Abstract
Canine kobuviruses (CaKVs) are newly recognized picornaviruses that have been recently detected in dogs in the U.S.A., Italy, U.K., the Republic of Korea and Tanzania. To trace the evolution of CaKV strains, a total of 201 fecal samples from rectal swabs of diarrheic dogs, which were obtained from May 2014 to April 2015 in northeast China, were detected by reverse transcription-PCR targeting a partial (504 bp) fragment of the 3D gene. Furthermore, a phylogenetic analysis of the CaKV strains identified in northeast China was conducted based on the partial 3D gene sequence. The results indicated that 36 fecal samples (17.91%, 36/201) were positive for CaKV, in which the co-infection rates of canine coronavirus, canine parvovirus-2 and canine bocavirus were 58.33%, 41.67%, and 11.11%, respectively. Sequence comparison of the partial 3D gene revealed nucleotide homologies of 94.4–100%, 95.6–98.6%, 94.3–97.6%, 94.4–96.3% and 93.3–95.1% within the 36 Chinese CaKV strains, and between the 36 Chinese CaKV strains and four CaKV reference strains from South Korea, Italy, U.S.A. and Tanzania, respectively. A phylogenetic tree revealed that the 36 Chinese CaKV strains formed one specific CaKV lineage with CaKVs that have recently been identified in other countries. The 36 Chinese CaKV strains were closely related to CaKV reference strains from Asia and Europe, but differed genetically from CaKV reference strains from North America and Africa. This study provides evidence that CaKVs circulate in diarrhoetic dogs in China and that they exhibit substantial genetic diversity and high co-infection rates with other enteric viruses.
Keywords: canine kobuvirus, epidemiology, genetic diversity, kobuvirus
The family Picornaviridae is highly diverse, currently comprising 17 genera, many of which consist of several species, subspecies and genotypes [1]. Kobuvirus is a newly recognized genus in the family Picornaviridae. Several studies have demonstrated that kobuviruses are transmitted through the fecal-oral route via the consumption of contaminated food or water, and they have a potential role in gastroenteritis in humans and animals [2, 8, 11]. Canine kobuviruses (CaKVs) have been recently discovered in dogs [10], demonstrating that pets can be infected with members of this genus. At present, epidemiological investigations of CaKVs in dogs are important for understanding their pathogenicity and given the interactions between pets and humans, their zoonotic potential.
To date, newly described CaKVs have been detected in healthy and diarrhoetic dogs in the US, Africa, Italy and the U.K. [4, 7, 10, 13, 15, 16]. However, the epidemiology of CaKVs in China remains unknown. To obtain information regarding the epidemiology of CaKVs, a surveillance study was conducted in diarrhoetic dogs in northeast China from May 2014 to April 2015. Furthermore, the phylogeny of the CaKV strains identified in China was analyzed. Our aim was to increase our understanding of the evolution of CaKVs.
MATERIALS AND METHODS
Sampling: In total, 201 rectal swab samples were collected in diarrhoetic dogs from animal hospitals in three districts—Harbin, Daqing and Mudanjiang—of Heilongjiang province in northeast China from May 2014 to April 2015 using 3.5-ml commercial virus sampling tubes (YOCON Biological Technology Co.. Ltd., Beijing, China). Of the 201 samples, 141 were collected from 2 animal hospitals in Harbin, 20 were collected from one animal hospital in Daqing, and 40 were collected from 1 animal hospital in Mudanjiang. All rectal swab samples were stored at −80°C.
RNA and DNA extraction: After 1 ml of fecal samples was centrifuged at 1,500 × g for 10 min at 4°C, the supernatant of each sample was transferred to a 1.5-ml Eppendorf (EP) tube. Viral RNA was extracted from each sample using the TIANamp Virus RNA Kit (Tiangen Biotech Co., Ltd., Beijing, China), according to the manufacturer’s instructions. Genomic DNA was extracted from the supernatant using a commercial TIANamp Stool DNA Kit (Tiangen Biotech Co., Ltd.), according to the manufacturer’s instructions. The extracted RNA and genomic DNAs were stored at −80°C.
Detection and sequence analysis of CaKVs: Molecular detection of CaKVs was conducted using reverse transcription-PCR (RT-PCR) targeting a 504-bp fragment of the 3D gene of CaKV described by Di Martino et al. [7]. The first cDNA was synthesized using random primers (6 nt) by Moloney murine leukemia virus (RNase H-) reverse transcriptase (Novoprotein Scientific Inc., Shanghai, China), according to the manufacturer’s instructions. PCR amplifications were conducted in a 50-µl reaction volume containing 0.1 µM of the forward primer, 0.1 µM of the reverse primer, 4 µl of cDNA, 25 µl of EmeraldAmp PCR Master Mix (2× Premix) (Takara Biotechnology Co., Ltd., Dalian, China) and an appropriate volume of double-distilled water. The mixtures were amplified by 35 cycles of 94°C for 45 sec, 54°C for 1 min and 72°C for 50 sec, followed by a final extension at 72°C for 10 min in an Applied Biosystems GeneAmp PCR System 9700 thermal cycler (Thermo Fisher Scientific, Waltham, MA, U.S.A.). After the amplification products were purified using the AxyPrep DNA Gel Extraction kit (Corning, Suzhou, China), the purified products were directly subjected to sequencing using the Sanger sequencing method. Each sample was sequenced three times. Thirty-six sequences were submitted to GenBank at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) under the accession numbers KT210390–KT210425.
Phylogenetic analysis of the 3D gene: For the phylogenetic analysis, partial sequences of the 3D gene of kobuviruses from canine and other species were retrieved from GenBank. To construct the phylogenetic trees, a multiple alignment of all target sequences was performed using ClustalX program version 1.83 [18]. Furthermore, phylogenetic trees of all target sequences were generated from the ClustalX-generated alignments by MEGA6.06 software using the neighbor-joining method [17]. Neighbor-joining phylogenetic trees were built with the p-distance model, 1,000 bootstrap replicates and, otherwise, the default parameters in MEGA 6.
Screening for canine enteric pathogens: All fecal samples that tested positive for CaKV were also screened for canine parvovirus type 2 (CPV-2), canine coronavirus (CCoV), canine astrovirus (CaAstV), canine norovirus (CNoV), canine bocavirus (CBoV) and Group A-Rotavirus (RV-A) by either PCR or RT-PCR, followed by sequencing, according to previously described protocols [3, 5, 6, 9, 12, 14].
RESULTS
Detection of CaKV: The characteristics of the CaKV-positive dogs and co-infections with other enteric viruses are shown in Table 1, and further analysis of the CaKV-positive samples is shown in Table 2. Of the 201 fecal samples, 17.91% (36/201) were positive for CaKV, and the CaKV-positive rates in Harbin, Daqing and Mudanjiang were 19.86% (28/141), 20% (4/20) and 10% (4/40), respectively. Co-infections with CCoV, CPV-2 and CBoV were found in the 36 CaKV-positive samples; 58.33% (21/36) were positive for CCoV; 41.67% (15/36) were positive for CPV-2; 11.11% (4/36) were positive for CBoV; 27.78% (10/36) were positive for CCoV and CPV-2; 5.56% (2/36) were positive for CPV-2 and CBoV; and 2.78% (1/36) were positive for CCoV, CPV-2 and CBoV.
Table 1. Characteristics of the CaKV positive dogs in northeast China and co-infection of CaKVs with other enteric viruses.
| No. | Strain | Accession No. | Collection date | Location | Breed | Gender | Age | Vaccined | Other enteric viruses |
|---|---|---|---|---|---|---|---|---|---|
| 1 | MDJ-10 | KT210390 | Nov-2014 | MDJ | MI | M | 4M | NO | CCoV (KT192642) |
| 2 | MDJ-13 | KT210391 | Oct-2014 | MDJ | LR | F | 2.5M | NA | CBoV (KR998481) |
| 3 | MDJ-27 | KT210392 | Apr-2015 | MDJ | NA | F | NA | NA | CBoV (KR998487), CPV-2 (KT074266) |
| 4 | MDJ-38 | KT210393 | Feb-2015 | MDJ | BF | M | 3.5M | Yes | — |
| 5 | HRB-a1 | KT210394 | Sep-2014 | HRB | NA | F | 3.5M | Yes | CBoV (KR998488), CCoV (KT192656), CPV-2 (KT074276) |
| 6 | HRB-a3 | KT210395 | Sep-2014 | HRB | GM | M | NA | No | CCoV (KT192658) |
| 7 | HRB-A4 | KT210396 | Sep-2014 | HRB | NA | F | 3M | NA | CCoV (KT192652), CPV-2 (KT074319) |
| 8 | HRB-A7 | KT210397 | Sep-2014 | HRB | SN | M | 3M | No | CCoV (KT192654) |
| 9 | HRB-A8 | KT210398 | Sep-2014 | HRB | NA | F | 1.5M | Yes | CCoV (KT192655), CPV-2 (KT074280) |
| 10 | DQ-alpha3 | KT210399 | Jan-2015 | DQ | CH | M | 2M | No | CCoV (KT192661) |
| 11 | DQ-alpha5 | KT210400 | Feb-2015 | DQ | PD | NA | 3M | NA | CCoV (KT192662) |
| 12 | DQ-alpha9 | KT210401 | Feb-2015 | DQ | CH | M | 1.5M | Yes | CBoV (KR998489) |
| 13 | DQ-beta3 | KT210402 | Nov-2014 | DQ | NA | F | 3M | No | CCoV (KT192675) |
| 14 | HRB-B1 | KT210403 | Sep-2014 | HRB | NA | M | NA | Yes | CCoV (KT192666) |
| 15 | HRB-B0 | KT210404 | Oct-2014 | HRB | PM | F | 3M | Yes | CCoV (KT192665), CPV-2 (KT074279) |
| 16 | HRB-B3 | KT210405 | Sep-2014 | HRB | PD | M | 3M | Yes | CCoV (KT192667) |
| 17 | HRB-b8 | KT210406 | Oct-2014 | HRB | SM | F | 2M | No | CCoV (KT192672), CPV-2 (KT074284) |
| 18 | HRB-B9 | KT210407 | Oct-2014 | HRB | NA | F | 3M | Yes | CCoV (KT192669) |
| 19 | HRB-bb1 | KT210408 | Oct-2014 | HRB | NA | M | 3M | Yes | — |
| 20 | HRB-bb3 | KT210409 | Oct-2014 | HRB | PM | NA | 2M | No | CCoV (KT192670), CPV-2 (KT074341) |
| 21 | HRB-C2 | KT210410 | Oct-2014 | HRB | TM | F | NA | No | CCoV (KT192678), CPV-2 (KT074285) |
| 22 | HRB-D1 | KT210411 | Oct-2014 | HRB | SN | M | 4M | Yes | CCoV (KT192682) |
| 23 | HRB-d4 | KT210412 | Nov-2014 | HRB | CO | F | 3M | Yes | — |
| 24 | HRB-D6 | KT210413 | Oct-2014 | HRB | GM | NA | NA | Yes | CCoV (KT192685) |
| 25 | HRB-D9 | KT210414 | Nov-2014 | HRB | NA | NA | 3.5M | Yes | CPV-2 (KT074296) |
| 26 | HRB-E8 | KT210415 | Nov-2014 | HRB | GM | M | 3M | Yes | CCoV (KT192687), CPV-2 (KT074300) |
| 27 | HRB-ee8 | KT210416 | Nov-2014 | HRB | BF | NA | NA | Yes | — |
| 28 | HRB-F1 | KT210417 | Nov-2014 | HRB | BF | M | 4M | Yes | — |
| 29 | HRB-F8 | KT210418 | Nov-2014 | HRB | PD | M | 3M | No | CCoV (KT192691), CPV-2 (KT074303) |
| 30 | HRB-G2 | KT210419 | Feb-2015 | HRB | SN | NA | 4M | No | — |
| 31 | HRB-G5 | KT210420 | Nov-2014 | HRB | NA | M | 4M | No | — |
| 32 | HRB-G8 | KT210421 | Nov-2014 | HRB | PM | NA | 4M | Yes | CPV-2 (KT074346) |
| 33 | HRB-G9 | KT210422 | Feb-2015 | HRB | NA | F | 3.5M | No | — |
| 34 | HRB-H2 | KT210423 | Dec-2014 | HRB | PD | F | 3M | NA | CPV-2 (KT074307) |
| 35 | HRB-H6 | KT210424 | Dec-2014 | HRB | PM | NA | 1.5M | NA | CCoV (KT192695) |
| 36 | HRB-K3 | KT210425 | Jan-2015 | HRB | NA | F | 3M | Yes | CPV-2 (KT074317) |
Note. For breed, GM: Golden Malinois, LR: Labrador Retriever, SM: Samoyed, PM: Pomeranian, SN: Schnauzer, BF: Bijon Frise, CO: Caucasian Owtcharka, TM: Tibetan Mastiff, PD: Poodle, CH: Chihuahua and MI: Manifold Intelligence; For gender, F: female and M: male; for age, M: month; for location, MDJ: Mudanjiang, HRB: Harbin, DQ: Daqing.
Table 2. Analysis of the CaKV positive samples and co-infection of other enteric viruses in CaKV positive samples.
| Location | Numbers of sample | Positive rate for CaKV | Other enteric viruses in the CaKV positive samples | |||||
|---|---|---|---|---|---|---|---|---|
| CCoV | CPV-2 | CBoV | CCoV+CPV-2 | CPV-2+CBoV | CCoV+CPV-2+CBoV | |||
| Harbin | 141 | 19.86% (28/141) | 60.71% (17/28) | 50% (14/28) | 3.57% (1/28) | 32.14% (9/28) | 3.57% (1/28) | 3.57% (1/28) |
| Daqing | 20 | 20% (4/20) | 75% (3/4) | — | 25% (1/4) | — | — | — |
| Mudanjiang | 40 | 10% (4/40) | 25% (1/4) | 25% (1/4) | 50% (2/4) | 25% (1/4) | 25% (1/4) | — |
| Total | 201 | 17.91% (36/201) | 58.33% (21/36) | 41.67% (15/36) | 11.11% (4/36) | 27.78% (10/36) | 5.56% (2/36) | 2.78% (1/36) |
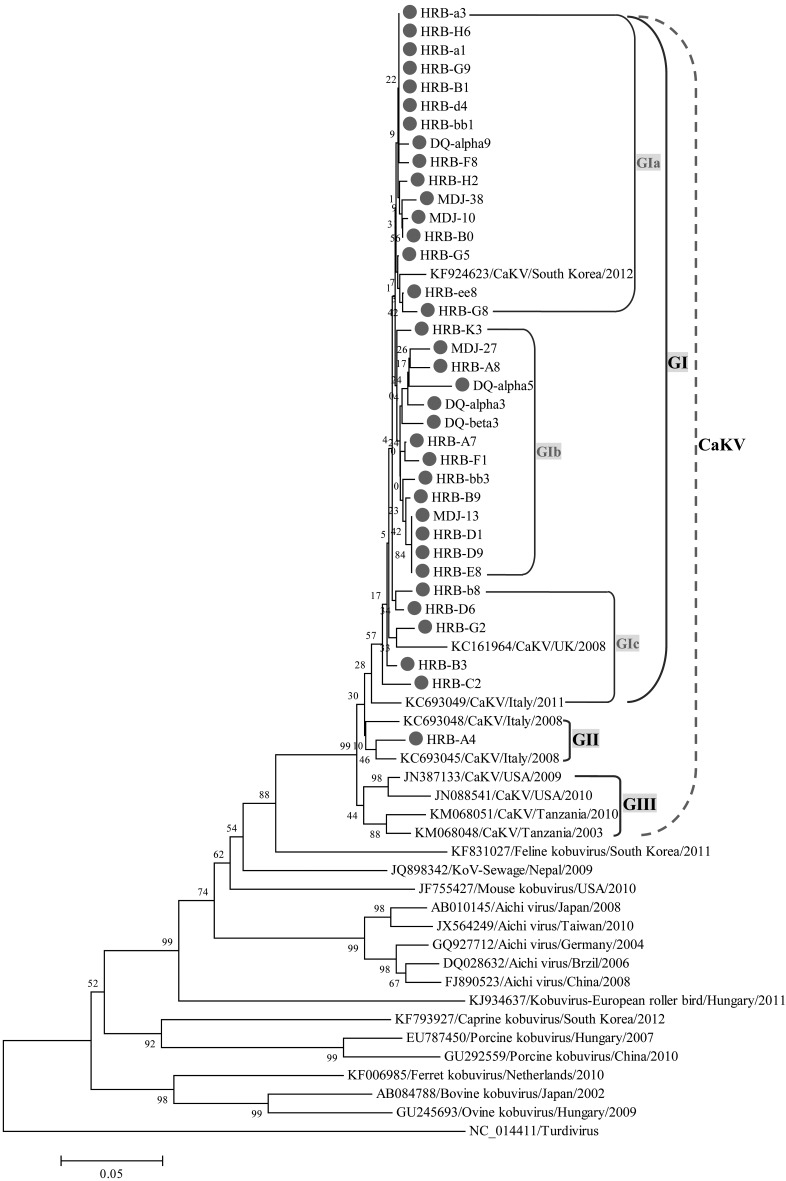
Sequence homology and phylogeny analyses of CaKV strains: Sequence comparisons of the partial 3D gene revealed 94.4–100%, 95.6–98.6%, 94.3–97.6%, 94.4–96.3%, 93.3–95.1%, 84.7–85.4%, 81.7–84%, 79.6–80.6%, 74.3–75.9%, 70.6–71.8%, 67.4–69% and 70.1–71.3% nucleotide identities within the 36 Chinese CaKV strains, between the 36 Chinese CaKV strains and four CaKV reference strains from South Korea, Italy, the USA and Tanzania, respectively, and between the 36 Chinese CaKV strains and seven kobuvirus reference strains from cats, mice, humans (Aichi virus), birds, goats, pigs and bovines, respectively (Table 3). Furthermore, the phylogenetic tree revealed that the 36 Chinese CaKV strains formed one specific CaKV lineage with the CaKVs that were recently identified in the other countries; the CaKV lineage was composed of three groups, GI, GII and GIII; one Chinese strain, HRB-A4, belonged to the GII group, while the other 35 Chinese strains belonged to the GI group, which was composed of three subgroups, GIa, GIb and GIc (Fig. 1).
Table 3. Sequence homology of the 3D gene of Chinese CaKV strains and other reference strains at nucleotide levels.
| Location | CaKV | Feline kobuvirus | Mouse kobuvirus | Human Aichi virus | Bird kobuvirus | Caprine kobuvirus | Porcine kobuvirus | Bovine kobuvirus | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strains identified in this study | Compared with 12D049 strain (KF924623) from South Korea | Compared with CKoV/19c/2012/IT strain (KC693045)/ from Italy | Compared with AN211D strain (JN387133) from USA | Compared with B103 strain (KM068051) from Tanzania | Compared with FK-13 strain (KF831027) from South Korea | Compared with M-5 strain (JF755427) from USA | Compared with Chshc7 strain (FJ890523) from China | Compared with SZAL6-KoV/2011/HUN strain (KJ934637) from Hungary | Compared with 12Q108 strain (KF793927) from South Korea | Compared with Y-1-CHI strain (GU292559) from China | Compared with U-1 strain (AB084788) from Japan | |
| Harbin | 95.4–100% | 95.6–98.6% | 95.5–97.6% | 94.9–96.3% | 93.8–95.1% | 84.5–85.4% | 81.7–84% | 79.9–80.8% | 74.5–75.9% | 70.8–71.1% | 67.4–69% | 70.1–71.3% |
| Daqing | 96.3–98.1% | 96.3–97.9% | 94.3–96.7% | 94.4–96.3% | 92.6–94.7% | 84.5–85.2% | 82.2–82.6% | 78.9–80.3% | 74.3–75.5% | 70.6–71.1% | 67.4–68.3% | 70.1–70.6% |
| Mudanjiang | 97.2–99.1% | 97.5–97.9% | 95.7–96.4% | 94.7–95.6% | 93.3–94.0% | 85.0–85.4% | 81.7–82.6% | 79.4–80.1% | 74.3–74.8% | 70.8–71.8% | 67.4–68.1% | 70.1–70.8% |
| Total | 94.4–100% | 95.6–98.6% | 94.3–97.6% | 94.4–96.3% | 93.3–95.1% | 84.7–85.4% | 81.7–84% | 79.6–80.6% | 74.3–75.9% | 70.6–71.8% | 67.4–69% | 70.1–71.3% |
Fig. 1.
Phylogenetic analysis of the CaKVs on the basis of the partial 3D gene. ●represents the CaKV strains identified in this study.
DISCUSSION
Kobuvirus is a relatively new and emerging genus of the Picornaviridae, with members infecting both humans and other animal species. In the current study, we conducted prevalence and phylogenetic analyses of canine kobuviruses in diarrhoetic dogs in northeast China. In our study, 36 of 201 (17.91%) fecal samples from diarrhoetic dogs were positive for CaKV, and the CaKV-positive rates in Harbin, Daqing and Mudanjiang were 19.86% (28/141), 20% (4/20) and 10% (4/40), respectively. In a previous study, a low prevalence of CaKV was reported by RT-PCR in the U.S.A. and Italy. Li et al. [13] reported that 10% (20/200) of both sick and healthy dogs tested positive for CaKV. Di Martino et al. [7] reported that 4.37% (6/137) of diarrhoetic dogs tested positive for CaKV. In contrast, a high prevalence of CaKV was reported in recent studies from the Republic of Korea. Oem et al. [15] reported that 19.04% (4/21) of diarrhoetic dogs tested positive for CaKV, while Choi et al. [5] reported that 50.55% (46/91) of diarrhoetic dogs tested positive for CaKV. The 17.91% CaKV-positive rate in our study is in agreement with that described by Oem et al. [15]. These data suggest that the prevalence of CaKVs may differ among different countries or regions.
These viruses have been implicated in diarrheal disease, but the evidence for a primary pathogenic role remains controversial. In our study, the CaKV-positive fecal samples were also screened for the leading causes of canine viral enteritis, including CPV-2, CCoV, CaAstV, CNoV, CaKV and RV-A. Co-infections with CPV-2, CCoV and CBoV were identified in the 36 CaKV-positive samples. Co-infection of CaKV with CCoV reached 58.33% (21/36), and co-infection of CaKV with CPV-2 reached 41.67% (15/36). These results are similar to those reported in a previous study of CaKV-positive samples from diarrhoetic dogs, in which the co-infection rates with CPV-2 and CCoV were 24.81% (34/137) and 22.62% (31/137), respectively [7]. In our study, CaKV was the only enteric virus identified in eight specimens, accounting for 22.22% (8/36) of all CaKV-positive samples. This result implies that CaKV alone may cause infections in the general population. However, the high co-infection rate of CaKV with other enteric viruses should be considered to be a composite factor for the occurrence of viral diarrhea in dogs in northeast China.
To investigate the genetic heterogeneity of CaKVs circulating in dogs in northeast China, sequence comparison and phylogeny analyses of the 36 CaKV-positive strains were conducted on the basis of the partial 3D gene sequence. Genetic analysis of the partial 3D region showed that the CaKVs identified in the Republic of Korea were more variable, sharing nucleotide sequence identities ranging from 85.1% to 99.8% [15]. The 36 CaKV-positive strains identified in China shared nucleotide sequence identities ranging from 94.4% to 100% and, thus, exhibited limited genetic diversity. The phylogenetic tree derived from the partial 3D gene from human and various animal species showed that CaKVs clustered into a single lineage. This finding supports the view that CaKVs from different countries are not restricted geographically and form a single lineage when compared with kobuviruses from other species. However, the lineage of the CaKV strains formed three groups—GI, GII and GIII—and the 36 Chinese CaKV strains were divided into the GI and GII groups. The 35 Chinese CaKV strains in the GI group were divided into the GIa, GIb and GIc subgroups. All 36 Chinese CaKV strains were closely related to the CaKV reference strains from Asia and Europe, and they differed genetically from the CaKV reference strains from North America and Africa. Thus, our data suggest that the lineages of CaKV strains may exhibit geographically differences. This conclusion needs to be validated by extensive epidemiological surveying in future studies.
In conclusion, the present study reports, for the first time, the prevalence and phylogenetic analysis of canine kobuviruses in diarrhoetic dogs in northeast China, and the CaKV strains circulating in northeast China exhibit genetic diversity and high co-infection rates with other enteric viruses. However, further investigations are needed to determine the relationship between CaKV infections and the ability of CaKVs to cause diarrhea in dogs.
Acknowledgments
This work was supported by the Program for New Century Excellent Talents in Heilongjiang Provincial University (grant no. 1252-NCET-016).
REFERENCES
- 1.Adams M. J., King A. M., Carstens E. B.2013. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2013). Arch. Virol. 158: 2023–2030. doi: 10.1007/s00705-013-1688-5 [DOI] [PubMed] [Google Scholar]
- 2.Ambert-Balay K., Lorrot M., Bon F., Giraudon H., Kaplon J., Wolfer M., Lebon P., Gendrel D., Pothier P.2008. Prevalence and genetic diversity of Aichi virus strains in stool samples from community and hospitalized patients. J. Clin. Microbiol. 46: 1252–1258. doi: 10.1128/JCM.02140-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buonavoglia D., Cavalli A., Pratelli A., Martella V., Greco G., Tempesta M., Buonavoglia C.2000. Antigenic analysis of canine parvovirus strains isolated in Italy. New Microbiol. 23: 93–96. [PubMed] [Google Scholar]
- 4.Carmona-Vicente N., Buesa J., Brown P. A., Merga J. Y., Darby A. C., Stavisky J., Sadler L., Gaskell R. M., Dawson S., Radford A. D.2013. Phylogeny and prevalence of kobuviruses in dogs and cats in the UK. Vet. Microbiol. 164: 246–252. doi: 10.1016/j.vetmic.2013.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi S., Lim S. I., Kim Y. K., Cho Y. Y., Song J. Y., An D. J.2014. Phylogenetic analysis of astrovirus and kobuvirus in Korean dogs. J. Vet. Med. Sci. 76: 1141–1145. doi: 10.1292/jvms.13-0585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costa E. M., de Castro T. X., Bottino Fde O., Garcia Rde C.2014. Molecular characterization of canine coronavirus strains circulating in Brazil. Vet. Microbiol. 168: 8–15. doi: 10.1016/j.vetmic.2013.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Martino B., Di Felice E., Ceci C., Di Profio F., Marsilio F.2013. Canine kobuviruses in diarrhoeic dogs in Italy. Vet. Microbiol. 166: 246–249. doi: 10.1016/j.vetmic.2013.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drexler J. F., Baumgarte S., de Souza Luna L. K., Eschbach-Bludau M., Lukashev A. N., Drosten C.2011. Aichi virus shedding in high concentrations in patients with acute diarrhea. Emerg. Infect. Dis. 17: 1544–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gómez M. M., de Mendonça M. C., Volotão Ede M., Tort L. F., da Silva M. F., Cristina J., Leite J. P.2011. Rotavirus A genotype P[4]G2: genetic diversity and reassortment events among strains circulating in Brazil between 2005 and 2009. J. Med. Virol. 83: 1093–1106. doi: 10.1002/jmv.22071 [DOI] [PubMed] [Google Scholar]
- 10.Kapoor A., Simmonds P., Dubovi E. J., Qaisar N., Henriquez J. A., Medina J., Shields S., Lipkin W. I.2011. Characterization of a canine homolog of human Aichivirus. J. Virol. 85: 11520–11525. doi: 10.1128/JVI.05317-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khamrin P., Maneekarn N., Okitsu S., Ushijima H.2014. Epidemiology of human and animal kobuviruses. Virusdisease 25: 195–200. doi: 10.1007/s13337-014-0200-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau S. K., Woo P. C., Yeung H. C., Teng J. L., Wu Y., Bai R., Fan R. Y., Chan K. H., Yuen K. Y.2012. Identification and characterization of bocaviruses in cats and dogs reveals a novel feline bocavirus and a novel genetic group of canine bocavirus. J. Gen. Virol. 93: 1573–1582. doi: 10.1099/vir.0.042531-0 [DOI] [PubMed] [Google Scholar]
- 13.Li L., Pesavento P. A., Shan T., Leutenegger C. M., Wang C., Delwart E.2011. Viruses in diarrhoeic dogs include novel kobuviruses and sapoviruses. J. Gen. Virol. 92: 2534–2541. doi: 10.1099/vir.0.034611-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mesquita J. R., Nascimento M. S.2012. Molecular epidemiology of canine norovirus in dogs from Portugal, 2007–2011. BMC Vet. Res. 8: 107. doi: 10.1186/1746-6148-8-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oem J. K., Choi J. W., Lee M. H., Lee K. K., Choi K. S.2014. Canine kobuvirus infections in Korean dogs. Arch. Virol. 159: 2751–2755. doi: 10.1007/s00705-014-2136-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olarte-Castillo X. A., Heeger F., Mazzoni C. J., Greenwood A. D., Fyumagwa R., Moehlman P. D., Hofer H., East M. L.2015. Molecular characterization of canine kobuvirus in wild carnivores and the domestic dog in Africa. Virology 477: 89–97. doi: 10.1016/j.virol.2015.01.010 [DOI] [PubMed] [Google Scholar]
- 17.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S.2013. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. doi: 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G.1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. doi: 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]