Abstract
Clinician attitudes towards multiplexed genomic testing may be vital to the success of translational programs. We surveyed clinicians at an academic medical center about their views on a large pharmacogenomics implementation, the PREDICT (Pharmacogenomic Resource for Enhanced Decisions in Care & Treatment) program. Participants were asked about test ordering, major factors influencing use of results, expectations of efficacy, and responsibility for applying results to patient care. Virtually all respondents (99%) agreed that pharmacogenomics variants influence patients’ response to drug therapy. The majority (92%) favored immediate, active notification when a clinically significant drug-genome interaction was present. However, clinicians were divided on which providers were responsible for acting on a result when a prescription change was indicated and whether patients should be directly notified of a significant result. We concluded genotype results were valued for tailoring prescriptions, but clinicians do not agree on how to appropriately assign clinical responsibility for actionable results from a multiplexed panel.
Introduction
The introduction of pharmacogenomics into clinical settings is accelerating as academic medical centers and integrated health systems have implemented pharmacogenomic testing, encouraged routine use within prescribing, and placed results within electronic health records (EHR).1–7 Many programs have adopted multiplexed, panel testing - in which multiple variants are tested simultaneously to inform prescribing- to leverage economies of scale and the potential for reuse of panel data over time.7–9 However, multiplexed testing may present challenges as clinicians may be expected to apply pharmacogenomic test results that were ordered in an unrelated clinical context and to consider genomic risks that are not relevant to their usual scope of practice.
Concerns about the ability of front line clinicians to manage genomic data are highlighted by surveys and qualitative studies of likely users.10–14 Nationally, fewer than one in eight primary care physicians has ordered a pharmacogenomic test or felt adequately informed to use the result. Within implementation programs, significant new educational efforts and clinical decision support strategies are designed to bridge this knowledge gap.1,3 However, little has been reported on the views of clinicians working in these new programs.
Education and implementation assistance from medical geneticists and knowledgeable pharmacists has been anticipated from the onset of genomic medicine.15 Even with the assistance of sophisticated EHR tools, clinicians’ understanding of pharmacogenomics and active engagement with pharmacogenomic testing is critical for test adoption and utilization. We report the outcomes of a survey administered to clinicians participating in a large pharmacogenomics program within an academic medical center. Those solicited had either requested or received results from a multiplexed pharmacogenomics panel performed for primary care and cardiovascular patient populations between 2010–2013.1 The present analysis focuses on clinicians’ perceptions of clinical utility, preparedness to effectively use pharmacogenomic test results, and questions of responsibility for disclosure and clinical use of multiplexed results over the course of patients’ care.
Materials and Methods
Pharmacogenomics Implementation
Clinicians solicited for this study participated in an institutional pharmacogenomics program launched in 2010. The program was designed to pre-emptively genotype patients, store actionable results as determined by the local pharmacy and therapeutics (P&T) committee, and provide program interpretations and recommendations at the point of care. During the initial implementation, program leaders gave educational seminars, distributed informational brochures, and conducted direct communications with clinicians through email and in-person meetings. The program created a web site summarizing the evidence for applying the tested variants to clinical care and linked pharmacogenomic results to the relevant page.1 Inpatient and outpatient clinical decision support provided guidance at the point of medication prescribing for five drugs, including clopidogrel and warfarin, during the survey period.2 A pharmacist-led active surveillance program focused on CYP2C19 and clopidogrel ensured key results were delivered to cardiology attending physicians following coronary stent placement.16 As part of program development, members of the patient population served by the institution gave feedback and guidance to program development as part of focus groups. Pharmacogenomic testing was performed under the terms of treatment consent similar to other laboratory testing within a health care environment.
Development of survey
Investigators conducting pharmacogenomics research developed the survey. Questions were based on a prior publication by Stanek, et al.,10 and contributions from authors. Two clinicians piloted the survey for clarity and completeness of the potential responses. The survey was designed to address the following domains: perception of clinical utility, preparedness to receive results, and assignment of clinical responsibility for communicating results and adjusting medications as warranted over the course of the patient’s care.
Sampling method
The survey was distributed by email between November 2012 and March 2013 to all clinicians within cardiology, primary care, and endocrinology who met the following criteria: 1) had ordered a panel-based test via the pharmacogenomics implementation or cared for a patient with a pharmacogenomic result in the previous one year and 2) held a position as an attending physician, specialty fellow physician, or nurse practitioner position with active prescribing privileges.
Collection of survey responses
Survey items were entered into REDCap, which features a secure environment for building and managing online surveys for research.17 A unique access code was created for each solicited subject, allowing the responses to be collected anonymously. After the initial email solicitation, non-responders were solicited with two additional emails. A modest incentive was provided for completing the survey. The Vanderbilt Institutional Research Board approved the study.
Data Analysis
Responses were included in the analysis if the majority of coded questions were answered including the key questions related to responsibility for results. Responses were tabulated as numeric counts and frequencies. Respondents were stratified by cardiology and non-cardiology specialty for the analysis related to questions about clinical responsibility for acting on pharmacogenomic results where a prescription change was indicated. All analyses were conducted in R version 3.0.1 (Vienna, Austria).
Results
Of 156 surveys distributed, 80 (52%) were returned with a complete response and 4 (3%) were returned incomplete. Respondents were evenly split between clinicians practicing cardiology (51%) and those practicing primary care or endocrinology (49%). Survey respondents were predominantly young with less than 15 years of practice (71%) and female (63%). Approximately half were attending faculty while the remainder consisted of nurse practitioners and fellows (Table 1).
Table 1.
Characteristics of Survey Respondents
| Cardiology N (%) |
Non-Cardiology N (%) |
Total N (%) |
|
|---|---|---|---|
| Age | |||
| 20–30 | 0 (0) | 0 (0) | 0 (0) |
| 31–40 | 22 (54) | 20 (51) | 42 (52) |
| 41–50 | 7 (17) | 12 (31) | 19 (24) |
| 51–60 | 7 (17) | 4 (10) | 11 (14) |
| 61–70 | 3 (7) | 2 (5) | 5 (6) |
| >71 | 2 (5) | 1 (3) | 3 (4) |
| Gender | |||
| Male | 11 (27) | 19 (49) | 30 (38) |
| Female | 30 (73) | 20 (51) | 50 (63) |
| Years of clinical practice | |||
| <5 | 9 (22) | 6 (15) | 15 (19) |
| 5–10 | 13 (32) | 14 (36) | 27 (34) |
| 11–15 | 5 (12) | 10 (26) | 15 (19) |
| 16–20 | 3 (7) | 5 (13) | 8 (10) |
| 21–25 | 0 (0) | 0 (0) | 0 (0) |
| >25 | 11 (27) | 4 (10) | 15 (19) |
| Practice Specialty | |||
| Internal Medicine: Primary Care Physician | 0 (0) | 21 (54) | 21(26) |
| Internal Medicine: Hospitalist | 0 (0) | 0 (0) | 0 (0) |
| Medical Specialty: Interventional Cardiology | 11 (27) | 0 (0) | 11(14) |
| Medical Specialty: General Cardiology | 30 (73) | 0 (0) | 30(38) |
| Medical Specialty: Endocrinologist | 0 (0) | 7 (18) | 7(9) |
| Pediatrics | 0 (0) | 1 (3) | 1(1) |
| Other | 0 (0) | 10 (26) | 10(13) |
| Position | |||
| Physician | 19 (46) | 32 (82) | 51 (64) |
| Fellow | 12 (29) | 1 (3) | 13 (16) |
| Resident physician | 0 (0) | 0 (0) | 0 (0) |
| Nurse practitioner | 8 (20) | 6 (15) | 14 (18) |
| Physician assistant | 0 (0) | 0 (0) | 0 (0) |
| Other | 2 (5) | 0 (0) | 2 (2.5) |
| Number of half-day outpatient sessions per week | |||
| 0–2 | 30 (73) | 18 (46) | 48 (60) |
| 3–4 | 8 (20) | 6 (15) | 14 (18) |
| 5–6 | 0 (0) | 4 1(0) | 4 (5) |
| 7–8 | 0 (0) | 9 (23) | 9 (11) |
| 9–10 | 3 (7) | 2 (5) | 5 (6) |
Preparedness to order pharmacogenomic testing
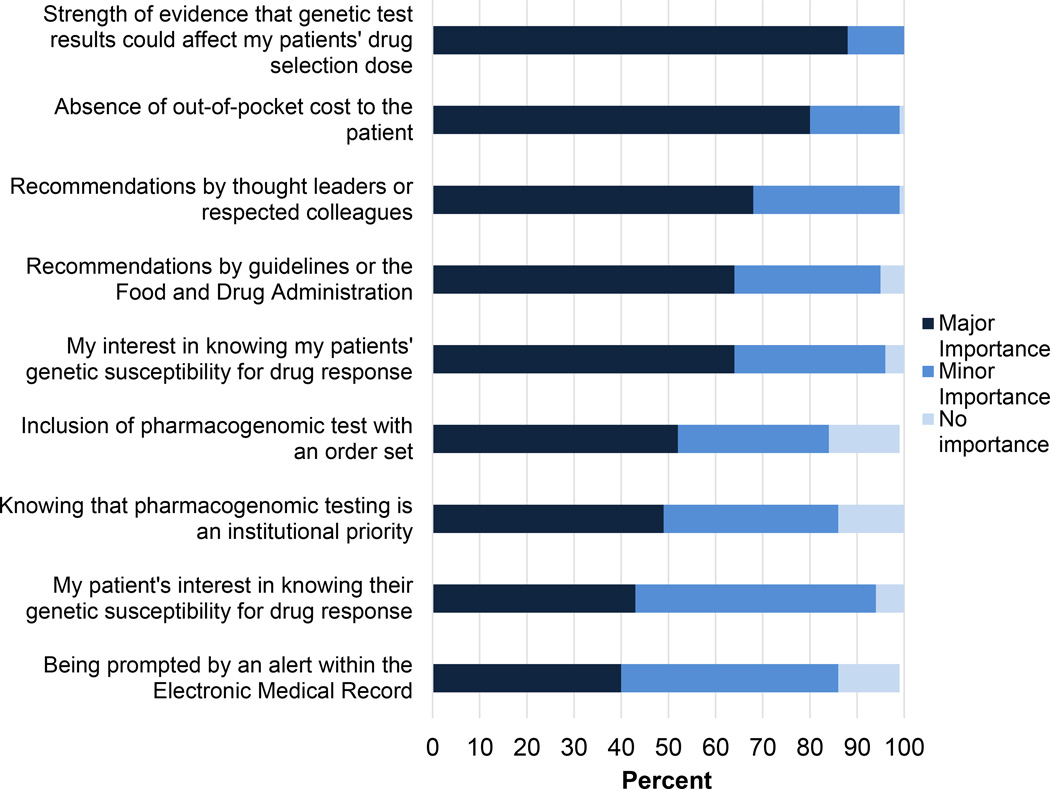
As expected based on study inclusion criteria, two-thirds (63%) had ordered or recommended a pharmacogenomic test in the prior six months. A high proportion of respondents (95%) were familiar with the institutional pharmacogenomics program. Nearly all (94%) prescribed at least one of the three drugs targeted by the program at the time of survey: simvastatin, clopidogrel, and warfarin. When deciding whether to order a pharmacogenomic test, the most important considerations reported were strength of evidence for the drug-gene interaction and patients’ out of pocket costs for testing (Figure 1).
Figure 1. Influential factors reported by clinicians when deciding whether to order a pharmacogenomics panel test.
Perception of clinical utility
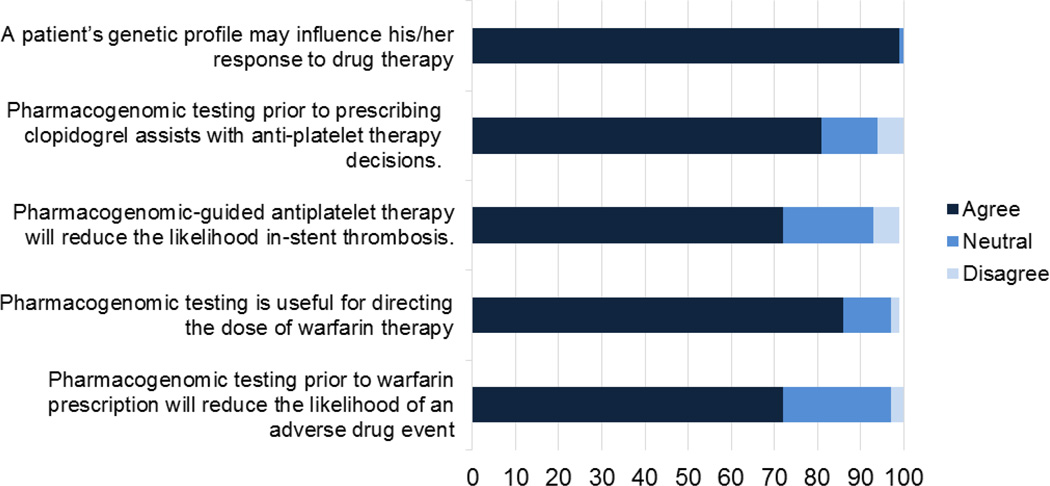
All but one respondent (99%) agreed that pharmacogenomic variants influence patients’ response to drug therapy. The majority agreed or strongly agreed with the clinical utility of CYP2C19 variants to tailor antiplatelet therapy following percutaneous coronary interventions (80%) and VKORC1 and CYP2C9 variants to tailor initial warfarin dosing (86%). The majority also agreed or strongly agreed that the variants affected patient outcomes, such as stent thrombosis and warfarin-related bleeding (Figure 2).
Figure 2. Attitudes towards clinical utility of genomic variants to tailor prescriptions.
Likert scale responses indicating strongly agree and strongly disagree are collapsed into agree and disagree categories.
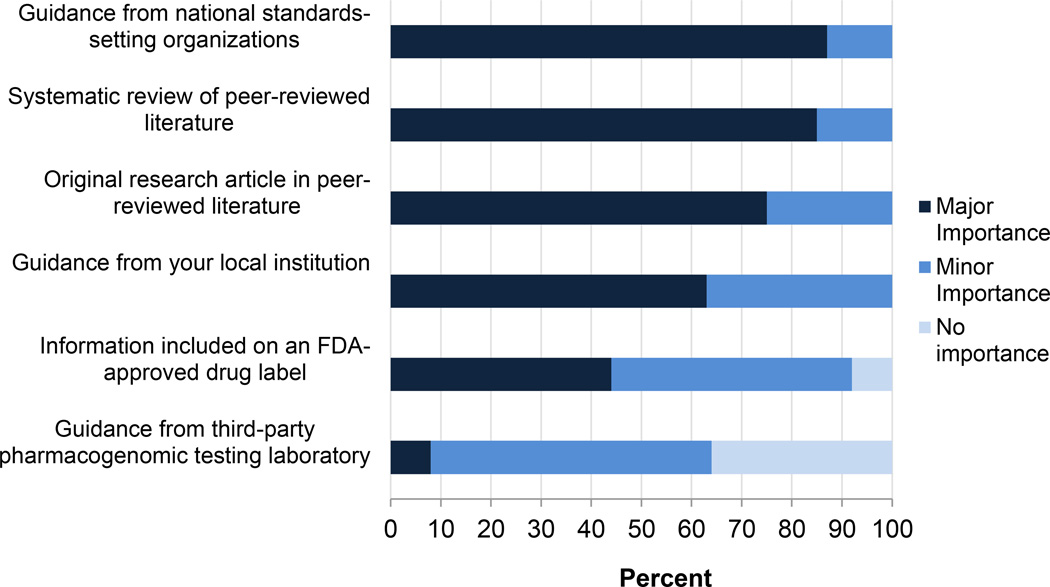
We asked participants to indicate which sources of information were of major, minor, or no importance to their perception of pharmacogenomic clinical utility. As indicated in Figure 3, respondents ranked published literature including systematic reviews and specialty society guidelines higher than guidance from the implementing institution, from a third party laboratory, or from Food and Drug Administration (FDA) approved medication labels.
Figure 3. Influential factors reported by clinicians when deciding to use pharmacogenomic variants to tailor therapy.
Preparedness to receive results
A minority of the respondents (19%) reported no prior instruction in pharmacogenomics; the remainder had completed undergraduate courses (11%), professional school instruction (31%), postgraduate coursework (23%), continuing medical education seminars (52%), or self-instruction by reading peer reviewed literature (51%). Overall, 70% of respondents felt they had adequate educational resources in the clinic to support clinical decision-making related to pharmacogenomics, a proportion that did not significantly differ between cardiology and non-cardiology providers (p=0.63). Many who did not feel adequate resources were available submitted free text responses requesting more point-of-care guidance with links to primary sources in the literature. Additionally, providers requested supplemental information geared towards patients to reduce the time required to educate patients about genomic variants and the rationale for tailoring therapy.
Responsibility for results
Survey subjects were asked to respond to two clinical scenarios applying pharmacogenomic results to clinical decision-making and to select which providers were responsible for clinical action (Table 2). The first scenario was modeled after the most common drug-genome interaction encountered at the time of the survey: prescribing clopidogrel in the setting of an intermediate or poor metabolism phenotype for CYP2C19. In the second scenario, new pharmacogenomic information pertinent to warfarin dosing became available months after the patient was initially tested. In both cases, respondents were asked to identify who should be notified and who should be responsible for acting on this result. Respondents could choose multiple selections.
Table 2.
Clinician attitudes regarding notification and responsibility for acting on a pharmacogenomics result
| Clinical scenario: A 65 year old patient with diabetes and hypertension experiences angina with brisk walking. Nuclear stress testing reveals evidence of cardiac ischemia. Upon referral to an interventional cardiologist, he is scheduled for elective angiography the following day and receives pharmacogenomic testing. He is prescribed aspirin and clopidogrel after successful placement of a drug-eluting stent and is discharged. One week later, the result of the pharmacogenomic test is returned and indicates that the patient is homozygous for the CYP2C19 *2 variant and thus is a poor metabolizer of clopidogrel. | ||
|
Cardiology N (%) |
Non- cardiology N (%) |
|
| In addition to including the results in the electronic medical record (EMR), who should be individually notified of the new pharmacogenomics result? (check all that apply) | ||
| Not necessary to notify any provider directly | 0 (0) | 0 (0) |
| Primary care provider | 22 (54) | 27 (69) |
| Specialist treating medical condition affected by test result | 37 (90) | 35 (90) |
| Provider who ordered pharmacogenomic test | 31 (76) | 27 (69) |
| Provider who prescribed drug therapy affected by test | 33 (80) | 37 (95) |
| Patient should be notified directly | 18(44) | 19 (49) |
| Which of the patient’s providers is responsible for acting on a pharmacogenomic result if a prescription change is indicated? (check all that apply) | ||
| Primary care provider | 3(7) | 7 (18) |
| Specialist treating medical condition affected by test result | 33 (80) | 29 (74) |
| Provider who ordered pharmacogenomic test | 23(56) | 20 (51) |
| Provider who previously prescribed drug therapy affected by test | 20 (49) | 23 (59) |
| When should providers be actively notified (e.g. with a reminder or prompt) if a prescription change based on the pharmacogenomic result is indicated? | ||
| As soon as results are available in the EMR | 37 (92) | 36 (92) |
| During the next appointment at Vanderbilt | 1 (2) | 1 (3) |
| Only when selecting antiplatelet medication using e-script | 2 (5) | 1 (3) |
| No reminder or prompt necessary | 0 (0) | 1 (3) |
| Continued scenario: Six months following the patient’s stent placement, the program begins reporting genetic results to guide warfarin therapy. Based on genetic and clinical variables, the patient is expected to have a stable therapeutic INR1 on a low dose of warfarin (<21mg/week) and increased risk of bleeding on standard or high doses of warfarin. Since his stent, the patient has resumed care with his primary care provider and cardiologist in his home town. | ||
| Who should be notified of the pharmacogenomic result? (check all that apply) |
Cardiology N (%) |
Non- cardiology N (%) |
| Vanderbilt provider who has seen the patient most recently | 11 (27) | 8 (21) |
| Primary care provider | 27 (66) | 30 (77) |
| Specialist treating medical condition affected by test result | 31 (76) | 28 (72) |
| Provider who ordered the pharmacogenomic test | 24 (59) | 22 (56) |
| Provider who will prescribe drug therapy affected by test | 35 (85) | 30 (77) |
| Patient should be notified directly | 19 (46) | 21 (54) |
| Who, within Vanderbilt, should take responsibility for following up with the patient or outside providers? (check all that apply) | ||
| Vanderbilt provider who has seen the patient most recently | 9 (22) | 4 (10) |
| Vanderbilt provider who ordered the pharmacogenomic test | 21 (51) | 24 (62) |
| PREDICT2 staff should contact the providers | 28 (68) | 28 (72) |
| PREDICT staff should contact the patient directly | 11 (27) | 12 (31) |
| What are your preferred methods of receiving notification of a pharmacogenomic result that may require you to take clinical action? | ||
| Standard laboratory reporting in EMR3 | 15 (37) | 19 (49) |
| Phone call from PREDICT staff | 4 (10) | 3 (8) |
| Electronic clinical message from PREDICT staff | 33 (80) | 27 (69) |
| Clinical decision support via e-prescribing and computerized physician order entry | 15 (37) | 16 (41) |
| Message to nursing staff or pharmacy directly | 1 (2) | 0 (0) |
INR = International Normalized Ratio
PREDICT is the name of the institutional pharmacogenomics program
EMR = Electronic Medical Records
No agreement emerged about which group of providers should be notified or who should take responsibility for clinical action when necessary. In the first scenario of an actionable result for clopidogrel and CYP2C19, most agreed that multiple providers should be notified; however, only 44%–49% agreed that the patient should also be directly notified (Table 1). The clinician most commonly selected by cardiologists for direct notification was the specialist treating the medical condition (90%) while non-cardiologists chose the provider who prescribed the drug affected by the result (95%). There was less agreement about which provider is responsible for acting on the result, but both groups most commonly chose the specialist treating the medical condition affected by the result (74–80%). Nearly all (92% in each group) wanted active notification as soon as the results were reported.
The second scenario also gave an actionable result, but this pharmacogenomic result for guiding initial warfarin dosing became available six months following the original testing. Again, about half of the respondents (46–54%) felt that the patient should be notified directly and the majority of respondents indicated that multiple clinicians should also be notified. While cardiologists most commonly selected the provider who would prescribe warfarin (85%), non-cardiologists selected both this option (77%) and the primary care provider (77%). Both cardiologists and non-cardiologists most commonly assigned responsibility for follow-up for this delayed result to the implementation program staff, to the provider who initially ordered the pharmacogenomic panel, or to both.
Discussion
Within an institutionally supported pharmacogenomics implementation program, clinicians expressed support for the concept that pharmacogenomic variants affect drug responses and more than 80% agreed that common drug-genome interactions for clopidogrel and warfarin reported by the program had clinical utility. The majority reported prior instruction in pharmacogenomics and felt adequately supported to use the results in clinical practice. National guidelines and published literature were favored as sources of guidance over local initiatives such as computerized prompts and institutional recommendations.
Several of these findings are distinct from those obtained from prior studies of physicians who practiced outside an implementation program. In one national survey, physicians reported near universal acceptance of the concept of pharmacogenomics, but had infrequently been educated on the topic and felt unprepared for test ordering and using the results.10 A second regional survey of primary care physicians and family practitioners yielded similar results.11 The differences highlight the importance of implementation programs to prepare end-users for ordering and interpreting pharmacogenomic results.
Our survey identified a lack of agreement regarding which clinician should be responsible for results with either immediate or potentially persistent value. Respondents assigned responsibility for long-term follow-up of genomic test results to an inconsistent array of providers, ranging from specialists to primary care providers to the administrative staff of the implementation program. In retrospect, this was not surprising since pharmacogenomics results can apply to a wide variety of clinical scenarios and survey respondents may not have felt comfortable with genetic information not directly related to their specialty. Nonetheless, this lack of agreement about who should act on pharmacogenomic results raises the risk that some patients may fall through the cracks. As a result, more work is needed to create systems for return of results that clearly assign responsibility for clinical action related to genomic variants.
Our study has several limitations. Subjects were selected within a tertiary care academic medical center program and survey responses may not be representative of the general practitioner population. Given the rapid changes in the evidence base for drug-genome interactions, survey responses related to specific clinical scenarios are expected to change over time. For example, the survey was performed prior to the publication of several large-scale studies of pharmacogenomic-guided dosing for warfarin,18,19 and thus, the physician responses presented here may not be fully indicative of current attitudes and practice related to that particular drug-gene interaction. Finally, clinicians may be influenced by a broader array of information sources than those indicated in the survey, and attitudes and preferences expressed by survey respondents may not always correspond with information-seeking behavior in the clinic.20
The growth of multiplexed pharmacogenomic testing is anticipated to occur both within and outside of the context of institutional implementation programs. Physicians operating within an implementation program report greater prior knowledge and educational support to order and use pharmacogenomic results than previously published results from a national sample. However, even in the context of a single health system, dilemmas persist related to assigning responsibility for pharmacogenomic results and require new strategies to ensure that patients receive the benefits of high-quality genome-informed care.
Acknowledgements
We thank Lisa Price for assisting with the survey. This project was funded by Vanderbilt University, the Centers for Disease Control and Prevention (U47CI000824), the National Heart, Lung, And Blood Institute (U01HL122904, U01HL105198), the National Institute for General Medical Sciences (U19HL065962), the National Human Genome Research Institute (U01HG006378), the National Center for Advancing Translational Sciences (UL1TR000445), KL2TR000446 and NICHD K23 HD000001. The analyses described herein are solely the responsibility of the authors alone and do not necessarily represent official views of the Centers for Disease Control and Prevention or the National Institutes of Health.
Additionally, the funding sources had no role in the study design, the collection, analysis, and interpretation of data, manuscript preparation, or the decision to submit the paper for publication.
Footnotes
Supplementary information is available at The Pharmacogenomics Journal’s website.
Conflict of Interest Disclosure: The authors declare no conflict of interest.
References
- 1.Pulley JM, Denny JC, Peterson JF, Bernard GR, Vnencak-Jones CL, Ramirez AH, et al. Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clin Pharmacol Ther. 2012;92:87–95. doi: 10.1038/clpt.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peterson JF, Bowton E, Field JR, Beller M, Mitchell J, Schildcrout J, et al. Electronic health record design and implementation for pharmacogenomics: a local perspective. Genet Med. 2013;15:833–841. doi: 10.1038/gim.2013.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shuldiner AR, Relling MV, Peterson JF, Hicks K, Freimuth RR, Sadee W, et al. The Pharmacogenomics Research Network Translational Pharmacogenetics Program: Overcoming Challenges of Real-World Implementation. Clinical Pharmacology & Therapeutics. 2013;94:207–210. doi: 10.1038/clpt.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rasmussen-Torvik LJ, Stallings SC, Gordon AS, Almoguera B, Basford MA, Bielinski SJ, et al. Design and Anticipated Outcomes of the eMERGE-PGx Project: A Multi-Center Pilot for Pre-Emptive Pharmacogenomics in Electronic Health Record Systems. Clin Pharmacol Ther. 2014 doi: 10.1038/clpt.2014.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell GC, Crews KR, Wilkinson MR, Haidar CE, Hicks JK, Baker DK, et al. Development and use of active clinical decision support for preemptive pharmacogenomics. J Am Med Inform Assoc. 2014;21:e93–e99. doi: 10.1136/amiajnl-2013-001993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bielinski SJ, Olson JE, Pathak J, Weinshilboum RM, Wang L, Lyke KJ, et al. Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time-using genomic data to individualize treatment protocol. Mayo Clin Proc. 2014;89:25–33. doi: 10.1016/j.mayocp.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffman JM, Haidar CE, Wilkinson MR, Crews KR, Baker DK, Kornegay NM, et al. PG4KDS: A model for the clinical implementation of pre-emptive pharmacogenetics. Am J Med Genet C Semin Med Genet. 2014;166:45–55. doi: 10.1002/ajmg.c.31391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Driest S, Shi Y, Bowton E, Schildcrout J, Peterson J, Pulley J, et al. Clinically Actionable Genotypes Among 10,000 Patients With Preemptive Pharmacogenomic Testing. Clin Pharmacol Ther. 2013 doi: 10.1038/clpt.2013.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schildcrout JS, Denny JC, Bowton E, Gregg W, Pulley JM, Basford MA, et al. Optimizing drug outcomes through pharmacogenetics: a case for preemptive genotyping. Clin Pharmacol Ther. 2012;92:235–242. doi: 10.1038/clpt.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stanek EJ, Sanders CL, Taber KAJ, Khalid M, Patel A, Verbrugge RR, et al. Adoption of Pharmacogenomic Testing by US Physicians: Results of a Nationwide Survey. Clin Pharmacol Ther. 2012;91:450–458. doi: 10.1038/clpt.2011.306. [DOI] [PubMed] [Google Scholar]
- 11.Haga S, Burke W, Ginsburg G, Mills R, Agans R. Primary care physicians’ knowledge of and experience with pharmacogenetic testing. Clin Genet. 2012;82:388–394. doi: 10.1111/j.1399-0004.2012.01908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haga SB, Carrig MM, O’Daniel JM, Orlando LA, Killeya-Jones LA, Ginsburg GS, et al. Genomic risk profiling: attitudes and use in personal and clinical care of primary care physicians who offer risk profiling. J Gen Intern Med. 2011;26:834–840. doi: 10.1007/s11606-011-1651-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haga SB, Tindall G, O’Daniel JM. Professional Perspectives About Pharmacogenetic Testing and Managing Ancillary Findings. Genet Test Mol Biomarkers. 2012;16:21–24. doi: 10.1089/gtmb.2011.0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hicks JK, Crews KR, Hoffman JM, Kornegay NM, Wilkinson MR, Lorier R, et al. A clinician-driven automated system for integration of pharmacogenetic interpretations into an electronic medical record. Clin Pharmacol Ther. 2012;92:563–566. doi: 10.1038/clpt.2012.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Varmus H. Getting ready for gene-based medicine. N Engl J Med. 2002;347:1526–1527. doi: 10.1056/NEJMe020119. [DOI] [PubMed] [Google Scholar]
- 16.Khan NA, Peterson JF. A surveillance tool to support quality assurance and research in personalized medicine. AMIA Annu Symp Proc. 2011;2011:701–708. PMCID: PMC3243202. [PMC free article] [PubMed] [Google Scholar]
- 17.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimmel SE, French B, Kasner SE, Johnson JA, Anderson JL, Gage BF, et al. A pharmacogenetic versus a clinical algorithm for warfarin dosing. N Engl J Med. 2013;369:2283–2293. doi: 10.1056/NEJMoa1310669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pirmohamed M, Burnside G, Eriksson N, Jorgensen AL, Toh CH, Nicholson T, et al. A Randomized Trial of Genotype-Guided Dosing of Warfarin. New England Journal of Medicine. 2013 doi: 10.1056/NEJMoa1311386. 131119084528003. [DOI] [PubMed] [Google Scholar]
- 20.Spurling GK, Mansfield PR, Montgomery BD, Lexchin J, Doust J, Othman N, et al. Information from pharmaceutical companies and the quality, quantity, and cost of physicians’ prescribing: a systematic review. PLoS Med. 2010;7:e1000352. doi: 10.1371/journal.pmed.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]