive proliferation in TNFα–treated organoids (Figure 6F). Therefore, despite being non-essential for normal tissue homeostasis, the miR-34a-mediated cell fate switch provides a safeguard against excessive ISC proliferation when stem cells regenerate under pro-inflammatory stress.
Figure 6. Loss of miR-34a inhibits asymmetric division and promotes ISC proliferation in organoids treated with TNFα.
(A) FACS analysis of Lgr5-EGFP-CreERT2 and Lgr5-EGFP-CreERT2/miR-34aflox/flox organoids with or without TNFα treatment. Percentage of Lgr5-GFP ISCs increased more dramatically in organoids from Lgr5-EGFP-CreERT2/miR-34aflox/flox mice. (B) RT-qPCR showing Lgr5 levels. (C and D) RT-qPCR (C) and Western blot (D) showing Ascl2 levels. (E) Representative images of organoids. Intestinal organoids from Lgr5-EGFP-CreERT2/miR-34aflox/flox mice grew into CCSC-like, undifferentiated spheres with high level of Lgr5-GPF upon TNFα treatment. (F) Cell proliferation measured by BrdU incorporation. (G and H) Representative images (G) and quantification (H) of symmetric and asymmetric division of Ascl2+ ISCs in Lgr5-EGFP-CreERT2 and Lgr5-EGFP-CreERT2/miR-34aflox/flox intestinal organoids with or without TNFα treatment. Tubulin staining indicates stages of mitosis. The anaphase/telophase images were taken from FACS-sorted doublets that were fixed and stained immediately without recovery. The cytokinesis images were taken from the pair-cell assay. Scale bar, 8μm. Error bars denote s.d. of triplicates. ***, p<0.001. p-value was calculated based on Student’s t-test. Also see Figure S4 and Figure S7.
Lgr5+ ISCs are thought to divide symmetrically in normal conditions (Lopez-Garcia et al., 2010; Snippert et al., 2010). We explored whether the presence of miR-34a has the capability to promote asymmetric division and differentiation to counter excessively proliferating ISCs. We first examined the division of intestinal organoid cells using both the pair-cell assay and direct immunofluorescence on Lgr5-GFP+ doublets freshly isolated by FACS, with antibodies against α- or β-tubulin to mark mitotic cells. Under normal organoid culture condition, only 4.6% of the Lgr5-GFP+ cells or 3.6% of Ascl2+ cells from Lgr5-EGFP-CreERT2 organoids divided asymmetrically, while asymmetric division was barely observed in miR-34a deficient Lgr5-GFP+ or Ascl2+ cells from Lgr5-EGFP-CreERT2/miR-34aflox/flox organoids. Remarkably, 3-day treatment of 10ng/ml TNFα caused 19% of Lgr5-GFP+ cells or 17.3% Ascl2+ cells from Lgr5-EGFP-CreERT2 organoids to divide asymmetrically. In contrast, miR-34a deficiency reduced such asymmetric division to less than 2% in Lgr5-EGFP-CreERT2/miR-34aflox/flox organoids (Figures S4J–S4K and 6G–6H).
miR-34a dependent asymmetric division in vivo
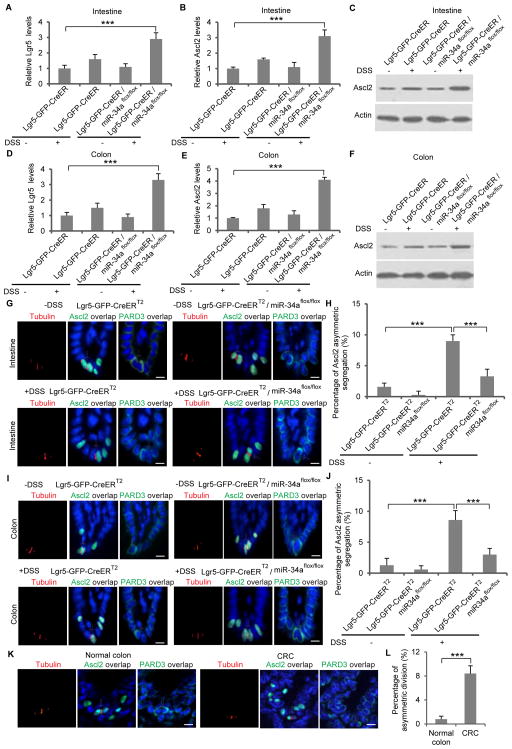
To examine whether inflammation also activates ISC asymmetric division in a miR-34a dependent manner in vivo, Lgr5-EGFP-CreERT2 and Lgr5-EGFP-CreERT2/miR-34aflox/flox mice were treated with 3% dextran sodium sulfate (DSS) in daily drinking water for 5 days, followed by 5 days of plain water supply for recovery. Tissues were then harvested and stained. Consistent with previous reports (Coste et al., 2007; Oh et al., 2014; Yan et al., 2009), DSS upregulated inflammatory factors TNFα, IL-1β and IL-6 in mouse intestine and colon inflammation (Figures S5D and S5E). Regeneration after DSS-induced tissue damage increased the number of Lgr5-GFP+ ISCs and Lgr5 and Ascl2 expression in the intestine and colon, which was further amplified by loss of miR-34a (Figures 7A–7F and S5F–S5G, 7A–7F). DSS treatment caused more proliferation in miR-34a deficient crypts, as shown by the number of cells incorporating BrdU (Figures S5H–S5I). Crypts were then stained for tubulin to identify dividing cell pairs with microtubule configuration consistent with telophase (the final phase of mitosis) – the midbody at the division plane during cytokinesis and asters at the poles. The cell polarity protein PARD3 was concurrently stained to validate division symmetry. Under stress, more ISCs switch to asymmetric division, from 2% to 13% of all Lgr5-GFP+ divisions and from 1.6% to 9% of all Ascl2+ divisions. Asymmetric division was remarkably decreased to 4% in miR-34a deficient mice (Figures 7G–7H and S6A–S6B). Notably, colon stem cells follow the same trend. During recovery from DSS treatment, Lgr5-GFP and Ascl2+ colon stem cells underwent more asymmetric division in a miR-34a dependent manner (Figures 7I–7J and S6C–S6D).
Figure 7. Loss of miR-34a inhibits asymmetric division and promotes ISC proliferation in crypts recovering from DSS treatment.
(A) RT-qPCR showing Lgr5 levels in mouse intestine. (B and C) RT-qPCR (B) and Western blot (C) showing Ascl2 levels in mouse intestine. (D) RT-qPCR showing Lgr5 levels in mouse colon. (E and F) RT-qPCR (E) and Western blot (F) showing Ascl2 levels in mouse colon. (G and H) Representative images (G) and quantification (H) of symmetric and asymmetric division of Ascl2+ intestinal stem cells in Lgr5-EGFP-CreERT2 and Lgr5-EGFP-CreERT2/miR-34aflox/flox mice with (+DSS) or without (-DSS) treatment. Cell polarity protein PARD3 was also stained. (I and J) Representative images (I) and quantification (J) of symmetric and asymmetric division of Ascl2+ colon stem cells in Lgr5-EGFP-CreERT2 and Lgr5-EGFP-CreERT2/miR-34aflox/flox mice with (+DSS) or without (-DSS) treatment. PARD3 established cell polarity. (K and L) Representative images (I) and quantification (J) of symmetric and asymmetric division of Ascl2+ cells in human normal colon and CRC tissue. Scale bar, 20μm. Error bars denote s.d. of triplicates. ***, p<0.001. p-value was calculated based on Student’s t-test. Also see Figures S5–S7.
We then tested whether asymmetric division can also be triggered by ISC proliferation due to genetic mutation. APC deficiency causes ISC proliferation and is an initiation step for adenomas and 90% of CRC (Schepers et al., 2012). We crossed transgenic mice carrying Lgr5-EGFP-CreERT2 and APCflox/flox alleles, and co-immunofluorescence for Lgr5-GFP and Tubulin confirmed that APC−/− intestinal tissues derived from Lgr5-EGFP-CreERT2/APCflox/flox mice induced with Tamoxifen in vivo contain asymmetric LGR5+/LGR5− or Ascl2+/Ascl2− division pairs (Figures S6E–S6H). Hence ISC proliferations in APC-deficient mouse adenomas can trigger asymmetric division.
To further validate the presence of asymmetric division in clinical samples, we examined 12 pairs of human normal colon and CRC samples. 10.6% of the Lgr5+ and 8.4% of the Ascl2+ dividing pairs were undergoing asymmetric division in CRC samples, in contrast to less than 1% in normal colon samples (Figures 7K–7L and S6I–S6J).
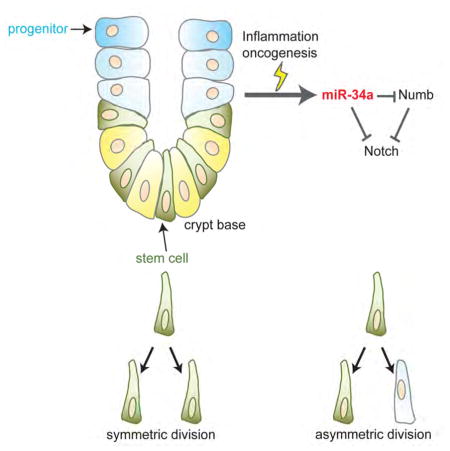
Taken together, the in vitro and in vivo data indicate that, despite being rare in normal tissue, the frequency of asymmetric division can be increased to rein in excessive stem cell proliferation during inflammation-induced regeneration/repair. Loss of miR-34a inhibits asymmetric division and promotes symmetric division that exacerbates stem cell proliferation (Figure S7).
DISCUSSION
Spatial imbalance of cell fate determinants can break symmetry and force bifurcation of cell fate. Here we show that the microRNA cell fate determinant miR-34a and canonical protein cell fate determinant Numb synergize to regulate self-renewal vs. differentiation of early-stage CCSC. miR-34a directly suppresses Numb to form an IFFL, which generates a robust binary switch response from Notch. This switch enhances bimodality of the population and separates CCSCs from non-CCSCs. Undermining this switch via Numb knockdown degrades bimodality and gives rise to an intermediate population of cells that have more ambiguous and plastic cell fate. We further showed that this cell fate determination switch likely provides a safeguard against excessive ISC self-renewal and proliferation in normal tissues. This safeguard mechanism can be triggered during tissue regeneration and repair after inflammation-induced damage, and its inactivation by miR-34a deletion exacerbates Lgr5+ ISC proliferation. The miR-34a-mediated asymmetric division is active in early-stage CCSCs, likely triggered by their excessive proliferation, and is eventually subverted by miR-34a silencing in late-stage CCSCs.
Like most microRNAs, miR-34a targets multiple genes. The level of free miR-34a available to bind Notch1 mRNA is subject to variation due to the expression of other miR-34a target genes. The IFFL may provide an additional benefit of buffering Notch and cell fate decision from such miR-34a copy number variation, because binary Notch level and its resulting bimodality is largely insensitive to precise miR-34a concentration as long as it does not cross the transition threshold.
miR-34a and Numb are lower in mouse Lgr5+ ISCs and higher in more differentiated non-ISCs, consistent with their roles of suppressing Notch. However, the fact that miR-34a deletion generates no obvious intestinal phenotype was puzzling initially. Why loss of miR-34a is so significant to CCSCs? The observation that miR-34a curbs excessive ISC proliferation under pro-inflammatory stress provides a potential answer: normal tissues possess seemingly non-essential or redundant mechanisms for robustness (Ebert and Sharp, 2012; Shen et al., 2008), and the importance of such mechanisms can become more prominent under stress or disease conditions. CCSCs in late-stage tumors eventually remove this barrier by silencing miR-34a and asymmetric division, contributing to more undifferentiated tumors (Bu et al., 2013a; Bu et al., 2013b). The concept of robustness may also provide insights into other microRNAs that are important tumor suppressors but not essential for normal tissue homeostasis.
The subject of ISC division symmetry has been intensely studied, which transformed our view of adult stem cell in mammalian tissue (McHale and Lander, 2014). Previously, ISCs were thought to undergo asymmetric division exclusively to protect their number and genomic integrity (Goulas et al., 2012; Potten et al., 2002; Quyn et al., 2010). However, Lgr5+ CBC cells were identified as actively cycling ISCs, and they perform symmetric division while competing with each other in a neutral drift process (Lopez-Garcia et al., 2010; Snippert et al., 2010).
Intriguingly, asymmetric division has been consistently observed in CCSCs, and its abrogation in favor of symmetric division increases their tumor initiating and proliferative capacity (Bu et al., 2013a; Hwang et al., 2014; O’Brien et al., 2012). Similar observations have been made in other types of cancer stem cells as well (Bajaj et al., 2015; Cicalese et al., 2009; Dey-Guha et al., 2011; Lathia et al., 2011; Pece et al., 2010; Pine et al., 2010; Sugiarto et al., 2011). Why do CCSCs activate asymmetric division, seemingly de novo, which curbs proliferation and promotes differentiation? Our data provide a potential explanation to this paradox: the mechanism of asymmetric division exists in ISC, but is largely silent during normal tissue homeostasis. The rate of asymmetric division is increased to rein in the number of proliferating Lgr5+ stem cells during tissue regeneration after inflammatory damages. It is plausible that asymmetric division may be activated to counter stem cell proliferation at the onset of oncogenesis and remains active in early-stage CCSCs, until being eventually silenced (e.g., through silencing miR-34a) by tumor progression.
EXPERIMENTAL PROCEDURES
CCSCs Isolation, Culture and Differentiation
CCSCs isolation, culture and differentiation were performed as described as previously (Bu et al., 2013a). These procedures are described in detail in the Supplemental Experimental Procedures.
Transgenic Mice and DSS treatment
Lgr5-EGFP-creERT2/miR-34aflox/flox mice were generated by interbreeding Lgr5-EGFP-creERT2 mice (Sato et al., 2009) and miR-34aflox/flox mice (Concepcion et al., 2012). Lgr5-EGFP-creERT2/APCflox/flox mice were generated by interbreeding Lgr5-EGFP-creERT2 mice with APCflox/flox mice (Shibata et al., 1997). Cre recombinase was induced by intraperitoneal injection of Tamoxifen (Sigma) dissolved in sterile corn oil for 5 consecutive days at a dose of 75mg/kg. For DSS treatment, 6–8 week old mice were treated with DSS (36,000–50,000 kDa; MP Biomedicals) in daily drinking water for 5 days, followed by plain water for 5 days. All animal experiments were approved by The Cornell Center for Animal Resources and Education (CARE) and followed the protocol (2009-0071 and 2010-0100).
Mouse intestinal organoid culture
Crypt isolation, cell dissociation, and organoid culturing were performed using previously described protocol (Sato et al., 2009). For TNFα treatment, organoid cells were cultured in medium containing 10ng/ml TNFα (R&D) for 72 hours.
Immunofluorescence
Pair-cell assay was used to investigate CCSC division. Disassociated single CCSC sphere cells were plated on an uncoated glass culture slide (Corning) and allowed to divide once. After fixed in cold methanol, the cells were blocked in 10% normal goat serum for 1 hour and then incubated with anti-ALDH1 (clone H-4, 1:100, Santa Cruz), anti-CK20 (clone H-70, 1:100, Santa Cruz), anti-Numb (1:100, Abcam) and anti-Notch1 (1:400, Abcam) antibody overnight at 4 °C. For the BrdU incorporation assay, the tissue sections were incubated in 1M HCl for 1 hour at 37 °C after fixation. The sections were then washed, and switched to 100 mM Na2B4O7 for 2 minutes. After blocked in 10% normal goat serum for 1 hour, the cells were then incubated with anti-BrdU (1:200, Sigma). The cells were then incubated with Rhodamine Red labeled secondary antibody (Invitrogen) for 1 hour at room temperature. After counterstained with DAPI (Invitrogen), the slides were observed under a fluorescent microscope (Olympus).
Divisions of Lgr5-EGFP ISCs were examined by three methods. First, Lgr5-GFP doublets were directly collected from intestinal organoids by FACS sorting based on GPF signal and cell size. The cells were then immediately fixed in 4% paraformaldehyde, permeabilized in 0.5% Triton-X and stained with anti-GFP-Alexa Fluor 488 (1:500, Abcam), anti-Ascl2 (1:100, Bioss) and anti-β-Tubulin-Cy3 antibodies (1:100, Sigma). In the second method, single Lgr5-GFP cells were plated in Matrigel and allowed to divide once. The cells were then fixed, permeabilized and stained with anti-GFP, anti-Ascl2 and anti-α-tubulin (1:500, Abcam) antibodies. In the third method, intestines from LGR5-EGFP-creERT2 and LGR5-EGFP-creERT2/APCflox/flox mice and human colon and CRC samples were fixed in 4% paraformaldehyde. Frozen sections were then prepared and stained with anti-GFP, anti-Ascl2, anti-PARD3 (1:200, Abcam) and anti-α-tubulin (1:500, Abcam) antibodies. After counterstaining with DAPI (Invitrogen), the slides were observed under a fluorescent microscope (Olympus).
RNA FISH
RNA FISH was performed as described as previously (Bu et al., 2013a). These procedures are described in detail in the Supplemental Experimental Procedures.
Flow Cytometry, RT-qPCR and Western blot
Flow Cytometry, RT-qPCR and Western blot were performed as described as previously (Bu et al., 2013a). These procedures are described in detail in the Supplemental Experimental Procedures.
Statistical Analysis
Data were expressed as mean ± standard deviation of three biological repeats. Student t-tests were used for comparisons, with p<0.05 considered significant.
Supplementary Material
Highlights.
miR-34a and Numb synergize to regulate asymmetric division of colon cancer stem cells
A miR-34a-Numb-Notch feedforward loop suppresses plasticity in cancer stem cells
Inflammation activates asymmetric division of intestinal and colon stem cells
Silencing miR-34a abolishes asymmetric division and promotes stem cell proliferation
Acknowledgments
This work was supported by NIH R01GM95990, NIH R01GM114254, NSF 1350659 career award, NSF 1137269, NYSTEM C029543, and DARPA 19-1091726.
Footnotes
Supplementary Information for this article includes seven figures, Supplementary Experimental Procedures and Supplementary References can be found with this article online.
AUTHOR CONTRIBUTIONS
P.B., L.W., and X.S. conceived the concept, designed the experiments, and co-wrote the manuscript. P.B., L.W. performed the experiments with the assistant from T.S. for the immunofluorescence, Y.A and S.K for immunohistochemistry. K.T. and A.K.V. created miR-34aflox/flox mice/Lgr5-GFP-IRES-CreERT2 mice. S.M.L. contributed the clinical samples. K.C and P.K.L.M performed computational simulation. C.J.C helped analyzed the data. P.B and L.W contributed equally to the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bader AG. miR-34 - a microRNA replacement therapy is headed to the clinic. Frontiers in genetics. 2012;3:120. doi: 10.3389/fgene.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj J, Zimdahl B, Reya T. Fearful Symmetry: Subversion of Asymmetric Division in Cancer Development and Progression. Cancer research. 2015;75:792–797. doi: 10.1158/0008-5472.CAN-14-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann J, Scheitza S, Wernet P, Fischer JC, Giebel B. Asymmetric cell division within the human hematopoietic stem and progenitor cell compartment: identification of asymmetrically segregating proteins. Blood. 2007;109:5494–5501. doi: 10.1182/blood-2006-11-055921. [DOI] [PubMed] [Google Scholar]
- Boon RA, Iekushi K, Lechner S, Seeger T, Fischer A, Heydt S, Kaluza D, Treguer K, Carmona G, Bonauer A, et al. MicroRNA-34a regulates cardiac ageing and function. Nature. 2013;495:107–110. doi: 10.1038/nature11919. [DOI] [PubMed] [Google Scholar]
- Bouchie A. First microRNA mimic enters clinic. Nature biotechnology. 2013;31:577. doi: 10.1038/nbt0713-577. [DOI] [PubMed] [Google Scholar]
- Bu P, Chen KY, Chen JH, Wang L, Walters J, Shin YJ, Goerger JP, Sun J, Witherspoon M, Rakhilin N, et al. A microRNA miR-34a-Regulated Bimodal Switch Targets Notch in Colon Cancer Stem Cells. Cell stem cell. 2013a;12:602–615. doi: 10.1016/j.stem.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu P, Chen KY, Lipkin SM, Shen X. Asymmetric division: a marker for cancer stem cells in early stage tumors? Oncotarget. 2013b;4:948–949. doi: 10.18632/oncotarget.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultje RS, Castaneda-Castellanos DR, Jan LY, Jan YN, Kriegstein AR, Shi SH. Mammalian Par3 regulates progenitor cell asymmetric division via notch signaling in the developing neocortex. Neuron. 2009;63:189–202. doi: 10.1016/j.neuron.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YJ, Lin CP, Ho JJ, He X, Okada N, Bu P, Zhong Y, Kim SY, Bennett MJ, Chen C, et al. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nature cell biology. 2011;13:1353–1360. doi: 10.1038/ncb2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicalese A, Bonizzi G, Pasi CE, Faretta M, Ronzoni S, Giulini B, Brisken C, Minucci S, Di Fiore PP, Pelicci PG. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell. 2009;138:1083–1095. doi: 10.1016/j.cell.2009.06.048. [DOI] [PubMed] [Google Scholar]
- Concepcion CP, Han YC, Mu P, Bonetti C, Yao E, D’Andrea A, Vidigal JA, Maughan WP, Ogrodowski P, Ventura A. Intact p53-dependent responses in miR-34-deficient mice. PLoS genetics. 2012;8:e1002797. doi: 10.1371/journal.pgen.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste A, Dubuquoy L, Barnouin R, Annicotte JS, Magnier B, Notti M, Corazza N, Antal MC, Metzger D, Desreumaux P, et al. LRH-1-mediated glucocorticoid synthesis in enterocytes protects against inflammatory bowel disease. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:13098–13103. doi: 10.1073/pnas.0702440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey-Guha I, Wolfer A, Yeh AC, Albeck JG, Darp R, Leon E, Wulfkuhle J, Petricoin EF, 3rd, Wittner BS, Ramaswamy S. Asymmetric cancer cell division regulated by AKT. Proceedings of the National Academy of Sciences of the United States of America; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fre S, Hannezo E, Sale S, Huyghe M, Lafkas D, Kissel H, Louvi A, Greve J, Louvard D, Artavanis-Tsakonas S. Notch lineages and activity in intestinal stem cells determined by a new set of knock-in mice. PLoS One. 2011;6:e25785. doi: 10.1371/journal.pone.0025785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goentoro L, Shoval O, Kirschner MW, Alon U. The incoherent feedforward loop can provide fold-change detection in gene regulation. Mol Cell. 2009;36:894–899. doi: 10.1016/j.molcel.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulas S, Conder R, Knoblich JA. The par complex and integrins direct asymmetric cell division in adult intestinal stem cells. Cell stem cell. 2012;11:529–540. doi: 10.1016/j.stem.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WL, Jiang JK, Yang SH, Huang TS, Lan HY, Teng HW, Yang CY, Tsai YP, Lin CH, Wang HW, et al. MicroRNA-146a directs the symmetric division of Snail-dominant colorectal cancer stem cells. Nature cell biology. 2014;16:268–280. doi: 10.1038/ncb2910. [DOI] [PubMed] [Google Scholar]
- Inaba M, Yamashita YM. Asymmetric stem cell division: precision for robustness. Cell stem cell. 2012;11:461–469. doi: 10.1016/j.stem.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Jackson HW, Waterhouse P, Sinha A, Kislinger T, Berman HK, Khokha R. Expansion of stem cells counteracts age-related mammary regression in compound Timp1/Timp3 null mice. Nature cell biology. 2015;17:217–227. doi: 10.1038/ncb3118. [DOI] [PubMed] [Google Scholar]
- Kaplan S, Bren A, Dekel E, Alon U. The incoherent feed-forward loop can generate non-monotonic input functions for genes. Mol Syst Biol. 2008;4:203. doi: 10.1038/msb.2008.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katajisto P, Dohla J, Chaffer CL, Pentinmikko N, Marjanovic N, Iqbal S, Zoncu R, Chen W, Weinberg RA, Sabatini DM. Stem cells. Asymmetric apportioning of aged mitochondria between daughter cells is required for stemness. Science. 2015;348:340–343. doi: 10.1126/science.1260384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Koo BK, Stange DE, Sato T, Karthaus W, Farin HF, Huch M, van Es JH, Clevers H. Controlled gene expression in primary Lgr5 organoid cultures. Nature methods. 2012;9:81–83. doi: 10.1038/nmeth.1802. [DOI] [PubMed] [Google Scholar]
- Lander AD, Kimble J, Clevers H, Fuchs E, Montarras D, Buckingham M, Calof AL, Trumpp A, Oskarsson T. What does the concept of the stem cell niche really mean today? BMC Biol. 2012;10:19. doi: 10.1186/1741-7007-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathia JD, Hitomi M, Gallagher J, Gadani SP, Adkins J, Vasanji A, Liu L, Eyler CE, Heddleston JM, Wu Q, et al. Distribution of CD133 reveals glioma stem cells self-renew through symmetric and asymmetric cell divisions. Cell Death Dis. 2011;2:e200. doi: 10.1038/cddis.2011.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine E, Zhang Z, Kuhlman T, Hwa T. Quantitative characteristics of gene regulation by small RNA. PLoS Biol. 2007;5:e229. doi: 10.1371/journal.pbio.0050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. The art of choreographing asymmetric cell division. Dev Cell. 2013;25:439–450. doi: 10.1016/j.devcel.2013.05.003. [DOI] [PubMed] [Google Scholar]
- Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Garcia C, Klein AM, Simons BD, Winton DJ. Intestinal stem cell replacement follows a pattern of neutral drift. Science. 2010;330:822–825. doi: 10.1126/science.1196236. [DOI] [PubMed] [Google Scholar]
- Mangan S, Itzkovitz S, Zaslaver A, Alon U. The incoherent feed-forward loop accelerates the response-time of the gal system of Escherichia coli. Journal of molecular biology. 2006;356:1073–1081. doi: 10.1016/j.jmb.2005.12.003. [DOI] [PubMed] [Google Scholar]
- McGill MA, McGlade CJ. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. The Journal of biological chemistry. 2003;278:23196–23203. doi: 10.1074/jbc.M302827200. [DOI] [PubMed] [Google Scholar]
- McHale PT, Lander AD. The protective role of symmetric stem cell division on the accumulation of heritable damage. PLoS Comput Biol. 2014;10:e1003802. doi: 10.1371/journal.pcbi.1003802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- Mukherji S, Ebert MS, Zheng GX, Tsang JS, Sharp PA, van Oudenaarden A. MicroRNAs can generate thresholds in target gene expression. Nat Genet. 2011 doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumuller RA, Knoblich JA. Dividing cellular asymmetry: asymmetric cell division and its implications for stem cells and cancer. Genes Dev. 2009;23:2675–2699. doi: 10.1101/gad.1850809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien CA, Kreso A, Ryan P, Hermans KG, Gibson L, Wang Y, Tsatsanis A, Gallinger S, Dick JE. ID1 and ID3 Regulate the Self-Renewal Capacity of Human Colon Cancer-Initiating Cells through p21. Cancer Cell. 2012;21:777–792. doi: 10.1016/j.ccr.2012.04.036. [DOI] [PubMed] [Google Scholar]
- Oh SY, Cho KA, Kang JL, Kim KH, Woo SY. Comparison of experimental mouse models of inflammatory bowel disease. Int J Mol Med. 2014;33:333–340. doi: 10.3892/ijmm.2013.1569. [DOI] [PubMed] [Google Scholar]
- Osella M, Bosia C, Cora D, Caselle M. The role of incoherent microRNA-mediated feedforward loops in noise buffering. PLoS Comput Biol. 2011;7:e1001101. doi: 10.1371/journal.pcbi.1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M, Ronzoni S, Bernard L, Viale G, Pelicci PG, Di Fiore PP. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140:62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Pine SR, Ryan BM, Varticovski L, Robles AI, Harris CC. Microenvironmental modulation of asymmetric cell division in human lung cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:2195–2200. doi: 10.1073/pnas.0909390107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popivanova BK, Kitamura K, Wu Y, Kondo T, Kagaya T, Kaneko S, Oshima M, Fujii C, Mukaida N. Blocking TNF-alpha in mice reduces colorectal carcinogenesis associated with chronic colitis. J Clin Invest. 2008;118:560–570. doi: 10.1172/JCI32453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potten CS, Owen G, Booth D. Intestinal stem cells protect their genome by selective segregation of template DNA strands. Journal of cell science. 2002;115:2381–2388. doi: 10.1242/jcs.115.11.2381. [DOI] [PubMed] [Google Scholar]
- Quyn AJ, Appleton PL, Carey FA, Steele RJ, Barker N, Clevers H, Ridgway RA, Sansom OJ, Nathke IS. Spindle orientation bias in gut epithelial stem cell compartments is lost in precancerous tissue. Cell stem cell. 2010;6:175–181. doi: 10.1016/j.stem.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, van de Wetering M, Clevers H. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–735. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- Schweisguth F. Regulation of notch signaling activity. Curr Biol. 2004;14:R129–138. [PubMed] [Google Scholar]
- Shen X, Collier J, Dill D, Shapiro L, Horowitz M, McAdams HH. Architecture and inherent robustness of a bacterial cell-cycle control system. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:11340–11345. doi: 10.1073/pnas.0805258105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata H, Toyama K, Shioya H, Ito M, Hirota M, Hasegawa S, Matsumoto H, Takano H, Akiyama T, Toyoshima K, et al. Rapid colorectal adenoma formation initiated by conditional targeting of the Apc gene. Science. 1997;278:120–123. doi: 10.1126/science.278.5335.120. [DOI] [PubMed] [Google Scholar]
- Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Sugiarto S, Persson AI, Munoz EG, Waldhuber M, Lamagna C, Andor N, Hanecker P, Ayers-Ringler J, Phillips J, Siu J, et al. Asymmetry-defective oligodendrocyte progenitors are glioma precursors. Cancer Cell. 2011;20:328–340. doi: 10.1016/j.ccr.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Flier LG, van Gijn ME, Hatzis P, Kujala P, Haegebarth A, Stange DE, Begthel H, van den Born M, Guryev V, Oving I, et al. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell. 2009;136:903–912. doi: 10.1016/j.cell.2009.01.031. [DOI] [PubMed] [Google Scholar]
- VanDussen KL, Carulli AJ, Keeley TM, Patel SR, Puthoff BJ, Magness ST, Tran IT, Maillard I, Siebel C, Kolterud A, et al. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development (Cambridge, England) 2012;139:488–497. doi: 10.1242/dev.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SE, Beronja S, Pasolli HA, Fuchs E. Asymmetric cell divisions promote Notch-dependent epidermal differentiation. Nature. 2011;470:353–358. doi: 10.1038/nature09793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Dalmasso G, Nguyen HT, Obertone TS, Sitaraman SV, Merlin D. Ste20-related proline/alanine-rich kinase (SPAK) regulated transcriptionally by hyperosmolarity is involved in intestinal barrier function. PLoS One. 2009;4:e5049. doi: 10.1371/journal.pone.0005049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.